Abstract
The ubiquitin E3 ligase gene related to anergy in lymphocytes (GRAIL) (Rnf128) is a type 1 transmembrane protein that induces T cell anergy through the ubiquitination activity of its cytosolic RING finger. GRAIL also contains an equally large luminal region consisting primarily of an uncharacterized protease-associated (PA) domain. Using two-hybrid technology to screen for proteins that bound the PA domain we identified CD151, a member of the tetraspanin family of membrane proteins. GRAIL bound to the luminal/extracellular portion of both CD151 and the related tetraspanin CD81 using its PA domain, which promoted ubiquitination of cytosolic lysine residues. GRAIL exhibited specificity for lysines only within the tetraspanin amino terminus even in the presence of other cytosolic lysine residues in the substrate. GRAIL-mediated ubiquitination promoted proteasomal degradation and cell surface down-regulation of tetraspanins via Lys-48 linkages. As a result, the juxtaposition of PA and RING finger domains across a lipid bilayer facilitates the capture of transmembrane substrates for subsequent ubiquitination. These findings identify for the first time a single subunit E3 ligase containing a substrate-binding domain spatially restricted by a membrane from its E2 recruitment domain as well as an E3 ligase for members of the tetraspanin family.
The ubiquitin proteasome pathway consists of a combination of proteins that determines with meticulous precision the post-translational fate of nearly every cellular protein. The sequential enzymes involved in conjugation of ubiquitin to a substrate, E12 activating enzymes, E2 transferases, and E3 ligases, provide a mechanism of increasing stringency to ensure the serial transfer of ubiquitin to its intended target (1). The majority of substrate specificity is conferred by a wide array of protein interaction motifs on the E3 ligase that are separate from the domains responsible for recruitment of the ubiquitination machinery (2). After covalent attachment of ubiquitin, other classes of ubiquitin-binding proteins decipher this highly informative post-translational modification by binding, editing, or removing the attached ubiquitin molecules (3–5).
E3 ligases are split into distinct families based on their E2 recruiting domains, including HECT, RING, and U box. RING finger proteins are the most numerous and can be further divided into multisubunit complexes (SCF and anaphase-promoting complex) or single subunit proteins with modular domains (Cbl and Mdm2). Multisubunit complexes partition function into individualized protein components with the substrate-binding region contained in the F box for the SCFs (6), whereas single subunit RING finger E3 ligases depend on substrate capture via protein-protein interaction domains particular to each (7). The regions critical for substrate binding among RING finger E3 ligases vary greatly, making prediction and identification of potential substrates difficult. In addition, the limited number of E3 ligases compared with the total number of proteins regulated by ubiquitination requires that each E3 ligase have flexibility to regulate multiple protein targets. How E3 ligases achieve selective substrate specificity among the pool of potential cellular substrates is currently not well understood.
The gene related to anergy in lymphocytes (GRAIL) is a type 1 transmembrane E3 ligase cloned following differential display screening to identify early factors that promoted T cell anergy (8). GRAIL expression has been correlated with a number of systems that prevent T cell proliferation and cytokine production, and its activity depends upon an intact cytosolic RING finger domain (9, 10). However, GRAIL mRNA expression is not exclusive to lymphocytes, suggesting that it could target substrates and affect cellular processes across numerous tissues. In addition, the amino terminus of GRAIL contains a protease-associated (PA) domain, which has been proposed to serve as a protein-protein or protein-sorting motif in plants, but its precise function is unknown (11, 12). Recent studies have also demonstrated that PA domain-containing proteins play a role in ubiquitin-mediated degradation of the transmembrane manganese transporter in yeast (13) and removal of misfolded endoplasmic reticulum glycoproteins in mammalian cells (14).
Of the few transmembrane E3 ligases currently known, a substantial number of their identified substrates are themselves membrane-resident proteins. Therefore, we asked whether the amino-terminal PA domain of GRAIL might serve as a substrate-binding motif to capture a luminal or extracellular domain of transmembrane proteins followed by ubiquitination of cytosolic residues by the GRAIL RING finger. Two-hybrid screening for proteins that interact with the amino terminus of GRAIL identified the tetraspanin family member CD151, a four-pass transmembrane protein that modulates cell-cell adhesion and integrin signaling (15, 16). Subsequent analyses demonstrated that GRAIL utilized its PA domain to bind directly extracellular regions of both CD151 and another tetraspanin, CD81, and this interaction promoted cytosolic ubiquitination of the tetraspanin substrate for proteasomal degradation. These results demonstrate a unique ubiquitination model in which a single subunit E3 ligase contains a substrate interaction domain for transmembrane proteins that is spatially restricted from its ubiquitination machinery across a lipid bilayer. Using this membrane juxtaposition of binding and ubiquitination, GRAIL might target additional transmembrane proteins for degradation.
EXPERIMENTAL PROCEDURES
Materials and Antibodies—Ammonium chloride, Brij96, CHAPS, chloroquine, Nonidet P-40, Triton X-100, mouse anti-FLAG-horseradish peroxidase (M2), and rabbit anti-V5 were purchased from Sigma. ALLN, MG132, and E64, were purchased from Calbiochem. DSP was purchased from Pierce. Mouse anti-HA-horseradish peroxidase (F-7), anti-HA-conjugated agarose (F-7), mouse anti-integrin β1 (P5D2), and mouse anti-CD81 (1.3.3.22) were purchased from Santa Cruz Biotechnology, Santa Cruz, CA. The mouse anti-CD58 (BRIC-5) antibody was from Southern Biotech, Birmingham, AL. Mouse anti-transferrin receptor (H68.4) and goat anti-rabbit-horseradish peroxidase were obtained from Zymed Laboratories Inc., San Francisco, CA. Mouse anti-human major histocompatibility complex class I was from eBioscience, San Diego, CA. Donkey anti-mouse IgG conjugated to Alexa Flour 488 and Alexa Fluor 594 was purchased from Molecular Probes, Eugene, OR. Goat anti-mouse conjugated to phycoerythrin was obtained from Pharmingen. Mouse anti-CD81 (5A6) and mouse anti-CD151 (1A5) were generous gifs from Dr. Shoshana Levy and Dr. James Quigley, respectively.
Bacterial Screening—The amino terminus of mouse GRAIL after the signal sequence (amino acids 40–204) was PCR-subcloned into the pBT vector from the BacterioMatch Two-Hybrid System (Stratagene, La Jolla, CA). This vector was screened against a liver cDNA library according to the manufacturer's standard protocol. Carbenicillin-resistant clones containing high β-galactosidase activity were sequenced, and open reading frames were identified by BLAST (basic local alignment search tool) search (NCBI).
Plasmids—All plasmids containing open reading frames for human and mouse tetraspanins were purchased from Open Biosystems, Huntsville, AL, except for human CD81, which was a generous gift from Dr. Shoshana Levy (Stanford University). All tetraspanins were PCR-subcloned into the pIRES-hrGFP-2a in frame with a carboxyl-terminal 3×HA tag (Stratagene). For all human tetraspanin constructs, humanized recombinant green fluorescent protein (GFP) was first removed from the expression vector.
Cell Culture—Human embryonic kidney 293T and HeLa cells were grown in Dulbecco's modified Eagle's medium + 5% heat-inactivated fetal bovine serum supplemented with l-glutamine and penicillin/streptomycin. Cells were transfected using TransIT-293 transfection reagent according to the manufacturer's standard protocol (Mirus Bio, Madison, WI). Cells were lysed with Brij96 lysis buffer to detect protein-protein interactions (20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% Brij96, protease inhibitors), and protein concentration was measured by Bradford assay (Bio-Rad). For ubiquitination assays, cells were incubated with ALLN (10 μg/ml) for 1.5–2 h at 37 °C prior to lysis. After lysis in 1% Brij96 buffer, lysates were denatured with 0.5% SDS at 100 °C for 10 min and then diluted to 0.1% SDS in Brij96 buffer for immunoprecipitation.
Immunoprecipitation and Western Blotting Analysis—Cell lysates were blocked for 1 h at 4°C with protein G-Sepharose (Amersham Biosciences) followed by immunoprecipitation overnight at 4 °C with the indicated agarose bound antibodies. After extensive washing in Brij96 buffer, proteins were eluted with 2× SDS sample buffer containing β-mercaptoethanol. Proteins were resolved by SDS-PAGE on 4–15% gradient gels (Bio-Rad) and transferred to polyvinylidene difluoride for analysis by Western blotting. Bands were illuminated using ECL Plus reagent (Amersham Biosciences). Filters were stripped using Restore buffer (Pierce).
Mutagenesis—Deletions and point mutations were made using the QuikChange II XL site-directed mutagenesis kit according to the manufacturer's standard protocol (Stratagene) and confirmed by DNA sequencing.
Immunofluorescence—Cells were grown on acid-treated, polylysine-coated No. 1 German coverslips for transfection. One hour prior to fixation, cells were incubated in serum-free medium containing 25 μm Alexa Fluor 633-labeled transferrin (Molecular Probes). Cells were fixed in 3.7% formaldehyde, permeabilized in 0.05% Triton X-100, and blocked in phosphate-buffered saline + 2% normal donkey serum (Jackson ImmunoResearch Laboratories, West Grove, PA). Cells were stained in phosphate-buffered saline + 1% normal donkey serum and mounted using VectaShield (Vector Laboratories, Burlingame, CA). Cells were imaged using an SP2 AOBS confocal microscope with a 63× oil immersion, 1.3 numerical aperture objective (Leica, Bannockburn, IL).
Flow Cytometry—Cells were dissociated 48 h following transfection and stained with appropriate primary and secondary antibodies in phosphate-buffered saline supplemented with 2% heat-inactivated bovine serum. Samples were run on a FACScan (BD Biosciences) and analyzed with FlowJo software (Tree-Star, Ashland, OR).
Cross-linking Assay—Transfected human embryonic kidney 293T cells were lysed in 1% Brij96 buffer. Cell lysates were split in half, and one set was cross-linked for 2 h on ice with 2 mm DSP followed by neutralization of free cross-linker with 50 mm Tris for 15 min on ice. SDS (0.1%) was added to both samples, and the tetraspanin was immunoprecipitated with anti-HA beads. Samples were washed, boiled in loading buffer with 10% β-mercaptoethanol for 10 min, and analyzed by Western blot.
RESULTS
Identification of Transmembrane Proteins That Bind the GRAIL PA Domain—The luminal portion of the type 1 transmembrane single unit E3 ubiquitin ligase GRAIL contains a PA domain, which is present in the sorting receptors of the plant vacuolar pathway and in the transferrin receptor in mammalian cells (17, 18). To identify potential PA-binding proteins, we screened a liver cDNA library in a bacterial two-hybrid system with the amino-terminal PA domain of mouse GRAIL, excluding the signal sequence, as “bait.” A bacterial system was used to avoid any secondary effects of the eukaryotic ubiquitin-proteasome system present in a yeast two-hybrid screen. Of the positive clones that contained regions of membrane-spanning proteins, the one chosen for further study contained the predominant protein-protein interaction motif, the large extracellular loop (LEL), of the transmembrane tetraspanin protein CD151 (Fig. 1A).
FIGURE 1.
The PA domain of GRAIL interacts with CD151. A, left, the region of mouse GRAIL used as bait in the bacterial screen, from amino acids 40 to 204. Right, the membrane topology of CD151, including the position of the small extracellular loop (SEL) and LEL. The portion of mouse CD151 identified from the liver cDNA library, from amino acids 117 to 253, is indicated by a dotted line. B, 293T cells were co-transfected with expression constructs for both V5-tagged mouse GRAIL and 3×HA-tagged mouse CD151 or empty vectors as control. Cell lysates were immunoprecipitated (IP) with anti-HA-conjugated beads and immunoblotted (IB) with anti-V5, anti-HA, and anti-transferrin receptor (TfnR). C, V5-tagged GRAIL and 3×HA-tagged CD151 were co-transfected into 293T cells, subjected to various lysis conditions followed by immunoprecipitation with anti-HA beads, and immunoblotted with anti-V5 and anti-HA. D, 293T cells were transfected with various V5-tagged expression constructs for wild-type GRAIL and GRAIL mutants along with 3×HA-tagged CD151. Cell lysates were immunoprecipitated with anti-HA blotted and immunoblotted with anti-V5 and anti-HA.
To verify the interaction between CD151 and full-length GRAIL in a eukaryotic system, transient co-expression of epitope-tagged constructs of these two proteins was performed in 293T cells. Immunoprecipitation of 3×HA-tagged CD151 resulted in co-immunoprecipitation of V5-tagged GRAIL (Fig. 1B), and reverse immunoprecipitation of GRAIL-V5 from separate lysates also pulled down CD151–3×HA (data not shown). This tetraspanin interaction was specific for the PA domain of GRAIL as another PA domain-containing protein with extensive intracellular co-localization with GRAIL, the transferrin receptor (8), did not co-immunoprecipitate with transfected 3×HA-tagged CD151 (Fig. 1B). One method to suggest a stoichiometric tetraspanin interaction is lysis with detergents of increasing hydrophobicity and ionic strength as was previously used to demonstrate the interaction between CD151 and the β1 and β3 integrins (16). GRAIL binding to CD151 survived solubilization with 1% Brij96 and with lower stoichiometry in 1% Triton X-100, implying a direct interaction between GRAIL and CD151 (Fig. 1C). Because Brij96 solubilization maintains the robust interaction between GRAIL and CD151 and avoids the formation of large tetraspanin complexes seen with CHAPS (19), this detergent was used for the remainder of the studies.
To further define the region of GRAIL that bound CD151, a series of V5-tagged GRAIL mutant constructs were co-expressed with 3×HA-tagged CD151 in 293T cells and assessed by co-immunoprecipitation for binding to CD151. As predicted, CD151 co-immunoprecipitated GRAIL constructs containing deletions or point mutations in the carboxyl-terminal domains (ΔCC, H2N2, and ΔZF), whereas a mutation removing the PA domain (ΔPA) prevented binding of CD151 to GRAIL (Fig. 1D).
The GRAIL PA Domain Directly Interacts with Transmembrane Tetraspanins—The predominant molecular characteristic of the tetraspanin family is the formation of an LEL domain. Although the sequence for the tetraspanin LEL regions diverge, a broad spectrum interaction of tetraspanin family members with proteins of the very late antigen family of β1 integrins has been reported (20). Therefore, we asked whether the PA domain of GRAIL might serve as an interaction domain for other members of the tetraspanin family in addition to CD151. To test our hypothesis, expression of V5-tagged GRAIL in 293T cells co-immunoprecipitated with 3×HA carboxyl-terminally tagged CD81 (Fig. 2A). Inclusion of an additional deletion mutant in an amino-terminal region of GRAIL (ΔVW) along with the ΔPA mutant in a co-immunoprecipitation assay with CD81 produced results similar to those seen with CD151 where an intact GRAIL amino terminus is required for binding (Fig. 2A). To demonstrate a direct interaction between GRAIL and tetraspanins, a covalent cross-linker (DSP) was added during lysis before supplementing with 0.1% SDS to prevent co-immunoprecipitation (Fig. 1C). The addition of DSP maintained the interaction between GRAIL and CD81, indicating that the GRAIL PA domain bound directly to the luminal/extracellular region of the transmembrane tetraspanins and acts as a bona fide protein-protein interaction motif in mammalian cells (Fig. 2B).
FIGURE 2.
The PA domain of GRAIL directly binds tetraspanins. A, left, 293T cells were transfected with expression constructs for V5-tagged GRAIL and 3×HA-tagged CD81 along with empty control vectors. Cell lysates were immunoprecipitated (IP) with anti-HA and immunoblotted (IB) with anti-V5 and anti-HA. Right, 293T cells were transfected with various V5-tagged expression constructs for wild-type GRAIL and GRAIL mutants along with 3×HA-tagged CD81. Cell lysates were immunoprecipitated with anti-HA and immunoblotted with anti-V5 or anti-HA. B, 293T cells were transfected with either 3×HA vector and V5-tagged GRAIL, 3×HA-tagged CD81 and V5 vector only, or 3×HA-tagged CD81 and V5-tagged GRAIL. Cells were lysed in 1% Brij96, lysates were divided in half, and one-half was cross-linked with DSP on ice for 2 h. Free DSP was neutralized with Tris, and 0.1% SDS was added to each sample. Cell lysates were then immunoprecipitated with anti-HA and immunoblotted with anti-GRAIL or anti-HA.
The GRAIL PA Domain Captures Tetraspanins for Cytosolic Ubiquitination by Its RING Finger—The cytosolic RING finger of GRAIL has been shown previously to confer E3 ligase activity against the cellular Rho family inhibitor RhoGDI (21). Therefore, we asked whether tetraspanin capture by the luminal PA domain of GRAIL could result in ubiquitination of one or more of the small cytosolic domains of these four-pass transmembrane proteins. Using a cellular ubiquitination assay in live cells, we determined whether GRAIL, compared with the enzymatically inactive H2N2 or nonbinding ΔPA mutants, could ubiquitinate tetraspanins. Because of the short half-life typical of polyubiquitinated proteins, a potent inhibitor of both proteasomes and lysosomal cathepsins, ALLN, was added to the cells no longer than 2 h prior to lysis to stabilize ubiquitinated proteins. In the presence of GRAIL, compared with LacZ, there was a dramatic increase in the amount of ubiquitinated CD81 and CD151 (Fig. 3A). To prevent detection of ubiquitinated proteins that could co-immunoprecipitate with tetraspanins, including autoubiquitinated GRAIL, cell lysates for the remaining cellular ubiquitination assays were first denatured in 0.5% SDS before immunoprecipitation. In these studies, transmembrane tetraspanin ubiquitination was dependent both on the ability of GRAIL to bind to the tetraspanin and its E3 ligase activity as demonstrated by reduced ubiquitination of CD81 and CD151 seen with the H2N2 or ΔPA mutants compared with wild-type GRAIL (Fig. 3B). The result of GRAIL ubiquitination was a decrease in the steady-state level of tetraspanin as seen when GRAIL was titrated into a fixed population of wild-type CD81 (Fig. 3C).
FIGURE 3.
GRAIL ubiquitination requires the PA and RING domains and results in tetraspanin degradation. A, following transfection with either V5-tagged GRAIL or lacZ; 3×HA-tagged CD81, CD151, or vector alone; and FLAG-tagged ubiquitin (Ub), 293T cells were incubated with ALLN for 2 h prior to lysis to enrich for ubiquitinated proteins. Cell lysates were immunoprecipitated (IP) with anti-HA beads and immunoblotted (IB) with anti-FLAG and anti-HA. B, 293T cells were transfected with V5-tagged GRAIL, ΔPA, H2N2, or vector alone constructs; 3×HA-tagged CD81 or CD151; and 3×FLAG-tagged ubiquitin. Cell lysates were denatured in SDS, diluted in lysis buffer, immunoprecipitated with anti-HA beads, and immunoblotted with anti-FLAG, anti-HA, and anti-V5. C, 293T cells were transfected with 0.4 μg of 3×HA-tagged CD81 and V5-tagged GRAIL at 0, 0.4, 0.8, 1.2, and 1.6 μg (empty V5 vector was added for a total of 1.6 μg of total V5 plasmid for each transfection). Cell lysates were separated by SDS-PAGE and blotted with the indicated antibodies.
GRAIL Ubiquitinates Residues Only within the Tetraspanin Cytosolic Amino Terminus—Ubiquitin molecules are predominantly added to lysine residues of target proteins. Previous alignments of human tetraspanins showed that the few conserved sequences were mainly centered in the transmembrane domains and cysteine motifs in the LEL along with a single lysine residue in the cytosolic amino terminus proximal to the first transmembrane region (22, 23). This orientation potentially provides access to the cytosolic RING finger of GRAIL for ubiquitin conjugation and suggested a conserved motif for tetraspanin ubiquitination. To test whether this shared lysine provided a preferred site for GRAIL-mediated ubiquitination, point mutants of the lysines in human CD81 and CD151 were constructed. Human CD81 (hCD81) contains two intracellular lysines only in the amino terminus at positions Lys-8 and conserved Lys-11, whereas human CD151 (hCD151) contains three lysines in the amino terminus at positions Lys-7, Lys-8, and conserved Lys-17 along with one lysine in each of the two other cytosolic domains (Fig. 4A). When GRAIL was co-expressed in cellular ubiquitination assays with hCD81 constructs containing single point mutations at either lysine 8 (K8R) or the conserved lysine at position 11 (K11R), the amount of hCD81 ubiquitination was reduced for both mutants roughly equally when compared with the ubiquitination density of wild-type hCD81 (Fig. 4B). In addition, an hCD81 protein devoid of both lysines (K8R,K11R) showed no GRAIL-mediated ubiquitination, whereas substituting a lysine for glutamic acid at position 4 in the double lysine-deleted hCD81 construct (K8R,K11R,E4K) restored the ability of GRAIL to ubiquitinate the tetraspanin. Mutating the conserved lysine (Lys-17) on hCD151 did not change the GRAIL-mediated ubiquitination seen compared with wild-type hCD151. However, the hCD151 mutant devoid of all three amino-terminal lysines (K7R,K8R,K17R) but still containing two additional non-amino-terminal cytosolic lysines was not ubiquitinated when co-expressed with GRAIL (Fig. 4B). Finally mutation of all the amino-terminal lysines conferred resistance to GRAIL-mediated degradation. Titrating in GRAIL to a fixed amount of the hCD81 K8R,K11R mutant, unlike the wild-type protein, did not change the steady-state levels of the tetraspanin (Fig. 4C).
FIGURE 4.
GRAIL ubiquitinates intracellular lysine residues only present in the tetraspanin amino terminus. A, ClustalW alignment of the amino terminus of human tetraspanins and diagram of CD81 and CD151 lysine point mutants constructed. * designates conserved lysine residue. B, 293T cells were transfected with either V5-tagged GRAIL or vector alone, 3×HA-tagged CD81 or CD151, and 3×FLAG-tagged ubiquitin (Ub). Cell lysates were denatured in SDS, immunoprecipitated (IP) with anti-HA beads, and immunoblotted (IB) with anti-HA and anti-FLAG. C, 293T cells were transfected with 0.5 μg of 3×HA-tagged CD81 K8R,K11R and V5-tagged GRAIL at 0, 0.4, 0.8, 1.2, 1.6, and 2 μg (empty V5 vector was added for a total of 2 μg of total V5 plasmid for each transfection). Cell lysates were separated by SDS-PAGE and blotted with the indicated antibodies.
These results demonstrate that, although the conserved lysines at position 11 of hCD81 and position 17 for hCD151 are not preferred sites for GRAIL-mediated ubiquitination, GRAIL can only ubiquitinate lysines in the amino terminus of these tetraspanins. In addition, results seen with the hCD81 K8R,K11R,E4K construct suggest that GRAIL can ubiquitinate lysines positioned anywhere in the amino terminus. Therefore, although the “conserved” amino-terminal lysine of tetraspanins does not serve as the primary site for ubiquitination, the GRAIL RING finger exhibits “regional specificity” for ubiquitinating lysine residues only within the amino-terminal, cytoplasmic domain of tetraspanins.
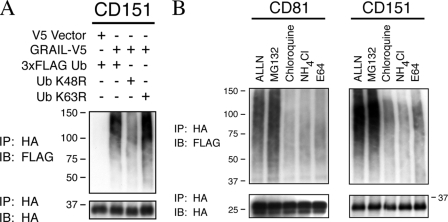
GRAIL Binding Promotes Lys-48-linked, Proteasomal Degradation of Tetraspanins—Ubiquitin itself contains seven lysine residues that can form isopeptide bonds with other ubiquitin molecules and result in polyubiquitin chains. The number of ubiquitin moieties added and the ubiquitin lysine used in the isopeptide bond between ubiquitins contribute to the ultimate fate of the ubiquitinated substrate. Polyubiquitin chains of four of more lysines joined via ubiquitin lysine 48 lead most often to proteasomal degradation, whereas ubiquitin lysine 63 conjugation can stabilize or activate the ubiquitinated substrate (24). To determine what type of polyubiquitin chains GRAIL formed on tetraspanins, 3×HA-tagged CD151 was co-expressed with V5-tagged GRAIL and either 3×FLAG-tagged wild-type ubiquitin or ubiquitin constructs containing arginine substitutions for lysine 48 (K48R) or lysine 63 (K63R). Transfection of K48R dramatically reduced ubiquitination of CD151, whereas the transfected K63R construct did not diminish CD151 ubiquitination, demonstrating that GRAIL polyubiquitinates tetraspanins using lysine 48 linkage (Fig. 5A). The presence of some ubiquitin conjugates in the presence of K48R ubiquitin is expected because this mutant competes with endogenous ubiquitin and can act as a “cap” to prevent elongation of lysine 48-linked polyubiquitin chains of varying lengths polymerized using endogenous ubiquitin.
FIGURE 5.
GRAIL polyubiquitinates tetraspanins via ubiquitin Lys-48 for proteasomal degradation. A, 293T cells were transfected with V5-tagged GRAIL or vector alone, 3×HA-tagged CD151, and either 3×FLAG-tagged wild-type ubiquitin (Ub), K48R ubiquitin, or K63R ubiquitin. Cell lysates were denatured in SDS, diluted in lysis buffer, immunoprecipitated (IP) with anti-HA beads, and immunoblotted (IB) with anti-FLAG and anti-HA. B, 293T cells were transfected with V5-tagged GRAIL, 3×HA-tagged CD81 or CD151, and 3×FLAG-tagged ubiquitin. Cells were incubated with various inhibitors affecting the proteasome or lysosome for 4 h at 37 °C prior to lysis. Cell lysates were denatured in SDS, immunoprecipitated with anti-HA beads, and immunoblotted with anti-FLAG and anti-HA.
To demonstrate that tetraspanin polyubiquitination led to proteasomal degradation, 293T cells were co-transfected with V5-tagged GRAIL, 3×FLAG-tagged wild-type ubiquitin, and either 3×HA-tagged CD81 or CD151. Four hours before lysis, cells were incubated with inhibitors of (a) both the proteasome and lysosomal cathepsins (ALLN), (b) only the proteasome (MG132), (c) vesicular trafficking to the lysosome (chloroquine and NH4Cl), or (d) lysosomal cysteine proteases (E64). The addition of both ALLN and MG132 stabilized ubiquitin conjugates on both CD81 and CD151, whereas inhibitors of lysosomal degradation or intracellular trafficking did not (Fig. 5B). These data demonstrate that GRAIL interaction promotes tetraspanin polyubiquitination via lysine 48-linked chains and leads to degradation at the proteasome.
Intracellular Co-localization Promotes Cell Surface Down-regulation of Tetraspanins—GRAIL has been shown to reside in perinuclear Rab5+ and Rab7+ vesicles of the recycling transferrin compartment, and tetraspanins have an extensive internal membrane distribution in addition to their localization on the cell surface (8, 25). To investigate where and the extent of GRAIL and tetraspanins overlap, HeLa cells were transfected with both V5-tagged GRAIL and 3×HA-tagged CD81 and CD151 followed by incubation of cells with Alexa 633-labeled transferrin. Co-localization of GRAIL and the tetraspanins was only seen in intracellular vesicles of the transferrin compartment (Fig. 6A). In addition, there were distinct locations of nonoverlap between the tetraspanins and GRAIL, indicating that intracellular populations of CD81 and CD151 exist that are independent of GRAIL regulation at the steady state.
FIGURE 6.
Intracellular co-localization with GRAIL results in down-regulation of cell surface tetraspanins. A, HeLa cells grown on coverslips were transfected with V5-tagged GRAIL and either 3×HA-tagged CD81 or CD151. One hour prior to fixation, cells were incubated with Alexa Fluor 633-conjugated transferrin. Cells were fixed and stained with anti-V5 and anti-HA. B, 293T cells were transfected with vector alone or the indicated GRAIL constructs, all expressing GFP from a downstream IRES. Cells were dissociated in trypsin-free buffer, stained for endogenous CD81 or CD151, and analyzed by flow cytometry. The percentages of cells in GFP+ quadrants are indicated along with the mean fluorescence intensity (MFI) of the tetraspanin stained channel from gated GFP+ cells. C, 293T cells were transfected with vector alone, wild-type GRAIL, GRAIL mutants, or Rnf130, all expressing GFP from a downstream IRES. Cells were dissociated in trypsin-free buffer; stained for endogenous CD81, CD58, or major histocompatibility complex class I; and analyzed by flow cytometry. The percentages of cells in both GFP+ quadrants are indicated.
Although the cellular ubiquitination assays indicated that GRAIL promoted tetraspanin ubiquitination, these studies did not test the effect of GRAIL on endogenous tetraspanins. To address the question of whether GRAIL-mediated ubiquitination could modulate endogenous tetraspanin expression, 293T cells were transfected with vector, GRAIL, ΔPA, or H2N2 constructs, all of which expressed GFP from a downstream internal ribosome entry site (IRES). Using GFP as a flow cytometry marker for GRAIL expression in live cells, cell surface expression of endogenous CD81 and CD151 were both reduced in GRAIL-expressing (GFP+) cells with CD81 reproducibly showing the greatest decrease in the mean fluorescence intensity of tetraspanin expression on GFP-gated cells (Fig. 6B). Expression of either the ΔPA or RING finger mutant failed to significantly reduce cell surface tetraspanin expression compared with vector alone, demonstrating that tetraspanin capture on the luminal side of the membrane followed by cytosolic ubiquitination was required for cell surface down-regulation by GRAIL. Using CD58 (LFA-3) and major histocompatibility complex class I as markers for expression of transmembrane proteins on the cell surface, we observed no nonspecific down-regulation of CD58 or major histocompatibility complex class I cell surface expression seen with wild-type GRAIL or any of the mutants (Fig. 6C). In addition, expression of the closest relative of GRAIL by sequence homology and similar domain layout, Rnf130 (26), did not down-regulate CD81 compared with wild-type GRAIL (Fig. 6C).
DISCUSSION
The number of transmembrane, single subunit RING finger E3 ligases characterized to date remains exceedingly small compared with the total number of RING finger E3 ligases. Examples include the K3 family from viruses and the orthologous MARCH family in mammals along with Tul1 from yeast (27–31). The type 1 transmembrane protein Tul1 binds membrane proteins that contain inappropriately exposed polar residues in or near their transmembrane domain, whereas the K3 and MARCH families are type II transmembrane proteins that ubiquitinate a variety of cell surface markers important in the immune system. For both of these groups of single unit E3 ligases, the transmembrane region serves as the primary substrate interaction motif.
We hypothesized that the newly characterized type 1 transmembrane single unit E3 ligase GRAIL contained a unique mechanism for binding and regulating transmembrane proteins. The presence of a PA domain could facilitate capture of a luminal or extracellular domain of a target transmembrane protein, thus allowing subsequent ubiquitination of a cytosolic domain by the RING finger of GRAIL. Screening for proteins that bound to the GRAIL luminal PA domain identified the LEL of the tetraspanin CD151. Co-immunoprecipitation experiments confirmed that an intact GRAIL PA domain was required for binding to not only CD151 (Fig. 1D) but also to another tetraspanin, CD81 (Fig. 2A). In addition, increasing the stringency of lysis (Fig. 1C) and cross-linking assays demonstrated that binding was direct (Figs. 2C). Thus the PA domain provides a protein-protein interaction domain to capture target transmembrane proteins as substrates. Although sequence alignments of the LEL of tetraspanins have revealed little primary sequence homology outside of small motifs necessary for disulfide bond formation and transmembrane regions, a common tertiary structure adopted by tetraspanins could serve as the site of interaction with the PA domain of GRAIL as suggested by recent modeling of the CD81 LEL region (32).
Co-expression of GRAIL and tetraspanins in cellular assays promoted ubiquitination of these four-pass transmembrane proteins in a GRAIL-specific fashion (Fig. 3A). Ubiquitin modification of tetraspanins required both a luminal substrate capture motif, the PA domain, and a RING finger for recruitment of cytosolic ubiquitination machinery (Fig. 3B). Titrating in GRAIL caused a decrease in steady-state tetraspanin from a fixed pool (Fig. 3C), suggesting that GRAIL can significantly reduce the half-life of substrate tetraspanins. GRAIL expression resulted in down-regulation of cell surface tetraspanins with varying degrees of potency (Fig. 6B), and this effect was not due to a more general effect on protein trafficking (Fig. 6C). This down-regulation could result from the overlap between GRAIL and tetraspanins in the recycling transferrin compartment, but complete elimination of tetraspanin expression was not likely because of distinct subcellular regions containing tetraspanins but lacking GRAIL (Fig. 6A). However, these data leave open the question of how GRAIL affects tetraspanins in an endogenous setting. GRAIL expression in naïve CD4+ T cells is low compared with the anergic state, and when combined with the long half-life shared among many tetraspanins, an increase in either tetraspanin duration or quantity after GRAIL knockdown was at present difficult to quantify.
Although there have been reports of “site-specific” ubiquitination (33, 34), E3 ligases can also ubiquitinate lysine residues that are in broad spatial alignment for ubiquitin transfer (35). Mutation of the conserved lysine in the amino terminus of human CD81 or CD151 had no effect on the ability of GRAIL to ubiquitinate either tetraspanin provided there was another lysine available in the amino terminus. However, mutation of all three lysines in the amino terminus of CD151 abrogated the ability of GRAIL to ubiquitinate CD151 even though one lysine was still present in each of the other two cytosolic domains (Fig. 4B). These data suggest that GRAIL contains regional specificity for tetraspanin ubiquitination where only the amino termini of the tetraspanins contain the proper spatial orientation for transfer of ubiquitin by GRAIL. This model would be difficult to predict a priori as the amino terminus of the tetraspanin is separated across a plasma membrane and not directly proximal to the LEL. A recent structural model for CD81 suggests that the amino terminus adopts an amphipathic membrane-parallel helix, which could be accessible to the cytosolic RING finger of GRAIL (36). How the cytosolic GRAIL RING finger recruits an E2 transferase with an activated ubiquitin molecule in the proper orientation for ubiquitination of only the amino terminus of tetraspanin molecules is an interesting question. The lack of ubiquitination or down-regulation of integrin β1, a highly stoichiometric CD151-binding protein, by GRAIL despite their co-localization further supports the specificity for tetraspanin ubiquitination (data not shown).
In summary, GRAIL is a unique single subunit transmembrane E3 ligase that completely separates its substrate recognition motif from its intrinsic ubiquitination machinery across a lipid bilayer. Both functional elements are required for substrate turnover as capture via the luminal or extracellular PA domain serves as a recruitment mechanism to preserve access to key cytosolic residues for ubiquitination via the RING finger. Because E3 ligases have multiple substrate targets, future studies will focus on elucidating how GRAIL utilizes this mechanism to regulate other single and multipass transmembrane proteins.
Acknowledgments
We are indebted to Cariel Taylor for excellent technical assistance. We gratefully acknowledge Dr. Shoshana Levy and the rest of the Levy laboratory for helpful discussions, antibodies, and plasmids. We also thank Drs. James Quigley and Andries Zijlstra for providing antibodies. We thank Drs. Patricia Jones, Gerald Crabtree, and Peter Jackson for critical discussion of the manuscript. Special thanks to Robyn Rajkovich and Lisa O'Brien for administrative support.
This work was supported, in whole or in part, by National Institutes of Health Grants CA 65237-17 and T32-AI07290-21 (both to N. L.). This work was also supported by the Tom and Susan Ford Fellowship (to N. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; Cbl, casitas B-lineage lymphoma; GRAIL, gene related to anergy in lymphocytes; PA, protease-associated; RING, really interesting new gene; SCF, skip-cullin-F box; LEL, large extracellular loop; Mdm2, mouse double minute 2; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; HA, hemagglutinin; HECT, homologous to E6-AP carboxyl terminus; GFP, green fluorescent protein; DSP, dithiobis(succinimidylpropionate); IRES, internal ribosome entry site; LFA-3, lymphocyte function-associated 3; MARCH, membrane-associated RING-CH; ALLN, N-acetyl-Leu-Leu-norleucine-CHO; h, human.
References
- 1.Hershko, A., Ciechanover, A., and Varshavsky, A. (2000) Nat. Med. 6 1073-1081 [DOI] [PubMed] [Google Scholar]
- 2.Jackson, P. K., Eldridge, A. G., Freed, E., Furstenthal, L., Hsu, J. Y., Kaiser, B. K., and Reimann, J. D. (2000) Trends Cell Biol. 10 429-439 [DOI] [PubMed] [Google Scholar]
- 3.Hicke, L., and Dunn, R. (2003) Annu. Rev. Cell Dev. Biol. 19 141-172 [DOI] [PubMed] [Google Scholar]
- 4.Richly, H., Rape, M., Braun, S., Rumpf, S., Hoege, C., and Jentsch, S. (2005) Cell 120 73-84 [DOI] [PubMed] [Google Scholar]
- 5.Kim, J. H., Park, K. C., Chung, S. S., Bang, O., and Chung, C. H. (2003) J. Biochem. (Tokyo) 134 9-18 [DOI] [PubMed] [Google Scholar]
- 6.Jackson, P. K., and Eldridge, A. G. (2002) Mol. Cell 9 923-925 [DOI] [PubMed] [Google Scholar]
- 7.Laney, J. D., and Hochstrasser, M. (1999) Cell 97 427-430 [DOI] [PubMed] [Google Scholar]
- 8.Anandasabapathy, N., Ford, G. S., Bloom, D., Holness, C., Paragas, V., Seroogy, C., Skrenta, H., Hollenhorst, M., Fathman, C. G., and Soares, L. (2003) Immunity 18 535-547 [DOI] [PubMed] [Google Scholar]
- 9.Heissmeyer, V., Macian, F., Im, S. H., Varma, R., Feske, S., Venuprasad, K., Gu, H., Liu, Y. C., Dustin, M. L., and Rao, A. (2004) Nat. Immunol. 5 255-265 [DOI] [PubMed] [Google Scholar]
- 10.Safford, M., Collins, S., Lutz, M. A., Allen, A., Huang, C. T., Kowalski, J., Blackford, A., Horton, M. R., Drake, C., Schwartz, R. H., and Powell, J. D. (2005) Nat. Immunol. 6 472-480 [DOI] [PubMed] [Google Scholar]
- 11.Bruinenberg, P. G., Doesburg, P., Alting, A. C., Exterkate, F. A., de Vos, W. M., and Siezen, R. J. (1994) Protein Eng. 7 991-996 [DOI] [PubMed] [Google Scholar]
- 12.Mahon, P., and Bateman, A. (2000) Protein Sci. 9 1930-1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stimpson, H. E., Lewis, M. J., and Pelham, H. R. (2006) EMBO J. 25 662-672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirao, K., Natsuka, Y., Tamura, T., Wada, I., Morito, D., Natsuka, S., Romero, P., Sleno, B., Tremblay, L. O., Herscovics, A., Nagata, K., and Hosokawa, N. (2006) J. Biol. Chem. 281 9650-9658 [DOI] [PubMed] [Google Scholar]
- 15.Yauch, R. L., Berditchevski, F., Harler, M. B., Reichner, J., and Hemler, M. E. (1998) Mol. Biol. Cell 9 2751-2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitter, S., Sincock, P. M., Jolliffe, C. N., and Ashman, L. K. (1999) Biochem. J. 338 61-70 [PMC free article] [PubMed] [Google Scholar]
- 17.Sanderfoot, A. A., Ahmed, S. U., Marty-Mazars, D., Rapoport, I., Kirchhausen, T., Marty, F., and Raikhel, N. V. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 9920-9925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence, C. M., Ray, S., Babyonyshev, M., Galluser, R., Borhani, D. W., and Harrison, S. C. (1999) Science 286 779-782 [DOI] [PubMed] [Google Scholar]
- 19.Hemler, M. E. (2001) J. Cell Biol. 155 1103-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berditchevski, F. (2001) J. Cell Sci. 114 4143-4151 [DOI] [PubMed] [Google Scholar]
- 21.Su, L., Lineberry, N., Huh, Y., Soares, L., and Fathman, C. G. (2006) J. Immunol. 177 7559-7566 [DOI] [PubMed] [Google Scholar]
- 22.Stipp, C. S., Kolesnikova, T. V., and Hemler, M. E. (2003) Trends Biochem. Sci. 28 106-112 [DOI] [PubMed] [Google Scholar]
- 23.Wright, M. D., and Tomlinson, M. G. (1994) Immunol. Today 15 588-594 [DOI] [PubMed] [Google Scholar]
- 24.Pickart, C. M., and Fushman, D. (2004) Curr. Opin. Chem. Biol. 8 610-616 [DOI] [PubMed] [Google Scholar]
- 25.Berditchevski, F., and Odintsova, E. (1999) J. Cell Biol. 146 477-492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guais, A., Siegrist, S., Solhonne, B., Jouault, H., Guellaen, G., and Bulle, F. (2006) Gene (Amst.) 374 112-120 [DOI] [PubMed] [Google Scholar]
- 27.Lehner, P. J., Hoer, S., Dodd, R., and Duncan, L. M. (2005) Immunol. Rev. 207 112-125 [DOI] [PubMed] [Google Scholar]
- 28.Bartee, E., Mansouri, M., Hovey Nerenberg, B. T., Gouveia, K., and Fruh, K. (2004) J. Virol. 78 1109-1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reggiori, F., and Pelham, H. R. (2002) Nat. Cell Biol. 4 117-123 [DOI] [PubMed] [Google Scholar]
- 30.Valdez-Taubas, J., and Pelham, H. (2005) EMBO J. 24 2524-2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohmura-Hoshino, M., Goto, E., Matsuki, Y., Aoki, M., Mito, M., Uematsu, M., Hotta, H., and Ishido, S. (2006) J. Biochem. (Tokyo) 140 147-154 [DOI] [PubMed] [Google Scholar]
- 32.Seigneuret, M., Delaguillaumie, A., Lagaudriere-Gesbert, C., and Conjeaud, H. (2001) J. Biol. Chem. 276 40055-40064 [DOI] [PubMed] [Google Scholar]
- 33.Scherer, D. C., Brockman, J. A., Chen, Z., Maniatis, T., and Ballard, D. W. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 11259-11263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geetha, T., Jiang, J., and Wooten, M. W. (2005) Mol. Cell 20 301-312 [DOI] [PubMed] [Google Scholar]
- 35.Pickart, C. M. (2001) Annu. Rev. Biochem. 70 503-533 [DOI] [PubMed] [Google Scholar]
- 36.Seigneuret, M. (2006) Biophys. J. 90 212-227 [DOI] [PMC free article] [PubMed] [Google Scholar]