Abstract
The ζ chain-associated 70-kDa protein (ZAP-70) of tyrosine kinase plays a critical role in T cell receptor-mediated signal transduction and the immune response. A high level of ZAP-70 expression is observed in leukemia, which suggests ZAP-70 as a logical target for immunomodulatory therapies. (-)-Epigallocatechin gallate (EGCG) is one of the major green tea catechins that is suggested to have a role as a preventive agent in cancer, obesity, diabetes, and cardiovascular disease. Here we identified ZAP-70 as an important and novel molecular target of EGCG in leukemia cells. ZAP-70 and EGCG displayed high binding affinity (Kd = 0.6207 μmol/liter), and additional results revealed that EGCG effectively suppressed ZAP-70, linker for the activation of T cells, phospholipase Cγ1, extracellular signaling-regulated kinase, and MAPK kinase activities in CD3-activated T cell leukemia. Furthermore, the activation of activator protein-1 and interleukin-2 induced by CD3 was dose-dependently inhibited by EGCG treatment. Notably, EGCG dose-dependently induced caspase-mediated apoptosis in P116.cl39 ZAP-70-expressing leukemia cells, whereas P116 ZAP-70-deficient cells were resistant to EGCG treatment. Molecular docking studies, supported by site-directed mutagenesis experiments, showed that EGCG could form a series of intermolecular hydrogen bonds and hydrophobic interactions within the ATP binding domain, which may contribute to the stability of the ZAP-70-EGCG complex. Overall, these results strongly indicated that ZAP-70 activity was inhibited specifically by EGCG, which contributed to suppressing the CD3-mediated T cell-induced pathways in leukemia cells.
For thousands of years, tea has been the most widely consumed beverage in the world after water. Historically, tea has been credited with various beneficial health effects, including medicinal efficacy in the prevention and treatment of numerous diseases. Thus, longevity and good health have often been associated with the habit of drinking tea (1). Four major polyphenolic catechins are found in green tea and include (-)-epicatechin (EC),3 (-)-epicatechin 3-gallate (ECG), (-)-epigallocatechin (EGC), and (-)-epigallocatechin 3-gallate (EGCG). A cup of green tea may contain 100–200 mg of EGCG (2). Several investigators have reported that green tea exerts cancer preventive activity at a variety of organ sites, including skin, lung, oral cavity, esophagus, stomach, small intestine, colon, pancreas, and mammary gland (1, 3, 4). However, the mechanisms explaining the cancer preventive activity of tea and tea polyphenols are still not clearly understood.
The ζ-associated 70-kDa protein (ZAP-70) is a Syk (spleen tyrosine kinase) family tyrosine kinase, which is associated with the ζ subunit of the T cell receptor (TCR). The ZAP-70 protein is primarily expressed in T cells and natural killer cells and plays an essential role in signaling through the T cell antigen receptor (5). The TCRs are associated with tyrosine phosphorylation of multiple proteins resulting in activation of various signaling pathways causing alterations in gene expression, increased T cell proliferation, and secretion of cytokines (6). CD3 (cluster of differentiation 3) stimulation of the T cell antigen receptor plays a role in tyrosine phosphorylation of a number of cellular substrates. An important substrate of ZAP-70 is the TCR ζ chain, which can mediate the transduction of extracellular stimuli into cellular effector functions (7, 8). ZAP-70 plays a critical role in cell surface expression of T cell antigen receptor-CD3 complex signaling during the early stages of T cell development and differentiation (9–13). The ZAP-70 tyrosine kinase is reported to play a critical role in T cell activation and the immune response, and therefore might be a logical target for immunomodulatory therapies (5). Crespo et al. (14) observed that among B cell and T cell lymphoproliferative disorders, a high level of ZAP-70 expression is found in T cell proliferative diseases, acute lymphoblastic leukemia, and a subgroup of chronic lymphocytic leukemia (CLL) (15, 16). These studies suggested that ZAP-70 could be an excellent prognostic biomarker in CLL.
Despite advances in T cell leukemia therapy, only a minority of patients achieve long term tumor-free survival with conventional chemotherapy but at the cost of significant, irreversible toxic side effects, which often limit effective treatment (17). Therefore, new therapeutic approaches with enhanced tumor selectivity and more favorable toxicity profiles are urgently needed.
EGCG, a major active constituent of green tea, has been shown to stimulate apoptosis and cell cycle arrest in various cancer cell lines, including prostate, colon, lung, leukemias, and lymphomas (3, 18). However, the anticancer mechanisms and molecular targets of EGCG are poorly understood, especially in T cell-mediated leukemias and lymphomas. Here we demonstrate that EGCG might be a potential immunomodulator for the management of ZAP-70-dependent T cell activation in human leukemias and lymphomas. The effects of EGCG were extensively investigated in a ZAP-70-deficient Jurkat T cell line (P116 cells) and a cell line in which ZAP-70 activity was recovered (P116.cl39 cells). Results suggested that ZAP-70 antagonists could be useful immunomodulatory therapeutic agents.
EXPERIMENTAL PROCEDURES
Materials—All media were obtained from American Type Culture Collection (Manassas, VA), and fetal bovine serum was from Gemini Bio-Products (Calabasas, CA). CNBr-Sepharose 4B, glutathione-Sepharose 4B, and [γ-32P]ATP were purchased from Amersham Biosciences. [3H]EGCG (13 Ci/mmol in ethanol containing 8 mg/ml ascorbic acid) was a gift from Dr. Yukihiko Hara (Food Research Laboratory, Mitsui Norin Co. Ltd., Fujieda, Shizuoka, Japan). Recombinant ZAP-70 and the poly-(Glu4-Tyr) peptide were from Upstate Biotechnology, Inc. (Charlottesville, VA). The following antibodies were used: anti-ZAP-70 (1E7.2), anti-linker for the activation of T cells (LAT) (FL-233), anti-phospholipase Cγ1 (PCLγ1) (E-12), anti-α-tubulin (TU-02) (all from Santa Cruz Biotechnology, Santa Cruz, CA), anti-β-actin (MOC, Sigma), anti-extracellular signaling-regulated kinase (ERK), anti-phospho-ERK (Thr202/Tyr204), anti-MAPK kinase (MEK), anti-phospho-MEK (Ser217/221), anti-phospho-PLCγ1 (Tyr783), and anti-phosphotyrosine 100 (Cell Signaling Technology, Inc. Charlottesville, VA). The ATP immobilized on agarose 4B was purchased from Fluka (St. Louis). The mouse anti-human CD3 monoclonal antibody, mouse IgG1κ, monoclonal immunoglobulin isotype control, anti-phospho-ZAP-70 (Tyr319), anti-phospho-ZAP-70 (Tyr493), anti-CD3ζ, and anti-phospho-CD3ζ (Tyr142) were purchased from Pharmingen™. The TnT® Quick Coupled transcription/translation system was from Promega (Madison, WI). EGCG, EC, ECG, and EGC were generous gifts from Dr. Chi-Tang Ho (Rutgers University, Piscataway, NJ).
Cell Culture—The Jurkat (clone E6–1) human T cell leukemia, ZAP-70-deficient Jurkat mutant P116, and ZAP-70 recovered P116 mutant P116.cl39 cells were kindly provided by Dr. Robert T. Abraham (Department of Immunology, Mayo Clinic, Rochester, MN) (19). Jurkat and P116 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, and the P116.cl39 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and 0.4 mg/ml G418.
Construction of Deletion Mutants and Site-directed Mutagenesis—A cDNA encoding ZAP-70 was generated by PCR and subcloned into the BamHI/EcoRI sites of the pcDNA4/HisMax vector (Amersham Biosciences) to produce the ZAP-70 protein. The deletion mutants of ZAP-70 were generated from ZAP-70 wild type and inserted in-frame into the BamHI/EcoRI sites of the pcDNA4/HisMax vector. The products were cut with BamHI and EcoRI and then subcloned into pcDNA4/HisMax, generating the constructs ZAP-70(1–256), ZAP-70(1–415), and ZAP-70-(1–466) according to the insert sizes. cDNAs encoding the Glu415, Lys369, Asp479, Glu386, and Arg465 mutants of ZAP-70 were generated using the QuikChange® multisite-directed mutagenesis kit (Stratagene, La Jolla, CA) and subcloned into the pcDNA4/HisMax vector as follows: ZAP-70 E415Q, ZAP-70 E415Q/K369R, ZAP-70 E415Q/D479N, ZAP-70 E415Q/K369R/D479N, and ZAP-70 E415Q/K369R/D479N/E386Q/R465K. The constructs were confirmed by DNA sequence analysis (GENEWIZ, South Plainfield, NJ).
In Vitro EGCG-Sepharose 4B and ATP-Agarose 4B Pulldown Assays—This method has been described previously (20). Briefly, the cells were transfected with 2 μg of each ZAP-70 point mutant using the jetPEI (Polyplus-Transfection SA, San Marcos, CA) reagent following the manufacturer's suggested protocol. The cellular supernatant fraction (Jurkat, P116, P116.cl39, or transfected cells), recombinant ZAP-70, or plasmids (pcDNA4/HIS/Max-ZAP-70 and deletion mutants of ZAP-70) were translated in vivo with l-[35S]methionine. Respective proteins were incubated with EGCG-Sepharose 4B beads or ATP-agarose 4B beads in reaction buffer (50 mm Tris, pH 7.5, 5 mm EDTA, 150 mm NaCl, 1 mm dithiothreitol, 0.01% Nonidet P-40, 2 μg/ml bovine serum albumin, 0.02 mm phenylmethylsulfonyl fluoride, 1× proteinase inhibitor). The beads were washed five times with buffer (50 mm Tris, pH 7.5, 5 mm EDTA, 150 mm NaCl, 1 mm dithiothreitol, 0.01% Nonidet P-40, 0.02 mm phenylmethylsulfonyl fluoride), and proteins bound to the beads were analyzed by autoradiography or immunoblotting with the appropriate antibodies.
Physical Binding and Kd Measurement—ZAP-70 binding assays were carried out as described (20) with some modifications. For analyzing concentration-dependent uptake, 1 nm to 10 μm concentrations of EGCG were applied. The Kd value was determined through nonlinear regression analysis using the Prizm 4.0 software program (Graphpad Inc., San Diego).
Molecular Modeling of the Interactions of EGCG with ZAP-70—Molecular docking studies were carried out using the Maestro software suite (Maestro, version 7.5, Schrödinger, New York). The EGCG molecule was drawn using the builder tool in Maestro and then optimized for docking in Ligprep. The ZAP-70 crystal structure complexed with staurosporine (Protein Data Bank code 1u59) was prepared for docking following the Glide standard procedure (21). Grids defining the protein receptor were generated considering the binding mode of staurosporine. Many protein-binding sites undergo structural rearrangements upon ligand binding, the so-called “induced fit” allows the binding site to follow the shape of the ligand with resulting better interactions. Consequently, molecular docking was performed with the induced fit docking protocol (Schrödinger Suite 2006). The induced fit docking is a part of the Maestro software suite that attempts to reproduce the protein conformational rearrangement upon binding. Initially, EGCG was docked with Glide in extra-precision mode using a softened potential: scaling the receptor (0.70) and the ligand (0.50) van der Waals radius. The best 20 EGCG poses were retained. Then the receptor side chains within 5 Å distance from the ligand were predicted and minimized for each protein-ligand complex, and then another round of minimizations was performed on each protein-ligand complex pose. Finally, Glide redocking in extra-precision mode of each protein-ligand complex within 30.0 kcal/mol of the lowest energy structure was performed. Herein the best-docked representative structure is presented.
Kinase Assay—The ZAP-70 kinase assay was performed at 30°C for 2 h in a 25-μl reaction mixture containing kinase buffer (50 mm Tris, pH 7.5, 10 mm MnCl2, 1 mm EGTA, 2 mm dithiothreitol, and 0.01% Brij), 1 μg of recombinant ZAP-70, 5 μg of the kinase substrate (poly(Glu4-Tyr) peptide, biotin conjugate), EGCG (0.5–20 μm), 100 μm ATP, and 1 μCi of [γ-32P]ATP. The reactions were stopped by adding 12.5 μl of termination buffer, and samples were quantified (counts/min) by scintillation counting.
Immunoprecipitation and Western Blotting—The P116 and P116.cl39 cells (5 × 105/ml) were grown in 75-cm3 flasks for 24 h and then serum-starved for 14 h at 37 °C. Where indicated, the cells were pretreated with EGCG (4 or 8 μm) for 2 h and then washed three times with phosphate-buffered saline. The cells were stimulated for 30 min at 37 °C with 2 μg/ml mouse IgG1κ monoclonal immunoglobulin isotype control and 2 μg/ml mouse anti-human CD3. The reactions were stopped by adding cold phosphate-buffered saline followed by centrifugation for subsequent lysis. Lysates were incubated overnight at 4 °C with the indicated antibodies and protein A/G Plus-agarose beads. After centrifugation, the beads were washed three times with washing buffer and were resuspended in SDS sample buffer. Eluted immunoprecipitates or whole-cell lysates were resolved by SDS-PAGE and transferred onto polyvinylidene difluoride membranes. The membranes were blocked with 5% skim milk and then incubated with appropriate antibodies. Immunocomplexes were detected by subsequent incubation with appropriate horseradish peroxidase-conjugated secondary IgG antibodies and visualized by ECL according to the manufacturer's instructions (Amersham Biosciences).
Electrophoretic Mobility Shift Assay—Nuclear extracts were prepared as described previously (22). The cells were treated and prepared as described above for Western blotting and then disrupted with a hypotonic buffer. The nuclei pellet was disrupted in a hypertonic buffer, and the nuclear extracts were retained for use in the DNA binding assay. A double-stranded deoxyoligonucleotide corresponding to AP (activator protein)-1 responsive elements (Santa Cruz Biotechnology) was end-labeled with [γ-32P]ATP using T4 kinase. Nuclear extracts (5 μg) were incubated in binding buffer for 15 min with poly(dI·dC) and the 32P-labeled DNA probe. The DNA binding activity was separated from free probe using a TBE Ready Gel Precast Gel (Bio-Rad). Following electrophoresis, the gel was dried and visualized by autoradiography.
Measurement of IL-2 Production—Quantification of interleukin (IL)-2 production from P116 and P116.cI39 cells was performed using a commercial enzyme-linked immunosorbent assay system (Pharmingen). The cells (5 × 105 cells/ml) were resuspended in fresh medium in a 48-well plate and incubated for 24 h. The cells were treated with various concentrations of EGCG and 2 μg/ml mouse IgG1κ monoclonal immunoglobulin isotype control or 2 μg/ml mouse anti-human CD3 for 24 or 48 h at 37 °C. The levels of IL-2 in the supernatant fractions were subsequently measured using an enzyme-linked immunosorbent assay kit (Pharmingen) according to the manufacturer's instructions.
MTS Assay—Cells (1 × 104) were seeded in a 96-well plate and then incubated for 24 h with different concentrations of EGCG (0, 0.5, 1, 2, or 4 μm) or EC, ECG, or EGC (4 μm) for 72 h. The effect of EGCG on viability was estimated using the Cell-Titer 96 AQueous One Solution cell proliferation assay kit (Promega, Madison, WI) according to the manufacturer's instructions. The assay solution was added to each well, and absorbance (492 nm and 690 background) was read with a 96-well plate reader (Labsystem Multiskan MS, Labsystem, Finland).
Annexin V Staining—Apoptosis was performed using the annexin V-FITC apoptosis detection kit as recommended by the manufacturer (MBL International Corp., Watertown, MA). Apoptosis was compared in P116 and P116.cl39 cells that were treated or not treated with 4 or 8 μm EGCG for 24, 48, 72, or 96 h. The cells were harvested and washed with phosphate-buffered saline and incubated for 5 min at room temperature with annexin V-FITC plus propidium iodide following the protocol included in the kit. Cells were analyzed on a FACSCalibur flow cytometer (BD Biosciences).
Statistical Analysis—Data are presented as means ± S.D. of triplicate samples from at least three independent experiments. Differences between means were assessed by one-way analysis of variance, and the minimum level of significance was set at p < 0.05.
RESULTS
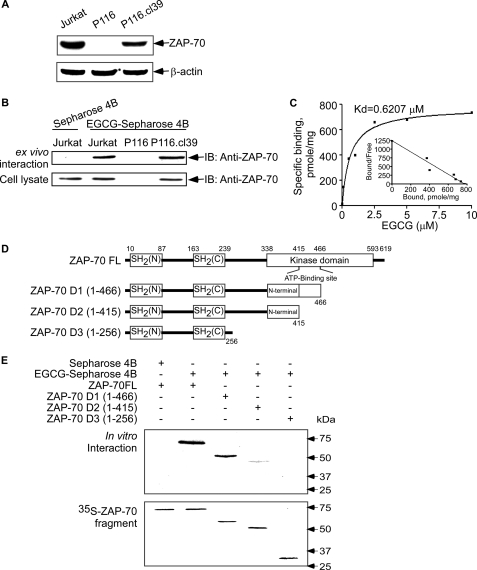
EGCG Binds with ZAP-70 ex Vivo and in Vitro—Expression of ZAP-70 was recently reported to be a reliable prognostic marker among patients with CLL (14). In a kinase screening experiment (kinase profiler specificity testing service, Upstate), EGCG (5 μm) was found to inhibit ZAP-70 kinase activity. We therefore studied the potential interaction of EGCG and ZAP-70 in wild type Jurkat cells, Jurkat cells (P116) that do not express ZAP-70, and in P116 cells (P116.cl39) in which ZAP-70 protein expression has been restored (Fig. 1A). The interaction of ZAP-70 and EGCG was determined in an EGCG-Sepharose 4B affinity chromatography experiment combined with immunoblotting with anti-ZAP-70. Results indicated that EGCG formed a complex with ZAP-70 in P116.cl39 cells but not in P116 cells (Fig. 1B). To characterize the physical binding between EGCG and ZAP-70, we measured the binding affinity (Kd) of the complex using a GST pulldown assay and 3H-labeled EGCG. The Kd value of ZAP-70 and EGCG binding was determined to be 0.6207 μmol/liter (Fig. 1C).
FIGURE 1.
Detection of ZAP-70 protein expression and EGCG binding with ZAP-70 and ZAP-70 deletion mutants. A, expression of ZAP-70 in Jurkat, P116 (ZAP-70-deficient), and P116.cl39 (ZAP-70-restored) cells. ZAP-70 was detected using specific antibodies as described under “Experimental Procedures.” Equal protein loading and protein transfer were confirmed with anti-β-actin. B, ZAP-70-EGCG binding ex vivo. EGCG-Sepharose 4B affinity chromatography was used to pull down ZAP-70 in lysates prepared from Jurkat, P116, or P116.cl39 cells. IB, immunoblot. C, specific binding assay for ZAP-70 and EGCG. The Kd (dissociation kinetic) value of the EGCG-ZAP-70 interaction (Kd = 0.6207 μm) was obtained by using a GST-ZAP-70 affinity-binding assay as described under “Experimental Procedures.” D, schematics of ZAP-70 full-length (ZAP-70 FL) and three deletion mutants (ZAP-70 D1, D2, and D3). A series of full-length and deletion mutants of ZAP-70 nucleotide constructs was created as indicated. E, in vitro identification of the EGCG-binding site of ZAP-70. The full-length and deletion mutants of ZAP-70 were translated in vivo with l-[35S]methionine using TnT and subjected to the EGCG-Sepharose 4B pulldown assay.
Identification of the EGCG-binding Site of ZAP-70—The ZAP-70 protein has a kinase domain, an SH2-kinase linker region, an inter-SH2 linker region, a C-terminal SH2 domain, and an N-terminal SH2 domain (6). To determine the binding region of ZAP-70 specific for EGCG, we created three serially deleted ZAP-70 mutants s follows: ZAP-70 D1 (amino acids 1–466), ZAP-70 D2 (amino acids 1–415), and ZAP-70 D3 (amino acids 1–256) (Fig. 1D). These mutants were used in combination with the EGCG-Sepharose 4B pulldown assay to determine the area of ZAP-70 to which EGCG binds. Results indicated that EGCG binds strongly with full-length ZAP-70 (ZAP-70FL) and the ZAP-70 D1 (amino acids 1–466) domain. On the other hand, ZAP-70 D2 (amino acids 1–415) showed weak binding with EGCG, whereas ZAP-70 D3 (amino acids 1–256) did not bind with EGCG, which suggested that EGCG binds primarily with the kinase domain (Fig. 1E).
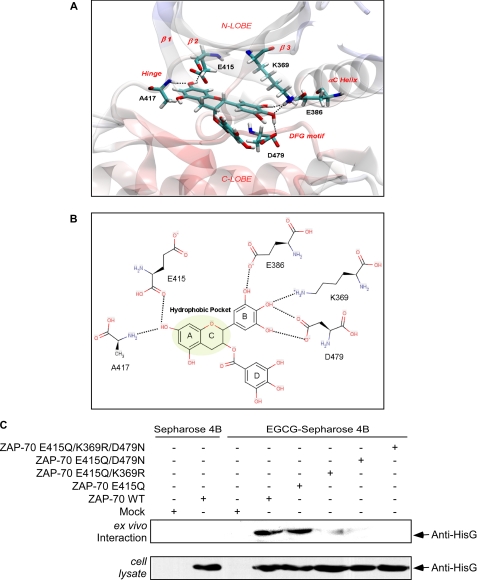
Structural Analysis of the Interaction between EGCG and ZAP-70—To better characterize the possible interaction between EGCG and the kinase/ATP binding domain of ZAP-70, we used the x-ray co-crystal structure of ZAP-70 complexed with staurosporine (Protein Data Bank code 1u59) as a starting point for a docking experiment. The accuracy of small molecule/protein docking increases when considering holo-proteins (23), and staurosporine is a rigid and flat molecule that creates an open conformation to the ZAP-70 ATP binding pocket. An open conformation is usually adopted by kinases when bound to small ligands, such as EGCG. In our docking model, EGCG presented a calculated binding affinity of -13.07 kcal/mol. One of the hydroxyl groups of the EGCG A ring, similar to other known kinase inhibitors, appears to form two hydrogen bonds with the kinase hinge region and, in particular, with the backbone carbonyl group of Glu415 and the amide group of Ala417 (Fig. 2A). The A–C ring system acts as an adenine mimic and likewise interacts with the front cleft hydrophobic pocket (Fig. 2B). The gallate moiety (D ring) occupies mainly the hydrophilic and solvent-exposed pocket covered by the G-loop, which is believed to be less important for ligand affinity (24). The side chains of lysine (Lys369) and aspartate (Asp479) may form a network of hydrogen bonds with the hydroxyl groups of the B ring. In addition, the side chain of Glu386 is expected to form a hydrogen bond with one of the hydroxyl groups of this ring (Fig. 2, A and B). This docking experiment suggested that disrupting specific hydrogen bonding networks by point mutation should affect the ability of EGCG to bind with ZAP-70 as modeled.
FIGURE 2.
Modeling of EGCG binding with the ZAP-70 protein. A, modeling of the three-dimensional spatial arrangement of EGCG within the ATP-binding site of ZAP-70. The amino acids involved in the hydrogen bonding interactions (dotted lines) with EGCG are depicted in atom-type stick representation, and the remaining part of the protein is shown as a transparent schematic. B, two-dimensional plot of the interactive network between EGCG and ZAP-70. The hydrophobic pocket occupied by the A-C ring system of EGCG is highlighted in green. C, ex vivo confirmation of the ZAP-70 amino acids that bind with EGCG. The point mutant plasmids of ZAP-70 were transfected into HEK 293 cells. Cell lysates were prepared and incubated with EGCG-Sepharose 4B beads in reaction buffer 2 h at 4 °C. The beads were then washed five times with buffer, and proteins bound to the beads were analyzed by immunoblotting with anti-HisG.
Therefore, to continue to assess the importance of the interaction between EGCG and the ZAP-70 kinase/ATP-binding site, we generated a variety of ZAP-70 point mutants, including ZAP-70 E415Q, ZAP-70 E415Q/K369R, ZAP-70 E415Q/D479N, ZAP-70 E415Q/K369R/D479N, and ZAP-70 E415Q/K369R/D479N/E386Q/R465K. The various ZAP-70 mutants were each transfected into HEK 293 cells to determine the effect on ZAP-70 binding with EGCG using EGCG-Sepharose 4B or Sepharose 4B combined with Western blot analysis (Fig. 2C). As predicted, the E415Q substitution did not affect the binding because EGCG forms hydrogen bonds with the main chain atoms in the hinge region. The binding seems to be dependent on the interactions of EGCG with Lys369 and Asp479, which we predicted would form a network of hydrogen bonds with the ligand B ring. The bulkier arginine in the K369R mutant very likely affects potential hydrogen bonding with EGCG but also may alter the shape of the cavity, preventing the ligand from assuming the correct orientation for satisfying all the necessary connections with the binding site. Replacing aspartate 479 with asparagine (D479N) also affects the ability of EGCG to bind. We predicted a hydrogen bond between the B ring and the aspartate side chain. The mutant, in which both Lys369 and Asp479 were substituted, consistently showed inhibition of EGCG binding. Thus, these data suggested that ZAP-70 Asp479 and Lys369 are required for the essential interactions of ZAP-70 with EGCG within the catalytic site.
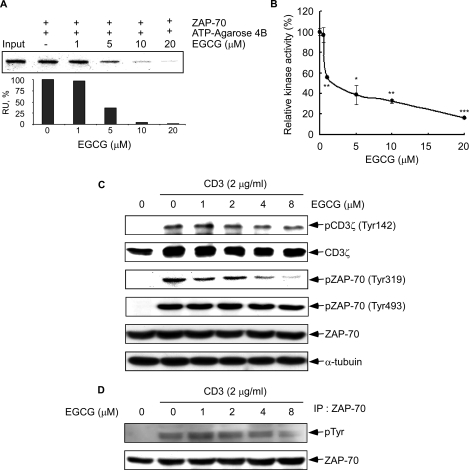
The information from the docking experiment suggested that EGCG might effectively inhibit ATP binding to active ZAP-70. Thus, we examined the effect of EGCG on ATP binding by pulldown assay using ATP-agarose 4B and the active ZAP-70 protein. Results confirmed that the binding of ATP with ZAP-70 decreased with increasing amounts of EGCG (Fig. 3A). This suggested that EGCG might inhibit ZAP-70 kinase activity by competing with ATP binding.
FIGURE 3.
Effect of EGCG on ZAP-70 kinase activity and CD3-induced phosphorylation of ZAP-70. A, EGCG binds to the ATPase catalytic domain of ZAP-70. Active ZAP-70 was incubated with various concentrations (0, 1, 5, 10, or 20 μm) of EGCG and ATP-agarose 4B beads in reaction buffer overnight at 4 °C. After washing, the proteins bound with ATP-agarose 4B beads were resolved by SDS-PAGE and analyzed by Western blot using a ZAP-70 antibody (top) and quantified (bottom) using the ImageJ software program (National Institutes of Health, Bethesda). B, EGCG inhibits ZAP-70 kinase activity in vitro. Kinase activity of ZAP-70 was assayed under the conditions described under “Experimental Procedures.” Data are represented as means ± S.D. of triplicate samples from three independent experiments. The asterisk (*, p < 0.05; **, p < 0.005; ***, p < 0.001) indicates a significant decrease in kinase activity compared with untreated control cells. C, phosphorylation of CD3ζ, pZAP-70 (Tyr319), and pZAP-70 (Tyr493) was detected by Western blot as described under “Experimental Procedures.” Equal protein loading and protein transfer were confirmed by stripping and incubating the same membrane with antibodies against α-tubulin, total CD3ζ, or ZAP-70. D, effect of EGCG on CD3-induced ZAP-70 total tyrosine phosphorylation. P116.cl39 cells were starved for 14 h followed by incubation for 1 h with EGCG at different concentrations (0, 1, 2, 4, or 8 μm). Cells were washed and then stimulated with 2 μg/ml CD3 for 30 min. Immunoprecipitation with anti-ZAP-70 was followed by anti-phosphotyrosine immunoblot analysis. Membranes were stripped and blotted with anti-ZAP-70.
We next determined whether EGCG could prevent ZAP-70 phosphorylation and/or suppress kinase activity. Kinase activity was analyzed using an in vitro kinase assay with active ZAP-70 and the preferred kinase substrate, poly(Glu4-Tyr) peptide. Results indicated that increasing concentrations of EGCG markedly suppressed ZAP-70 kinase activity, and 3 μm EGCG significantly inhibited ZAP-70 kinase activity by 50% (Fig. 3B). ZAP-70 is recruited to the phosphorylated CD3 and ζ subunits after TCR stimulation (25). The immunoreceptor tyrosine-based activation motifs (ITAM) of the signal-transducing antigen receptor subunit (CD3 and ζ) are phosphorylated by Src PTK thus allowing the Syk family PTK ZAP-70 to bind to the ITAM (26). EGCG had no effect on CD3-induced phosphorylation of CD3ζ (Fig. 3C). TCR-mediated Lck activity leads to phosphorylation of ZAP-70 on Tyr493 in the regulatory loop of the PTK domain resulting in the up-regulation of ZAP-70 kinase activity (27). Our data showed that EGCG had no effect on phosphorylation of ZAP-70 (Tyr493) (Fig. 3C). Tyrosine 319 is a key phosphorylation site of ZAP-70 and is phosphorylated upon recruitment of ZAP-70 to T cell antigen receptor signaling or by ZAP-70 itself (28–31). Experiments were performed to compare total tyrosine phosphorylation of ZAP-70 and autophosphorylation of Tyr319. EGCG inhibited autophosphorylation of ZAP-70 at Tyr319 in a dose-dependent manner (Fig. 3C). Notably, EGCG at4or8 μm inhibited Tyr319 autophosphorylation by 76 or 93%, respectively, compared with cells treated with only CD3 (Fig. 3C). On the other hand, total tyrosine phosphorylation was inhibited at about 55% by EGCG (Fig. 3D). The percent inhibition was determined by densitometric analysis of EGCG-treated cells compared with CD3-treated cells after normalization to ZAP-70 total levels (Fig. 3D). Overall, these results indicated that EGCG does not affect ZAP-70 recruitment to the TCR activation complex, but specifically inhibits ZAP-70.
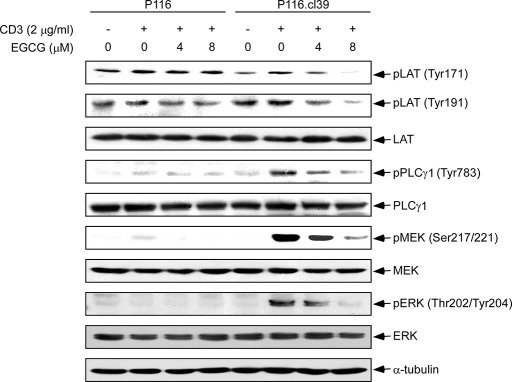
Effects of EGCG on the CD3-induced Phosphorylation of Downstream Kinases—Activation of T cells can be initiated through cell surface molecules in addition to the TCR-CD3 complex (7, 32, 33). Expression of the TCR-CD3 complex is associated with acute-type adult T cell leukemia and is observed in lymphoma-type adult T cell leukemia patients (34, 35). TCR activates signaling by recruiting and activating PTK of the Src, Syk, and Tec families (8, 36–39). Activated ZAP-70 phosphorylates adapter LAT at multiple conserved tyrosine residues (Tyr171 and Tyr191) within the SH2-binding motifs, exposing these motifs as the docking sites for PLCγ1 (40, 41). Precisely how the formation of the LAT-associated signaling complex leads to ERK1/2 activation is unclear. ERK1/2 and MEK both need to be activated in order for TCR engagement to result in T cell activation (42). We examined the effect of EGCG on the phosphorylation of several of these downstream kinases in ZAP-70 expressing (P116.cl39) or deficient (P116) cells. The cell lines were treated with increasing concentrations of EGCG (0–8 μm) for 2 h followed by stimulation with CD3 (2 μg/ml) for 30 min. An EGCG-induced dose-dependent decrease in phosphorylation of LAT (Tyr171, Tyr191), PLCγ1, MEK, and ERK1/2 (Tyr202/Tyr204) was observed in P116cl.39 cells compared with untreated control cells (Fig. 4). On the other hand, EGCG had no effect on phosphorylation of these various kinases in ZAP-70-deficient P116 cells.
FIGURE 4.
The effect of EGCG on CD3-induced phosphorylation of downstream kinases. Cells (P116 and P116.cl39) were starved for 14 h and then incubated for 1 h with EGCG at various concentrations (0, 4, or 8 μm). Cells were washed and then stimulated with 2 μg/ml of CD3 for 30 min. Thirty μg of cellular extract were separated in each lane on a 10% SDS-polyacrylamide gel as described under “Experimental Procedures.” Equal protein loading and transfer of proteins were confirmed by stripping and incubating the same membrane with anti-α-tubulin.
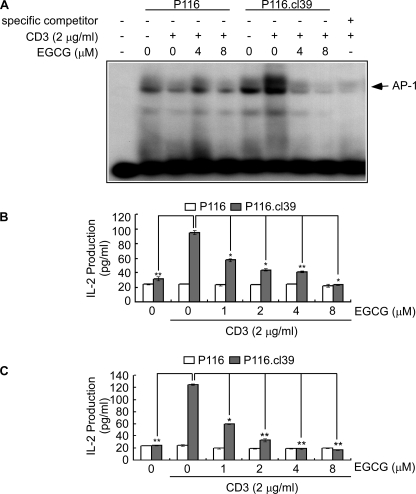
Effect of EGCG on CD3-induced IL-2 Production and DNA Binding Activity of AP-1—Activated ERK1/2 translocates to the nucleus and directly regulates various transcription factors. Multiple transcription factors regulate the IL-2 gene, including AP-1, nuclear factor-κB (NF-κB), Oct-1, and nuclear factor of activated T cells. We hypothesized that the inhibition of ZAP-70 by EGCG might decrease CD3-induced IL-2 secretion and DNA binding activity of AP-1. We therefore examined the effect of EGCG on IL-2 secretion and DNA binding activity of AP-1 in supernatant fractions from P116 and P116.cl39 cells. Results indicated that EGCG suppressed CD3-induced AP-1 DNA binding (Fig. 5A) and IL-2 production at 24 h (Fig. 5B) or 48 h (Fig. 5C) in P116.cl39 (ZAP-70 expressing) cells but had no effect on P116 ZAP-70-deficient cells.
FIGURE 5.
The effect of EGCG on CD3-mediated AP-1 DNA binding and IL-2 cytokine release. A, gel-shift assay. Nuclear extracts were obtained from cells pretreated with different concentrations of EGCG (0, 4, or 8 μm) for 1 h and then exposed to mouse IgG1κ monoclonal immunoglobulin isotype control (2 μg/ml) or mouse anti-human CD3 (2 μg/ml) for 24 h. The nuclear extracts were loaded onto a 5% polyacrymide gel and probed with γ-32P-labeled AP-1. Specific binding of nuclear proteins to the AP-1-responsive element was analyzed by the electrophoretic mobility shift assay. B and C, IL-2 secretion was measured from cells pretreated for 1 h with various concentrations of EGCG and then exposed to mouse IgG1κ monoclonal immunoglobulin isotype control (2 μg/ml) or mouse anti-human CD3 (2 μg/ml) for 24 (B) or 48 h (C). IL-2 production was measured in supernatant fractions by an enzyme-linked immunosorbent assay according to the manufacturer's instructions. Data are represented as the average of triplicate samples from three independent experiments. The asterisks indicate a significant change relative to P116.cl39 cell stimulated with CD3 (*, p < 0.005; **, p < 0.001).
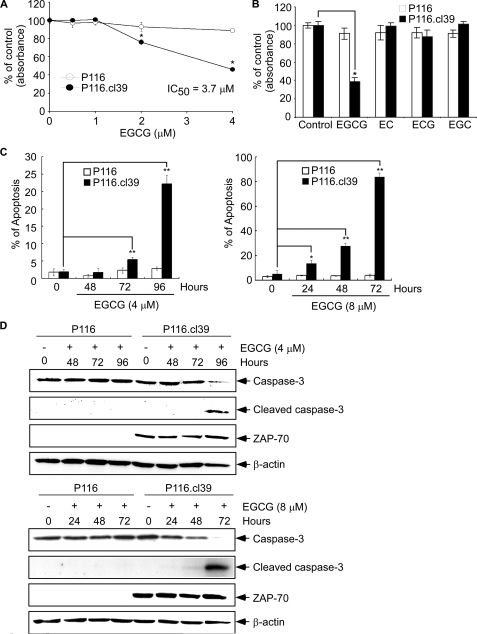
EGCG Inhibits Cell Viability and Induces Apoptosis in ZAP-70-expressing Cells but Has No Effect on ZAP-70-deficient Cells—Results thus far suggested that ZAP-70 induces phosphorylation of pLAT, pPLCγ1, pMEK, and pERK and DNA binding activity of AP-1. These signaling molecules are critical for cell survival (43, 44), and the blocking of these signals by EGCG may at least partially explain the ability of EGCG to induce apoptosis. ZAP-70-deficient P116 cells will be more resistant to the effect of EGCG on viability or apoptosis compared with the ZAP-70-expressing P116.cl39 cells. P116 and P116.cl39 cells were cultured for 72 h in a 96-well culture plate with increasing concentrations of EGCG, and then EGCG cytotoxicity was measured using the MTS assay. Treatment with EGCG (2 μm) resulted in about a 25% decrease in the viability of P116.cl39 cells (Fig. 6A). Only about 40% of P116.cl39 cells survived treatment with 4 μm EGCG, whereas more than 95% of the P116 cells remained viable after treatment with 4 μm EGCG. Treatment with the EGCG analogues, EC, ECG, or EGC, had no effect on P116 or P116.cl39 cells (Fig. 6B), suggesting a specific effect for EGCG. To determine whether the effect of EGCG on cell viability was because of apoptosis, flow cytometry was used in combination with Western blot analysis of caspase-3 and cleaved caspase-3. Caspase-3 is used as a marker for apoptosis in these experiments. Results indicated that the percentage of annexinV-stained positive cells (early apoptotic and late apoptotic/necrotic cells) increased substantially in P116.cl39 cells treated for 72 or 96 h with 4 or 8 μm EGCG (Fig. 6C). Very little apoptosis was observed in P116 ZAP-70-deficient cells. The key components of the biochemical pathways of caspase activation play a central role in the execution of apoptosis (45, 46). We therefore examined the effect of EGCG on caspase-3 and cleaved caspase-3 in P116 and P116.cl39 cells by Western blot analysis. Caspase-3 was detected in P116 cells but did not change with EGCG treatment, whereas capase-3 was substantially decreased in P116.cl39 cells following EGCG treatment (Fig. 6D). Cleaved caspase-3 was detected in the EGCG-treated P116.cl39 cells but not in the EGCG-treated P116 ZAP-70-deficient cells (Fig. 6D). These results suggested that EGCG treatment induces apoptosis only in ZAP-70-expressing leukemia cells through a caspase-3-dependent pathway. In addition, results confirmed that ZAP-70-deficient P116 cells are more resistant to the effects of EGCG on viability or apoptosis than the ZAP-70-expressing P116.cl39 cells. Overall, these results suggested that EGCG suppressed cell viability and induced cell apoptosis through a ZAP-70-mediated mechanism.
FIGURE 6.
The effect of EGCG and EGCG analogs on cell viability and apoptosis in P116 and P116. cl39 cells. A, effect of EGCG on cell viability. P116 or P116.cl39 cells (1 × 103 cells/200 μl) were each seeded in a 96-well microtiter plate and incubated for 72 h with increasing concentrations of EGCG. The effect of EGCG on cell viability was estimated with the MTS assay kit, as described under “Experimental Procedures.” B, specific effect of EGCG on cell viability was compared with EGCG analogues in P116 and P116.cl39 cells by the MTS assay. C, analysis of apoptosis by flow cytometry after 4 μm EGCG (left) or 8 μm EGCG (right) treatment of cells for various times. Cells were labeled with annexin V-FITC and propidium iodide, and the distribution pattern of live and apoptotic cells was determined by flow cytometry. The graphs indicate percentage of early apoptotic and late apoptotic/necrotic cells. EGCG-treated cells were compared with untreated cells, and data are shown as the average of triplicate samples from three independent experiments. The asterisks (*, p < 0.005; **, p < 0.0001) indicate a significant increase in apoptosis in EGCG-treated cells compared with untreated cells. D, effect of EGCG on expression of caspase-3 and cleaved caspase-3 in P116 and P116.cl39 cells as determined by Western blotting.
DISCUSSION
The ability of tea to prevent tumorigenesis in various in vitro and in vivo systems has been reported. However, the mechanisms of action to explain the effects are not clearly understood. In particular, although EGCG inhibits cellular viability and induces apoptosis in various leukemias, the molecular mechanism(s) by which this occurs in T cell-mediated leukemia is not clear. The biological activities of EGCG and other components need to be characterized more fully to enhance our understanding of the biological effects of tea consumption. Cancer is a multistep process, with accumulation of mutations in tumor-suppressor genes and the dominant expression of oncogenes. Tyrosine kinases include the largest group of oncoproteins (47). The recent development of a series of relatively specific protein-tyrosine kinase (PTK) inhibitors with the ability to suppress the proliferation of tumor cells expressing the target PTK in vivo shows that inhibition of deregulated, dominant oncogenic PTKs is often enough to slow tumor progression (48). Research results suggest that targeted molecular cancer therapies can potentially deliver treatment directly to a specific protein or gene target and optimize efficacy and thus reduce adverse side effects often associated with traditional chemotherapy. Many research groups have reported that the ZAP-70 family of tyrosine kinases are selectively retained and expressed in T cell proliferative diseases (49). A subgroup of CLL was reported to overexpress ZAP-70 thus enabling T cells to develop and differentiate through CD3-TCR-mediated signaling pathways (16). Therefore, ZAP-70 has been suggested to be an excellent biomarker for prognosis in CLL (9–11) and might be a logical target for the development of potent immunosuppressive agents for T cell-mediated diseases (5, 50). We showed that the green tea polyphenol, EGCG, directly interacts with and suppresses ZAP-70 activity. Molecular modeling and biochemical experiments confirmed that EGCG interacted with the ATP-binding pocket and effectively competed with ATP for binding.
Activation of the TCR induces numerous downstream signaling events, including the activation of various transcription factors, IL-2 promoter-driven transcription, protein tyrosine phosphorylation, intracellular calcium mobilization, MAPK activation, and phosphorylation of multiple important signaling molecules (51). To understand the effect of tea consumption on leukemias with ZAP-70 overexpression, we used ZAP-70-deficient Jurkat (P116) mutant cells and ZAP-70-recovered (P116.c139) cells as a model system to investigate the involvement of EGCG in T cell activation. Our results showed that EGCG impaired CD3-induced activation of TCR signaling through the inhibition of ZAP-70 kinase activity and autophosphorylation of ZAP-70 at Tyr319 for T cell antigen receptor-dependent signaling. The mutation of ZAP-70 Tyr319 site dramatically impaired TCR signaling (28). EGCG had no effect on phosphorylation of ZAP-70 on Tyr493. ZAP-70 is a protein-tyrosine kinase thought to play a critical role in T cell receptor (TCR) signal transduction. During T cell activation, ZAP-70 binds to a conserved signaling motif known as the ITAM and becomes phosphorylated at Tyr493 (27, 52). This result indicated that EGCG did not affect ZAP-70 recruitment to the TCR activation complex and that EGCG specifically inhibited ZAP-70. EGCG inhibited CD3-induced phosphorylation of several ZAP-70 downstream signaling molecules, including LAT (Tyr171/191), PLCγ1 (Tyr783), MEK (Ser217/221), and ERK (Thr202/Tyr204). EGCG also suppressed CD3-induced IL-2 production and activation of transcription factor AP-1. ZAP-70 was required for inhibition of T cell leukemia proliferation and induction of apoptosis by EGCG because EGCG had no effect on ZAP-70-deficient P116 cells, which also display severe defects in TCR signaling. These results indicated that EGCG specifically targets ZAP-70. Others have reported that EGCG induces apoptosis in human T cell acute lymphoblastic leukemia Jurkat cells (53), peripheral blood T lymphocytes, or leukemic blast cells of adult T cell leukemia patients (54, 55). However, how the formation of this signaling complex leads to apoptosis is unclear. Our results provide strong evidence that ZAP-70 is a direct mediator of EGCG-induced apoptosis in leukemia cells and suggest that EGCG suppresses CD3-TCR-mediated signaling (pLAT, pPLCγ1, pMEK, and pERK, and DNA binding activity of AP-1 and IL-2) in leukemia through its inhibition of ZAP-70.
Acknowledgments
We thank Dr. Robert T. Abraham for P116 and P116.cl39 cell lines and A. Hansen for secretarial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants CA81064, CA77646, CA111536, and CA120388. This work was also supported by The Hormel Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: EC, (-)-epicatechin; ECG, (-)-epicatechin 3-gallate; EGC, (-)-epigallocatechin; EGCG, (-)-epigallocatechin 3-gallate; TCR, T cell receptor; IL, interleukin; PTK, protein-tyrosine kinase; CLL, chronic lymphocytic leukemia; MAPK, mitogen-activated protein kinase; MEK, MAPK kinase; ERK, extracellular signaling-regulated kinase; PLC, phospholipase C; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium); ITAM, immunoreceptor tyrosine-based activation motif; FITC, fluorescein isothiocyanate; SH2, Src homology domain 2; LAT, anti-linker for the activation of T cells.
References
- 1.Yang, C. S., Maliakal, P., and Meng, X. (2002) Annu. Rev. Pharmacol. Toxicol. 42 25-54 [DOI] [PubMed] [Google Scholar]
- 2.Zaveri, N. T. (2006) Life Sci. 78 2073-2080 [DOI] [PubMed] [Google Scholar]
- 3.Yang, C. S., and Wang, Z. Y. (1993) J. Natl. Cancer Inst. 85 1038-1049 [DOI] [PubMed] [Google Scholar]
- 4.Yang, D. J., and Hwang, L. S. (2006) J. Chromatogr. A 1119 277-284 [DOI] [PubMed] [Google Scholar]
- 5.Jin, L., Pluskey, S., Petrella, E. C., Cantin, S. M., Gorga, J. C., Rynkiewicz, M. J., Pandey, P., Strickler, J. E., Babine, R. E., Weaver, D. T., and Seidl, K. J. (2004) J. Biol. Chem. 279 42818-42825 [DOI] [PubMed] [Google Scholar]
- 6.Deindl, S., Kadlecek, T. A., Brdicka, T., Cao, X., Weiss, A., and Kuriyan, J. (2007) Cell 129 735-746 [DOI] [PubMed] [Google Scholar]
- 7.Cantrell, D. (1996) Annu. Rev. Immunol. 14 259-274 [DOI] [PubMed] [Google Scholar]
- 8.Cantrell, D. A. (2002) Immunology 105 369-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamblin, T. J. (2007) Best Pract. Res. Clin. Haematol. 20 455-468 [DOI] [PubMed] [Google Scholar]
- 10.Gibbs, G., Bromidge, T., Howe, D., and Johnson, S. (2007) Int. J. Lab. Hematol. 29 225-227 [DOI] [PubMed] [Google Scholar]
- 11.Zanotti, R., Ambrosetti, A., Lestani, M., Ghia, P., Pattaro, C., Remo, A., Zanetti, F., Stella, S., Perbellini, O., Prato, G., Guida, G., Caligaris-Cappio, F., Menestrina, F., Pizzolo, G., and Chilosi, M. (2007) Leukemia (Baltimore) 21 102-109 [DOI] [PubMed] [Google Scholar]
- 12.Chan, A. C., Irving, B. A., Fraser, J. D., and Weiss, A. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 9166-9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan, A. C., Iwashima, M., Turck, C. W., and Weiss, A. (1992) Cell 71 649-662 [DOI] [PubMed] [Google Scholar]
- 14.Crespo, M., Bosch, F., Villamor, N., Bellosillo, B., Colomer, D., Rozman, M., Marce, S., Lopez-Guillermo, A., Campo, E., and Montserrat, E. (2003) N. Engl. J. Med. 348 1764-1775 [DOI] [PubMed] [Google Scholar]
- 15.Buggins, A. G., Hirst, W. J., Pagliuca, A., and Mufti, G. J. (1998) Br. J. Haematol. 100 784-792 [DOI] [PubMed] [Google Scholar]
- 16.Khan, I. H., Mendoza, S., Rhyne, P., Ziman, M., Tuscano, J., Eisinger, D., Kung, H. J., and Luciw, P. A. (2006) Mol. Cell. Proteomics 5 758-768 [DOI] [PubMed] [Google Scholar]
- 17.Bremer, E., ten Cate, B., Samplonius, D. F., de Leij, L. F., and Helfrich, W. (2006) Blood 107 2863-2870 [DOI] [PubMed] [Google Scholar]
- 18.Nakazato, T., Ito, K., Miyakawa, Y., Kinjo, K., Yamada, T., Hozumi, N., Ikeda, Y., and Kizaki, M. (2005) Haematologica 90 317-325 [PubMed] [Google Scholar]
- 19.Williams, B. L., Schreiber, K. L., Zhang, W., Wange, R. L., Samelson, L. E., Leibson, P. J., and Abraham, R. T. (1998) Mol. Cell. Biol. 18 1388-1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ermakova, S., Choi, B. Y., Choi, H. S., Kang, B. S., Bode, A. M., and Dong, Z. (2005) J. Biol. Chem. 280 16882-16890 [DOI] [PubMed] [Google Scholar]
- 21.Friesner, R. A., Banks, J. L., Murphy, R. B., Halgren, T. A., Klicic, J. J., Mainz, D. T., Repasky, M. P., Knoll, E. H., Shelley, M., Perry, J. K., Shaw, D. E., Francis, P., and Shenkin, P. S. (2004) J. Med. Chem. 47 1739-1749 [DOI] [PubMed] [Google Scholar]
- 22.Shim, J. H., Cho, K. J., Lee, K. A., Kim, S. H., Myung, P. K., Choe, Y. K., and Yoon, D. Y. (2005) Proteomics 5 2112-2122 [DOI] [PubMed] [Google Scholar]
- 23.McGovern, S. L., and Shoichet, B. K. (2003) J. Med. Chem. 46 2895-2907 [DOI] [PubMed] [Google Scholar]
- 24.Fabbro, D., Ruetz, S., Buchdunger, E., Cowan-Jacob, S. W., Fendrich, G., Liebetanz, J., Mestan, J., O'Reilly, T., Traxler, P., Chaudhuri, B., Fretz, H., Zimmermann, J., Meyer, T., Caravatti, G., Furet, P., and Manley, P. W. (2002) Pharmacol. Ther. 93 79-98 [DOI] [PubMed] [Google Scholar]
- 25.Kersh, E. N., Shaw, A. S., and Allen, P. M. (1998) Science 281 572-575 [DOI] [PubMed] [Google Scholar]
- 26.Neumeister, E. N., Zhu, Y., Richard, S., Terhorst, C., Chan, A. C., and Shaw, A. S. (1995) Mol. Cell. Biol. 15 3171-3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan, A. C., Dalton, M., Johnson, R., Kong, G. H., Wang, T., Thoma, R., and Kurosaki, T. (1995) EMBO J. 14 2499-2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Bartolo, V., Mege, D., Germain, V., Pelosi, M., Dufour, E., Michel, F., Magistrelli, G., Isacchi, A., and Acuto, O. (1999) J. Biol. Chem. 274 6285-6294 [DOI] [PubMed] [Google Scholar]
- 29.Watts, J. D., Affolter, M., Krebs, D. L., Wange, R. L., Samelson, L. E., and Aebersold, R. (1994) J. Biol. Chem. 269 29520-29529 [PubMed] [Google Scholar]
- 30.Brill, L. M., Salomon, A. R., Ficarro, S. B., Mukherji, M., Stettler-Gill, M., and Peters, E. C. (2004) Anal. Chem. 76 2763-2772 [DOI] [PubMed] [Google Scholar]
- 31.Williams, B. L., Irvin, B. J., Sutor, S. L., Chini, C. C., Yacyshyn, E., Bubeck Wardenburg, J., Dalton, M., Chan, A. C., and Abraham, R. T. (1999) EMBO J. 18 1832-1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alberola-Ila, J., and Hernandez-Hoyos, G. (2003) Immunol. Rev. 191 79-96 [DOI] [PubMed] [Google Scholar]
- 33.Iwashima, M. (2003) Immunol. Rev. 191 196-210 [DOI] [PubMed] [Google Scholar]
- 34.Suzushima, H., Hattori, T., Asou, N., Wang, J. X., Nishikawa, K., Okubo, T., Anderson, P., and Takatsuki, K. (1991) Cancer Res. 51 6084-6088 [PubMed] [Google Scholar]
- 35.van Dongen, J. J., Quertermous, T., Bartram, C. R., Gold, D. P., Wolvers-Tettero, I. L., Comans-Bitter, W. M., Hooijkaas, H., Adriaansen, H. J., de Klein, A., and Raghavachar, A. (1987) J. Immunol. 138 1260-1269 [PubMed] [Google Scholar]
- 36.Acuto, O., and Cantrell, D. (2000) Annu. Rev. Immunol. 18 165-184 [DOI] [PubMed] [Google Scholar]
- 37.Lin, J., and Weiss, A. (2001) J. Cell Sci. 114 243-244 [DOI] [PubMed] [Google Scholar]
- 38.van Leeuwen, J. E., and Samelson, L. E. (1999) Curr. Opin. Immunol. 11 242-248 [DOI] [PubMed] [Google Scholar]
- 39.Wange, R. L., Malek, S. N., Desiderio, S., and Samelson, L. E. (1993) J. Biol. Chem. 268 19797-19801 [PubMed] [Google Scholar]
- 40.Martelli, M. P., Lin, H., Zhang, W., Samelson, L. E., and Bierer, B. E. (2000) Blood 96 2181-2190 [PubMed] [Google Scholar]
- 41.Houtman, J. C., Houghtling, R. A., Barda-Saad, M., Toda, Y., and Samelson, L. E. (2005) J. Immunol. 175 2449-2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dumont, F. J., Staruch, M. J., Fischer, P., DaSilva, C., and Camacho, R. (1998) J. Immunol. 160 2579-2589 [PubMed] [Google Scholar]
- 43.Freund-Michel, V., and Frossard, N. (2008) Pharmacol. Ther. 117 52-76 [DOI] [PubMed] [Google Scholar]
- 44.Junttila, M. R., Li, S. P., and Westermarck, J. (2008) FASEB J. 22 954-965 [DOI] [PubMed] [Google Scholar]
- 45.Budihardjo, I., Oliver, H., Lutter, M., Luo, X., and Wang, X. (1999) Annu. Rev. Cell Dev. Biol. 15 269-290 [DOI] [PubMed] [Google Scholar]
- 46.Earnshaw, W. C. (1999) Nature 397 387, 389 [DOI] [PubMed] [Google Scholar]
- 47.Blume-Jensen, P., and Hunter, T. (2001) Nature 411 355-365 [DOI] [PubMed] [Google Scholar]
- 48.Rodrigues, G. A., and Park, M. (1994) Curr. Opin. Genet. Dev. 4 15-24 [DOI] [PubMed] [Google Scholar]
- 49.Bene, M. C. (2006) Cytometry B. Clin. Cytom. 70 204-208 [DOI] [PubMed] [Google Scholar]
- 50.Wiestner, A., Rosenwald, A., Barry, T. S., Wright, G., Davis, R. E., Henrickson, S. E., Zhao, H., Ibbotson, R. E., Orchard, J. A., Davis, Z., Stetler-Stevenson, M., Raffeld, M., Arthur, D. C., Marti, G. E., Wilson, W. H., Hamblin, T. J., Oscier, D. G., and Staudt, L. M. (2003) Blood 101 4944-4951 [DOI] [PubMed] [Google Scholar]
- 51.Brdicka, T., Kadlecek, T. A., Roose, J. P., Pastuszak, A. W., and Weiss, A. (2005) Mol. Cell. Biol. 25 4924-4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iwashima, M., Irving, B. A., van Oers, N. S., Chan, A. C., and Weiss, A. (1994) Science 263 1136-1139 [DOI] [PubMed] [Google Scholar]
- 53.Nakagawa, H., Hasumi, K., Woo, J. T., Nagai, K., and Wachi, M. (2004) Carcinogenesis 25 1567-1574 [DOI] [PubMed] [Google Scholar]
- 54.Li, H. C., Yashiki, S., Sonoda, J., Lou, H., Ghosh, S. K., Byrnes, J. J., Lema, C., Fujiyoshi, T., Karasuyama, M., and Sonoda, S. (2000) Jpn. J. Cancer Res. 91 34-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Asano, Y., Okamura, S., Ogo, T., Eto, T., Otsuka, T., and Niho, Y. (1997) Life Sci. 60 135-142 [DOI] [PubMed] [Google Scholar]