Abstract
Methionine is an essential amino acid in mammals at the junction of methylation, protein synthesis, and sulfur pathways. However, this amino acid is highly susceptible to oxidation, resulting in a mixture of methionine-S-sulfoxide and methionine-R-sulfoxide. Whether methionine is quantitatively regenerated from these compounds is unknown. Here we report that SK-Hep1 hepatocytes grew on methionine-S-sulfoxide and consumed this compound by import and methionine-S-sulfoxide reductase (MsrA)-dependent reduction, but methionine-R-sulfoxide reductases were not involved in this process, and methionine-R-sulfoxide could not be used by the cells. However, SK-Hep1 cells expressing a yeast free methionine-R-sulfoxide reductase proliferated in the presence of either sulfoxide, reduced them, and showed increased resistance to oxidative stress. Only methionine-R-sulfoxide was detected in the plasma of wild type mice, but both sulfoxides were in the plasma of MsrA knock-out mice. These results show that mammals can support methionine metabolism by reduction of methionine-S-sulfoxide, that this process is dependent on MsrA, that mammals are inherently deficient in the reduction of methionine-R-sulfoxide, and that expression of yeast free methionine-R-sulfoxide reductase can fully compensate for this deficiency.
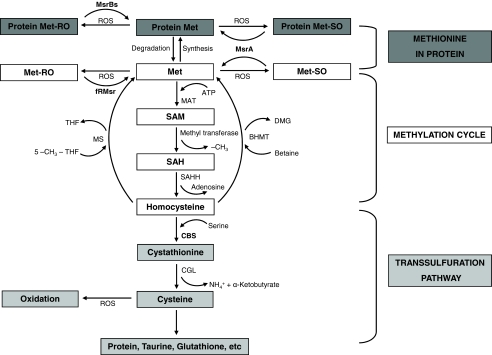
Methionine (Met) is an essential amino acid in mammals. Besides its utilization for protein synthesis, Met supports the cellular S-adenosylmethionine-dependent methylation cycle and is used for biosynthesis of cysteine and other compounds (Fig. 1) (1–3). As a central metabolite in the global sulfur and methylation pathways, Met homeostasis is subject to exquisite regulation that tightly controls fluxes of Met-derived metabolites and Met itself (4, 5).
FIGURE 1.
Metabolism of Met and Met sulfoxide in mammalian cells. Protein synthesis and methylation and trans-sulfuration pathways represent major uses of free Met. The current study adds information on an additional Met cycle wherein Met is oxidized to Met-RO and Met-SO, and only Met-SO can be reduced to Met. Expression of yeast fRMsr allows utilization of Met-RO, compensating for deficiency in this function in mammals. MAT, methionine adenosyltransferase; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; SAHH, S-adenosylhomocysteine hydrolase; CGL, cystathionine γ-lyase; MS, methionine synthase; THF, tetrahydrofolate; DMG, dimethyl glycine; BHMT, betaine:homocysteine methyltransferase.
Despite extensive information on Met regulation, one pathway critical to Met availability and homeostasis did not receive sufficient attention to date. Being a sulfur-containing amino acid, Met, along with Cys, is highly susceptible to oxidation by reactive oxygen species (ROS)2 generated during oxidative stress and normal cellular metabolism. ROS can oxidize both free Met and Met residues in proteins, resulting in a diastereomeric mixture of Met sulfoxides: Met-S-sulfoxide (Met-SO) and Met-R-sulfoxide (Met-RO) (6, 7).
To repair Met-SO and Met-RO residues in proteins, cells have evolved two families of enzymes known as Met sulfoxide reductases: MsrA that is specific for Met-SO and MsrB that specifically reduces Met-RO (8). The active sites of proteins in both Msr families are better adapted for binding Met sulfoxide residues than free Met sulfoxides (6). Although both MsrA and MsrB are capable of reducing free Met sulfoxides (9), a contribution of these enzymes to the reduction of these amino acids is not known (10).
Mammals have one MsrA and three MsrB isozymes, with selenoprotein MsrB1 localized to cytosol and nucleus, MsrB2 to mitochondria, and MsrB3 to the endoplasmic reticulum (9). A single MsrA is partitioned into various cellular compartments by alternative first exon splicing and folding-dependent cytosolic retention of the protein containing a mitochondrial targeting signal (11–14). MsrA and MsrB are thought to both repair oxidatively damaged Met residues and serve as antioxidant proteins, thus supporting the role of Met residues in scavenging ROS (15). Consistent with these ideas, deletion of Msrs reduces life span in yeast and mice (16, 17), whereas their overexpression can increase life span in fruit flies and yeast cells (17–19).
In comparison with the repair of oxidized Met residues, reduction of free Met sulfoxides in mammals received little attention. On the other hand, several cellular fractions with proteins distinct from MsrA and MsrB were found to reduce free Met sulfoxides in bacteria (20). Furthermore, by analyzing bacterial cells deficient in MsrA and MsrB, two enzymes were identified that acted exclusively on free Met sulfoxides. A molybdoprotein-containing biotin sulfoxide reductase BisC (21) was found to reduce free Met-SO, whereas a GAF domain-containing protein fRMsr was specific for free Met-RO (22).
In this work, we characterized the ability of mammals to reduce free Met sulfoxides. Whereas Met-SO was reduced by MsrA, mammalian cells were deficient in the reduction of free Met-RO. However, cells expressing a yeast fRMsr homolog could efficiently utilize Met-RO. Thus, this study both revealed an inherent deficiency of mammalian cells in an important anti-oxidant repair process and identified a protein that can compensate for this deficiency.
EXPERIMENTAL PROCEDURES
Preparation of Free Met-SO and Met-RO—Free Met-SO and Met-RO were prepared from mixed l-Met-R,S-sulfoxide (Sigma) according to the method of Lavine (23). To obtain diastereomers of higher purity, we repeated the separation process twice for each sulfoxide. Purity of Met-SO and Met-RO was assessed by an HPLC procedure using o-phthalaldehyde (OPA) (Sigma)-derivatized amino acids (7) and found to exceed 98%.
Cell Culture—SK-Hep1 (ATCC: HTB-52™) and fRMsr-transfected SK-Hep1 cells were cultured in DMEM or Met-free DMEM (Invitrogen) supplemented with 0.1 mm Met, 0.1 mm Met-RO, 0.1 mm Met-SO, or 0.1 mm Met-RSO. The medium also contained 10% dialyzed fetal bovine serum and an antibiotics-antimycotic mixture (Invitrogen) of 100 units/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, and 0.25 μg/ml amphotericin B. In initial experiments, we examined media containing different amounts of Met and found that 0.1 mm Met was optimal and avoided Met deficiency. Thus, 0.1 mm Met sulfoxides were used in further experiments. Cell culture experiments, with the exception of a proliferation assay, were carried out in 6-well plates, and the cells were maintained at 37 °C in a 5% CO2 atmosphere.
Growth of SK-Hep1 cells was analyzed in modified DMEM containing Met, Met-SO, Met-RO, or Met-RSO (0.1 mm of each amino acid) or with no addition of these compounds. In addition, the media were supplemented or not with 100 nm sodium selenite. The cells were analyzed at 0, 24, 48, and 72 h. Another experiment involving SK-Hep1 cells was done with cells grown in serum-free modified DMEM, containing insulin (5 μg/ml) and transferrin (10 μg/ml), Met or Met-RSO (0.1 mm of each amino acid) and 100 nm sodium selenite (or with no addition of this compound), and separately in 10% dialyzed fetal bovine serum-containing DMEM containing 0.1 mm Met with 100 nm sodium selenite (or not). The cells were analyzed at 0, 24, 48, and 72 h. Cell growth assays were carried out as described below.
Proliferation Assay—Cell growth was quantified using colorimetric MTS assay (Promega). The cells in regular DMEM were plated in 96-well plates at 5 × 103 cells/well, washed with PBS, and specialized DMEM were added that contained Met, Met-SO, Met-RO, or Met-RSO 24 h after plating. To assay for cell proliferation, a tetrazolium compound (inner salt; MTS) and an electron coupling reagent (phenazine methosulfate) were mixed according to the manufacturer's protocol, and then 20 μl of the mixture were added to 100 μl of phenol red-excluded medium that further replaced cell culture medium. After 90 min of incubation at 37 °C in the atmosphere of 5% CO2, cell proliferation was measured at indicated time periods from 0 to 96 h at 450 nm in a plate reader. Direct counting of viable cells using 0.2% trypan blue was done in 6-well plates.
An HPLC Analysis of Media Samples—The cells were plated in 6-well plates at 7.5 × 104 cells/well in 1.5 ml of regular DMEM. After 24 h, the cells were washed with PBS, and the medium was changed to modified DMEM containing Met, Met-SO, Met-RO, or Met-RSO (0.1 mm of each amino acid). 90 μl of medium from each well was collected at 0, 24, 48, 72, and 96 h and mixed with 10 μl of 100% trichloroacetic acid. After incubation at 4 °C for 10 min and centrifugation at 13,000 rpm for 15 min, the supernatant (50 μl) was diluted 10 times with distilled water and prepared for OPA derivatization. OPA derivatization of amino acids and HPLC analysis were performed as described (7) with minor modifications. The derivatization reagent was freshly prepared as a stock solution (40 mg of o-phthalaldehyde, 1 ml of methanol, 50 μl of 2-mercaptoehanol, 5 ml of 0.1 m Na2B4O7, pH 9.5) at room temperature in a capped amber vial. Sample solutions (2–5 μl) were mixed with the OPA derivatization reagent to a 100-μl final volume. Following a 2-min reaction at room temperature, the mixture was fractionated on a Zorbax Eclipse XDB-C8 column (4.6×150 mm). Changes in Met levels in the fRMsr activity assay were similarly quantified. Detection was by fluorescence of Met derivatives using a Waters 474 scanning fluorescence detector with excitation at 330 nm and emission at 445 nm.
Knockdown of MsrA in SK-Hep1 Cells by Small Interfering RNA—Double-stranded small interfering RNAs (Dharmacon) for targeting human MsrA mRNA were used to knockdown this gene in SK-Hep1 cells. The cells were transfected with small interfering RNA by using DharmaFECT™ 1 Transfection Reagent (Dharmacon) according to the manufacturer's protocol. Proliferation of these cells was assayed 60 h after transfection in comparison with control transfected cells.
Transfection of Yeast fRMsr Gene into SK-Hep1 Cells—A His6 tag sequence was cloned at the C terminus of yeast fRMsr, the sequence was inserted into a mammalian expression vector, pCI-neo (Promega), and the resulting construct was verified by DNA sequencing. SK-Hep1 cells were transfected or cotransfected with the expression vector coding for fRMsr and pEGFP-N1 (Clontech) vector or with an empty pCI-neo vector and pEGFP-N1 vector using FuGENE 6 transfection reagent according to the manufacturer's suggestion. To establish a stable cell line, the cells were selected in the presence of 800 μg/ml G418 sulfate (Promega) and further maintained in the presence of 400 μg/ml G418 sulfate. Expression of recombinant fRMsr was verified by Western blotting using anti-His tag antibodies, and Msr activities were assayed by an HPLC method using OPA derivatization.
Cloning and Expression of Yeast fRMsr—Characterization of S. cerevisiae fRMsr (YKL069W) is described in a separate paper.3 Briefly, a yeast fRMsr cDNA, which encodes a protein homologous to fRMsr from Escherichia coli, was amplified from Saccharomyces cerevisiae genomic DNA and subcloned into pET21b vector (Novagen) using primers 5′-AAACATATGATGGGCTCATCAACCGGGTTTC-3′ (sense) and 5′-AAGCGGCCGCGACACATGATTTATTAATTAATTTAGCAAG-3′ (antisense). After transformation into BL21 (DE3) E. coli, the subcloned sequence was verified by DNA sequencing. Cells with the plasmid in 500 ml of LB medium containing 100 μg/ml ampicillin were grown until A600 reached 0.6–0.8, followed by the addition of isopropyl β-d-thiogalactopyranoside to 0.3 mm. Protein expression was at 30 °C for 4 h, followed by harvesting cells by centrifugation at 4,000 rpm for 5 min. The cells were washed with PBS and stored at -70 °C until use.
To purify the protein, cell pellet was dissolved in resuspension buffer (Tris-HCl, pH 7.5, 15 mm imidazole, 300 mm NaCl), and phenylmethylsulfonyl fluoride was added to a final concentration of 0.5 mm. After sonication, supernatant was collected by centrifugation at 8,000 rpm for 30 min. The supernatant was loaded onto a cobalt Talon resin (Clontech) pre-equilibrated with resuspension buffer. Following washing with the same buffer, the protein was eluted with elution buffer (Tris-HCl, pH 7.5, 300 mm imidazole, 300 mm NaCl). Fractions containing yeast fRMsr were pooled together and dialyzed overnight against PBS in a dialysis cassette (Pierce).
Activity Assays of fRMsr—Reaction mixture (40 μl) included 50 mm dithiothreitol, 1 mm substrate (Met-RO or Met-SO), and purified enzyme or cell lysate prepared by treatment with CelLytic™M cell lysis reagent (Sigma). 20 μl of the reaction mixture was mixed with 2 μl of trichloroacetic acid and subjected to OPA derivatization as described above and then injected onto the column to measure endogenous Met level in the samples. An additional 20 μl from the same original reaction mixture were incubated at 37 °C for 30 min and then subjected to OPA derivatization by the same procedure using trichloroacetic acid. After derivatization of the sample (2–5 μl) by adding OPA solution to 100 μl as a final volume, 50 μl of the mixture were subjected to an HPLC separation as described above.
Analysis of SK-Hep1 and fRMsr-transfected SK-Hep1 Cells—SK-Hep1 and fRMsr-transfected SK-Hep1 cells were plated in 6-well plates at 7.5 × 104 cells/well in regular DMEM. The medium was changed to Met-free DMEM supplemented with either Met or individual Met sulfoxides at 24 h after plating. The cells were then collected every 24 h until 96 h. Protein concentration was measured by the Bradford assay, and the samples were probed by standard immunoblot assays with polyclonal MsrA (kindly provided by Bertrand Friguet), cystathionine β synthase (CBS) (kindly provided by Ruma Banerjee), and activating transcription factor 3 (ATF3) (Santa Cruz Biotechnology) antibodies.
Microscopy—A Nikon TE-300 microscope was used to prepare images of SK-Hep1 cells grown on different Met and Met sulfoxide media. Microscopy analyses were carried out in the University of Nebraska-Lincoln Microscopy Core Facility.
75Se Metabolic Labeling—SK-Hep1 cells were plated in 6-well plates at 7.5 × 104 cells/well in 1.5 ml of regular DMEM. After 24 h, the cells were washed with PBS, and the medium was changed to modified DMEM containing Met, Met-SO, Met-RO, or Met-RSO (0.1 mm of each amino acid). 0.01 mCi of freshly neutralized [75Se]selenious acid (specific activity, 1,000 Ci/mmol; University of Missouri Research Reactor) was added in each well at 48 h after changing the medium, and the cells were incubated at 37 °C for 24 h in 5% CO2 atmosphere, followed by harvesting the cells. The cell extracts (30 μg of total protein) were electrophoresed on 10% Bis-Tris gels and transferred onto polyvinylidene difluoride membranes (Invitrogen). The 75Se radioactivity pattern was visualized by using a PhosphorImager (GE Healthcare).
Resistance of fRMsr-expressing and Control SK-Hep1 Cells to Oxidative Stress—SK-Hep1 cells stably transfected with either an empty pCI-neo vector and fRMsr construct were plated in 96-well plates at 3.0 × 104 cells/well. The medium was changed to a serum-free medium, and H2O2 was added to each well at indicated concentrations (0–1000 μm). Cell viability was measured by using colorimetric MTS assay.
Mice—MsrA knock-out mice (16) were kindly provided by Drs. Rodney Levine and Geumsoo Kim (National Institutes of Health). These mice and control C57BL6 mice were used for blood sampling. In addition, C57BL6 mice, which were subjected to selenium deficiency or a control diet containing 0.4 ppm Se in the form of sodium selenite for 8 months, were used.
HPLC Analysis of Met-RO and Met-SO Level in Mouse Plasma—Mouse blood was centrifuged at 13,000 rpm for 15 min. The supernatant was prepared for OPA derivatization without dilution following trichloroacetic acid precipitation and analyzed at isocratic flow rate of 2 min/ml at 89:11 (v/v) of 20 mm sodium acetate, pH 5.8 (solvent A) and methanol (solvent B) as described in the supplemental materials.
RESULTS
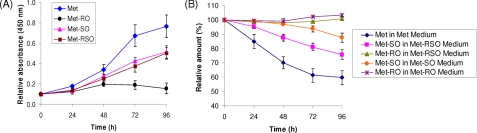
SK-Hep1 Cells Grow in the Presence of Met and Met-SO, but Not in the Presence of Met-RO—To examine the capacity of mammalian cells to provide Met for cellular metabolism by Met sulfoxide reduction, human hepatoma SK-Hep1 cells were grown in Met-free DMEM supplemented with 0.1 mm Met, 0.1 mm Met-SO, 0.1 mm Met-RO, or 0.1 mm mixed Met-RO/Met-SO (Met-RSO) (Fig. 2A). SK-Hep1 hepatocytes grew best in the presence of Met and could also proliferate in Met-SO and Met-RSO media, although at a reduced rate. In contrast, Met-RO did not support the growth of SK-Hep1 cells. Morphology of cells maintained on Met-SO and Met-RO media was also different (see below). After 96 h on the Met-RO medium, SK-Hep1 hepatocytes were long and narrow, similar in shape to Met-restricted cells, whereas the cells grown on Met-SO resembled those maintained in the presence of Met. These data suggested that SK-Hep1 cells have a system for import and reduction of Met-SO, which provides them with Met, whereas these cells are unable to utilize free Met-RO.
FIGURE 2.
Growth and consumption of free Met sulfoxides by SK-Hep1 cells. A, growth of SK-Hep1 hepatocytes on Met and Met sulfoxide media. The cells were grown in Met-free DMEM supplemented with 0.1 mm Met (diamonds), 0.1 mm Met-RO (triangles), 0.1 mm Met-SO (circles), or 0.1 mm Met-RSO (squares). Cell growth was quantified in a cell proliferation assay at 0, 24, 48, 72, and 96 h. The error bars represent the standard deviations from five independent experiments. B, consumption of Met in Met medium (diamonds), Met-SO in Met-RSO medium (squares), Met-RO in Met-RSO medium (triangles), Met-SO in Met-SO medium (circles), and Met-RO in Met-RO medium (stars) at 0, 24, 48, 72, and 96 h. The initial concentration of each amino acid was 0.1 mm. The error bars represent the standard deviations from three independent experiments.
To determine whether Met, Met-RO, and Met-SO were consumed by SK-Hep1 hepatocytes, levels of these compounds were determined in each growth medium at 0, 24, 48, 72, and 96 h. For this purpose, we developed an HPLC procedure that utilized OPA derivatization of free amino acids following trichloroacetic acid precipitation of medium components (Fig. 2B and supplemental Fig. S1). We found this method to be superior to the currently available procedures for quantitative analysis of free Met sulfoxides and Met. It was recently reported (25) that many Met sulfoxide derivatives, such as widely used dabsyl chloride-reacted forms, are subject to nonenzymatic Met sulfoxide reduction at high temperatures, but the corresponding derivatization procedures themselves require heating. In contrast, the OPA derivatization process is completed at room temperature after 2–3 min, thus avoiding a heating step and associated nonenzymatic Met sulfoxide reduction.
With this procedure, we found that relative amounts of Met in the Met-supplemented medium decreased to ∼60% of the initial level at 96 h. In contrast, Met-RO levels were not changed in Met-RO and Met-RSO media at any time points. Met-SO was decreased to 88% in the Met-SO-supplemented medium and to 77% in the Met-RSO medium (Fig. 2B). These data show that SK-Hep1 cells could import and consume both Met-SO and Met from the medium to support cell proliferation. On the other hand, consistent with the inability of SK-Hep1 cells to grow on Met-RO, this compound was not utilized. Moreover, when cells were grown on Met-RSO, only Met-SO was consumed.
SK-Hep1 Cell Extracts Are Active in the Reduction of Met-SO but Do Not Reduce Met-RO—Because both MsrA and MsrB were reported to support low level reduction of free Met sulfoxides in in vitro assays (9), it was of interest to examine the contribution of these enzymes to Met sulfoxide reduction under physiological conditions in mammalian cells. We measured specific activities of SK-Hep1 cell extracts for the reduction of free Met-RO and Met-SO. No free Met-RO reductase activity was detected (it was within background), whereas the activity for free Met-SO reduction was ≈38.6 pmol/min/mg of protein (Table 1).
TABLE 1.
Specific activities of SK-Hep1 cells towards free Met sulfoxides The table shows the values ± standard deviations from three independent experiments.
|
Specific activity for each Met-O
|
|||
|---|---|---|---|
| Met-RO | Met-SO | ||
| pmol/min/mg | |||
| yfRMsra | 30015 ± 2652 | NDb | |
| SK-Hep1 | NDb | 38.6 ± 3.6 | |
| yfRMsr transfected SK-Hep1 | 46.4 ± 5.0 | 42.7 ± 5.9 | |
yfRMsr is yeast-free methionine-R-sulfoxide reductase
A signal corresponding to specific activity less than 5 pmol/min/mg is shown as ND (not detectable)
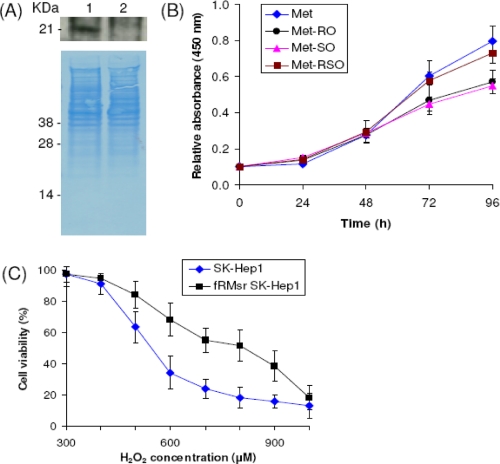
Role of MsrA in the Reduction of Free Met-SO—To examine the role of MsrA in providing cells with Met by reduction of free Met-SO, we knocked down its expression by small interfering RNA. Decreased MsrA expression was verified by Western blot assays (Fig. 3A). We found that MsrA-deficient cells grew neither in Met-SO nor Met-RO medium, whereas Met still supported their growth. Thus, MsrA is responsible for the reduction of Met-SO acquired from the media by SK-Hep1 cells. These data also indicate that none of the three mammalian MsrB isozymes contributes significantly to the enzymatic reduction of free Met sulfoxides and to providing Met to support cell growth.
FIGURE 3.
Characterization of MsrA knockdown SK-Hep1 cells. A, Western blot analysis of MsrA in MsrA-knockdown SK-Hep1 cells (lane 1) and control SK-Hep1 cells (lane 2). B, MsrA knockdown SK-Hep1 cells were grown in media containing 0.1 mm Met (diamonds), Met-RO (circles), Met-SO (triangles), or Met-RSO (squares) for 96 h. Cell growth was measured by an MTS cell proliferation assay at 0, 24, 48, 72, and 96 h. The error bars represent the standard deviations from three independent experiments.
A Yeast Enzyme Specific for Free Met-RO—A recent study identified a bacterial enzyme specific for free Met-RO (22). This GAF domain-containing protein was designated as fRMsr. We analyzed sequence databases for homologs of this protein and found that mammals and other animals lack fRMsr. However, homologs of this protein were detected in many lower eukaryotes (supplemental Fig. S2).3 We cloned fRMsr homolog from S. cerevisiae and expressed it in E. coli as a His-tagged protein. The recombinant protein was soluble and had the expected molecular weight as determined by SDS-PAGE and mass spectrometry, whereas the native yeast protein functioned as fRMsr in S. cerevisiae cells.3 The recombinant yeast fRMsr protein exhibited high activity toward free Met-RO (≈33 nmol/min/mg of protein), whereas it was inactive with Met-SO as well as with dabsyl-Met-RO and dabsyl-Met-SO.
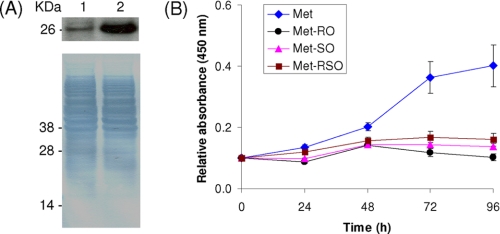
Yeast fRMsr-expressing SK-Hep1 Cells Grow on Met-RO—Yeast fRMsr was stably expressed in SK-Hep1 cells in the form of His-tagged protein, and its expression was verified in Western blot assays with antibodies specific for His tag (Fig. 4A). Specific activity of fRMsr-transfected cells for Met-RO was ≈46.4 pmol/min/mg of protein, and the activity of these cells toward Met-SO was ≈42.7 pmol/min/mg of protein (Table 1). The transfected cells grew on Met-SO or Met-RO medium, and their growth was essentially indistinguishable from that of cells grown in the presence of Met (Fig. 4B). Morphology of fRMsr expressing cells on the Met-RO-supplemented medium was similar to that of cells grown on Met (Fig. 5). Thus, yeast fRMsr expressed in SK-Hep1 cells could reduce free Met-RO in quantities sufficient to compensate for Met deficiency. These data also indicated that Met-RO could be imported into SK-Hep1 cells from the medium.
FIGURE 4.
Characterization of SK-Hep1 cells expressing yeast fRMsr. A, Western blot analysis of SK-Hep1 cells stably expressing yeast His-tagged fRMsr (lane 1) and control SK-Hep1 cells (lane 2) with anti-His antibodies. B, yeast fRMsr-transfected SK-Hep1 cells were grown in media containing 0.1 mm Met (diamonds), 0.1 mm Met-RO (circles), 0.1 mm Met-SO (triangles), or 0.1 mm Met-RSO (squares) until 96 h. Cell growth was measured by an MTS cell proliferation assay at 0, 24, 48, 72, and 96 h. C, resistance of fRMsr-expressing SK-Hep1 cells to hydrogen peroxide treatment. Control (diamonds) and fRMsr-expressing SH-Hep1 (squares) cells were treated with the indicated concentrations of hydrogen peroxide, and their viability was assayed at 24 h. The error bars show the standard deviations from three independent experiments.
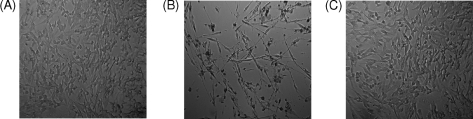
FIGURE 5.
Morphology of cells grown on sulfoxide media. A, an image of SK-Hep1 cells grown in the Met-SO medium for 96 h. B, an image of SK-Hep1 cells grown in Met-RO medium for 96 h. C, an image of SK-Hep1 cells stably expressing yeast fRMsr grown in Met-RO medium for 96 h.
Increased Resistance of fRMsr-expressing SK-Hep1 Cells to Oxidative Stress—SK-Hep1 cells expressing yeast fRMsr were further examined for resistance to oxidative stress by subjecting them (and control cells) to hydrogen peroxide treatment. At higher concentrations of hydrogen peroxide (above 400 μm), fRMsr-expressing cells showed significantly higher viability than control cells (Fig. 4C). Thus, yeast fRMsr protected SK-Hep1 cells from oxidative stress caused by hydrogen peroxide treatment. The increased resistance of transfected cells to oxidative stress is likely because of reduction of free Met-RO formed by Met oxidation in the presence of hydrogen peroxide. In addition, these data indicate that reversible oxidation and reduction of free Met-RO (and by analogy free Met-SO) provides mammalian cells with an antioxidant defense system.
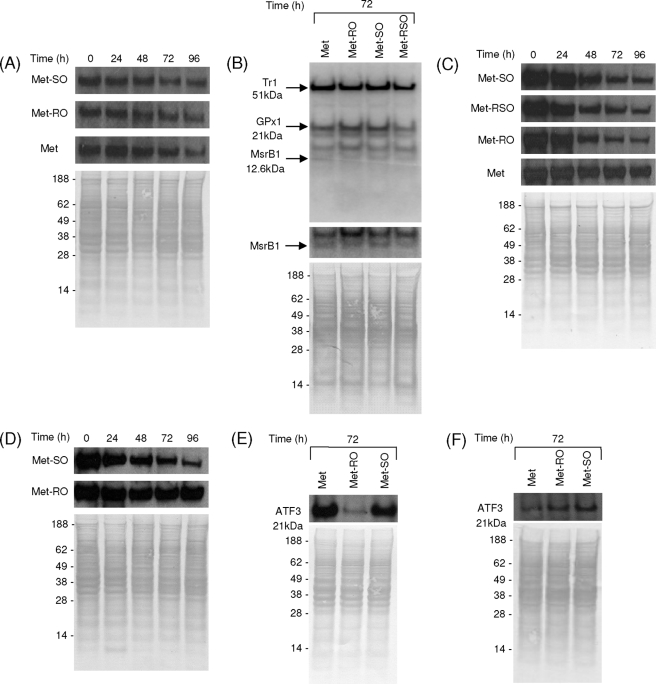
MsrA and Selenoprotein Expression in Cells Grown on Met and Met Sulfoxide Media—We tested whether expression of MsrA is regulated by availability of Met or Met sulfoxides by maintaining cells in the corresponding growth media (Fig. 6A). Although levels of this enzyme slightly decreased after 96 h growth of SK-Hep1 cells, this change was observed in all samples, and we did not find significant differences in MsrA expression among samples examined. Thus, MsrA expression was not influenced by Met sulfoxide levels in cell culture media. We also tested whether the addition of Met sulfoxides regulates expression of selenoproteins, many of which are important antioxidant enzymes or redox regulators (Fig. 6B). We metabolically labeled SK-Hep1 cells with 75Se and examined selenoprotein patterns. No difference was observed among cells grown on Met, Met-RO, Met-SO, and Met-RSO media. The addition of selenium (100 nm sodium selenite) also did not influence cell growth on Met and Met sulfoxide in fetal bovine serum-supplemented and insulin/transferrin (supplemental Fig. S3) media. These data further argue against the role of selenoprotein MsrB1, which is a major peptide Met-RO reductase in mammals, in the reduction of free Met sulfoxides.
FIGURE 6.
CBS, ATF3, MsrA, and selenoprotein expression in SK-Hep1 cells grown in Met and Met sulfoxide media. A, Western blot analysis of MsrA in SK-Hep1 cells grown in media containing 0.1 mm Met, Met-RO, or Met-SO at 0, 24, 48, 72, and 96 h. B, metabolic labeling of SK-Hep1 cells grown in media containing Met, Met-RO, Met-SO, or Met-RSO, with 75Se. Cell extracts were analyzed by SDS-PAGE, and the 75Se pattern was visualized with a PhosphorImager (upper panel). Migration of major selenoproteins, thioredoxin reductase1 (Tr1), and glutaredoxin peroxidase1 (GPx1) is indicated. Migration of MsrB1 is also shown, and the corresponding area of the gel is enlarged in the middle panel. The bottom panel shows protein loading. C, Western blot analysis of CBS in SK-Hep1 cells grown in media with 0.1 mm Met, Met-RO, Met-SO, or Met-RSO at 0, 24, 48, 72, and 96 h. D, Western blot analysis of CBS in fRMsr-expressing SK-Hep1 cells grown in media containing 0.1 mm Met-RO or Met-SO at 0, 24, 48, 72, and 96 h. E, Western blot analysis of ATF3 in SK-Hep1 cells grown in media with 0.1 mm Met, Met-RO, or Met-SO at 72 h. F, Western blot analysis of ATF3 in fRMsr-expressing SK-Hep1 cells grown in media containing 0.1 mm Met, Met-RO or Met-SO at 72 h.
CBS Expression in Cells Grown on Met and Met Sulfoxide Media—Met restriction in SK-Hep1 cells is known to reduce CBS levels by decreasing the availability of S-adenosylmethionine, which stabilizes CBS (4). Thus, this protein can be viewed as a marker of S-adenosylmethionine status (and therefore Met availability). We examined expression of CBS in cells grown on Met and Met sulfoxide media (Fig. 6, C and D). In the presence of Met-RO, SK-Hep1 cells reduced expression of this protein, whereas the cells grown on Met maintained stable CBS levels. In cells grown on the Met-SO medium, CBS levels were decreased, suggesting that although MsrA supports proliferation of SK-Hep1 cells by providing them with Met, these cells still suffer from Met deficiency (Fig. 6C). Interestingly, fRMsr-expressing cells grown on Met-RO had normal CBS levels (Fig. 6D). Thus, fRMsr can fully compensate for the lack of Met-RO reductase activity in SK-Hep1 hepatocytes and provide these cells with the full amount of Met needed for cellular metabolism.
ATF3 Expression in Cells Grown on Met and Met Sulfoxide Media—ATF3 is known to be expressed at low levels in normal and quiescent cells but is rapidly induced by various stresses, such as amino acid deprivation, oxidative stress, and ER stress (26, 27). ATF3 is also relevant to the control of cell proliferation and pathway cross-talk regulation (24). Interestingly, SK-Hep1 cells grown on Met-RO for 72 h showed low expression levels of ATF3 as compared with cells on Met or Met-SO (Fig. 6E), suggesting that the cells grown on Met-RO had metabolic and proliferative defects caused by Met limitation. Importantly, SK-Hep1 cells being quiescent on Met-RO recovered normal metabolic function and the ability to proliferate when transfected with a construct coding for yeast fRMsr (Figs. 4B and 6F).
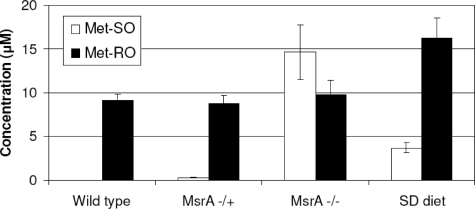
Met-SO and Met-RO Levels in Mouse Plasma—To test whether differences in Met-SO and Met-RO consumption and reduction that were evident in cell culture experiments are also observed in an animal system, we analyzed Met-SO and Met-RO levels in mouse plasma. Met-SO was not detected in plasma of wild type mice, whereas Met-RO concentration was ≈9.1 ± 0.7 μm (quantified by providing known amounts of Met-RO and Met-SO standards to plasma samples) (Fig. 7 and supplemental Fig. S4). To test whether MsrA is responsible for the low systemic Met-SO in mouse blood, we further examined samples from MsrA knock-out mice. These mice had similar levels of Met-SO and Met-RO in plasma (Fig. 7 and supplemental Fig. S4). Met-SO concentration was found to be ≈14.6 ± 3.1 μm, and Met-RO concentration was ≈9.8 ± 1.6 μm (Fig. 7 and supplemental Fig. S4). These data show that deletion of MsrA caused a remarkable increase specifically in Met-SO levels, indicating that MsrA is responsible for the low levels of Met-SO in plasma of wild type mice. We also subjected wild type mice to selenium deficiency, which reduced MsrB1 to almost undetectable levels in liver (data not shown). Under these conditions, plasma Met-SO was ≈3.7 ± 0.6 μm, and Met-RO was ≈16.2 ± 2.4 μm (Fig. 7 and supplemental Fig. S4), indicating that both sulfoxides were slightly elevated. Besides MsrB1, several selenoproteins serve as antioxidant proteins and show reduced expression in selenium deficiency, resulting in increased oxidative stress. Thus, slight elevation in both Met-SO and Met-RO under conditions of low dietary selenium is due to an overall increased oxidative stress caused by systemic selenoprotein deficiency.
FIGURE 7.
Analysis of Met-SO and Met-RO in mouse plasma. Concentrations of Met-SO and Met-RO were determined in plasma of wild type, heterozygous MsrA knock-out (+/-), homozygous MsrA knock-out (-/-), and selenium-deficient (SD) mice. The error bars represent the standard deviations from four independent experiments.
DISCUSSION
Prior to this study, there was little understanding of the pathways for reduction of free Met-SO and Met-RO in mammalian cells. Although both Met residues and free Met are known to be susceptible to oxidation by ROS, and the reduction of Met sulfoxide residues is carried out by four enzymes in mammals, whether free Met sulfoxides are reduced or wasted was not known. Surprisingly, analysis of human SK-Hep1 hepatocytes grown on media containing Met, Met-RSO or individual Met sulfoxide diastereomers showed a significant difference in cell proliferation and Met sulfoxide consumption between Met-SO and Met-RO (Fig. 2). Met-SO, but not Met-RO, supported the growth of SK-Hep1 cells and was consumed from the medium. In addition, extracts of SK-Hep1 cells showed high free Met-SO reductase activity, whereas free Met-RO activity was not detected (Table 1). Certainly, MsrA was an obvious candidate for the role in free Met-SO reduction, and further experiments confirmed this idea because MsrA-deficient SK-Hep1 cells were no longer able to utilize free Met-SO and could not grow on either Met sulfoxide. These findings, as well as experiments with selenium deficiency and supplementation in SK-Hep1 hepatocytes, also demonstrated that none of the three mammalian MsrBs had a role in providing these cells with Met by enzymatic free Met sulfoxide reduction.
We were able to extend these observations from cell culture to an animal model. Plasma samples from wild type mice had undetectable Met-SO, whereas the bulk of Met sulfoxide occurred in the form Met-RO (∼9 μm). Moreover, MsrA knock-out mice had both diastereomers at similar levels, indicating a specific elevation in Met-SO. Thus, not only hepatocytes, but also mice were deficient in Met-RO reduction and accumulated this compound in plasma. In addition, low levels of Met-SO were clearly caused by reduction of this compound by MsrA. We also found that subjecting wild type mice to selenium deficiency slightly elevated both diastereomers, likely because of increased oxidative stress caused by systemic selenoprotein deficiency. This finding again excluded the role of MsrB1 in the reduction of Met-RO in mice.
A recent report by Lowther and co-workers (22) identified an enzyme, fRMsr, that catalyzes the reduction of free Met-RO in E. coli. However, our sequence analyses revealed that mammals lack this protein. Nevertheless, fRMsr homologs were detected in a variety of prokaryotes as well as in many single-celled eukaryotes. We characterized an fRMsr homolog from S. cerevisiae, which showed robust free Met-RO reductase activity. Furthermore, SK-Hep1 cells expressing this protein grew equally well on Met and Met-RSO media (Fig. 4B). Thus, fRMsr could fully compensate for inability of SK-Hep1 cells to reduce free Met-RO and could provide these cells with Met in quantities sufficient for cell growth. In addition, we observed increased resistance of fRMsr-expressing cells to hydrogen peroxide treatment, suggesting that reduction of free Met sulfoxides provides cells with antioxidant defense as previously proposed (15).
In view of such an important function, it is puzzling why mammals lack fRMsr. Perhaps, environment at the time when animals evolved could have played a role. However, once the enzyme is lost, the ability for Met-RO reduction may be difficult to replace, and mammalian cells are now faced with this defect. In this regard, the finding that a yeast reductase fRMsr can fully compensate for the deficiency in Met-RO reduction could provide avenues for correcting this shortcoming in mammals and other animals.
Met is an essential amino acid in mammals, which is exclusively derived from food sources and linked to many metabolic processes, including protein synthesis, methylation, sulfur metabolism, and ROS scavenging. In particular, Met metabolism has an important role in the synthesis of antioxidant compounds, such as cysteine, taurine, and GSH, through the transsulfuration pathway in liver, kidney, and small intestine. Thus, free Met plays a role in antioxidant defense not only by scavenging ROS (through reversible oxidation) but also as precursor of other antioxidant compounds.
In conclusion, this work revealed an ability of mammalian cells to utilize free Met sulfoxides; however, of the two stereoisomers, only Met-SO is reduced in sufficient quantities, and this occurs in a MsrA-dependent manner. In contrast, mammals are deficient in the reduction of free Met-RO because of a loss of fRMsr in animals during evolution. Importantly, we found that this potential Achilles' heel of the mammalian anti-oxidant repair system could be addressed by expression of a yeast fRMsr, suggesting strategies for improved protection of mammals from oxidative stress and disease and for influencing the aging process.
Supplementary Material
Acknowledgments
We thank Drs. Ruma Banerjee and Bertrand Friguet for providing antibodies and Drs. Rodney Levine and Geumsoo Kim for providing MsrA knock-out mice.
This work was supported, in whole or in part, by National Institutes of Health Grant AG021518 (to V. N. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: ROS, reactive oxygen species; ATF3, activating transcription factor 3; CBS, cystathionine β-synthase; fRMsr, free methionine-R-sulfoxide reductase; OPA, o-phthalaldehyde; Met-SO, methionine-S-sulfoxide; Met-RO, methionine-R-sulfoxide; MsrA, methionine sulfoxide reductase A; MsrB, methionine sulfoxide reductase B; HPLC, high pressure liquid chromatography; DMEM, Dulbecco's modified Eagle's medium; PBS, phosphate-buffered saline; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium.
D. T. Le, B. C. Lee, Y. Zhang, D. E. Fomenko, S. M. Marino, E. Hacioglu, G. H. Kwak, A. Koc, H. Y. Kim, and V. N. Gladyshev, submitted for publication.
References
- 1.Baker, D. H. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 17897-17902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosnan, J. T., and Brosnan, M. E. (2006) J. Nutr. 136 1636S-1640S [DOI] [PubMed] [Google Scholar]
- 3.Stipanuk, M. H. (2004) Annu. Rev. Nutr. 24 539-577 [DOI] [PubMed] [Google Scholar]
- 4.Prudova, A., Bauman, Z., Braun, A., Vitvitsky, V., Lu, S. C., and Banerjee, R. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 6489-6494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elmore, C. L., and Matthews, R. G. (2007) Antioxid. Redox Signal. 9 1911-1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boschi-Muller, S., Olry, A., Antoine, M., and Branlant, G. (2005) Biochim. Biophys. Acta 1703 231-238 [DOI] [PubMed] [Google Scholar]
- 7.Sharov, V. S., Ferrington, D. A., Squier, T. C., and Schöneich, C. (1999) FEBS Lett. 455 247-250 [DOI] [PubMed] [Google Scholar]
- 8.Weissbach, H., Resnick, L., and Brot, N. (2005) Biochim. Biophys. Acta 1703 203-212 [DOI] [PubMed] [Google Scholar]
- 9.Kim, H. Y., and Gladyshev, V. N. (2004) Mol. Biol. Cell 15 1055-1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oien, D. B., and Moskovitz, J. (2007) Biochem. Biophys. Res. Commun. 352 556-559 [DOI] [PubMed] [Google Scholar]
- 11.Haenold, R., Wassef, R., Hansel, A., Heinemann, S. H., and Hoshi, T. (2007) Free Radic. Res. 28 1-13 [DOI] [PubMed] [Google Scholar]
- 12.Vougier, S., Mary, J., and Friguet, B. (2003) Biochem. J. 373 531-537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, H. Y., and Gladyshev, V. N. (2006) BMC Mol. Biol. 7 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, H. Y., and Gladyshev, V. N. (2005) Biochemistry 44 8059-8067 [DOI] [PubMed] [Google Scholar]
- 15.Stadtman, E. R., Moskovitz, J., Berlett, B. S., and Levine, R. L. (2002) Mol. Cell Biochem. 234–235, 3-9 [PubMed] [Google Scholar]
- 16.Moskovitz, J., Bar-Noy, S., Williams, W. M., Requena, J., Berlett, B. S., and Stadtman, E. R. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12920-12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koc, A., Gasch, A. P., Rutherford, J. C., Kim, H. Y., and Gladyshev, V. N. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 7999-8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wassef, R., Haenold, R., Hansel, A., Brot, N., Heinemann, S. H., and Hoshi, T. (2007) J. Neurosci. 27 12808-12816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruan, H., Tang, X. D., Chen, M. L., Joiner, M. L., Sun, G., Brot, N., Weissbach, H., Heinemann, S. H., Iverson, L., Wu, C. F., and Hoshi, T. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2748-2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Etienne, F., Spector, D., Brot, N., and Weissbach, H. (2003) Biochem. Biophys. Res. Commun. 300 378-382 [DOI] [PubMed] [Google Scholar]
- 21.Ezraty, B., Bos, J., Barras, F., and Aussel, L. (2005) J. Bacteriol. 187 231-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, Z., Johnson, L. C., Weissbach, H., Brot, N., Lively, M. O., and Lowther, W. T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 9597-9602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavine, T. F. (1947) J. Biol. Chem. 169 477-491 [PubMed] [Google Scholar]
- 24.Tamura, K., Hua, B., Adachi, S., Guney, I., Kawauchi, J., Morioka, M., Tamamori-Adachi, M., Tanaka, Y., Nakabeppu, Y., Sunamori, M., Sedivy, J. M., and Kitajima, S. (2005) EMBO J. 24 2590-2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukushima, E., Shinka, Y., Fukui, T., Atomi, H., and Imanaka, T. (2007) J. Bacteriol. 189 7134-7144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan, Y., Chen, H., Siu, F., and Kilberg, M. S. (2003) J. Biol. Chem. 278 38402-38412 [DOI] [PubMed] [Google Scholar]
- 27.Okamoto, A., Iwamoto, Y., and Maru, Y. (2006) Mol. Cell. Biol. 26 1087-1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.