Abstract
Two groups of tau, 3R- and 4R-tau, are generated by alternative splicing of tau exon 10. Normal adult human brain expresses equal levels of them. Disruption of the physiological balance is a common feature of several tauopathies. Very early in their life, individuals with Down syndrome (DS) develop Alzheimer-type tau pathology, the molecular basis for which is not fully understood. Here, we demonstrate that Dyrk1A, a kinase encoded by a gene in the DS critical region, phosphorylates alternative splicing factor (ASF) at Ser-227, Ser-234, and Ser-238, driving it into nuclear speckles and preventing it from facilitating tau exon 10 inclusion. The increased dosage of Dyrk1A in DS brain due to trisomy of chromosome 21 correlates to an increase in 3R-tau level, which on abnormal hyperphosphorylation and aggregation of tau results in neurofibrillary degeneration. Imbalance of 3R- and 4R-tau in DS brain by Dyrk1A-induced dysregulation of alternative splicing factor-mediated alternative splicing of tau exon 10 represents a novel mechanism of neurofibrillary degeneration and may help explain early onset tauopathy in individuals with DS.
The microtubule-associated protein tau plays an important role in the polymerization and stabilization of neuronal microtubules. Tau is thus crucial to both the maintenance of the neuronal cytoskeleton and the maintenance of the axonal transport. Abnormal hyperphosphorylation and accumulation of this protein into neurofibrillary tangles (NFTs)2 in neurons, first discovered in Alzheimer disease (AD) brain (1, 2), is now known to be a characteristic of several related neurodegenerative disorders called tauopathies (3). Several different etiopathogenic mechanisms lead to development of NFTs (4).
Adult human brain expresses six isoforms of tau from a single gene by alternative splicing of its pre-mRNA (5, 6). Inclusion or exclusion of exon 10 (E10), which codes for the second microtubule-binding repeat, divides tau isoforms into two main groups, three (3R)- or four (4R)-microtubule-binding repeat tau. They show key differences in their interactions with tau kinases as well as their biological function in the polymerization and stabilization of neuronal microtubules. In the adult human brain, 3R-tau and 4R-tau are expressed at similar levels (5, 7). Several specific mutations in the tau gene associated with frontotemporal dementias with Parkinsonism linked to chromosome 17 (FTDP-17) cause dysregulation of tau E10 splicing, leading to a selective increase in either 3R-tau or 4R-tau. It has therefore been suggested that equal levels of 3R-tau and 4R-tau may be critical for maintaining optimal neuronal physiology (8).
Down syndrome (DS), caused by partial or complete trisomy of chromosome 21, is the most common chromosomal disorder and one of the leading causes of mental retardation in humans. Individuals with DS develop Alzheimer-type neurofibrillary degeneration as early as the fourth decade of life (9). The presence of Alzheimer-type amyloid pathology in DS is attributed to an extra copy of APP gene. However, the molecular basis of neurofibrillary pathology remains elusive.
Alternative splicing of tau E10 is tightly regulated by complex interactions of splicing factors with cis-elements located mainly at E10 and intron 10. Serine/arginine-rich (SR) proteins are a superfamily of conserved splicing factors in metazoans and play very important roles in alternative splicing (10). One such protein, splicing factor 2, also called alternative splicing factor (ASF), plays essential and regulatory roles in alternative splicing of E10 by binding to a polypurine enhancer of exonic splicing enhancer located at tau E10 (11, 12). The function and subcellular localization of ASF is tightly regulated by phosphorylation (13-16).
Dyrk1A (dual-specificity tyrosine phosphorylation-regulated kinase 1A) lies at the Down syndrome critical region of chromosome 21 and contributes to several phenotypes of DS in transgenic mice (17, 18). Multiple biological functions of Dyrk1A are suggested by its interaction with a myriad of cellular proteins including transcription and splicing factors (19). It is distributed throughout the nucleoplasm with a predominant accumulation in nuclear speckles (20, 21), the storage site of inactivated SR proteins, including ASF. Because of its overexpression in DS brain and its predominant localization in nuclear speckles, we hypothesized that Dyrk1A could affect phosphorylation of ASF, and in doing so, disturb ASF-regulated alternative splicing of tau E10, leading to the apparent dysregulation of the balance of 3R-tau and 4R-tau.
In the present study, we provide direct evidence that Dyrk1A can phosphorylate ASF at Ser-227, Ser-234, and Ser-238, driving it into nuclear speckles. By preventing its association with nascent transcripts, phosphorylation of ASF by Dyrk1A causes exclusion of tau E10, leading to an increase in 3R-tau level and an imbalance of 3R-tau and 4R-tau in DS brain. Dysregulation of alternative splicing of tau E10 represents a novel mechanism of neurofibrillary degeneration in DS and offers a unique therapeutic target.
EXPERIMENTAL PROCEDURES
Human Brain Tissue—Tissue from the temporal cortices of six DS and six control brains (see Table 1) for biochemical studies was obtained from the Brain Bank for Developmental Disabilities and Aging of the New York State Institute for Basic Research in Developmental Disabilities. Diagnosis of all human cases was genetically and histopathologically confirmed, and the brain tissue samples were stored at -70 °C until used.
TABLE 1.
Down syndrome and normal control brains used in this study
| Group | Case number | Gender | Age at death | PMIa |
|---|---|---|---|---|
| years | h | |||
| DS | 69 | M | 65 | 4.5 |
| 1139 | F | 58 | 5 | |
| 1162 | F | 55 | 5 | |
| 1238 | M | 55 | 6 | |
| 1283 | F | 59 | 6 | |
| 1342 | M | 61 | 3 | |
| Mean ± DS | 58.8 ± 3.8 | 4.9 ± 1.1 | ||
| Control | 241 | F | 67 | 2.5 |
| 244 | M | 86 | 1.5 | |
| 248 | F | 61 | 7 | |
| 252 | F | 68 | 3 | |
| 255 | F | 67 | 4 | |
| 256 | M | 59 | 6 | |
| Mean ± DS | 68.0 ± 9.5 | 4.0 ± 2.1 |
PMI, postmortem interval.
Plasmids, Proteins, and Antibodies—Recombinant rat Dyrk1A and mammalian expression vector pcDNA3 containing either rat Dyrk1A or kinase-dead Dyrk1AK188R were prepared as described previously (22). pCEP4-ASF-HA, pCEP4-9G8-HA, pCEP4-SC35-HA, and pCEP4-SRp55-HA were gifts from Dr. Tarn of the Institute of Biomedical Sciences, Academia Sinica, Taiwan. pCI-SI9/LI10 containing a tau minigene, SI9/LI10, comprising tau exons 9, 10, and 11, part of intron 9, and the full length of intron 10 has been described (23). Monoclonal antibody 8D9 was raised against a histidine-tagged protein containing the first 160 residues of rat Dyrk1A (24). The monoclonal anti-HA, anti-α-tubulin, and anti-β-actin were bought from Sigma. Monoclonal anti-3R-tau and anti-4R-tau were from Upstate Biotechnology (Lake Placid, NY). Monoclonal anti-tau (tau-5) was from Chemicon International, Inc. (Pittsburgh, PA). Monoclonal anti-human tau (43D) and polyclonal anti-tau (R134d) were described previously (25). Peroxidase-conjugated anti-mouse and anti-rabbit IgG were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA); monoclonal anti-SR protein (1H4) and anti-ASF antibodies, tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-rabbit IgG, and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG were from Santa Cruz Biotechnology (Santa Cruz, CA). The ECL kit was from Amersham Biosciences, and [γ-32P]ATP and [32P]orthophosphate were from MP Biomedicals (Irvine, CA).
Plasmid Construction and DNA Mutagenesis—pGEX-2T-ASF was constructed by PCR amplification from pCEP4-ASF and subcloning into pGEX-2T to express GST-ASF fusion protein. Mutation of Ser-227 to Ala of ASF was achieved by using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) with primers (forward, 5′-caggagtcgcagttacgccccaaggagaagcagag-3′, and reverse, 5′-ctctgcttctccttggggcgtaactgcgactcctg-3′). The mutation of Ser-234 or Ser-238 to Ala was generated by using PCR from vector pGEX-2T-ASF with the same forward primer as described above and different reverse primers (5′-cggaattcttatgtacgagagcgagatctgctatgacggggagaatagcgtggtgctcctctgc-3′ for Ser-234 to Ala and 5′-cggaattcttatgtacgagagcgagatctgctatgacggggagcatagc-3′ for Ser-238 to Ala). The mutations at Ser-227, Ser-234, and Ser-238 to three Ala (ASFS3A) were produced by using PCR to pGEX-2T-ASFS227A with reverse primer (5′-cggaattcttatgtacgagagcgagatctgctatgacggggagcatagcgtggtgctcctctgc-3′). All the PCR products were digested and inserted into pGEX-2T. The mutations were confirmed by DNA sequencing analysis. For mammalian vectors, ASF mutants were constructed by digestion of those ASF mutants in pGEX-2T vector and inserting them into pCEP4 to generate pCEP4-ASFS3A. By using a similar strategy, we also mutated these three Ser residues, 227, 234, and 238, to aspartic acid to generate plasmid pCEP4-ASFS3D.
Cell Culture and Transfection—COS-7, HEK-293T, SH-SY5Y, and HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen) at 37 °C. Normal human neuronal progenitor cells (Lonza, Walkersville, MD) were maintained in Neurobasal supplemented with 2% B27 (Invitrogen), 20 ng/ml fibroblast growth factor 2 (FGF-2), 20 ng/ml epidermal growth factor, and 10 ng/ml leukemia inhibitory factor and differentiated with 10 μm retinoid acid in the maintenance medium for 6 days. All transfections were performed in triplicate with Lipofectamine 2000 (Invitrogen) in 12-well plates. The cells were transfected with 2.4 μg of plasmid DNA and 5 μl of Lipofectamine 2000 in 1 ml Opti-MEM (Invitrogen) for 5 h at 37 °C (5% CO2), after which 1 ml Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum was added. For co-expression experiments, 2.4 μg of total plasmid was used containing 0.8 μg of E10 splicing vector, 0.8 μg of ASF vectors, and 0.8 μg of Dyrk1A vector or their control vectors.
In Vitro Phosphorylation of ASF by Dyrk1A—For in vitro ASF phosphorylation by Dyrk1A, GST-ASF or GST (0.2 mg/ml) was incubated with various concentrations of Dyrk1A in a reaction buffer consisting of 50 mm Tris-HCl (pH 7.4), 10 mm β-mercaptoethanol, 0.1 mm EGTA, 10 mm MgCl2, and 0.2 mm [γ-32P]ATP (500 cpm/pmol). After incubation at 30 °C for 30 min, the reaction was stopped by adding an equal volume of 2× Laemmli sample buffer and boiling. The reaction products were separated by SDS-PAGE. Incorporation of 32P was detected by exposure of the dried gel to phosphorimaging system (BAS-1500, Fujifilm).
GST Pull Down—GST or GST-ASF was purified by affinity purification with glutathione-Sepharose as described above but without elution from the beads. These beads coupled GST or GST-ASF were incubated with crude extract from rat brain homogenate in buffer (50 mm Tris-HCl, pH 7.4, 8.5% sucrose, 50 mm NaF, 1 mm Na3VO4, 0.1% Triton X-100, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 10 μg/ml pepstatin). After a 4-h incubation at 4 °C, the beads were washed with washing buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 1 mm dithiothreitol) six times, the bound proteins were eluted by boiling in Laemmli sample buffer, and the samples were subjected to Western blot analysis.
Co-immunoprecipitation—HEK-293T cells were co-transfected with pCEP4-ASF-HA and pcDNA3-Dyrk1A for 48 h as described above. The cells were washed twice with phosphate-buffered saline (PBS) and lysed by sonication in lysate buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 50 mm NaF, 1 mm Na3VO4, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, 2 μg/ml leupeptin, and 2 μg/ml pepstatin). Insoluble materials were removed by centrifugation; the supernatants were preabsorbed with protein G-conjugated agarose beads and incubated with anti-HA or anti-Dyrk1A antibody 8D9 overnight at 4 °C, and then protein G beads were added. After a 4-h incubation at 4 °C, the beads were washed with lysate buffer twice and with Tris-buffered saline twice, and bound proteins were eluted by boiling in Laemmli sample buffer. The samples were subjected to Western blot analysis with the indicated primary antibodies.
Co-localization Study—HeLa cells were plated in 12-well plates onto coverslips 1 day prior to transfection at 50-60% confluence. HA-tagged ASF constructs were singly transfected or co-transfected with Dyrk1A or Dyrk1Ak188R constructs as described above. Two days after transfection, the cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. After washing with PBS, the cells were blocked with 10% goat serum in 0.2% Triton X-100/PBS for 2 h at 37 °C and incubated with rabbit anti-HA antibody (1:200) and mouse anti-Dyrk1A (8D9, 1:10,000) overnight at 4 °C. After washing and incubation with secondary antibodies (TRITC-conjugated goat anti-rabbit IgG and FITC-conjugated goat anti-mouse IgG, 1:200), the cells were washed extensively with PBS and incubated with 5 μg/ml Hoechst 33342 for 15 min at room temperature. The cells were washed with PBS, mounted with Fluoromount-G, and revealed with a Leica TCS-SP2 laser-scanning confocal microscope.
RNA Interference—For inhibition of ASF and Dyrk1A expression, HEK-293T cells cultured in 12-well plates were transfected with various amounts of short interfering RNA (siRNA) using Lipofectamine 2000. After a 48-h transfection, cells were lysed, and protein and RNA were extracted as described above. siRNA target sequences to ASF were 5′-GTAGAACCCATGTTGTATA-3′, 5′-GTTCCAATGTATTGGTGTA-3′, and 5′-GGAGCTGGATCATTGGATT-3′ (Santa Cruz Biotechnology). Dyrk1A SMARTpool target sequences to Dyrk1A were 5′-TAAGGATGCTTGATTATGA-3′, 5′-GCTAATACCTTGGACTTTG-3′, 5′-GAAAACAGCTGATGAAGGT-3′, and 5′-AAACTCGAATTCAACCTTA-3 (Dharmacon, Lafayette, CO). Both strands of siRNAs had two uridines at 3′. The same amount of scramble siRNA was used for controls.
Quantitation of Tau E10 Splicing by Reverse Transcription-PCR (RT-PCR)—Total cellular RNA was isolated from cultured cells by using an RNeasy mini kit (Qiagen GmbH). One microgram of total RNA was used for first-strand cDNA synthesis with oligo(dT)15-18 by using an Omniscript reverse transcription kit (Qiagen GmbH). PCR was performed by using Prime-STAR™ HS DNA Polymerase (Takara Bio Inc., Otsu, Shiga, Japan) with primers (forward 5′-GGTGTCCACTCCCAGTTCAA-3′ and reverse 5′-CCCTGGTTTATGATGGATGTTGCCTAATGAG-3′) to measure alternative splicing of tau E10 under conditions: at 98 °C for 3 min, at 98 °C for 10 s, and at 68 °C for 40 s for 30 cycles and then at 68 °C for 10 min for extension. The PCR products were resolved on 1.5% agarose gels and quantitated using the Molecular Imager system (Bio-Rad).
Immunohistochemical Staining—The serial sections from the temporal inferior gyrus of six cases of DS and six control cases (see Table 1) were examined. For comparison, brain sections from the same region of three AD cases with a moderately severe stage of the disease (global deterioration scale, stage 6) and three cases with a severe stage of the disease (global deterioration scale, stage 7) were examined by immunohistochemical staining. One brain hemisphere was fixed in 10% buffered formalin, dehydrated in ethanol, and infiltrated and embedded in polyethylene glycol. The tissue blocks were cut serially into 50-μm-thick sections. Monoclonal antibody RD3 (diluted 1:500) was used for detection of 3R-tau, and RD4 (diluted 1:200) was used to detect 4R-tau. The endogenous peroxidase in the sections was blocked with 0.2% hydrogen peroxide in methanol. The sections were then treated with 10% fetal bovine serum in PBS for 30 min to block nonspecific binding. The antibodies were diluted in 10% fetal bovine serum in PBS and were incubated with sections overnight at 4 °C. The sections were washed and treated for 30 min with biotinylated sheep anti-mouse IgG antibody diluted 1:200. The sections were treated with an extravidin peroxidase conjugate (1:200) for 1 h, and the product of reaction was visualized with diaminobenzidine (0.5 mg/ml with 1.5% hydrogen peroxide in PBS). After immunostaining, the sections were lightly counterstained with hematoxylin.
Sarkosyl-insoluble Tau Preparation—Homogenates of control and DS brains prepared in 10 volumes of buffer (50 mm Tris-HCl, 8.5% sucrose, 50 mm NaF, 10 mm β-mercaptoethanol, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 50 nm okadaic acid, and 10 μg/ml aprotinin, leupeptin, and pepstatin) were centrifuged at 16,000 × g for 10 min. The supernatant was adjusted to 1% N-lauroylsarcosine and 1% β-mercaptoethanol and incubated for 1 h at room temperature. After the incubation, supernatant was spun at 100,000 × g for 1 h at 25 °C. The resulting pellet was dissolved in Laemmli sample buffer and subjected to Western blot analysis.
Phosphorylation of ASF in Cultured Cells—COS7 cells were transfected with pCEP4-ASF-HA and cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. After a 45-h transfection, the medium was replaced with [32P]orthophosphate (10 mCi) in Dulbecco's modified Eagle's medium (-phosphate) supplemented with 10% fetal bovine serum plus 10 μm Tg003 and/or 50 μm EGCG. After a 3-h incubation, the cells were harvested in lysate buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 50 NaF, 1 mm Na3VO4, 50 mm okadaic acid, 0.1% Triton X-100, 0.1% Nonidet P-40, 0.25% sodium deoxychalate, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and 10 μg/ml aprotinin, leupeptin, and pepstatin). Insoluble materials were removed by centrifugation, and the supernatant was used for immunoprecipitation as described above. Immunoprecipitated ASF-HA by anti-HA was analyzed by immunoblotting and autoradiography.
Mass Spectrometry—GST-ASF fusion protein was phosphorylated by Dyrk1A as described above. To maximize the yield of the phosphorylated protein, the reaction was carried out for 1 h with high amounts of the kinase (1:6 molar ratio of GST-ASF and GST-Dyrk1A). The phosphorylated products were separated in SDS-PAGE and stained by Coomassie Blue. The gel piece was in-gel tryptic-digested. Proteolytic peptides were extracted from the gel followed by TiO2-based immobilized metal affinity chelate enrichment for the phosphopeptide. The resulting fraction was concentrated and reconstituted in 10 μl of 5% formic acid for liquid chromatography-tandem mass spectrometry analysis separately.
Statistical Analysis—Where appropriate, the data are presented as the means ± S.D. Data points were compared by the unpaired two-tailed Student's t test, and the calculated p values are indicated in Figs. 4, 5 and 6. For the analysis of the correlation between Dyrk1A level and 3R-tau or 4R-tau levels in human brain homogenates, the Pearson product-moment correlation coefficient r was calculated.
FIGURE 4.
Dyrk1A inhibits the role of ASF in tau exon 10 inclusion. a, overexpression of Dyrk1A inhibited the role of ASF in tau exon 10 inclusion. Mini-tau gene pCI-SI9/LI10 was co-transfected with ASF or Dyrk1A into COS7 cells for 48 h, and the total RNA was extracted and subjected for measurement of tau exon 10 splicing by using RT-PCR. Con, control. b, knock down of Dyrk1A by its siRNA elevated exon 10 inclusion induced by ASF. Mini-tau gene was co-transfected into HEK-293T cells with ASF and siRNA of Dyrk1A or its scrambled form, and then tau exon 10 splicing was analyzed by RT-PCR after a 48-h transfection. Expression of endogenous Dyrk1A was decreased by siRNA of Dyrk1A dose-dependently (inset panel). Transfection of Dyrk1A siRNA significantly increased the 4R-tau expression when compared with the scrambled form. c, inhibition of Dyrk1A increased 4R-tau expression in differentiated human neuronal progenitor cells. Human neuronal progenitor cells were differentiated with retinoid acid for 6 days and then treated with 12.5 μm EGCG or 10 μm Harmine for 24 h to inhibit Dyrk1A. The cell lysates were objected to Western blots with anti-3R-tau and anti-4R-tau antibodies. The ratio of 4R-tau and 3R-tau was calculated. The data are presented as mean ± S.D. d, Dyrk1A drove ASF into speckles. HA-ASF and/or Dyrk1A or Dyrk1AK188R were co-transfected into HeLa cells. After a 48-h transfection, the cells were fixed and immunostained by anti-HA and anti-Dyrk1A and followed by TRITC-anti-rabbit IgG or FITC-anti-mouse IgG, respectively. Hoechst was used for nucleus staining. **, p < 0.01 versus control group, #, p < 0.05 versus ASF group.
FIGURE 5.
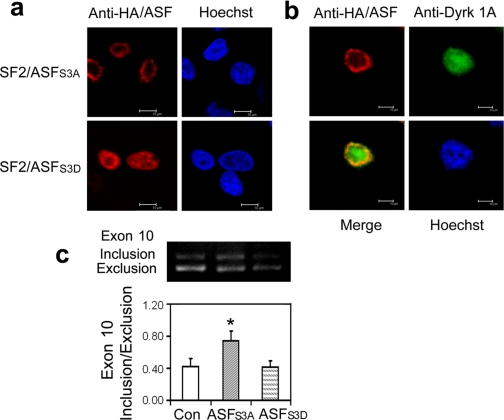
Mutations of ASF affect its subcellular location and role in tau exon 10 inclusion. a, subcellular location of ASF mutants. HA-tagged ASFS3A or ASFS3D was overexpressed in HeLa cells, and then the cells were fixed and immunostained with anti-HA and FITC-labeled secondary antibody. Hoechst was used for nucleus staining. b, Dyrk1A could not drive ASFS3A into speckles. HA-tagged ASFS3A (red) was co-expressed with Dyrk1A (green) in HeLa cells and immunostained as described above. c, effects of ASF mutations on tau exon 10 splicing. The mini-tau gene was co-transfected with ASFS3A or ASFS3D into COS7 cells, and tau exon 10 splicing was analyzed by RT-PCR after a 48-h transfection. *, p < 0.05 versus control (Con) group.
FIGURE 6.
Increased 3R-tau in DS brain correlates with overexpresson of Dyrk1A. a, Dyrk1A level was increased by ∼50% in DS brain. Immuno-dot blots of temporal cortical homogenates from six DS and six control (Con) cases were developed with monoclonal antibody 8D9 to Dyrk1A and quantitated by densitometry. b, total tau was increased in DS brain. Total tau level in temporal cortical homogenates from six DS and six control cases was detected by Western blots with polyclonal antibody R134d and quantitated by densitometry. c and d, the levels of 3R-tau and 4R-tau were altered in DS brain. 3R-tau and 4R-tau in the same samples were measured by Western blots with anti-3R-tau (RD3) and anti-4R-tau (RD4), respectively, quantitated by densitometry, and normalized by total tau level. For the Western blots shown in panels b-d, the amount of proteins applied in homogenates of DS cases was 30% of that in the control cases. **, p < 0.01, and *, p < 0.05. e, NFTs in DS brain were mainly 3R-tau-positive. Series sections from the temporal inferior gyrus of a 58-year-old DS subject (case number 1139) (DS) and AD subject with comparable severe AD (global deterioration scale, stage 7) (AD) were immunostained with anti-3R-tau or anti-4R-tau. f, sarcosyl-insoluble tau in DS brain was dominantly 3R-tau. DS brain extracts were adjusted to 1% N-lauroylsarcosine and 1% β-mercaptoethanol and incubated for 1 h at room temperature. Sarcosyl-insoluble tau was collected from the pellet of 100,000 × g centrifugation at 25 °C and dissolved in Laemmli sample buffer. The brain extracts and the sarcosyl-insoluble protein were analyzed proportionally for the levels of 3R-tau and 4R-tau by Western blots. The levels of 3R-tau and 4R-tau in the sarkosyl-insoluble fraction were normalized with taus in the brain extracts, and the level of 4R-tau was designated as 100%. The data are presented as mean ± S.D. **, p < 0.01, g, correlation of 3R-tau or 4R-tau level (x axis) with Dyrk1A level (y axis). The levels of 3R (left) or 4R-tau (right) measured in c or d were plotted against the level of Dyrk1A measured in e. Open circles are control cases, and closed circles are DS cases.
RESULTS
ASF Promotes Tau E10 Inclusion—3R-tau and 4R-tau are generated by alternative splicing of tau E10, an event known to be regulated by a host of SR family splicing factors, including ASF, 9G8, SC35, and SRp55 (11, 12, 26-28). To compare the E10 splicing efficiency of these SR proteins, we co-transfected mini-tau gene pCI-I9/LI10, consisting of tau exons 9-11, part of intron 9 (SI9), and the full length of intron 10 (LI10) (23), together with each of the four known E10 splicing factors, into HEK-293T cells and detected E10 splicing by RT-PCR. We found that of the four SR proteins, ASF promoted E10 inclusion most effectively (Fig. 1a). This effect was not cell type-specific, as overexpression of ASF in cells including HEK-293T, COS7, HeLa, and SH-SY5Y, also up-regulated 4R-tau production (Fig. 1b).
FIGURE 1.
Overexpression of ASF promotes tau exon 10 inclusion. a, overexpression of SR proteins affected tau E10 splicing. the pCI-SI9/LI10 mini-tau-gene was co-transfected with the same amount of pCEP4-SR proteins into HEK239-T cells. Total RNA was subjected to RT-PCR for measurement of tau exon 10 splicing after a 48-h transfection. b, expression of ASF promoted tau exon 10 inclusion. pCI-SI9/LI10 was co-transfected with pCEP4-ASF into various cell lines indicated under each panel. Tau exon 10 splicing was measured by RT-PCR after a 48-h transfection. c, knock-down of ASF by siRNA inhibited tau exon 10 inclusion. pCI-SI9/LI10 was co-transfected with ASF siRNA or scramble siRNA into HEK-293T cells. The expression of ASF was measured by Western blot (upper panel), and the tau exon 10 splicing was measured by RT-PCR after a 48-h transfection (lower panel). Con, control.
To establish the requirement of ASF in E10 inclusion in vivo, we next knocked down endogenous ASF by transfecting HEK-293T cells with ASF-specific siRNA (Fig. 1c). Decreased ASF expression by siRNA significantly decreased 4R-tau production (Fig. 1c), further supporting an essential role of ASF in tau E10 inclusion.
Dyrk1A Phosphorylates ASF at Ser-227, Ser-234, and Ser-238—The biological activity of ASF is regulated by its phosphorylation (13-15). To study whether Dyrk1A phosphorylated ASF, we phosphorylated GST-ASF in vitro by Dyrk1A. We found that GST-ASF (Fig. 2b), but not GST (Fig. 2d), was phosphorylated by Dyrk1A in an enzyme concentration-dependent manner (Fig. 2b). To study the phosphorylation of ASF by Dyrk1A in vivo, we overexpressed HA-ASF in COS7 cells and labeled the cells with [32P]orthophosphate. ASF is phosphorylated heavily in vivo. To better learn the effect of inhibition of Dyrk1A on ASF phosphorylation, we lowered down the phosphorylation of ASF by Tg003, a Clk/Sty inhibitor, and then added Dyrk1A inhibitor EGCG. We found that Clk/Sty inhibitor significantly decreased 32P incorporation into ASF and increased ASF mobility shift (Fig. 2c). When compared with Tg003 treatment, EGCG further inhibited the 32P incorporation. However, EGCG did not further change the mobility shift (Fig. 2c). These results indicate that ASF is phosphorylated by Dyrk1A at different sites from that phosphorylated by Clk/Sty in cultured cells.
FIGURE 2.
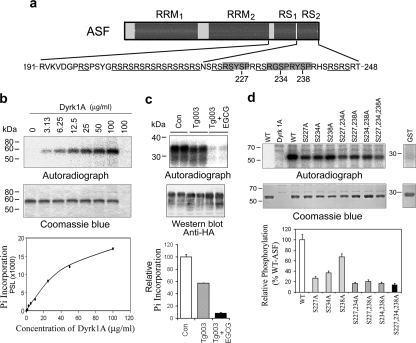
Dyrk1A phosphorylates ASF in vitro at Ser-227, Ser-234, and Ser-238. a, schematic of ASF domain structures and the serine residues (227, 234, and 238) in three conserved motifs (gray box) of Dyrk1A phosphorylation sites that were identified in this study. b, autoradiography of ASF phosphorylation by Dyrk1A in vitro. Recombinant GST-ASF was incubated with various concentrations of Dyrk1A indicated above each lane for 30 min at 30 °C and separated by SDS-PAGE and visualized with Coomassie Blue staining (middle panel). The last lane is Dyrk1A alone, without GST-ASF. After drying the gel, the 32P incorporated into ASF was measured by using a phosphorimaging device (BAS-1500, Fuji) (upper panel). The level of 32P was normalized by the protein level detected by Coomassie Blue staining was plotted to Dyrk1A concentration (lower panel). c, inhibition of Dyrk1A by EGCG decreased the phosphorylation of ASF in cultured cells. COS7 cells were transfected with pCEP4-ASF-HA for 45 h and then treated with 10 μm Tg003 and 50 μm EGCG to inhibit Clk and Dyrk1A, respectively. At the same time, [32P]orthophosphate was added to label the phosphoproteins. After a 3-h treatment and phospho-labeling, the cells were harvested, and the cell lysates were subjected to immunoprecipitation with anti-HA. The immunoprecipitated ASF-HA was analyzed by autoradiography and Western blot with anti-HA. The 32P incorporated into ASF-HA was normalized by ASF-HA level detected with anti-HA. Con, control; Pi, phosphate group. d, mutation of Ser to Ala at Ser-227, Ser-234, and/or Ser-238 inhibited ASF phosphorylation by Dyrk1A. Mutants of GST-ASF at the Ser residue indicated above each lane were phosphorylated by Dyrk1A (10 μg/ml) in vitro for 30 min. 32P incorporation into GST-ASFs was measured by using phosphorimaging analysis after the phosphorylation products were separated by SDS-PAGE. The 32P incorporated was normalized by protein level detected with Coomassie Blue staining. WT, wild type.
To map the putative phosphorylation sites on ASF, we in vitro phosphorylated GST-ASF using a high concentration of Dyrk1A (130 μg/ml, an enzyme/substrate molar ratio of ∼1/6) for 60 min followed by SDS-PAGE separation of the phosphoproducts. A slower mobility of phospho-GST-ASF was evident by Coomassie Blue staining (data not shown), indicating that ASF was phosphorylated by Dyrk1A. Phospho-GST-ASF was then subjected to mass spectrometry after in-gel trypsin digestion. Surprisingly, no phosphorylated peptides were detected (data not shown). Peptide recovery data from mass spectrometry showed the absence of C-terminal 40 amino acid residues, including part of RS1 and all RS2 (Fig. 2a and supplemental Fig. 1). This finding suggested that the phosphorylation sites were probably located within this region.
Dyrk1A is a proline-arginine-directed Ser/Thr protein kinase and prefers an RX(X)(S/T)P motif. ASF contains three such motifs, all of them within 22 amino acid residues of the C-terminal RS2 domain (Fig. 2a). To determine whether Dyrk1A-mediated phosphorylation of ASF occurred within the consensus regions, we mutated three Ser residues within the motifs to Ala, either individually or in combination followed by in vitro phosphorylation by Dyrk1A. We found that any given single mutation decreased ASF phosphorylation, which was further compounded by double or triple mutations (Fig. 2d, lowest panel). This finding suggested that Dyrk1A phosphorylated ASF mainly at Ser-227, Ser-234, and Ser-238 in vitro.
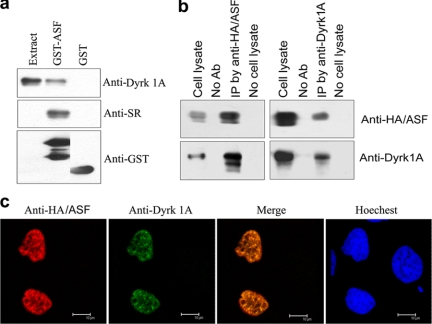
ASF Interacts with Dyrk1A—The in vitro phosphorylation of ASF by Dyrk1A led us to investigate whether the two proteins interact with each other in vitro and in vivo. Employing GST-pulldown assay and immunoprecipitation studies, we found that only GST-ASF, but not GST, could pull down Dyrk1A from rat brain extract (Fig. 3a), and Dyrk1A and ASF could be co-immunoprecipitated by antibodies to each protein (Fig. 3b). These results confirmed the interaction between ASF and Dyrk1A in vitro.
FIGURE 3.
ASF interacts with Dyrk1A. a, Dyrk1A was pulled down from rat brain extract by GST-ASF. GST-ASF or GST coupled onto glutathione-Sepharose was incubated with rat brain extract. After washing, bound proteins were subjected to Western blots by using anti-GST, anti-SR protein, and anti-Dyrk1A. Only GST-ASF, but not GST, pulled down Dyrk1A. All these lanes are from the same blot from which unrelated lanes between the second and third lanes were removed. b, ASF and Dyrk1A could be co-immunoprecipitated by each other's antibodies. ASF tagged with HA and Dyrk1A were co-expressed in HEK-293T cells for 48 h. The cell extract was incubated with anti-HA or anti-Dyrk1A, and then protein G beads were added into the mixture. The bound proteins were subjected to Western blots by using antibodies indicated at the right of each blot. Dyrk1A and HA-ASF were co-immunoprecipitated by each other's antibodies, respectively. No Ab, no antibody; IP, immunoprecipitation. c, co-localization of ASF with Dyrk1A in nucleus. HA-ASF and Dyrk1A were co-transfected into HeLa cells. After a 48-h transfection, the cells were fixed and immunostained by anti-HA or anti-Dyrk1A and followed by TRITC-anti-rabbit IgG or FITC-anti-mouse IgG. Hoechst was used for nucleu staining.
To study their interaction in vivo, we co-expressed HA-tagged ASF (HA-ASF) and Dyrk1A in HeLa cells and established their subcellular localization by using confocal microscopy. Both ASF and Dyrk1A were co-localized in the nucleus and enriched in speckles (Fig. 3c), giving further evidence to their possible interaction in cultured cells.
Dyrk1A Inhibits Tau E10 Inclusion Induced by ASF—To study whether Dyrk1A affects the biological activity of ASF, we determined ASF-mediated tau E10 splicing by overexpressing both Dyrk1A and ASF in COS 7 cells. Splicing products of 3R- and 4R-tau were quantitated by RT-PCR. We found that Dyrk1A significantly inhibited ASF-mediated tau E10 inclusion, and co-expression of ASF with kinase-dead Dyrk1A (Dyrk1AK188R) had no effect on tau E10 splicing (Fig. 4a). Furthermore, knockdown of endogenous Dyrk1A with siRNA (Fig. 4b, inset) increased 4R-tau (Fig. 4b), confirming that ASF-mediated tau E10 splicing is regulated by Dyrk1A.
Differentiated human neuronal progenitor cells with retinoid acid express both 3R-tau and 4R-tau. In these cells, inhibition of Dyrk1A with either harmine or EGCG elevated 4R-tau expression (Fig. 4c, upper panel) and resulted in the increase in the ratio of 4R-tau to 3R-tau (Fig. 4c, lower panel), indicating that Dyrk1A regulates endogenous tau exon 10 splicing.
ASF shuttles between the cytoplasm and the nucleus, as well as within the nucleus, a process dependent on its state of phosphorylation. To determine whether Dyrk1A-induced phosphorylation of ASF also affected its subcellular localization, we overexpressed ASF alone or in combination with either Dyrk1A or kinase-dead Dyrk1AK188R in HeLa cells. Employing laser confocal microscopy, ASF was found to be localized primarily in the nuclear periphery of ASF-only transfected cells (Fig. 4d). When co-expressed with Dyrk1A, ASF translocated into nuclear speckles and co-localized with Dyrk1A (Figs. 3c and 4d), whereas co-expression with Dyrk1AK188R had no effect on ASF localization (Fig. 4d). Together, these results suggest that Dyrk1A drives ASF from the nuclear periphery into speckles, preventing the association of ASF with nascent transcripts and resulting in an increase in 3R-tau production.
ASF promoted tau exon 10 splicing in all tested cell lines (Fig. 1b). However, the promoting effect of ASF on tau exon 10 splicing was not to the same extent in different experiments by using the same cell line (Figs. 1, a and c and 4, a and b), suggesting that basal activities of endogenous splicing factors or their regulators are different in cells with different conditions, which was further confirmed by the study that HEK-293T cells with different passage showed different tau splicing level (supplemental Fig. 2).
ASF Pseudophosphorylated at Ser-227, Ser-234, and Ser-238 Mainly Localizes in Speckles and Does Not Promote Tau E10 Inclusion—To evaluate the central role of Ser-227, Ser-234, and Ser-238 in conferring biological activity to ASF, we mutated these sites to either alanine (ASFS3A) or aspartic acid (ASFS3D) and studied the localization and biological activity of ASF in transiently transfected HeLa cells. We observed that ASFS3A was mainly localized in the nuclear periphery (Fig. 5a), and when co-overexpressed with Dyrk1A, it failed to translocate to the nuclear speckles (Fig. 5b). In contrast, ASFS3D, in which serine residues at the three sites were mutated to aspartate to mimic phosphorylation (pseudophosphorylation), was enriched in speckles (Fig. 5a), a situation reminiscent of the subnuclear localization of ASF upon phosphorylation by Dyrk1A (Figs. 3c and 4d). We further studied whether phosphorylation-resistant ASFS3A promoted tau E10 inclusion in transfected cells. As expected, ASFS3A promoted tau E10 inclusion, whereas pseudophosphorylated ASFS3D did not change the tau E10 splicing pattern in tau transcripts (Fig. 5c). Together, these results strongly suggest that phosphorylation of ASF at Ser-227, Ser-234, and Ser-238 changes its localization from the nuclear periphery, a location of active splicing, to speckles, and in so doing, inhibits ASF-mediated tau E10 inclusion, thereby altering the production of 3R-tau and 4R-tau.
3R-tau Is Increased and Is Correlated to Dyrk1A Overexpression in DS—In DS brain, as expected due to three copies of the Dyrk1A gene, the protein level of Dyrk1A is increased by 50% (Fig. 6a). To study whether the increased dosage of Dyrk1A in DS brain affects the proportion of 3R- and 4R-tau, we subjected brain homogenates from temporal cortices of six DS and six age- and postmortem interval-matched control cases (Table 1) to Western and immuno-dot blots (data not shown) using antibodies against 3R-tau (RD3), 4R-tau (RD4), or total tau (R134d, or a mixture of 43D and tau 5). We found that the level of tau protein was elevated in DS brains to ∼3-fold of control cases (Fig. 6b). This increase was due to a selective increase in 3R-tau (Fig. 6c). The level of 3R-tau (normalized to total-tau) was 4-fold increases, whereas that of 4R-tau (normalized to total-tau) was decreased by ∼25% in DS when compared with control brains (Fig. 6, c and d). Dephosphorylation of tau by alkaline-phosphatase did not affect the labeling by these antibodies (data not shown), confirming that the antibodies were phosphorylation-independent. These results suggest that the proportion of 3R-tau is markedly increased and that the balance of 3R-tau and 4R-tau is disturbed in DS when compared with control brain.
To determine whether increased 3R-tau was associated with neurofibrillary pathology, we compared 3R- and 4R-tau-specific immunostaining in temporal cortices from DS and control cases employing RD3 and RD4 antibodies (Fig. 6e). In control brain, RD3- or RD4-positive NFTs in the temporal cortex were rare (data not shown). However, NFTs in DS cases showed a predominant anti-3R-tau immunoreactivity (Fig. 6e, left panel). In comparable AD cases, the 3R-tau-positive NFTs were 2-3 times less than in similar area from subjects with DS (Fig. 6e, right panel). To learn whether the 3R-tau is predominant in sarkosyl-insoluble tau, we prepared a sarcosyl-insoluble fraction from control and DS brain extracts. In normal human brain, sarcosyl-insoluble tau was not detectable (data not shown). In DS brain, 3R-tau in the sarcosyl-insoluble fraction showed 3.1-fold enrichment when compared with 4R-tau (Fig. 6f), indicating that 3R-tau is predominant in sarcosyl-insoluble tau. These results suggest that neurofibrillary degeneration in the DS brain is primarily associated with 3R-tau.
To study whether overexpression of Dyrk1A correlated with an imbalance of 3R- and 4R-tau, we plotted Dyrk1A levels with 3R-tau or 4R-tau levels in brain homogenate of individual case. We found a strong correlation between 3R-tau and Dyrk1A levels and an inverse correlation between 4R-tau and Dyrk1A levels (Fig. 6g), indicating that the overexpression of Dyrk1A in DS brain may contribute to an increase in 3R-tau/4R-tau ratio.
DISCUSSION
The amyloid pathology in DS brain is believed to be due to an extra copy of the APP gene. However, the presence of NFTs in trisomy 21 and the etiology of the neurofibrillary degeneration seen in DS remain elusive. A close look at the neurofibrillary degeneration of DS reveals a pattern of tau pathology reminiscent of several tauopathies constituting frontotemporal dementia. These tauopathies are caused by dysregulation of alternative splicing of tau E10, causing a shift in the ratio of 3R/4R tau. In the present study, we propose a novel pathogenic mechanism of tau pathology that clearly explains how the imbalance in 3R-tau and 4R-tau develops, which might cause neurofibrillary degeneration, along with brain amyloidosis, leading to early onset Alzheimer-type dementia in individuals with DS. We propose Dyrk1A, a kinase located in the Down syndrome critical region, as a key regulator of tau alternative splicing. By being overexpressed in the DS brain, Dyrk1A alters the nuclear distribution of splicing factor ASF by phosphorylating at Ser-227, Ser-234, and Ser-238, making it unavailable to the tau transcript, and causing a selective increase in 3R-tau level. The abnormal 3R/4R tau ratio resulting from defective alternative splicing of tau E10 is primarily responsible for the Alzheimer-type neurofibrillary degeneration seen in DS (Fig. 7).
FIGURE 7.
The model for the inhibitory role of Dyrk1A in tau exon 10 inclusion via phosphorylation of ASF. ASF binds to polypurine enhancer cis-element of exonic splicing enhancer (ESE) at tau E10 and promotes E10 inclusion. Overexpression of Dyrk1A phosphorylates ASF at Ser-227, Ser-234, and Ser-238, which drives it into speckles from nascent transcripts and leads to E10 exclusion and increase of 3R-tau production. Increased 3R-tau disrupts the balance of 3R-/4R-tau required for normal function of adult human brain and aggregates in affected neurons, which initiates and/or accelerates the formation of NFTs and develops tauopathy in DS brain. ESS, exonic splicing silencer.
Tau E10 splicing is regulated by several SR or SR-like proteins, including ASF, SC35, 9G8, Tra2β, and SRp55 (11, 12, 26-28). We found ASF to be the most effective tau E10 splicing factor when compared with other SR proteins. ASF is known to bind to a polypurine enhancer on tau E10 and has been reported to play essential and regulatory roles in tau E10 inclusion (12). Its activity and subcellular localization are tightly regulated by its degree of phosphorylation. To date, four kinases, including SR protein kinase (SRPK) 1, SRPK2, Clk/Sty, and DNA topoisomerase-I, have been reported to phosphorylate ASF (14, 29-31). Phosphorylation by SRPK1 drives ASF from cytosol into the nucleus, and phosphorylation by Clk/Sty causes release of ASF from speckles, the storage compartments of inactive SR proteins (16, 32). Thus, both SRPK1 and Clk/Sty phosphorylate ASF and recruit it into nascent transcripts, resulting in enhancement of its role in regulation of alternative splicing. Our data identify Dyrk1A as a novel ASF kinase. We have found that Dyrk1A phosphorylates ASF mainly at three sites (Ser-227, Ser-234, and Ser-238) within the consensus motif (RX(X)(T/S)P) of Dyrk1A, none of which are known to be phosphorylated by the other four ASF kinases. In addition to ASF, Dyrk1A also phosphorylates other splicing factors and regulates their activity (20, 21). However, whether phosphorylation of these factors plays any role in the alternative splicing of tau is not known yet. In the present study, we further show that upon phosphorylation by Dyrk1A, ASF is recruited from the nuclear periphery to the speckles, an event that corresponds to a dramatic decrease in tau E10 inclusion. However, the level of ASF remains unchanged in the DS brain (see supplemental Fig. 3). Therefore, we believe that it is the phosphorylation of ASF at Ser-227, Ser-234, and Ser-238 by Dyrk1A that causes dysregulation of tau E10 alternative splicing, leading to an imbalance in 3R-tau and 4R-tau, ultimately laying foundations for the development of neurofibrillary degeneration in DS.
Although hyperphosphorylation of tau plays a fundamental role in the development of Alzheimer-type neurofibrillary degeneration, imbalance in the cellular levels of 3R- and 4R-tau is emerging as an important concept in this pathology. Several lines of evidence, from transgenic mouse models to human tauopathies, emphasize the importance of a critical 3R/4R tau ratio in the cell, which, if disturbed for a given species, may lead to the characteristic neurofibrillary pathology. For example, overexpression of human 3R-tau, but not 4R-tau (33) in mice (where 4R-tau is the only isoform expressed in adult life), produces age-dependent tauopathy (34), whereas in adult Ts65Dn mouse, a transgenic mouse model of DS, there is no evidence of neurofibrillary pathology (35), although tau phosphorylation is increased at several sites (36). Because E10 becomes a constitutively expressed exon in the adult mouse, overexpression of six human tau isoforms in the transgenic mouse 8c model does not produce neurofibrillary degeneration; 3R/4R balance in the Tg 8c mouse is maintained by endogenous 4R-tau (37). However, when the same 8c mouse is crossed with a tau knock-out mouse, the resultant htau mouse now has more 3R-tau than 4R-tau and thus displays abundant neurofibrillary pathology (38). We see a similar pattern of a disturbed 3R/4R balance in several human tauopathies. Close to 50% of all mutations in the tau gene causing human FTDP-17 affect tau exon E10 splicing and alter 3R/4R tau ratio (10). Most of these mutations (N279K, L284L, ΔN296, N296N, N296H, P301S, S305N, S303S, E10 + 3, E10 + 11, E10 + 12, E10 + 13, E10 + 14, and E10 + 16) increase tau exon E10 inclusion and raise the normal 4R/3R ratio from 1 to 2-3 (39). Other tau gene mutations such as ΔK280, E10 + 19, and E10 + 29 decrease E10 inclusion and reduce the 4R/3R ratio (40-42). In addition to FTDP-17, dysregulation of E10 splicing may also contribute to other human tauopathies, such as Pick disease (with a predominant increase in 3R-tau), progressive supranuclear palsy (4R-tau up-regulation), and corticobasal degeneration (4R-tau up-regulation) (43).
4R- and 3R-taus have inherent differences in regulating microtubule dynamics, and the pathology triggered by a 3R/4R tau imbalance may be differentially affected by the particular tau isoform. For example, 3R-tau is known to bind to microtubules with lesser efficiency than the 4R-tau (44-46). We observed an ∼4-fold increase in 3R-tau in the DS brain and believe that the resultant free 3R-tau becomes a readily available substrate for hyperphosphorylation by tau kinases. In the case of human tauopathies with a low 3R/4R ratio, 3R-tau probably becomes free due to the high affinity binding of 4R-tau to microtubules. Free tau has been shown to be a favorable substrate for abnormal hyperphosphorylation than its microtubule-bound counterpart (47). In DS brain, the situation is further compounded by the fact that Dyrk1A not only causes a 3R/4R imbalance, but being a tau kinase, also leads to hyperphosphorylation of tau at many sites including Thr-212. Phosphorylation of tau by Dyrk1A can prime tau for further abnormal hyperphosphorylation by glycogen synthase kinase-3β (36). The high levels of Aβ in DS brain due to an extra dose of the APP gene may also significantly accelerate tau pathology by activating tau kinases including glycogen synthase kinase-3β (GSK-3β) (48). It has been shown that infusion of Aβ in tauP301L transgenic mice causes neurofibrillary degeneration, and the double transgenic mice generated by crossing Tg2567 (swAPP) animals with tauP301L animals have significantly increased neurofibrillary pathology (49, 50). Thus, a multifaceted effect of an extra copy of Dyrk1A and APP gene in DS may help explain the occurrence of neurofibrillary degeneration ∼20 years earlier than in AD. Apart from AD-like pathology, 50% of DS patients have one or more forms of congenital cardiac anomalies. Interestingly, a recent report suggested the fundamental role of ASF in programming excitation-contraction coupling in the cardiac muscle (51). Moreover, ASF was recently identified as an onco-protein in a study examining a number of solid organ tumors, which may provide insight into the 20-fold increase in the incidence of leukemia in DS patients when compared with the general population (52). Thus, Dyrk1A-induced changes in ASF activity may elucidate the molecular biology underlying some of the tell tale features of DS.
In summary, the present study provides novel insights into the etiology of neurofibrillary pathology and presents intriguing data about the molecular basis of the 3R/4R imbalance seen in the DS brain. We show that Dyrk1A level is increased in DS brain due to an extra copy of the Dyrk1A gene located in the Down syndrome critical region. We further show that Dyrk1A phosphorylates the splicing factor ASF at three sites, causing a shift in its nuclear localization and making it unavailable for tau E10 splicing. This results in severe up-regulation of 3R-tau mRNA and is directly responsible for 3R/4R tau imbalance in the DS brain. Dyrk1A may further accelerate neurofibrillary degeneration by contributing to tau hyperphosphorylation. Regulation of ASF-mediated tau E10 alternative splicing by Dyrk1A opens new avenues for research and development of therapeutics for tauopathies.
Supplementary Material
Acknowledgments
We thank Dr. A. Alonso for critical reading, M. Marlow for editorial suggestions, and J. Murphy for secretarial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant Grants AG027429 (to C.-X. G.), AG019158 (to K. I.), and HD43960 (to J. W.). This work was also supported by grants from Nantong University, the New York State Office of Mental Retardation and Developmental Disabilities, and the University of Massachusetts Medical School and by grants from the National Natural Science Foundation of China (Grants 30572076 and 30770468 to F. L.) and Alzheimer's Association (NIRG-08-91126 to F. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains three supplemental figures.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: NFTs, neurofibrillary tangles; DS, Down syndrome; AD, Alzheimer disease; APP, amyloid precursor protein; ASF alternative splicing factor; Dyrk1A, dual-specificity tyrosine phosphorylation-regulated kinase 1A; E10, exon 10; FTDP-17, frontotemporal dementias with Parkinsonism linked to chromosome 17; 3R-tau, three-repeat tau; 4R-tau, four-repeat tau; SR protein, serine/arginine-rich proteins; SRPK, SR protein kinase; TRITC, tetramethyl rhodamine isothiocyanate; FITC, fluorescein isothiocyanate; PBS, phosphate buffered saline; GST, glutathione S-transferase; HA, hemagglutinin; siRNA, small interfering RNA; RT-PCR, reverse transcription-PCR; EGCG, epigallocatechin gallate.
References
- 1.Grundke-Iqbal, I., Iqbal, K., Quinlan, M., Tung, Y. C., Zaidi, M. S., and Wisniewski, H. M. (1986) J. Biol. Chem. 261 6084-6089 [PubMed] [Google Scholar]
- 2.Grundke-Iqbal, I., Iqbal, K., Tung, Y. C., Quinlan, M., Wisniewski, H. M., and Binder, L. I. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 4913-4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballatore, C., Lee, V. M., and Trojanowski, J. Q. (2007) Nat. Rev. Neurosci. 8 663-672 [DOI] [PubMed] [Google Scholar]
- 4.Iqbal, K., Alonso, A. C., Chen, S., Chohan, M. O., El-Akkad, E., Gong. C.-X., Khatoon, S., Li, B., Liu, F., Rahman, A., Tanimukai, H., and Grundke-Iqbal, I. (2005) Biochim. Biophys. Acta 1739 198-210 [DOI] [PubMed] [Google Scholar]
- 5.Goedert, M., Spillantini, M. G., Jakes, R., Rutherford, D., and Crowther, R. A. (1989) Neuron 3 519-526 [DOI] [PubMed] [Google Scholar]
- 6.Andreadis, A., Brown, W. M., and Kosik, K. S. (1992) Biochemistry 31 10626-10633 [DOI] [PubMed] [Google Scholar]
- 7.Kosik, K. S., Orecchio, L. D., Bakalis, S., and Neve, R. L. (1989) Neuron 2 1389-1397 [DOI] [PubMed] [Google Scholar]
- 8.Goedert, M., and Jakes, R. (2005) Biochem. Biophy. Acta 1739 240-250 [DOI] [PubMed] [Google Scholar]
- 9.Wisniewski, K. E., Wisniewski, H. M., and Wen, G. Y. (1985) Ann. Neurol. 17 278-282 [DOI] [PubMed] [Google Scholar]
- 10.D'Souza, I., and Schellenberg, G. D. (2005) Regulation of tau isoform expression and dementia. Biochim. Biophys. Acta 1739 104-115 [DOI] [PubMed] [Google Scholar]
- 11.Kondo, S., Yamamoto, N., Murakami, T., Okumura, M., Mayeda, A., and Imaizumi, K. (2004) Genes Cells 9 121-130 [DOI] [PubMed] [Google Scholar]
- 12.D'Souza, I., and Schellenberg, G. D. (2006) J. Biol. Chem. 281 2460-2469 [DOI] [PubMed] [Google Scholar]
- 13.Gui, J. F., Lane, W. S., and Fu, X.-D. (1994) Nature 369 678-682 [DOI] [PubMed] [Google Scholar]
- 14.Colwill, K., Pawson, T., Andrews, B., Prasad, J., Manley, J. L., Bell, J. C., and Duncan, P. I. (1996) EMBO J. 15 265-275 [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao, S. H., and Manley, J. L. (1998) EMBO J. 17 6359-6367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ngo, J. C. K., Chakrabarti, S., Ding, J.-H., Velazquez-Dones, A., Nolen, B., Aubol, B. E., Adams, J. A., Fu, X.-D., and Ghosh, G. (2005) Mol. Cell 20 77-89 [DOI] [PubMed] [Google Scholar]
- 17.Arron, J. R., Winslow, M. M., Polleri, A., Chang, C. P., Wu, H., Gao, X., Neilson, J. R., Chen, L., Heit, J. J., Kim, S. K., Yamasaki, N., Miyakawa, T., Francke, U., Graef, I. A., and Crabtree, G. R. (2006) Nature 441 595-600 [DOI] [PubMed] [Google Scholar]
- 18.Gwack, Y., Sharma, S., Nardone, J., Tanasa, B., Iuga, A., Srikanth, S., Okamura, H., Bolton, D., Feske, S., Hogan, P. G., and Rao, A. (2006) Nature 441 646-650 [DOI] [PubMed] [Google Scholar]
- 19.Galceran, J., de Graaf, K., Tejedor, F. J., and Becker, W. (2003) J. Neural Transm. Suppl. 67 139-148 [DOI] [PubMed] [Google Scholar]
- 20.Alvarez, M., Estivill, X., and de la Luna, S. (2003) J. Cell Sci. 116 3099-3107 [DOI] [PubMed] [Google Scholar]
- 21.De Graaf, K., Hekerman, P., Spelten, O., Herrmann, A., Packman, L. C., Bussow, K., Muller-Newen, G., and Becker, W. (2006) BMC Biochem. 7 716512921 [Google Scholar]
- 22.Chen-Hwang, M. C., Chen, H. R., Elzinga, M., and Hwang, Y. W. (2002) J. Biol. Chem. 277 17597-17604 [DOI] [PubMed] [Google Scholar]
- 23.Yu, Q., Guo, J., and Zhou, J. (2004) J. Neurochem. 90 164-172 [DOI] [PubMed] [Google Scholar]
- 24.Wegiel, J., Kuchna, I., Nowicki, K., Frackowiak, J., Dowjat, K., Silverman, W. P., Reisberg, B., DeLeon, M., Wisniewski, T., Adayev, T., Chen-Hwang, M. C., and Hwang, Y. W. (2004) Brain Res. 1010 69-80 [DOI] [PubMed] [Google Scholar]
- 25.Liu, F., Iqbal, K., Grundke-Iqbal, I., Hart, G. W., and Gong, C.-X. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10804-10809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao, L., Wang, J., Wang, Y., and Andreadis, A. (2007) Mol. Cell Neurosci. 34 48-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez, F., Perez, M., Lucas, J. J., Mata, A. M., Bhat, R., and Avila, J. (2004) J. Biol. Chem. 279 3801-3806 [DOI] [PubMed] [Google Scholar]
- 28.Wang, Y., Wang, J., Gao, L., Lafyatis, R., Stamm, S., and Andreadis, A. (2005) J. Biol. Chem. 280 14230-14239 [DOI] [PubMed] [Google Scholar]
- 29.Gui, J. F., Tronchère, H., Chandler, S. D., and Fu, X.-D. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 10824-10828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, J., Xiao, S. H., and Manley, J. L. (1998) Genes Dev. 12 2222-2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossi, F., Labourier, E., Forne, T., Divita, G., Derancourt, J., Riou, J. F., Antoine, E., Cathala, G., Brunel, C., and Tazi, J. (1996) Nature 381 80-82 [DOI] [PubMed] [Google Scholar]
- 32.Koizumi, J., Okamoto, Y., Onogi, H., Mayeda, A., Krainer, A. R., and Hagiwara, M. (1999) J. Biol. Chem. 274 11125-11131 [DOI] [PubMed] [Google Scholar]
- 33.Götz, J., Probst, A., Spillantini, M. G., Schäfer, T., Jakes, R., Bürki, K., and Goedert, M. (1995) EMBO J. 14 1304-1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishihara, T., Hong, M., Zhang, B., Nakagawa, Y., Lee, M. K., Trojanowski, J. Q., and Lee, V. M. T. (1999) Neuron 24 751-762 [DOI] [PubMed] [Google Scholar]
- 35.Holtzman, D. M., Santucci, D., Kilbridge, J., Chua-Couzens, J., Fontana, D. J., Daniels, S. E., Johnson, R. M., Chen, K., Sun, Y., Carlson, E., Alleva, E., Epstein, C. J., and Mobley, W. C. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 13333-13338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, F., Liang, Z., Wegiel, J., Hwang, Y. W., Iqbal, K., Grundke-Iqbal, I., Ramakrishna, N., and Gong, C.-X. (2008) FASEB J., in press [DOI] [PMC free article] [PubMed]
- 37.Duff, K., Knight, H., Refolo, L. M., Sanders, S., Yu, X., Picciano, M., Malester, B., Hutton, M., Adamson, J., Goedert, M., Burki, K., and Davies, P. (2000) Neurobiol. Dis. 7 87-98 [DOI] [PubMed] [Google Scholar]
- 38.Andorfer, C., Kress, Y., Espinoza, M., de Silva, R., Tucker, K. L., Barde, Y. A., Duff, K., and Davies, P. (2003) J. Neurochem. 86 582-590 [DOI] [PubMed] [Google Scholar]
- 39.Hutton, M., Lendon, C. L., Rizzu, P., Baker, M., Froelich, S., Houlden, H., Pickering-Brown, S., Chakraverty, S., Isaacs, A., Grover, A., Hackett, J., Adamson, J., Lincoln, S., Dickson, D., Davies, P., Petersen, R. C., Stevens, M., de Graaff, E., Wauters, E., van Baren, J., Hillebrand, M., Joosse, M., Kwon, J. M., Nowotny, P., Che, L. K., Norton, J., Morris, J. C., Reed, L. A., Trojanowski, J., Basun, H., Lannfelt, L., Neystat, M., Fahn, S., Dark, F., Tannenberg, T., Dodd, P. R., Hayward, N., Kwok, J. B., Schofield, P. R., Andreadis, A., Snowden, J., Craufurd, D., Neary, D., Owen, F., Oostra, B. A., Hardy, J., Goate, A., van Swieten, J., Mann, D., Lynch, T., and Heutink, P. (1998) Nature 393 702-705 [DOI] [PubMed] [Google Scholar]
- 40.D'Souza, I., Poorkaj, P., Hong, M., Nochlin, D., Lee, V. M., Bird, T. D., and Schellenberg, D. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 5598-5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanford, P. M., Shepherd, C. E., Halliday, G. M., Brooks, W. S., Schofield, P. W., Brodaty, H., Martins, R. N., Kwok, J. B., and Schofield, P. R. (2003) Brain 126 814-826 [DOI] [PubMed] [Google Scholar]
- 42.van Swieten, J. C., Bronner, I. F., Azmani, A., Severijnen, L. A., Kamphorst, W., Ravid, R., Rizzu, P., Willemsen, R., and Heutink, P. (2007) J. Neuropathol. Exp. Neurol. 66 17-25 [DOI] [PubMed] [Google Scholar]
- 43.Yoshida, M. (2006) Neuropathology 26 457-470 [DOI] [PubMed] [Google Scholar]
- 44.Lu, M., and Kosik, K. S. (2001) Mol. Biol. Cell 12 171-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goode, B. L., Chau, M., Denis, P. E., and Stuart, C. (2000) J. Biol. Chem. 275 38182-38189 [DOI] [PubMed] [Google Scholar]
- 46.Alonso, A. D., Zaidi, T., Novak, M., Barra, H. S., Grundke-Iqbal, I., and Iqbal, K. (2001) J. Biol. Chem. 276 37967-37973 [DOI] [PubMed] [Google Scholar]
- 47.Sengupta, A., Novak, M., Grundke-Iqbal, I., and Iqbal, K. (2006) FEBS Lett. 580 5925-5933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takashima, A., Noguchi, K., Sato, K., Hoshino, T., and Imahori, K. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 7789-7793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Götz, J., Chen, F., van Dorpe, J., and Nitsch, R. M. (2001) Science 293 1491-1495 [DOI] [PubMed] [Google Scholar]
- 50.Lewis, J., Dickson, D. W., Lin, W. L., Chisholm, L., Corral, A., Jones, G., Yen, S. H., Sahara, N., Skipper, L., Yager, D., Eckman, C., Hardy, J., Hutton, M., and McGowan, E. (2001) Science 293 1487-1491 [DOI] [PubMed] [Google Scholar]
- 51.Xu, X., Yang, D., Ding, J. H., Wang, W., Chu, P. H., Dalton, N. D., Wang, H. Y., Bermingham, J. R., Jr., Ye, Z., Liu, F., Rosenfeld, M. G., Manley, J. L., Ross, J., Jr., Chen, J., Xiao, R. P., Cheng, H., and Fu, X.-D. (2005) Cell 120 59-72 [DOI] [PubMed] [Google Scholar]
- 52.Karni, R., de Stanchina, E., Lowe, S. W., Sinha, R., Mu, D., and Krainer, A. R. (2007) Nat. Struct. Mol. Biol. 14 185-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.