Abstract
To ensure inheritance by daughter cells, many low-copy number bacterial plasmids, including the R1 drug-resistance plasmid, encode their own DNA segregation systems. The par operon of plasmid R1 directs construction of a simple spindle structure that converts free energy of polymerization of an actin-like protein, ParM, into work required to move sister plasmids to opposite poles of rod-shaped cells. The structures of individual components have been solved, but little is known about the ultrastructure of the R1 spindle. To determine the number of ParM filaments in a minimal R1 spindle, we used DNA-gold nanocrystal conjugates as mimics of the R1 plasmid. We found that each end of a single polar ParM filament binds to a single ParR/parC-gold complex, consistent with the idea that ParM filaments bind in the hollow core of the ParR/parC ring complex. Our results further suggest that multifilament spindles observed in vivo are associated with clusters of plasmids segregating as a unit.
In eubacteria and archaea, many biologically important processes are carried out by genes encoded on large, low-copy number plasmids. These include genes conferring resistance to antibiotic and heavy metal toxicity, as well as genes involved in host cell invasion and pathogenicity. These large plasmids face a difficult challenge. To reduce the metabolic load they impose on the host cell, their copy numbers must be kept to a minimum (one to two per chromosome equivalent) (1). At such low-copy numbers, they can no longer rely on chance for their maintenance in the bacterial population. To be stably maintained, these low-copy number plasmids must, like chromosomes, be actively segregated to daughter cells before division. Two classes of plasmid segregation systems (Types I and II) have been known and studied for years (2), and recent work has uncovered several more (3, 4). Each system appears to be encoded on a single operon and to be composed of three pieces: 1) a centromeric DNA sequence, 2) a DNA-binding protein, and 3) a polymer-forming protein. The most well understood of these systems is the Type II segregation machinery encoded by the R1 drug-resistance plasmid. Segregation of R1 is driven by the par operon, which consists of a 150-bp centromeric sequence (parC), a repressor protein (ParR) that binds to the centromeric sequence, and a divergent actin-like protein (ParM) that polymerizes into dynamically unstable filaments in the presence of ATP (5). Binding of the ParR/parC complex to ParM filaments stabilizes them against catastrophic disassembly and promotes their elongation via insertional polymerization at the interface with the ParR/parC complex. When both ends of a ParM filament are bound to ParR/parC complexes, elongation of the filament pushes the attached plasmids in opposite directions.
High-resolution structures of components of the R1 spindle are available (6, 7), but provide little information about the ultrastructure of the R1 spindle itself. The atomic structure of ParM monomers reveals a basic similarity to conventional actin, whereas electron microscopy reveals that ParM and actin filaments differ significantly (8). Like actin, ParM filaments are structurally polarized, with distinct ends corresponding to the barbed and pointed ends of a conventional actin filament. Unlike actin, the long-pitch, two-start helix of ParM filaments is left-handed rather than right-handed, and the orientation of the ParM monomer with respect to the filament axis is significantly different from that of actin. Both free and ParR/parC-bound filaments elongate at equal rates from each end. This fact and the polar nature of ParM filaments place constraints on the mechanism of the interaction with the ParR/parC complex as follows. (a) ParR/parC contains two distinct ParM-binding sites, one for “pointed” ends and one for “barbed” ends; (b) spindles are composed of bundles of antiparallel filaments, and the complex recognizes an identical surface at each end of the bundle; or (c) ParR/parC interacts with a surface present at or near both filament ends.
X-ray crystallography of the ParR/parC complex reveals that multiple ParR dimers assemble into a helical ring with parC DNA running around the outside edge. This “segrosome” ring is hollow, with a 6-nm hole through the center. The size of the hole suggests that a single ParM filament (also 6 nm in diameter) might bind the inside of the ParR/parC ring complex (6, 7). By forming a collar around the end of a filament, the ParR/parC complex would pose no barrier to elongation, and if the affinity of the complex for ParM depends on the nucleotide bound to the filament, hydrolysis of ATP within the filament could promote tracking of the complex with the elongating filament end. This is similar to the way in which the Dam1 complex is thought to interact with the dynamic ends of microtubules (9). In addition, a collar encircling a ParM filament might prevent monomer dissociation, providing an attractive explanation for how the ParR/parC complex stabilizes filaments against catastrophic disassembly. This model appears to be at odds with in vivo studies of R1 spindles that suggest plasmids are segregated by spindles composed of multiple filaments (10). However, R1 plasmids are also observed to cluster in vivo, and it is unclear whether multifilament spindles observed in vivo are assembled by pairs of individual plasmids or by pairs of plasmid clusters.
To better understand the coupling of the ParR/parC complex to ParM filaments, we determined the maximum number of filaments that can simultaneously associate with an individual ParR/parC complex. To do this, we attached defined numbers of parC-containing DNA molecules to gold nanoparticles and counted the number of filaments associated with each particle. Preparation of such discrete DNA-Au particles is well established (11, 12). We initiated ParM filament formation in vitro in the presence of DNA-Au nanoparticle conjugates and visualized associated ParM filaments by transmission electron microscopy. The high contrast of gold nanoparticles enabled us to identify the location of ParR/parC complexes relative to ParM filaments.
EXPERIMENTAL PROCEDURES
Preparation of Gold Colloid—Samples of 5- and 10-nm gold colloid were prepared according to a literature procedure (13), described in detail by Claridge et al. (14). The gold particles were used as an electron-dense tag for parC strands.
Hybridization of Thiolated Double-stranded parC DNA—The 150-bp parC was composed of four oligonucleotides (synthesized and purified by polyacrylamide gel electrophoresis, Integrated DNA Technologies, Coralville, IA) as shown in Fig. 1A. The following oligonucleotide sequences were used (cpc1 denotes the strand containing the cp and c1 regions; p2 denotes the strand containing the p and 2 regions): c2, 5′-(thiol)-TTT TTG GTG TTT TTT TGG TGT GTG TTT GGG TAT GTT TTG GGT TTT AAA TGG GTT TGT TTG-3′; cpc1, 5′-TCA AGT TTA CCC CAT TTC AAC CAT CAA TCA ATG ATT ATT TGT CTT GTT TTG GTG TTT TAT TGG GTT GGG TAT GGG TTG TTT TTG GGT TTT GTT TAA AAA-3′; 1, 5′-TTT TTA AAC AAA ACC CAA AAA CAA CCC ATA CCC AAC CCA ATA AAA CAC CAA AAC AAG ACA-3′; and p2, 5′-AAT AAT CAT TGA TTG ATG GTT GAA ATG GGG TAA ACT TGA CAA ACA AAC CCA CTT AAA ACC CAA AAC ATA CCC AAA CAC ACA CCA AAA AAA CAC C-3′.A solution with 6 μm of each of the four oligonucleotides in 100 mm NaCl was heated at 80 °C for 2 min and then cooled to room temperature over 1 h to allow the strands to hybridize.
FIGURE 1.
Gold particles conjugated to defined numbers of parC-containing, double-stranded DNA molecules are prepared by covalent thiol-Au attachment and separated by gel electrophoresis. A, the 150-bp parC construct consists of two regions (1 and 2), with a promoter (p) region in-between; c1, cp, and c2 denote the corresponding regions on the complementary strand. To incorporate a thiol group at one end of the construct, we used synthesized DNA. Each parC strand was divided into two parts (shown schematically as gray and black), for a total of four synthesizable oligonucleotides. The four oligonucleotides were hybridized together before conjugation to gold particles. B, conjugation of thiolated parC to gold particles resulted in a mixture of particles with different numbers of parC attached. The mixture was run in a 3% agarose gel to separate different types of conjugates. Particles conjugated to different numbers of parC sequences were then excised from the gel and extracted. Left lane, gold particles alone. Right lane, gold particles conjugated with DNA. Separation of gold particles with zero, one, two, and three DNA strands attached is clearly shown.
Synthesis of parC-Au Conjugates—DNA-Au conjugates were prepared and isolated according to Zanchet et al. (11) and Loweth et al. (13). A 3% agarose gel was used to separate different types of conjugates.
Protein Purification—Untagged ParM and ParR were purified following procedures described by Garner et al. (15).
Mixing of parC-Au Conjugates with Other Par Components and ATP—ParM and ParR were diluted to 23 and 2 μm, respectively, in Buffer F (100 mm KCl, 30 mm Tris-HCl (pH 7.5), 1 mm MgCl2, and 1 mm Tris(2-carboxyethyl)phosphine). A 20-μl solution of 15–30 nm parC-Au, 350–700 nm ParR, and 2.3 μm ParM in Buffer F/FA (1% (w/v) 400-centipoise methylcellulose, 100 mm KCl, 30 mm Tris-HCl (pH 7.5), 1 mm MgCl2, and 500 μm Tris(2-carboxyethyl)phosphine HCl) (15) was mixed and incubated on ice for 10–20 min while transmission electron microscopy grids were plasma-ionized. The sample was pipetted onto a strip of Parafilm, and 1.5 μl of ATP (100 mm) was added for a total of 15–60 s to initiate ParM polymerization. Experiments without ParR were performed similarly, but with Buffer F in place of ParR to maintain similar sample volume and salt concentration. Control experiments of parC-Au particles alone, all components without ParM, and all components without ATP were performed, with no filaments observed in any of these cases (supplemental Fig. 1). Occasionally gray wisps of ∼50 nm in length attached to the gold particles were observed, all in control experiments, leading us to hypothesize that the wisps are parC-Au strands. These occasional observations were not surprising; uranyl acetate is a known stain for large DNA sequences, but DNA on the order of hundreds of base pairs (such as parC) is more difficult to visualize.
Preparation and Visualization of Samples by Transmission Electron Microscopy—Carbon-coated copper transmission electron microscopy grids (Ted Pella) were ionized for 30 s in a Harrick Plasma PDC-32G oxygen plasma cleaner to increase surface hydrophilicity; the grids were then floated carbon-side down for 1–10 s on the 15-μl sample + 1.5 μl of ATP (10 mm) reaction. The sample was wicked off and allowed to partially air-dry. Once grids were nearly dry, 10 μl of 2% uranyl acetate was added to each grid and incubated for 1 min. The uranyl acetate stain was wicked off, and the grids were then immersed sequentially in two 50-ml tubes of deionized water for a total of 50 s. The water was wicked off, and grids were allowed to air-dry prior to analysis. Transmission electron microscopy was performed using a Phillips Tecnai G2 20.
Determination of Normalized Structure Occurrence—Wide-field images (magnification × 19,500–38,000) for experiments with and without ParR were analyzed, and the observed structures on each image (ParM structures capped on both ends with a gold particle (Au-M-Au), gold particles associated with one end of a ParM filament structure (Au-M), ParM with gold close to end, ParM with gold close to side, ParM filaments alone) were counted manually. The normalized structure occurrence was obtained by dividing the number of that structure type by the total number of structures observed; normalized occurrences for experiments with and without ParR were tabulated separately.
Measurement of ParM Filament Width in Au-M and Au-M-Au Structures—Images (magnification × 38,000–97,000) of Au-M structures (53 total) and Au-M-Au structures (27 total) were analyzed using NIH ImageJ image analysis software (rsb.info.nih.gov/ij/). A line scan perpendicular to the length of each ParM filament yielded a plot of pixel intensity versus distance; the distance between the darkest points of the scan was taken to be the filament width. Five line scans for each filament were taken at different points along the filament, and the mean of the resulting widths was taken to be the average width of the filament. The histograms for each type of structure were obtained by binning the widths: widths of 4.5 < x ≤ 5.5 nm were binned as 5 nm, and so on, up to 8 nm.
RESULTS
Garner et al. (15) found that, in the presence of ParR, polystyrene particles conjugated to hundreds of copies of parC DNA-stabilized bundles of up to 100 ParM filaments. To determine the number of ParM filaments that interact with a single ParR/parC complex, we prepared gold particles conjugated to single copies of parC-containing DNA, an established and well characterized procedure (11, 12). Briefly, we incubated gold particles with thiolated parC-containing DNA to obtain particles conjugated to zero, one, two, or three parC sequences. We separated different sized conjugates by gel electrophoresis (Fig. 1B), excised bands containing individual, conjugated species from the gel, and extracted and purified the monoconjugated particles.
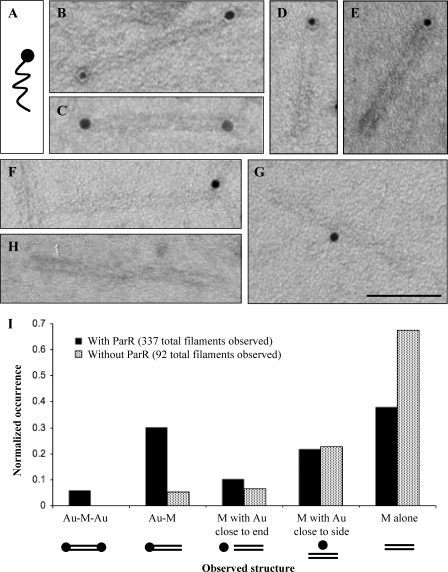
We mixed monoconjugated parC-Au (Fig. 2A) with purified ParR and ParM, added ATP to induce filament formation, and observed the resulting structures by transmission electron microscopy. We then quantitated the specific association between ParM filaments and parC-Au monoconjugates by comparing and counting the types of structures observed in the absence and presence of the DNA-binding protein ParR (Fig. 2I). In the absence of ParR, we rarely observed gold particles associated with ParM filaments (Fig. 2H), whereas in the presence of ParR, we often observed gold particles closely associated with one or both ends of a ParM structure, with no observable gap between the filament and the gold particle. Addition of ParR increased the frequency of Au-M (Fig. 2, D and E) by >5-fold (Fig. 2I). Likewise, in the presence of ParR, we observed Au-M-Au (Fig. 2, B and C), but never observed such Au-M-Au structures in the absence of ParR (Fig. 2I). These data indicate that the majority of the Au-M and all of the Au-M-Au structures seen in the presence of ParR represent parC-Au nanoparticles bound specifically to ParM filaments via ParR.
FIGURE 2.
Individual ParR/parC-Au complexes attach to one or both ends of a single ParM filament. A, monoconjugated parC-Au (schematic) was used as an R1 plasmid mimic to investigate the number of ParM filaments that would bind to a single parC. B and C, single Au-M-Au filaments are shown. D and E, single Au-M filaments are shown. F, ParM filaments with a parC-Au particle close to but separated from the filament end (M with gold close to end) were observed with near equal probability with and without the binding protein ParR. G, ParM filaments with a parC-Au particle close to or touching the filament side (M with gold close to side) were observed with near equal probability with and without ParR. Scale bar = 50 nm. H, uncapped ParM filaments with no gold particles close by (M alone) were observed both with and without ParR. I, normalized histograms (occurrence divided by total number of filaments counted) indicate the numbers of observed structures when monoconjugated parC-Au, ParM, and ATP were combined with and without ParR.
We also observed gold particles in proximity to the end or side of ParM filaments but separated by an observable gap (Fig. 2, F and G). We scored all such cases in which gold particles were within 15 nm of a ParM filament and found that they occurred with equal frequency in the absence and presence of ParR, suggesting that they represent chance proximity between filaments and particles (Fig. 2I). The majority of ParM filaments in the absence and presence of ParR were not colocalized with gold particles (Fig. 2H). This is reasonable because we used 2.3 μm ParM, a concentration at which filaments can form spontaneously (15).
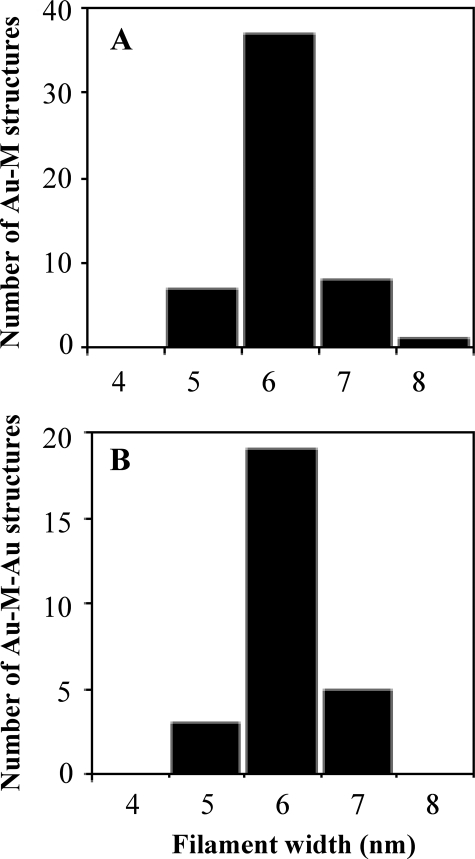
To determine the number of filaments bound to an individual ParR/parC complex, we measured the width of the ParM filaments in Au-M and Au-M-Au structures. In both cases, we measured an average width of 6.0 ± 0.6 nm (Fig. 3), in good agreement with the width of single ParM filaments (16, 17). We never observed ParM structures >8 nm in width, indicating that none of the observed structures (n = 80) contained two filaments.
FIGURE 3.
Measurement of the widths of ParM structures in various relationships with ParR/parC-Au particles demonstrates that the binding capacity of the ParR/parC complex is a single ParM filament. Filament widths are shown for Au-M structures, 53 total measured (A), and Au-M-Au structures, 27 total measured (B). In both types of structures, an average width of 6 nm was observed, in agreement with the accepted width for single ParM filaments.
We observed singly capped filaments more frequently than doubly capped, consistent with a lower probability that a ParM filament will encounter two ParR/parC-Au complexes compared with one. The optimal ParM concentration for electron microscopy was ∼2.5 μm. At higher concentrations (7–15 μm, near the ParM concentration estimated inside R1-containing bacterial cells), the filament density was too high to accurately distinguish capped from uncapped filaments. We also tried assembling spindles at high concentration and diluting them immediately prior to visualization, but found that the filaments were sheared and broken by pipetting. At lower concentrations (approaching 0.6 μm, the critical concentration for ATP ParM filaments (15)), we observed capped filaments but with significantly reduced frequency.
Attempts to increase the encounter frequency by increasing the concentration of ParR/parC-Au particles in solution resulted in a density of gold particles too high to distinguish individual Au-M and Au-M-Au structures.
To determine whether our failure to observe more than one ParM filament per gold particle reflects the binding capacity of the ParR/parC complex or an experimental artifact, we mixed ParM with gold nanoparticles conjugated to four or more parC sequences (Fig. 4A). In the presence of ParR and ATP, the multiconjugated gold particles associated with the ends of bundles composed of up to six ParM filaments (Fig. 4, B–E). These data argue strongly that our results with monoconjugated gold particles reflect the binding capacity of the ParR/parC complex. The occurrence of multifilament spindles under these conditions is consistent with our previous observation of multifilament spindles formed by multiconjugated particles (15) and suggests that, once a single filament spindle is formed, the probability of the tethered particles capturing additional filaments is relatively high.
FIGURE 4.
Multiconjugated ParR/parC-Au particles (four or more parC DNAs per gold particle) interact with bundles containing multiple ParM filaments. A, multiconjugated parC-Au (schematic) was used to investigate filament binding behavior to multiple parC. B–E, ParM bundles with a ParR/parC-Au complex at one or both ends are shown. Scale bar = 50 nm.
DISCUSSION
DNA-Au nanoparticle conjugates have been used for a variety of purposes, including nanoparticle self-assembly (14) and as probes of biomolecular dynamics (18). For small gold nanoparticles (<15 nm in diameter) and DNA oligonucleotides longer than 100 bp, it is straightforward to tune the number of DNA molecules attached to each particle (11, 12). Our in vitro results using DNA-Au monoconjugates to mimic the R1 plasmid demonstrate that a single ParR/parC complex binds either end of a single ParM filament. This implies that, in vivo, pairs of plasmids are segregated by polymerization of single ParM filaments. This is consistent with what is known about the physical properties of ParM filaments. Theoretical considerations and measurements of force produced by polymerizing actin filaments suggest that, in vivo, a growing ParM filament can generate forces on the order of 1 piconewton (19). Stiffness measurements2 indicate that ParM filaments have a persistence length (∼10 μm) several times longer than the length of a bacterial cell (1–3 μm) and that, at bacterial length scales, ParM filaments require piconewton forces to buckle. These forces are several orders of magnitude greater than those required to push plasmids through bacterial cytoplasm (20).
Taken together with recent structural studies of the ParR/parC complex (6, 7), our results suggest that a single 6-nm diameter ParM filament might fit within the ParR segrosome ring. This arrangement would explain three important experimental observations regarding ParM polymer dynamics. By forming a collar around the ParM filament rather than binding the end of the filament “face on,” the ParR/parC ring 1) could interact with identical surfaces at either end of a ParM filament, 2) would not be expected to affect the elongation rate of bound ParM filaments, and 3) could interact with no more than one filament at a time. One hypothesis for how the ParR/parC rings surf on the growing ends of ParM filaments would thus be that the ring has high affinity for ATP-bound portions of the filament and lower affinity for portions of the filament that have hydrolyzed bound ATP (15). An appealing analogy for this situation is the way in which Dam1 complexes are thought to encircle microtubules. Dam1 rings appear to prefer the plus-end of a microtubule, but by applying force to the complex, it can be moved away from the tip and slide along the entire length of the microtubule (21). Additional single molecule experiments will be required to determine whether this is also true for ParR/parC rings.
Finally, we previously found that, in vivo, R1 spindles are occasionally composed of at least two ParM filaments (10). Our results using monoconjugated gold particles suggest that these multifilament spindles contain at least two ParR/parC complexes at each end. This implies that clusters composed of multiple plasmids (22) can segregate together as a single unit in vivo. This is also consistent with our previous observations of plasmid dynamics in vivo. Using plasmids labeled with fluorescently tagged DNA-binding proteins, we often observed segregation of plasmid foci with dramatically different fluorescence intensities (10), suggesting that the two foci contained different numbers of plasmids. Plasmid clustering is not well understood, but we suggest that it may play a previously unsuspected role in replication and segregation in vivo.
Supplementary Material
Acknowledgments
We thank C. S. Campbell for protein preparation and helpful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant PN2 EY016546[ to the University of California San Francisco/University of California Berkeley Nanomedicine Development Center as administered through the University of California San Francisco. This work was also supported by grants from the Sandler Family Supporting Foundation, National Institutes of Health Grant 5R01GM079556-03, and the Director, Office of Science, Office of Basic Energy Sciences, of the United States Department of Energy under Contract DE-AC02-05CH11231 (to R. D. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
C. L. Choi, S. A. Claridge, E. C. Garner, A. P. Alivisatos, and R. D. Mullins, unpublished data.
References
- 1.Nordström, K., Ingram, L. C., and Lundbäck, A. (1972) J. Bacteriol. 110 562–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerdes, K., Møller-Jensen, J., and Bugge Jensen, R. (2000) Mol. Microbiol. 37 455–466 [DOI] [PubMed] [Google Scholar]
- 3.Larsen, R. A., Cusumano, C., Fujioka, A., Lim-Fong, G., Patterson, P., and Pogliano, J. (2007) Genes Dev. 21 1340–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, E., Herrera, N. C., Gunderson, F. Q., Derman, A. I., Dance, A. L., Sims, J., Larsen, R. A., and Pogliano, J. (2006) EMBO J. 25 5919–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Møller-Jensen, J., Jensen, R. B., Löwe, J., and Gerdes, K. (2002) EMBO J. 21 3119–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Møller-Jensen, J., Ringgaard, S., Mercogliano, C. P., Gerdes, K., and Lowe, J. (2007) EMBO J. 26 4413–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumacher, M. A., Glover, T. C., Brzoska, A. J., Jensen, S. O., Dunham, T. D., Skurray, R. A., and Firth, N. A. (2007) Nature 450 1268–1272 [DOI] [PubMed] [Google Scholar]
- 8.Orlova, A., Garner, E. C., Galkin, V. E., Heuser, J., Mullins, R. D., and Egelman, E. H. (2007) Nat. Struct. Mol. Biol. 14 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westermann, S., Wang, H. W., Avila-Sakar, A., Drubin, D. G., Nogales, E., and Barnes, G. (2006) Nature 440 565–569 [DOI] [PubMed] [Google Scholar]
- 10.Campbell, C. S., and Mullins, R. D. (2007) J. Cell Biol. 179 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zanchet, D., Micheel, C. M., Parak, W. J., Gerion, D., and Alivisatos, A. P. (2001) Nano Lett. 1 32–35 [Google Scholar]
- 12.Claridge, S. A., Liang, H. W., Basu, S. R., Frechet, J. M. J., and Alivisatos, A. P. (2008) Nano Lett. 8 1202–1206 [DOI] [PubMed] [Google Scholar]
- 13.Loweth, C. J., Caldwell, W. B., Peng, X. G., Alivisatos, A. P., and Schultz, P. G. (1999) Angew. Chem. Int. Ed. Engl. 38 1808–1812 [DOI] [PubMed] [Google Scholar]
- 14.Claridge, S. A., Goh, S. L., Frechet, J. M. J., Williams, S. C., Micheel, C. M., and Alivisatos, A. P. (2005) Chem. Mater. 17 1628–1635 [Google Scholar]
- 15.Garner, E. C., Campbell, C. S., Weibel, D. B., and Mullins, R. D. (2007) Science 315 1270–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garner, E. C., Campbell, C. S., and Mullins, R. D. (2004) Science 306 1021–1025 [DOI] [PubMed] [Google Scholar]
- 17.van den Ent, F., Møller-Jensen, J., Amos, L. A., Gerdes, K., and Löwe, J. (2002) EMBO J. 21 6935–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sönnichsen, C., Reinhard, B. M., Liphardt, J., and Alivisatos, A. P. (2005) Nat. Biotechnol. 23 741–745 [DOI] [PubMed] [Google Scholar]
- 19.Dye, N. A., and Shapiro, L. (2007) Trends Cell Biol. 17 239–245 [DOI] [PubMed] [Google Scholar]
- 20.Giardini, P. A., Fletcher, D. A., and Theriot, J. A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 6493–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashbury, C. L., Gestaut, D. R., Powers, A. F., Franck, A. D., and Davis, T. N. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pogliano, J., Ho, T. Q., Zhong, Z., and Helinski, D. R. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 4486–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.