Abstract
In polycystic kidney disease (PKD), polycystin-2 (PC2) is frequently mutated or truncated in the C-terminal cytoplasmic tail (PC2-C). The currently accepted model of PC2-C consists of an EF-hand motif overlapping with a short coiled coil; however, this model fails to explain the mechanisms by which PC2 truncations C-terminal to this region lead to PKD. Moreover, direct PC2 binding to inositol 1,4,5-trisphosphate receptor, KIF3A, and TRPC1 requires residues in PC2-C outside this region. To address these discrepancies and investigate the role of PC2-C in PC2 function, we performed de novo molecular modeling and biophysical analysis. De novo molecular modeling of PC2-C using the ROBETTA server predicts two domains as follows: an EF-hand motif (PC2-EF) connected by a linker to a previously unidentified C-terminal coiled coil (PC2-CC). This model differs substantially from the current model and correlates with limited proteolysis, matrix-assisted laser desorption/ionization mass spectroscopy, N-terminal sequencing, and improved coiled coil prediction algorithms. PC2-C is elongated and oligomerizes through PC2-CC, as measured by analytical ultracentrifugation and size exclusion chromatography, whereas PC2-EF is globular and monomeric. We show that PC2-C and PC2-EF have micromolar affinity for calcium (Ca2+) by isothermal titration calorimetry and undergo Ca2+-induced conformational changes by circular dichroism. Mutation of predicted EF-hand loop residues in PC2 to alanine abolishes Ca2+ binding. Our results suggest that PC2-CC is involved in PC2 oligomerization, and PC2-EF is a Ca2+-sensitive switch. PKD-associated PC2 mutations are located in regions that may disrupt these functions, providing structural insight into how PC2 mutations lead to disease.
Polycystic kidney disease (PKD)4 is among the most common life-threatening inherited disorders, with clinical consequences characterized by renal and hepatic cysts (1). Most cases of PKD (>95%) are linked with mutations in the genes Pkd1 or Pkd2, which encode the membrane protein polycystin-1 (PC1) and the calcium (Ca2+)-permeable channel, polycystin-2 (PC2), respectively (2, 3). PC2 belongs to the TRP channel family and is expressed in most tissues (4–8). It has six transmembrane spans, and both C and N termini are cytoplasmic. PC1 and PC2 interact directly and co-localize to primary cilia where they are hypothesized to be necessary for a mechanosensory or chemosensory response that triggers a rise in intracellular Ca2+ (9). Many aspects of PC2 function are mediated by the cytoplasmic C-terminal tail (PC2-C), including co-assembly with PC1 through the PC1 C-terminal cytoplasmic tail (10–13). Two-thirds of pathogenic mutations in PC1 and >90% of pathogenic mutations in PC2 result in truncations of their C-terminal cytoplasmic regions and are predicted to abrogate interaction between the proteins. There are no structural data describing PC2, and current descriptions of PC2-C lack functional and biochemical information.
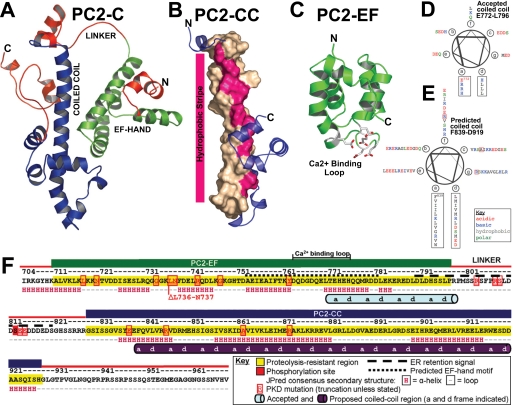
The currently accepted domain model of PC2-C consists of an EF-hand motif overlapping with a short coiled coil (6, 12). Helical wheel projection of this purported coiled coil (Glu772–Leu796) shows that residues in positions “a” and “d” of the helix (i.e. the coiled coil interface) are hydrophilic or charged and would be unfavorable in a coiled coil interface (Fig. 1D). PKD-associated truncations have been identified outside of these proposed domains in PC2-C (Fig. 1F), suggesting that residues important for PC2 function are located in the truncated regions. Moreover, PC2 binds directly to several proteins, including the inositol 1,4,5-trisphosphate receptor (14), TRPC1 (15), tropomyosin-1 (16), Id2 (17), troponin-1 (18), KIF3A (19), and PC1 (9, 20). All of these interactions are dependent on residues C-terminal to the currently accepted domain organization of PC2-C. To address these discrepancies and to aid future studies into the molecular basis of PKD pathogenesis and progression, we constructed a validated structural model of PC2-C.
FIGURE 1.
De novo modeling of PC2-C predicts a two-domain structure connected by a flexible linker. A, model of PC2-C (Ile704–Val968) obtained from ROBETTA showing PC2-EF (green), PC2-CC (blue), and proteolytically sensitive regions identified by limited proteolysis (red). B, ROBETTA model of PC2-CC (Gly828–His927) shown as schematic with the central helix (Phe839–Met870) depicted as surface. Coiled coil interface a and d residues predicted by MARCOIL are highlighted pink. C, ROBETTA model of PC2-EF alone (Lys719–Met800) showing putative Ca2+-binding residues (sticks). D, helical wheel projection of PC2-C (Glu772–Leu796) previously reported to contain a coiled coil, and E, of (Phe839–Asp919) proposed in this study to be the actual coiled coil domain. F, schematic of PC2-C domain structure. PC2-EF (green), PC2-CC (blue) and proteolytically sensitive regions (red) are colored accordingly. Mutations listed are from the Autosomal Dominant Polycystic Kidney Disease (ADPKD) Database (PKDB). Our proposed coiled coil domain (purple cylinder) is shown with a and d coiled coil interface residues labeled.
We have determined a de novo molecular model of PC2-C using the ROBETTA server (21–23) and validated characteristics of this model using biophysical and biochemical analysis. We show that PC2-C contains two domains, a single EF-hand motif (PC2-EF) connected by a linker to a coiled coil domain (PC2-CC). We propose that PC2-CC is the actual coiled coil domain that has been assumed in the literature. Our results suggest that PC2-CC is involved in PC2 oligomerization and that PC2-EF acts as a Ca2+-sensitive switch. PKD-associated PC2 truncation mutations are located in regions that would disrupt these functions, providing insight into how PC2 mutations may lead to disease.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—Fragments of polycystin-2 are as follows. PC2-C (Ile704–Val968), PC2-EF (Asn720–Pro797), and PC2-CC (Gly828–His927) were PCR-amplified from human PC2 cDNA (obtained from S. Somlo, Yale University), cloned into pET-28 (a+) (Novagen), and transformed into BL21(DE3) CodonPlus RIL (Stratagene) for bacterial expression. Mutant variants of PC2-EF (PC2-EF-X-Z T771A/E774A) and PC2-CC (L842P, V846E, M849K, I853P, I856K, V846E/I856K, and M849K/V863E) were created using QuikChange multiple site-directed mutagenesis kit (Stratagene). Cells were grown to OD595 ∼0.4 at 37 °C and then shifted to 18 °C after induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside at ∼18 h. Cells were harvested and resuspended in Buffer A (20 mm Tris, 500 mm NaCl, pH 8.0), lysed by freeze thaw/sonication with lysozyme (∼1 mg/ml) (Sigma), and clarified by centrifugation. Supernatant was loaded onto a 1-ml HisTrap column (GE Healthcare). The column was washed with 30 column volumes of Buffer A and then with 10 column volumes of Buffer A + 50 mm imidazole. Purified polycystin-2 fragments were eluted in Buffer A + 500 mm imidazole and applied to a Superdex 200 SEC column equilibrated in Buffer A. PC2-CC was treated with thrombin (1 unit/mg) for all experiments to remove the His tag. PC2-C and PC2-EF contain an N-terminal His tag with the sequence “MGSSHHHHHHSSGLVPRGSHM.”
Molecular Modeling—PC2-C (Ile704–Val968) and PC2-EF (Asn720–Pro797) were submitted to the full-chain structure prediction server ROBETTA (21–23). Models were analyzed for secondary structure content using iMolTalk (24) and visualized using PyMOL. Coordinates for the ROBETTA models are included as supplemental data. The ROBETTA model for PC2-C-(801–968) and PC2-EF-(719–800) were also ligated to create a theoretical model of PC2-C (supplemental Fig. 2). Coordinates for this theoretical model are available in the supplemental data file.
Limited Proteolysis Domain Mapping—PC2-C was treated with trypsin (1:100 w/w) or proteinase K (1:10,000 w/w) in 25 mm Tris, 250 mm NaCl, pH 8.0. Reactions were quenched using proteinase inhibitor mixture (Roche Applied Science). Products were resolved on SDS-PAGE, transferred to polyvinylidene difluoride, and bands sent for N-terminal sequencing (Tufts University Core Facility). Identical reaction products were sent for MALDI-MS (Tufts University Core Facility). MALDI-MS results were analyzed using FindPept (25).
Circular Dichroism—CD spectra (260–185 nm) were recorded on an AVIV 215 spectrophotometer (Lakewood, NJ) at 25 °C with 1-nm step size, 1-nm bandwidth, 3 s averaging time, and 1-mm path length. PC2-C and PC2-EF were stripped of Ca2+ with 100 mm EDTA. Protein concentrations were measured by A280 (extinction coefficients: 9970 m-1 cm-1 (PC2-C), 1490 m-1 cm-1 (PC2-EF), and 6990 m-1 cm-1 (PC2-CC)), and Bradford assay (Bio-Rad). Samples were diluted in double distilled H2O and spectra recorded at 0.17 mg/ml (PC2-C), 0.10 mg/ml (PC2-EF), and 0.05 mg/ml (PC2-CC). PC2-C (0.17 mg/ml) and PC2-EF (0.10 mg/ml) spectra were also recorded in the presence of 5 mm Ca2+. Three spectra were averaged before processing. CD spectra were converted to mean residue ellipticity [θ] (degrees cm2 dmol-1 residue-1) using IgorCD in IgorPro. Spectra were base-line corrected by subtracting averaged data (250–260 nm). Secondary structure calculations were performed using DICHROWEB and CDSSTR (26–28). PC2-C and PC2-EF spectra (260–190 nm) in the presence and absence of Ca2+ were fit with reference set 7, whereas PC2-CC spectra (260–185 nm) were fit with reference set 6 (26). Secondary structure values were compared with the secondary structure content of de novo models of PC2-C (Ile704–Val968), PC2-EF (Lys719–Met800), or PC2-CC (Gly827–His927), respectively.
Isothermal Titration Calorimetry—Measurements were done on a VP ITC (MicroCal, Northampton, MA) instrument; PC2-C (208 μm) was in the cell, and Ca2+ (2.5 mm) was in the syringe. For PC2-EF (188 μm) and PC2-EF-X-Z (T771A/E774A) (180 μm), Ca2+ was 5 mm. Proteins were stripped of Ca2+ with 100 mm EDTA and desalted in 25 mm Tris, 250 mm NaCl, pH 7.5. 79 injections of 3 μl were made at 210-s intervals. Data were fit to a 1-site model using ORIGIN 7.0 (MicroCal), and dissociation constant (Kd), stoichiometry (n), and enthalpy (ΔH) of binding determined.
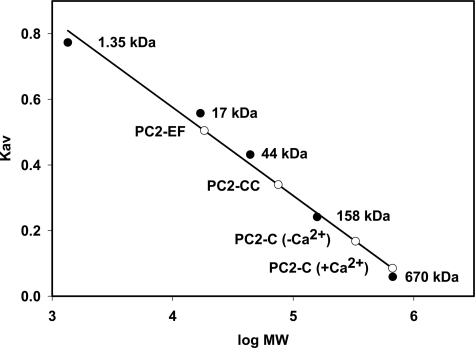
Size Exclusion Chromatography—PC2-C, PC2-EF, or PC2-CC (0.5 ml each) was applied to a Superdex 200 analytical grade SEC column (GE Healthcare) equilibrated in 25 mm Tris, 250 mm NaCl, pH 8.0, at 0.1 ml/min. The column was calibrated using the following: thyroglobulin 670 kDa, bovine γ-globulin kDa 158, chicken ovalbumin 44 kDa, equine myoglobin 17 kDa, and vitamin B12 1.35 kDa (Bio-Rad). The distribution coefficient, KAV, was measured and plotted versus log (molecular weight): KAV = (Ve - Vo)/(Vt - Vo) where Ve = elution volume; Vo = column void volume; Vt = total volume. KAV was calculated for PC2-C (+Ca2+), PC2-C (-Ca2+) (following treatment with 100 mm EDTA), PC2-EF, and PC2-CC.
Analytical Ultracentrifugation—Sedimentation equilibrium experiments were performed with a Beckman Optima XL-I at the Center for Analytical Ultracentrifugation of Macromolecular Assemblies (University of Texas Health Science Center, San Antonio, Dept. of Biochemistry). AUC experiments were analyzed with UltraScan version 9.5 (29). Hydrodynamic corrections were made according Laue et al. (30), as implemented in UltraScan. The partial specific volumes of PC2-EF, PC2-CC, and PC2-C were estimated according to Cohn and Edsall (31), as implemented in UltraScan yielding 0.722, 0.732, and 0.717 cm3/g, respectively. Equilibrium data were fit to multiple models, and the best chosen based on fitting statistics and visual inspection of the residual run patterns. 2DSA and Monte Carlo analyses were calculated on a Linux Beowulf cluster at the Bioinformatics Core Facility, Department of Biochemistry, University of Texas, Health Science Center, San Antonio, and on Lonestar/Teragrid (Texas Advanced Computing Center, University of Texas, Austin). Samples were analyzed in a buffer containing 300 mm NaCl. Sedimentation velocity experiments were performed at 20 °C and 60,000 rpm. Absorbance samples were spun in two-channel Epon/charcoal centerpieces in the AN-60-TI rotor. Loading concentrations of 1.0 OD (optical density at 280 nm) for PC2-CC, 0.25 OD for PC2-EF, and 1.0 OD for PC2-C were scanned at 280 nm with 0.001-cm step size setting and no averaging, corresponding to 120 μm for PC2-CC (ε = 6970 OD280 mol-1 cm-1), 163 μm for PC2-EF (ε = 1280 OD280 mol-1 cm-1), and 87 μm for PC2-C (ε = 9530 OD280 mol-1 cm-1). Extinction coefficients were estimated according to Gill and von Hippel (32). Velocity data were analyzed with 2DSA (33) combined with Monte Carlo analysis (34), direct nonlinear least squares fitting of finite element solutions of the Lamm equation (35) combined with the enhanced van Holde-Weischet method as implemented in UltraScan (36).
RESULTS AND DISCUSSION
Molecular Modeling of the C-terminal Tail of Polycystin-2 Predicts Two Domains Connected by a Flexible Linker—De novo structural models of PC2-C (Ile704–Val968) and PC2-EF (Lys719–Met800) were obtained as output from the ROBETTA server (coordinate files are available as supplemental material). The PC2-C model predicts an α-helical, two-domain elongated structure connected by a linker containing a known PC2 phosphorylation site (Ser812) (37). Domain 1 (∼Ile704–Ser794) consists of a globular α-helical bundle (PC2-EF), whereas domain 2 (∼Gly828–Val968) contains a striking ∼40-residue-long central α-helix (Tyr836–Lys876) characteristic of coiled coil-containing proteins (PC2-CC) (Fig. 1A). Unexpectedly, domain 1 was not modeled as an EF-hand motif in the top theoretical structure received as output from the ROBETTA server. Because residues believed to participate in Ca2+ binding were improperly positioned for Ca2+ coordination in this model, and the de novo protocol utilized in ROBETTA is optimized for single domain proteins, we submitted the sequence for the predicted EF-hand domain alone (Lys719–Met800) for de novo modeling to the ROBETTA server. Modeling the smaller PC2-EF fragment resulted in a canonical single EF-hand motif with a well defined Ca2+-binding site (Fig. 1C). The ROBETTA protocol does not attempt to predict or model Ca2+-binding sites; thus the presence of a credible Ca2+ coordination loop in our model is independent evidence in support of the presence of a Ca2+-binding EF-hand motif in PC2-C. Interestingly, a structural domain with similarities to a coiled coil (domain 2) was found C-terminal to the position of the purported coiled coil domain in the currently accepted model of PC2-C. To investigate the possibility that a previously undiscovered coiled coil motif is present in the C-terminal cytoplasmic tail of PC2, the sequence of PC2-C was analyzed for coiled coil propensity using the program MARCOIL (38) which contains an improved, more stringent prediction algorithm. The resulting output from MAR-COIL predicts that residues Phe839–Asp919 have a high probability to form a coiled coil (supplemental Fig. 3). Heptad repeat residues (a–d) were mapped and found to correspond to a hydrophobic stripe on the long α-helix in the PC2-CC model (Fig. 1, B and E and supplemental Fig. 1). Typically, a–d residues are hydrophobic and make up the coiled coil interface (39).
The results of our de novo modeling for the cytoplasmic tail of PC2 suggest that PC2-CC is the actual coiled coil domain assumed in the literature. Residues within PC2-CC are necessary for direct binding to PC2-interacting proteins (PC1, TRPC1, and KIF3A) and PC2-PC2 oligomerization (9, 14–20). Furthermore, PC2-CC includes the most C-terminal pathogenic PKD-associated truncation variant (R872X) (40). This truncation product is unable to interact with the C terminus of PC1 (12), leading to a plausible hypothesis for PKD pathogenesis. These modeling studies identify a previously unreported coiled coil domain within the C-terminal cytoplasmic tail of PC2, which may serve as an oligomerization interface ablated by PKD-associated truncations.
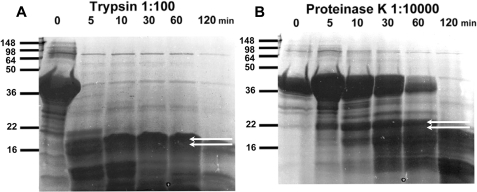
Domain Mapping Using Limited Proteolysis Validates the Presence and Relative Positions of Two Domains within the C-terminal Tail of Polycystin-2—To test the validity of our de novo structural model, we mapped the domain structure of PC2-C using limited proteolysis experiments coupled with MALDI-MS and N-terminal sequencing. PC2-C (Ile704–Val968) was treated with trypsin or proteinase K to identify proteolytically resistant fragments under single hit kinetics (Fig. 2). Two proteolytically resistant bands from each reaction were analyzed by N-terminal sequencing and correspond to fragments that begin with the sequences: “ALVKL” (Ala711–Leu715) and “NTVDD” (Asn720–Asp724) from trypsin digestion and “SFPRS” (Ser804–Ser808) and “SSRRR” (Ser823–Arg827) from proteinase K digestion. Notably, both proteinase K cleavage sites occur within the predicted linker region of our PC2-C model. Given the broad substrate specificity of proteinase K, these results support the presence of an interdomain linker between residues Arg803 and His822. Identical proteolysis reaction products were analyzed by MALDI-MS and matched with fragments of the PC2-C sequence using the N-terminal sequencing results as a constraint, corresponding to Ala711–Pro797 and Gly828–His927. These results were obtained before modeling was complete because full chain automated structure prediction using the ROBETTA server can take several months. The residues corresponding to these PC2-C fragments were mapped onto our de novo model of PC2-C and overlap with the positions of the predicted domains, validating our two domain model and the position of the linker region (Fig. 1D). Using these results we generated PC2-EF (Asn720–Pro797) and PC2-CC (Gly828–His927) for biophysical analysis.
FIGURE 2.
Limited proteolysis identifies two stable domains, PC2-EF and PC2-CC. Time course of limited proteolysis of PC2-C using (A) trypsin (1:100 w/w) or (B) proteinase K (1:10,000 w/w) as analyzed by SDS-PAGE. Proteolytically resistant bands analyzed by N-terminal sequencing are indicated with white arrows and correspond to cleavage at ALVKL and NTVDD for trypsin and SFPRS and SSRRR for proteinase K. Identical limited proteolysis reaction products were analyzed by MALDI-MS to identify fragments corresponding to PC2-EF and PC2-CC.
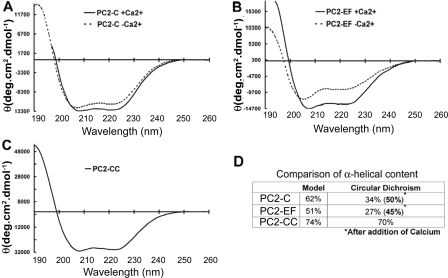
Circular Dichroism Shows That PC2-C Has a Highly α-Helical Fold and Undergoes Ca2+-induced Conformational Changes—The de novo model of PC2-C predicts an all α-helical protein. To test this, we compared the secondary structure content of the PC2-C model with values obtained by CD spectroscopy. CD spectra were recorded for PC2-C, PC2-EF, and PC2-CC and show each to contain high α-helical content in agreement with our modeling results (Fig. 3). Secondary structure content, calculated using DICHROWEB (27), was compared with percentage α-helicity from our structural model of PC2-C for the entire model and for individual domains. Because the model of PC2-C suggested a Ca2+ binding domain, we also investigated the Ca2+ sensitivity of the conformation of PC2-C. PC2-C and PC2-EF spectra were recorded in the absence and presence of Ca2+. PC2-EF shows an increase from ∼27 to ∼45% α-helicity after Ca2+ addition, and PC2-C increases from ∼34 to ∼50% α-helicity with Ca2+. These values for Ca2+-bound PC2-C (∼50%) and PC2-EF (∼45%) correspond with our structural models of PC2-C (62%) and PC2-EF (51%). Secondary structure content calculated from CD spectra for PC2-CC (∼70% α-helix) is also in agreement with our structural model of the coiled coil domain (74%). CD thus validates the α-helical fold predicted by our model of PC2-C and suggests that Ca2+ binding induces a conformational change within the EF-hand domain.
FIGURE 3.
CD of PC2-C, PC2-EF, and PC2-CC indicate high α-helical secondary structure content. CD spectra were recorded for PC2-C, PC2-EF, and PC2-CC, and secondary structure values were compared with the secondary structure content of the de novo models of PC2-C (Ile704–Val968) or PC2-EF (Lys719–Met800). CD spectra of PC2-C (A) or PC2-EF (B) were recorded in the presence (smooth line) and absence (dotted line) of Ca2+. C, CD spectrum of PC2-CC. D, α-helical content of PC2-C, PC2-EF, and PC2-CC estimated from CD spectra using DICHROWEB in comparison with values obtained from de novo models of PC2-C (Ile704–Val968) or PC2-EF (Lys719–Met800).
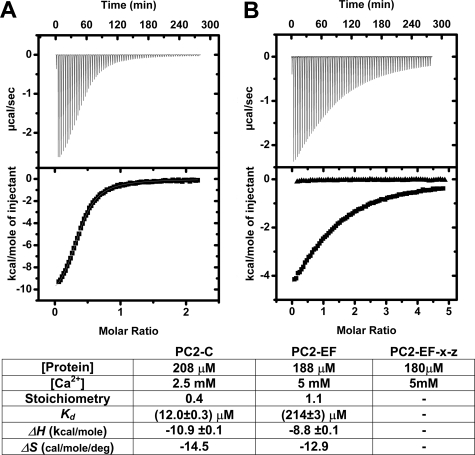
PC2-C Contains a Ca2+-binding EF-hand Domain by Isothermal Titration Calorimetry That May Be Regulated by Residues Outside of the EF-hand Fold—The presence of an EF-hand motif within the C-terminal cytoplasmic tail of PC2 has been reported (41), but not experimentally confirmed. Our structural model and CD analysis suggest that this EF-hand domain binds Ca2+. To verify and quantify this Ca2+ binding, we conducted isothermal titration calorimetry (ITC) (Fig. 4). Fitting ITC isotherms with a one-site binding model indicates that PC2-C binds Ca2+ with a Kd of ∼12 μm and a stoichiometry of 1:0.4, whereas PC2-EF binds Ca2+ with lower affinity (Kd ∼ 214 μm) and 1:1 stoichiometry. To verify that residues predicted by our ROBETTA model of the EF-hand domain of PC2 are directly involved in Ca2+ binding, we created PC2-EF-X-Z (T771A/E774A) by site-directed mutagenesis. Thr771 and Glu774 are predicted to be directly involved in Ca2+ coordination according to our model (supplemental Fig. 4), and the resulting ITC isotherms following Ca2+titration indicate a complete loss of affinity for Ca2+ (Fig. 4B).
FIGURE 4.
Isothermal titration calorimetry of PC2-C and PC2-EF identifies a functional Ca2+-binding site. A, PC2-C (208 μm); B, PC2-EF (188 μm) or PC2-EF-X-Z (T771A/E774A) (180 μm, filled triangles) was titrated with Ca2+, and the resulting isotherm was fit to a one-site binding model. Bottom, ITC fitting results and derived thermodynamic parameters for PC2-C and PC2-EF Ca2+ titrations.
Interestingly, our attempts to prepare PC2-C in a Ca2+-free state by treatment of proteins with EDTA resulted in a binding stoichiometry of 1:0.4, indicating that a significant fraction of PC2-C remained in the Ca2+-bound state. This result contrasts with PC2-EF, where identical treatment seems to have removed all bound Ca2+. The reduced effectiveness of EDTA treatment correlates with the 17-fold lower affinity of the isolated EF-hand domain. The stoichiometry values for Ca2+ binding do not affect the final Kd values obtained from fitting procedures. The differential affinity for Ca2+ of PC2-EF alone versus PC2-C implies that regions of PC2-C outside of PC2-EF are involved in stabilization or regulation of the Ca2+-binding site and support the presence of a physiologically relevant Ca2+-binding site within PC2.
Oligomerization of the C-terminal Tail of Polycystin-2 Is Mediated by an Extended Coiled Coil Domain as Shown by Size Exclusion Chromatography—Our structural model and MAR-COIL analysis predict that PC2-CC contains a coiled coil domain with a hydrophobic stripe. We investigated whether this region of PC2-C is important for oligomerization. Analytical SEC was used to analyze the conformation and oligomerization states of PC2-C, PC2-EF, and PC2-CC (Fig. 5). PC2-C (32 kDa) elutes at ∼670 kDa, suggesting both oligomerization and an elongated molecular conformation. Stripping Ca2+ from PC2-C reduces the apparent molecular weight by 50%, implying a change in oligomerization state or molecular conformation driven by Ca2+. PC2-EF (12 kDa) elutes at ∼19 kDa, consistent with the calculated molecular weight of a monomer, whereas PC2-CC (12 kDa) elutes at ∼75 kDa, indicating both oligomerization and an elongated conformation. To test the possibility that residues corresponding to positions a and d in the PC2-CC coiled coil interface are directly mediating PC2 oligomerization, we created PC2-CC variants with substitutions of hydrophobic a or d residues to either glutamate, lysine, or proline. (PC2-CC L842P, V846E, M849K, I853P, I856K, V846E/I856K, or M849K/V863E). All point mutations tested resulted in complete insolubility of recombinant PC2-CC suggesting the importance of the integrity of the coiled coil interface for proper folding and possibly oligomerization. (supplemental Fig. 5) It is possible that the extended hydrophobic interface created by these a and d residues was disrupted by the insertion of charged residues or putative helix breaking proline residues resulting in nonspecific aggregation and misfolding. Interestingly, although mutations resulting in truncation of the entire coiled coil segment have been identified in PKD patients within our proposed coiled coil domain, no point mutations resulting in amino acid substitutions have been found. These findings support our structural model of PC2-C and further show that PC2-C is an extended oligomeric macromolecule with PC2-CC responsible for this oligomerization.
FIGURE 5.
Size exclusion chromatography of PC2-C, PC2-EF, and PC2-CC shows the coiled coil motif in PC2-CC may mediate PC2 oligomerization. A Superdex 200 analytical grade SEC column was calibrated using thyroglobulin 670 kDa, bovine γ-globulin kDa 158, chicken ovalbumin 44 kDa, equine myoglobin 17 kDa, vitamin B12 1.350 kDa. KAV was determined for each standard (closed circles) and plotted versus log molecular weight. KAV was determined (open circles) for PC2-C (+Ca2+), PC2-EF, and PC2-CC and their apparent molecular weights determined. KAV for PC2-C (-Ca2+) was measured following treatment with EDTA. PC2-C elutes at ∼670 kDa, but this shifts to ∼330 kDa following removal of Ca2+. PC2-EF elutes at ∼19 kDa consistent with a monomeric state. PC2-CC elutes at ∼75 kDa consistent with an extended conformation and oligomerization.
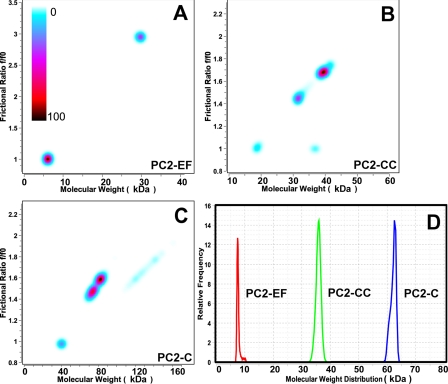
Analytical Ultracentrifugation Shows That the C-terminal Tail of Polycystin-2 Has an Extended Oligomeric Structure with Oligomerization Mediated by the Coiled Coil Domain—To further examine the oligomerization state and molecular conformation of PC2-C, PC2-EF, and PC2-CC, we performed high speed sedimentation velocity ultracentrifugation experiments. In a velocity experiment, sample composition can be determined and multiple components resolved. The van Holde-Weischet analysis provides a model-independent view of the sedimentation coefficient distribution (42), whereas 2DSA resolves heterogeneity in the solution according to both shape and molecular weight. Combining the 2DSA with a Monte Carlo analysis reduces the effects of stochastic noise on the appearance of false-positive peaks and enhances the inherent signal in the data (34). We used a color gradient indicating the relative concentration of solutes to create pseudo-three-dimensional plots (Fig. 6). Van Holde-Weischet analysis showed that all samples were mostly free of aggregates and degradation products, and 2DSA analysis showed one predominant species with narrow confidence bands, and several minor species with little signal. Velocity sedimentation results suggest that PC2-CC is mostly present as an oligomer (trimer or tetramer) with an elongated shape, and that PC2-EF is monomeric with globular shape (Fig. 6, A and B). PC2-C appears to be present in a monomer-dimer equilibrium in velocity sedimentation experiments, which at the concentration tested (87 μm) appears to be mostly shifted to the dimer state (Fig. 6C). These results are also in agreement with our SEC analysis, and support a model of PC2-C as an oligomeric elongated macromolecule whose oligomerization is mediated by the C-terminal coiled coil domain, PC2-CC, identified in this study.
FIGURE 6.
Velocity sedimentation analysis indicates PC2-CC mediates oligomerization of PC2-C and has an elongated molecular shape. Velocity data were analyzed with 2DSA combined with Monte Carlo analysis (A–C), and results are shown in pseudo-three-dimensional plots versus frictional coefficient (f/f0). f/f0 >1.0 indicate an extended molecular shape. The relative concentration of solutes is shown using a color gradient (light blue to black with black centered peaks corresponding to the highest concentration). A, PC2-EF exists as a monomer with globular shape. B, PC2-CC is present as an elongated oligomeric species (trimer or tetramer). C, PC2-C appears to be elongated and present in a monomer-dimer equilibrium. D, van Holde-Weischet differential sedimentation distributions plotted as molecular weight distributions. All proteins appear narrowly distributed in the sedimentation coefficient.
Conclusion—In PKD, PC2 is frequently truncated in PC2-C. The currently accepted domain model of PC2-C fails to explain the mechanisms by which truncations in PC2-C lead to PKD. Moreover, direct PC2 binding to proteins involved in PKD progression requires residues outside this region of PC2-C. To address these discrepancies and provide a framework to investigate the role of PC2-C in PC2 function and PKD progression, we performed de novo molecular modeling and validated this model through biochemical and biophysical analysis.
Structural modeling using the ROBETTA server predicts an extended α-helix C-terminal to the currently accepted coiled coil domain of PC2. The correlation of our structural model with limited proteolysis and MARCOIL results suggests that there is a previously unidentified coiled coil domain C-terminal to the EF-hand motif, and that this is the actual coiled coil domain assumed in the literature. Using AUC and SEC, we found the EF-hand motif domain to be monomeric, whereas the coiled coil domain forms oligomers. Mutagenesis of a and d heptad repeat residues predicted to form the PC2 coiled coil interface result in complete insolubility of recombinant PC2-CC, suggesting their importance in folding and possibly oligomerization. This is in agreement with findings for TRP channels where a C-terminal coiled coil region mediates channel oligomerization (43), but it remains to be tested on active full-length PC2 ion channels.
Ca2+ binding to the EF-hand motif domain was investigated using ITC, CD, and site-directed mutagenesis. This is the first experimental data supporting the presence of a Ca2+-binding site within PC2. EF-hand motif proteins bind Ca2+ with affinities ranging from nanomolar to millimolar (44), and our ITC experiments for the EF-hand domain of PC2 suggest one Ca2+-binding site with micromolar affinity. Given the relatively high concentration of Ca2+ predicted at the mouth of the PC2 channel pore and locally because of opening of associated ryanodine or inositol 1,4,5-trisphosphate receptor channels at the endoplasmic reticulum surface, a micromolar affinity for Ca2+ may allow PC2-EF to sense local changes in Ca2+ concentration. This result, coupled with the conformational changes observed upon Ca2+ binding, suggests a functional role for Ca2+ in PC2 activation.
We report here the first molecular model of the C-terminal cytoplasmic tail of PC2. This model provides a framework to guide experiments probing normal PC2 function and the molecular basis of PKD pathogenesis and progression. Our results validate the presence of a functional Ca2+-binding site and identify a previously unreported coiled coil domain in PC2. These analyses further suggest that the coiled coil domain may serve as a PC2 oligomerization interface. Additionally, the presence of a coiled coil encompassing ∼Tyr836–Lys876 may provide a structural explanation for previous reports that show the importance of this region for PC2 binding to PC1 and other proteins. Significantly, all known PKD-associated PC2 truncations disrupt this new PC2 coiled coil domain. Our results thus provide a plausible molecular mechanism for PKD pathogenesis.
Supplementary Material
Acknowledgments
We thank E. Folta-Stogniew of the Keck Facility, Yale University, for help conducting the ITC experiments and S. Somlo, L. Cohen, and S. Jordt for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants DK57328 (to B. E. E.), CA009085 (training grant to E. T. P.), RR022200 (UltraScan software development), and CA054174 (to the Center for Analytical Ultracentrifugation of Macromolecular Assemblies facility). This work was also supported in part by a grant from the PKD Foundation, UltraScan software development was also supported by the Robert J. Kleberg Jr., and Helen C. Kleberg Foundation, and AUC calculations were supported by National Science Foundation Grant TGMCB070038 (to B. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and coordinates for the ROBETTA models and the theoretical model PC2-C.
Footnotes
The abbreviations used are: PKD, polycystic kidney disease; PC2, polycystin-2; PC2-C, polycystin-2 C-terminal cytoplasmic domain; PC2-EF, polycystin-2 EF-hand domain; PC2-CC, polycystin-2 coiled coil domain; ITC, isothermal titration calorimetry; AUC, analytical ultracentrifugation; MALDI-MS, matrix-assisted laser desorption/ionization mass spectroscopy; 2DSA, two-dimensional spectrum analysis; SEC, size exclusion chromatography.
References
- 1.Wu, G., D'Agati, V., Cai, Y., Markowitz, G., Park, J. H., Reynolds, D. M., Maeda, Y., Le, T. C., Hou, H., Jr., Kucherlapati, R., Edelmann, W., and Somlo, S. (1998) Cell 93(2), 1, 7-188 [DOI] [PubMed] [Google Scholar]
- 2.Somlo, S., and Ehrlich, B. (2001) Curr. Biol. 11(9), R356-360 [DOI] [PubMed] [Google Scholar]
- 3.Clapham, D. E. (2003) Nature 426 517-524 [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez-Perrett, S., Kim, K., Ibarra, C., Damiano, A. E., Zotta, E., Batelli, M., Harris, P. C., Reisin, I. L., Arnaout, M. A., and Cantiello, H. F. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 1182-1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koulen, P., Cai, Y., Geng, L., Maeda, Y., Nishimura, S., Witzgall, R., Ehrlich, B. E., and Somlo, S. (2002) Nat. Cell Biol. 4 191-197 [DOI] [PubMed] [Google Scholar]
- 6.Mochizuki, T., Wu, G., Hayashi, T., Xenophontos, S. L., Veldhuisen, B., Saris, J. J., Reynolds, D. M., Cai, Y., Gabow, P. A., Pierides, A., Kimberling, W. J., Breuning, M. H., Deltas, C. C., Peters, D. J., and Somlo, S. (1996) Science 272 1339-1342 [DOI] [PubMed] [Google Scholar]
- 7.Anyatonwu, G. I., and Ehrlich, B. E. (2005) J. Biol. Chem. 280 29488-29493 [DOI] [PubMed] [Google Scholar]
- 8.Luo, Y., Vassilev, P. M., Li, X., Kawanabe, Y., and Zhou, J. (2003) Mol. Cell. Biol. 23 2600-2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Somlo, S., and Markowitz, G. S. (2000) Curr. Opin. Nephrol. Hypertens. 9 385-394 [DOI] [PubMed] [Google Scholar]
- 10.Tsiokas, L., Kim, E., Arnould, T., Sukhatme, V. P., and Walz, G. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 6965-6970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu, G. M., and Arnaout, M. A. (2002) Genomics 79 87-94 [DOI] [PubMed] [Google Scholar]
- 12.Qian, F., Germino, F. J., Cai, Y., Zhang, X., Somlo, S., and Germino, G. G. (1997) Nat. Genet. 16 179-183 [DOI] [PubMed] [Google Scholar]
- 13.Hanaoka, K., Qian, F., Boletta, A., Bhunia, A. K., Piontek, K., Tsiokas, L., Sukhatme, V. P., Guggino, W. B., and Germino, G. G. (2000) Nature 408 990-994 [DOI] [PubMed] [Google Scholar]
- 14.Li, Y., Wright, J. M., Qian, F., Germino, G. G., and Guggino, W. B. (2005) J. Biol. Chem. 280 41298-41306 [DOI] [PubMed] [Google Scholar]
- 15.Tsiokas, L., Arnould, T., Zhu, C., Kim, E., Walz, G., and Sukhatme, V. P. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 3934-3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Q., Dai, Y., Guo, L., Liu, Y., Hao, C., Wu, G., Basora, N., Michalak, M., and Chen, X. Z. (2003) J. Mol. Biol. 325 949-962 [DOI] [PubMed] [Google Scholar]
- 17.Li, X., Luo, Y., Starremans, P. G., McNamara, C. A., Pei, Y., and Zhou, J. (2005) Nat. Cell Biol. 7 1202-1212 [DOI] [PubMed] [Google Scholar]
- 18.Li, Q., Shen, P. Y., Wu, G., and Chen, X. Z. (2003) Biochemistry 42 450-457 [DOI] [PubMed] [Google Scholar]
- 19.Li, Q., Montalbetti, N., Wu, Y., Ramos, A., Raychowdhury, M. K., Chen, X. Z., and Cantiello, H. F. (2006) J. Biol. Chem. 281 37566-37575 [DOI] [PubMed] [Google Scholar]
- 20.Delmas, P. (2004) Biol. Res. 37 681-691 [DOI] [PubMed] [Google Scholar]
- 21.Chivian, D., Kim, D. E., Malmstrom, L., Schonbrun, J., Rohl, C. A., and Baker, D. (2005) Proteins 61 Suppl. 7, 157-166 [DOI] [PubMed] [Google Scholar]
- 22.Chivian, D., Kim, D. E., Malmstrom, L., Bradley, P., Robertson, T., Murphy, P., Strauss, C. E., Bonneau, R., Rohl, C. A., and Baker, D. (2003) Proteins 53 Suppl. 6, 524-533 [DOI] [PubMed] [Google Scholar]
- 23.Kim, D. E., Chivian, D., and Baker, D. (2004) Nucleic Acids Res. 32 W526-W531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diemand, A. V., and Scheib, H. (2004) Nucleic Acids Res. 32 W512-W516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gattiker, A., Bienvenut, W. V., Bairoch, A., and Gasteiger, E. (2002) Proteomics 2 1435-1444 [DOI] [PubMed] [Google Scholar]
- 26.Sreerama, N., and Woody, R. W. (2000) Anal. Biochem. 287 252-260 [DOI] [PubMed] [Google Scholar]
- 27.Whitmore, L., and Wallace, B. A. (2004) Nucleic Acids Res. 32 W668-W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobley, A., Whitmore, L., and Wallace, B. A. (2002) Bioinformatics (Oxf.) 18 211-212 [DOI] [PubMed] [Google Scholar]
- 29.Demeler, B. (2005) in Modern Analytical Ultracentrifugation: Techniques and Methods (Scott, D. J., Harding, S. E., and Rowe, A. J., eds) pp. 210-229, Royal Society of Chemistry, Cambridge
- 30.Laue, T. M., Shah, B. D., Ridgeway, T. M., and Pelletier, S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding, S. E., Rowe, A. J., and Horton, J. C., eds) pp. 90-125, Royal Society of Chemistry, Cambridge
- 31.Cohn, E. J., and Edsall, J. T. (1943) Proteins, Amino Acids, and Peptides as Ions and Dipolar Ions, Reinhold, New York
- 32.Gill, S. C., and von Hippel, P. H. (1989) Anal. Biochem. 182 319-326 [DOI] [PubMed] [Google Scholar]
- 33.Brookes, E., Boppana, R. V., and Demeler, B. (2006) Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, November 11–17, 2006, Association for Computing Machinery, New York
- 34.Demeler, B., and Brookes, E. (2007) Colloid Polym. Sci. 286 129-137 [Google Scholar]
- 35.Demeler, B., and Saber, H. (1998) Biophys. J. 74 444-454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demeler, B., and van Holde, K. E. (2004) Anal. Biochem. 335 279-288 [DOI] [PubMed] [Google Scholar]
- 37.Cai, Y., Anyatonwu, G., Okuhara, D., Lee, K. B., Yu, Z., Onoe, T., Mei, C. L., Qian, Q., Geng, L., Wiztgall, R., Ehrlich, B. E., and Somlo, S. (2004) J. Biol. Chem. 279 19987-19995 [DOI] [PubMed] [Google Scholar]
- 38.Delorenzi, M., and Speed, T. (2002) Bioinformatics (Oxf.) 18 617-625 [DOI] [PubMed] [Google Scholar]
- 39.Lupas, A. N., and Gruber, M. (2005) Adv. Protein Chem. 70 37-78 [DOI] [PubMed] [Google Scholar]
- 40.Reynolds, D. M., Hayashi, T., Cai, Y., Veldhuisen, B., Watnick, T. J., Lens, X. M., Mochizuki, T., Qian, F., Maeda, Y., Li, L., Fossdal, R., Coto, E., Wu, G., Breuning, M. H., Germino, G. G., Peters, D. J., and Somlo, S. (1999) J. Am. Soc. Nephrol. 10 2342-2351 [DOI] [PubMed] [Google Scholar]
- 41.Pei, Y., Wang, K., Kasenda, M., Paterson, A. D., Liang, Y., Huang, E., Lian, J., Rogovea, E., Somlo, S., and St. George-Hyslop, P. (1998) Kidney Int. 53 1127-1132 [DOI] [PubMed] [Google Scholar]
- 42.Demeler, B., Saber, H., and Hansen, J. C. (1997) Biophys. J. 72 397-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuruda, P. R., Julius, D., and Minor, D. L., Jr. (2006) Neuron 51 201-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gifford, J. L., Walsh, M. P., and Vogel, H. J. (2007) Biochem. J. 405 199-221 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.