Abstract
Renal tubulo-interstitial inflammation is frequently associated with polyuria and urine concentration defects. This led us to investigate the effects of the major pro-inflammatory nuclear factor κB (NF-κB) pathway on aquaporin 2 (AQP2) expression by the collecting duct. Using immortalized collecting duct principal cells (mpkCCDcl4), we found that, acting independently of vasopressin, activation of NF-κBby lipopolysaccharide (LPS) decreased AQP2 mRNA and protein levels in a time- and dose-dependent manner but did not decrease AQP2 mRNA stability. Consistently, constitutively active IκB kinase β decreased AQP2 expression. The LPS-induced decrease in AQP2 mRNA levels was confirmed in rat kidney slices and was reproduced both under conditions of elevated cAMP concentration and V2 receptor antagonism. Computer analysis of the AQP2 promoter revealed two putative κB elements. Mutation of either κB element abolished the LPS-induced decrease of luciferase activity in cells expressing AQP2 promoter-luciferase plasmid constructs. Chromatin immunoprecipitation revealed that LPS challenge decreased p65, increased p50 and p52, and had no effect on RelB and c-Rel binding to κB elements of the AQP2 promoter. RNA-mediated interference silencing of p65, p50, and p52 confirmed controlled AQP2 transcription by these NF-κB subunits. We additionally found that hypertonicity activated NF-κB in mpkCCDcl4 cells, an effect that may counteract the Tonicity-responsive enhancer binding protein (TonEBP)-dependent increase in AQP2 gene transcription. Taken together, these findings indicate that NF-κB is an important physiological regulator of AQP2 transcription.
Selective transcellular water permeability across the renal epithelium is facilitated by the aquaporin 1 (AQP1)3 water channel, expressed in the proximal tubule and thin descending limb of Henle's loop (1, 2) and AQP2, -3, and -4, expressed in collecting duct (CD) principal cells (3). AQP2 inserted in the apical plasma membrane enhances CD apical water permeability. Water exits these cells via basolateral AQP3 and AQP4. By controlling both short term (plasma membrane insertion) and long term (transcriptional and post-transcriptional) AQP2 expression, the antidiuretic hormone [8-arginine]vasopressin (AVP) plays a major role in regulating water reabsorption (3–7). In addition to AVP, AQP2 expression is influenced by numerous factors that act independently of AVP including other hormones, i.e. aldosterone and insulin (8, 9), extracellular calcium (10), and environmental tonicity (11–13).
The NF-κB family of transcription factors consists of five members (p65, p50/p105, p52/p100, RelB, and c-Rel) that contain a Rel-homology domain required for the formation of various combinations of homo- and heterodimers and for DNA binding. Generally, dimers that contain p65 or c-Rel are transcriptional activators, whereas p50 and p52 homodimers act as repressors. The differential dimerization that occurs between NF-κB family members allows for cell-specific transcriptional modulation of target genes in response to a large number of stimuli. This accounts for the role played by NF-κB in the expressional regulation of genes involved in numerous physiological functions including the immune and inflammatory response, environmental stress response, cell survival, cell growth, proliferation and cycling, and neuronal signaling (14, 15). Rel-homology domain is also involved in NF-κB interaction with the family of IκB inhibitory proteins that prevent NF-κB from translocating to the nucleus. In the canonical NF-κB signaling pathway, activation of IκB kinases (IKK), predominantly by IKKβ, leads to phosphorylation, polyubiquitination, and subsequent proteasomal degradation of IκB (predominately IκBα), allowing translocation of released NF-κB (mostly p50-p65 and p50-c-Rel dimers) to the nucleus and transcriptional activation of target genes (15).
Renal tubulo-interstitial inflammation is frequently associated with polyuria and urinary concentration defects (16, 17). Urinary concentration defects associated with decreased AQP2 expression have been documented in rat experimental models of glomerular disease, septic kidney, and post-obstructive kidney (18–21). Computer-assisted analysis of the first 2 kilobases of the AQP2 promoter revealed two putative κB binding elements. Taken together these observations may suggest that tubulo-interstitial inflammation leads to activation of NF-κB in CD principal cells and, thus, down-regulation of AQP2 expression. The aim of the present study was, therefore, to investigate the effects of altered NF-κB activity on AQP2 transcription. Results revealed that LPS treatment of cultured mouse CD principal cells or rat kidney slices decreased both basal and cAMP-stimulated AQP2 expression. Combined silencing of NF-κB subunits, site-directed mutagenesis of the AQP2 promoter, and the chromatin immunoprecipitation assay revealed that reduced AQP2 transcription under conditions of elevated NF-κB activity is largely mediated by decreased p65 and increased p50 and p52 binding to κB elements of the AQP2 promoter. These observations highlight the importance of down-regulated transcriptional AQP2 activity in the physiological control of AQP2 expression and suggest a role for AQP2 in polyuria associated with renal tubulo-interstitial diseases.
MATERIALS AND METHODS
Cell Culture and Transfection—mpkCCDcl4 or mCCDcl1 cells (passages 22–34) were seeded on permeable filters (Transwell®, Corning Costar, Cambridge, MA) and grown in culture medium supplemented with 2% fetal calf serum (11) (22). Cells were grown to confluence and then in serum- and hormone-free medium for 48 h before performing experiments. Iso-osmotic medium (300 mosmol/kg) was made hypertonic (400 or 500 mosmol/kg) by replacing a fraction of apical and basal medium (75–150 μl/600 μl of apical medium and 150–300 μl/1200 μl of basal medium) with NaCl-enriched medium. Medium osmolality was checked using an osmometer.
Transfection was performed by electroporating cells as previously described (23) in the presence of either 1.2 nmol of Stealth RNAi against p65, RelB, p105/p50, or p100/p52 (Invitrogen), 8 pmol of plasmid containing either eGFP, constitutively active IKKβ (24), or super-repressor IκBα (25) mutants, or 8 pmol of plasmid containing luciferase constructs. We have previously estimated that ∼70% of cells are efficiently transfected by electroporation (26). Constitutively active IKKβ, obtained by mutating Ser-176 and Ser-180 residues to Glu, and IκBα super-repressor mutant, obtained by mutating serine phosphorylation sites (Ser-32 and Ser-36) to alanine, were generous gifs from Dr. S. Gosh (Yale University School of Medicine, New Haven, CT). RNAi sense primer against p65 was 5′-GGAACAGUUCGAAUCUCCCUGGUCA-3′, and that for scrambled p65 RNAi was 5′-GCACCUGCAGUUUGAUGCUGAUGAA-3′. RNAi sense primer against p50 was 5′-GGGAGGAGAUUUACCUUCUCUGUGATT-3′, and that for scrambled p50 RNAi was 5′-CAGCCGGAUACUUCCAAACUUAAUUTT-3′. RNAi sense primer against p52 was 5′-GGACAUGACUGCUCAAUUUTT-3′, and that for scrambled p52 RNAi was 5′-GUAGGAUGCACUAUCUCUATT-3′. RNAi sense primer against RelB was 5′-AGAAACCAGUGUUCUUGGATT-3′, and that for scrambled RelB RNAi was 5′-GAACCUAGUUAAUACAUCATT-3′. Transfected cells were seeded on permeable filters, grown in culture medium supplemented with 2% fetal calf serum for 2 days and then in serum- and hormone-free medium for 48 h (24 h for cells transfected with RNAi) before performing experiments.
Preparation of Rat Kidney Slices and RNA Isolation—Male Wistar rats (150–200 g body weight; Taconic Farms, Hudson, NY) were anesthetized with an intraperitoneal injection of sodium pentobarbital (65 mg/kg body weight). Both kidneys were removed, cut into 2–3-mm-thick slices using a razor blade, and quickly placed in Hanks' balanced salt solution (110 mm NaCl, 5 mm KCl, 1.2 mm MgSO4, 1.8 mm CaCl2, 4 mm sodium acetate, 1 mm sodium Citrate, 6 mm d-glucose, 6 mm l-alanine, 1 mm NaH2PO4, 3 mm Na2HPO4, 25 mm NaHCO3, pH 7.4) at 37 °C equilibrated with 5% CO2, 95% O2. Slices 0.5-mm in thickness were cut with a Stadie-Riggs microtome (Thomas Scientific, Swedesboro, NJ) as previously described (27). The slices (3 slices/experimental condition) were then incubated in Hanks' balanced solution at 37 °C for 6 h alone or in the presence of various stimuli. Bathing solutions were continuously bubbled and changed every 2 h. The slices were then placed overnight at 4 °C in RNAlater RNA stabilization reagent (Qiagen, Venlo, The Netherlands). The following day, 25 mg of cortical tissue was excised, snapfrozen in liquid nitrogen, and ground with a mortar and pestle. The tissue powder was then homogenized, and total RNA was purified using RNeasy Mini kit (Qiagen) following the manufacture's instructions.
Western Blot Analysis—Preparation of total cell lysate was performed as previously described (11). Equal amounts of protein (50 μg) were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Immobilon-P, Millipore, Bedford, MA) as previously described (4). Na+, K+-ATPase α-subunit and AQP2 were detected by Western blotting using polyclonal rabbit antibodies (Refs. 28) and 29, respectively) at a 1:2000 dilution, and NF-κB p65, p50, p100, p52, RelB, c-Rel, IκBα, and α-tubulin were detected by Western blotting using monoclonal rabbit or mouse antibodies purchased from Cell Signaling (Danvers, MA) diluted 1:2000. The antigen-antibody complexes were detected using Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore, Billerica, MA). Bands were quantified using a video densitometer and ImageQuant software (GE Healthcare).
Real-time PCR Analysis—RNA extraction, reverse transcription, and real-time PCR analysis was performed as previously described (11). Primers used for detection of mouse acidic ribosomal phosphoprotein P0 were 5′-AATCTCCAGAGGCACCATTG-3′ and 5′-GTTCAGCATGTTCAGCAGTG-3′, those for mouse TNFα were 5′-GACCCTCACACTCAGATCATCTTCT-3′ and 5′-CCACTTGGTGGTTTGCTACGA-3′, those for mouse MCP-1 were 5′-GGCTCAGCCAGATGCAGTTAA-3′ and 5′-CCTACTCATTGGGATCATCTTGCT-3′, those for mouse IκBα were 5′-CGGAGGACGGAGACTCGTT-3′ and 5′-TTCACCTGACCAATGACTTCCA-3′, those for mouse p65 were 5′-GGAGTTCCAGTACTTGCC-3′ and 5′-GTCCTTTTGCGCTTCTCT-3′, those for mouse p50/105 were 5′-CATCCCGGAGTCACGAAATC-3′ and 5′-GCACAATCTTTAGGGCCATTTT-3′, those for mouse p52/p100 were 5′-CGTTCATAAACAGTATGCCATTGTG-3′ and 5′-CCCACGCTTGCGTTTCAG-3′, those for mouse RelB were 5′-GCTGGGAATTGACCCCTACA-3′ and 5′-CATGTCGACCTCCTGATGGTT-3′, those for mouse AQP2 were 5′-CTTCCTTCGAGCTGCCTTC-3′ and 5′-CATTGTTGTGGAGAGCATTGAC-3′, those for rat AQP2 were 5′-GGCCACCTCCTTGGG-3′ and 5′-GGAGCGGGCTGGATTCA-3′, those for rat interleukin-1β were 5′-CAGGATGAGGACCCAAGCA-3′ and 5′-CACAGAGGACGGGCTCTTCT-3′, and those for rat actin were 5′-TGTTGCCCTAGACTTCGAGCA-3′ and 5′-GGACCCAGGAAGGAAGGCT-3′. Mouse P0 and rat actin were used as internal standards, and data were analyzed as previously described (5).
Firefly Luciferase Assay—Luciferase plasmid constructs used for transfection were NF-κB-driven luciferase plasmid containing three κB enhancer elements (p(κB)3 IFN-Luc plasmid) (31), nucleotides –517 to +109 of the mouse AQP2 gene in front of luciferase using pGL3-Basic (Promega, Madison, WI), nucleotides –2043 to +109 of the mouse AQP2 gene in front of luciferase using pGL3-Basic, mutant AQP2 promoter constructs made by changing TGGAA of TonE to TTTAA, mutant κB constructs made by changing GGGAGTTCCT of the κB binding site located at –666 to GCCAGTTCCT, and/or changing GGGGTTTTCC of the κB binding site located at –2030 to GCCGTTTTCC. The numbering was based on the genomic sequence from NCBI. We confirmed the transcription start site by primer extension and ribonuclease protection assay (data not shown). Luciferase activity was measured using the Luciferase Assay System (Promega) according to the manufacturer's instructions. The light produced was measured using a Lumat LB 9507 luminometer (EG&G Berthold).
Chromatin Immunoprecipitation Assay—Chromatin immunoprecipitation assay was performed as previously described (32). Briefly, after stimulation, cells were rapidly fixed in ice-cold cross-linking solution (1% formaldehyde, 9 mm NaCl, 4.5 mm HEPES, pH 8) before cell lysis (0.5% Non-idet P-40, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 50 mm NaF, 1 mm orthovanadate, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 10 mm Tris, pH 8). Nuclei were then isolated by centrifugation, lysed (1% Triton X-100, 0.5% SDS, 0.5% Sarkosyl, 0.5 m NaCl, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 50 mm NaF, 1 mm orthovanadate, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 10 mm Tris, pH 8), and pelleted chromatin was resuspended (100 mm NaCl, 1 mm EDTA, 10 mm Tris, pH 8) before fragmentation (<1 kilobase) by sonication. For immunoprecipitation, chromatin extracts (100 μg) were precleared for 1 h with protein A-Sepharose beads (Amersham Biosciences) and salmon sperm DNA in radioimmune precipitation assay buffer (1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 140 mm NaCl, 1 mm EDTA, 10 mm Tris, pH 8) and then incubated overnight at 4 °C with rabbit polyclonal anti-p65, anti-p50, or anti-52 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). Immune complexes were precipitated with protein A-Sepharose beads for 3 h at 4 °C. Reversal of cross-link was achieved by overnight incubation at 65 °C with proteinase K and 1% SDS. DNA was extracted with phenol-chloroform and precipitated with ethanol. Real-time PCR was performed as described above. Primers flanking the κB site of the AQP2 promoter located 2030 bp upstream of the start codon were 5′-TCACCAAAGGTCACTCAGGA-3′ and 5′-ATCAAGATTGGGCCTTGATG-3′, and those flanking the κB site of the AQP2 promoter located 666 bp upstream of the start codon were 5′-AAGTTGACCTTGGTGACTCTATCA-3′ and 5′-GGAAGAGGATGGAAAACAACT-3′. Primers flanking the κB sites of the IκBα promoter were 5′-GCTTCTCAGTGGAGGACGAG-3′ and 5′-CTGGCTGAAACATGGCTGT-3′.
Statistics—Results are given as the mean ± S.E. from n independent experiments. Each experiment was performed on mpkCCDcl4 cells from the same passage. Statistical differences were assessed using the Mann-Whitney U test or the Kruskal-Wallis test for comparison of two groups or more, respectively. A p value <0.05 was considered significant.
RESULTS
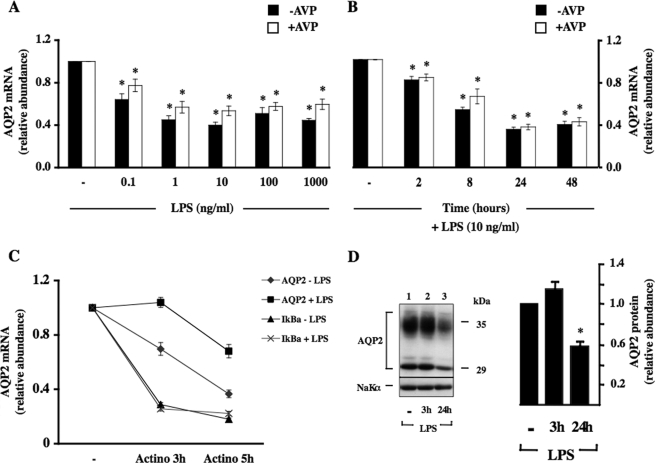
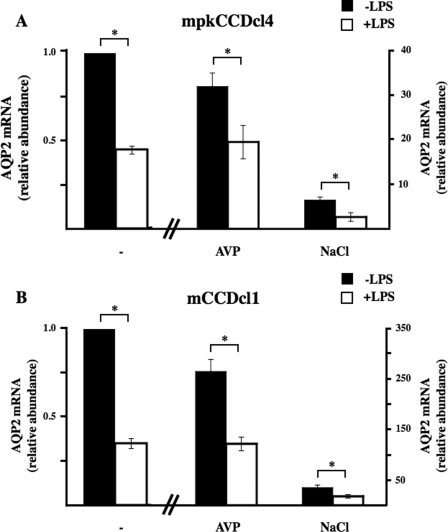
NF-κB Decreases AQP2 Transcription—We first analyzed the effects of increased NF-κB activity on AQP2 expression by stimulating mpkCCDcl4 cells with lipopolysaccharide (LPS) from Escherichia coli (O55:B5; Calbiochem). As previously described (5), 10–10 m AVP increased AQP2 mRNA expression by 30-fold after 24 h of stimulation. LPS decreased AQP2 mRNA expression in both a dose- and time-dependent manner with a maximal effect achieved at 10 ng/ml LPS (Fig. 1A) after 24 h of stimulation (Fig. 1B). The extent of decreased AQP2 mRNA expression induced by LPS was similar in the presence or absence of AVP. To determine whether down-regulated AQP2 mRNA expression in response to LPS was due to decreased AQP2 gene transcription or to decreased AQP2 mRNA stability, mpkCCDcl4 cells were incubated or not with 100 ng/ml LPS for 3 or 5 h in the presence of the transcriptional inhibitor actinomycin D (Fig. 1C). As previously reported (11) AQP2 mRNA expression was reduced in cells treated with actinomycin D alone. Unexpectedly, whereas LPS had no effect on actinomycin d-induced IκBα mRNA degradation, it increased AQP2 mRNA stability (Fig. 1C), indicating that reduced AQP2 mRNA expression induced by LPS is not due to altered AQP2 mRNA stability. Western blot analysis revealed that reduced AQP2 mRNA expression after LPS stimulation was accompanied by reduced AQP2 protein expression (Fig. 1D). We have previously shown that long term (≥24 h) exposure of mpkCCDcl4 cells to hypertonic (≥350 mosmol/kg) medium increases AQP2 expression (11). Enhanced AQP2 mRNA expression after 24 h of hypertonic (≥500 mosmol/kg) challenge (11) was reduced to a similar extent in non-stimulated cells and in AVP-treated cells after 24 h of LPS stimulation (Fig. 2A). We compared our observations made in mpkCCDcl4 cells to those obtained in a recently established, physiologically relevant mouse cortical collecting duct cell line, mCCDcl1 (22). Similar to mpkCCDcl4 cells, LPS reduced AQP2 mRNA expression to similar extents in non-stimulated and AVP- or hypertonic-challenged mCCDcl1 cells (Fig. 2B).
FIGURE 1.
Increased NF-κB activity decreases AQP2 transcription in cultured collecting duct cells. A and B, cells were preincubated or not with 10–10 m AVP for 24 h and then challenged for an additional 24 h with 0–1000 ng/ml LPS (A) or for 0–48 h with 10 ng/ml LPS (B) before RNA extraction. Real-time PCR was performed using primers specific for AQP2. Results are expressed relative to control values determined in the absence of LPS and in the absence (filled bars) or presence (open bars) of AVP. Bars are the mean ± S.E. from four independent experiments. *, p < 0.05. C, cells were immediately lysed or were preincubated with 5 × 10–6 m actinomycin D for 30 min before 3 or 5 h of incubation in the absence or presence of 100 ng/ml LPS (in the continuous presence of actinomycin D) before RNA extraction. Real-time PCR was performed using primers specific for AQP2 (diamonds and squares) or IκBα (triangles and ×'s). Results are expressed as the percentage of control values for each gene determined in the absence of actinomycin D. Data are the mean ± S.E. from three independent experiments. D, cells were preincubated for 24 h with 10–10 m AVP and then for an additional 3 or 24 h with 10 ng/ml LPS before protein extraction. Western blot analysis was performed on AQP2, and Na+, K+-ATPaseα1 subunit (NaKα) was used as a loading control. A representative image is shown. AQP2 abundance is expressed as the ratio of optical density values measured in the presence of LPS and that measured in the absence of LPS. Values are the mean ± S.E. from three independent experiments. *, p < 0.05.
FIGURE 2.
LPS decreases both AVP-induced and hypertonicity-enhanced AQP2 expression in cultured collecting duct cells. Cultured mpkCCDcl4 (A) or mCCDcl1 (B) cells were preincubated 24 h or not with either 10–10 m AVP or hypertonic (NaCl, 500 mosmol/kg) medium and were then stimulated for an additional 24 h with 100 ng/ml LPS before RNA extraction. Real-time PCR was performed using primers specific for AQP2. Results are expressed relative to control values determined in the absence of stimuli. Bars are the mean ± S.E. from three independent experiments. *, p < 0.05.
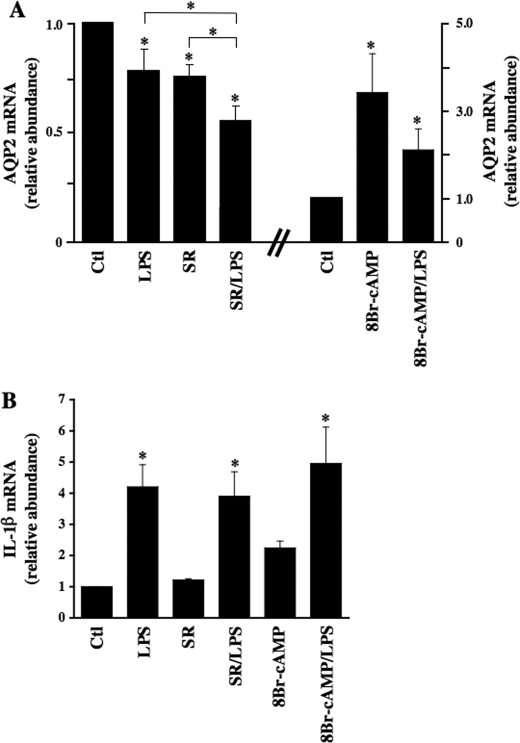
To confirm the physiological relevance of our in vitro findings in cultured CD principal cells, we assessed the effect of LPS on AQP2 expression ex vivo in rat kidney slices. In agreement with results obtained in cultured cells, 100 ng/ml LPS alone significantly decreased AQP2 mRNA levels after 6 h of incubation (Fig. 3A, 0.78 ± 0.11 as compared with control tissue standardized to 1). AQP2 expression was reduced by the addition of 10–6 m of the V2 receptor antagonist SR121463B alone (0.76 ± 0.06). Reduced levels of AQP2 mRNA expression, however, were greatest in tissue incubated with both LPS and SR121463B (0.56 ± 0.08). Conversely, increased AQP2 mRNA expression induced by 10–3 m 8-bromo-cAMP (3.43 ± 0.89) was reduced in tissue simultaneously incubated with 8-bromo-cAMP and LPS (2.12 ± 0.48). LPS was found to induce a similar increase of interleukin-1β mRNA expression when applied alone or together with SR121463B or 8-bromo-cAMP indicating that NF-κB activation by LPS was similar under all three conditions (Fig. 3B).
FIGURE 3.
LPS decreases AQP2 expression independently of the V2 receptor and intracellular cAMP in rat kidney. Wistar rat kidney slices were incubated for 6 h in the absence or presence of 100 ng/ml LPS with or without 10–6 m SR121463B (SR) or 10–3 m 8-bromo-cAMP before mRNA extraction. Real-time PCR was performed using primers specific for AQP2 (A) and interleukin-1β (IL-1β)(B). Results are expressed relative to control values determined in the absence of stimuli. Bars are the mean ± S.E. from triplicate measurements performed in three different animals. *, p < 0.05. Ctl, control.
Altogether, our results suggest that LPS inhibits both basal and enhanced AQP2 transcriptional activity. Moreover, our results suggest that the effect of LPS is at least partly independent of V2 receptor activity.
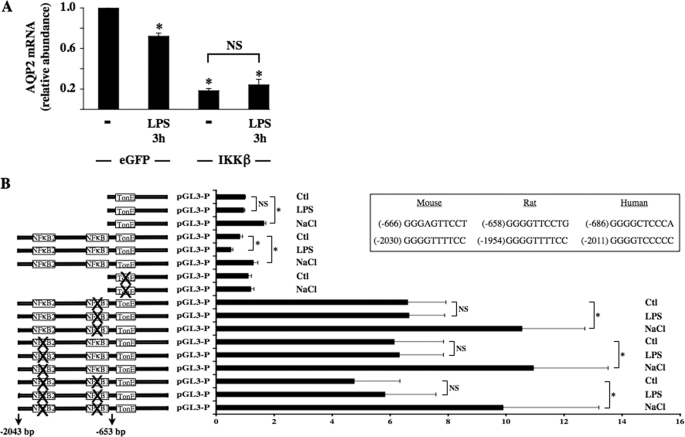
Characterization of Two κB Binding Elements Located in the AQP2 Promoter—Because LPS is a potent activator of the so-called classical NF-κB pathway, which relies on activation of IKKβ, we assessed the effect of overexpression of a constitutively active IKKβ mutant on AQP2 expression levels by mpkCCDcl4 cells. Cells transfected with constitutively active IKKβ mutant expressed significantly less AQP2 mRNA as compared with cells transfected with eGFP, and LPS did not further reduce AQP2 expression in cells expressing mutant IKKβ (Fig. 4A). These results indicate that decreased AQP2 transcription after LPS treatment at least partly relies on increased NF-κB activity.
FIGURE 4.
Two κB binding elements located in the AQP2 promoter mediate the decrease of AQP2 transcriptional activity in response to NF-κB activation. A, cells transfected with an expression plasmid that contained either eGFP or constitutively active IKKβ and were stimulated or not for 3 h with 100 ng/ml LPS before RNA extraction. Real-time PCR was performed using primers specific for AQP2. Results are expressed relative to control values determined in cells transfected with eGFP-containing plasmid in the absence of LPS. Bars are the mean ± S.E. from four independent experiments. *, p < 0.05. NS, no significant difference. B, cells were transfected with a luciferase reporter gene 5′-flanked by either the first 517 or 2043 bp of mouse AQP2 promoter, the first 517 bp of mouse AQP2 promoter of which the TonE sequence was mutated, or the first 2043 bp of mouse AQP2 promoter of which either or both putative κB binding sites were mutated. Transfected cells were challenged with either hypertonic (NaCl, 500 mosmol/kg) medium or LPS (100 ng/ml) for 24 h before measurement of luciferase activity. Results are expressed relative to control values determined in cells transfected with luciferase reporter gene 5′-flanked by the first 517 bp of mouse AQP2 promoter and incubated for 24 h in isotonic medium. Bars are the mean ± S.E. from three independent experiments. *, p < 0.05; NS, no significant difference. The positions, relative to the start codon, and sequences of putative κB binding sites between different species are shown in the inset. Ctl, control.
Based on structural and genetic studies, κB binding sites display a loose consensus sequence, often cited as 5′-GGGRNNYYCC-3′ (N, nucleotide; Y, pyrimidine nucleoside) (33). We have previously shown that a highly conserved TonE sequence, located 625 bp upstream of the ATG start codon, is a key element mediating increased AQP2 transcription after hypertonic challenge (26). In the present study we investigated the functional roles of two putative κB binding sites present in the first 2043 bp of the AQP2 promoter. In mouse AQP2 promoter, these sites are located 666 and 2030 bp upstream of the ATG start codon (see the inset of Fig. 4B). We investigated the functional importance of both κB binding sites by performing a luciferase gene reporter assay (Fig. 4B). As previously shown (26), 24 h of hypertonic stimulation (500 mosmol/kg) increased luciferase activity in cells transfected with a reporter construct encoding luciferase 5′-flanked by either the first 653 (AQP2 653) or 2043 (AQP2 2043) bp upstream of the ATG start codon of the AQP2 gene (Fig. 4B) containing an intact TonE enhancer element. 24 h of LPS (100 ng/ml) stimulation significantly decreased luciferase activity in cells transfected with AQP2 2043 but not AQP2 653. Mutation of either or both κB binding sites increased basal luciferase activity as compared with cells transfected with wild-type AQP2 2043. In addition, although hypertonicity increased luciferase activity in cells expressing mutant κB binding sites, mutation of either κB binding site abolished the LPS-induced decrease of luciferase activity. Altogether, these results reveal that an increase of NF-κB activity reduces AQP2 transcription and that this event requires two intact κB binding sites present in the first 2043 bp upstream of the ATG start codon.
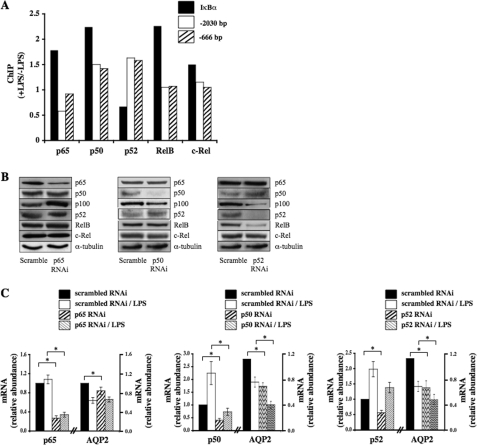
We next investigated which members of the NF-κB family bind to the AQP2 promoter by comparing the ability of p65, RelB, c-Rel, p50, and p52 to bind to either κB site in response to LPS (Fig. 5A). LPS increased p65, RelB, c-Rel, and p50, but not p52, binding to the IκBα promoter, used as a control for chromatin immunoprecipitation analysis. Anti-p65 immunoprecipitation of DNA fragments of the AQP2 promoter containing κB binding sites located either 2030 or 666 bp upstream of the start codon revealed decreased p65 binding to the proximal κB site (–2030) in response to LPS challenge. On the other hand, LPS increased the ability of both p50 and p52 to bind to either κB binding site of the AQP2 promoter. Binding of RelB and c-Rel to either κB element was not affected by LPS challenge. These results strongly indicate that decreased AQP2 transcriptional activity in response to an increase of NF-κB activity principally arises from decreased p65 binding and increased binding of p50 and p52 homodimers and/or p50/p52 heterodimers to κB elements of the AQP2 promoter.
FIGURE 5.
The effect of LPS is dependent on altered p65, p52, and p50 binding to the AQP2 promoter. A, cells were challenged or not with 100 ng/ml LPS for 3 h before DNA fragmentation and immunoprecipitation using anti-p65, p50, p52, RelB, or c-Rel antibodies. Real-time PCR was performed using primers flanking either the NF-κB binding element of the AQP2 promoter (–2030, open box; –666, hatched box) or using primers flanking NF-κB binding element of the IκBα promoter (closed box). Results are expressed as the ratio of values obtained in the presence and in the absence of LPS. Shown are results of one of three similar experiments. ChIP, chromatin immunoprecipitation. B and C, cells were transfected with either scrambled RNAi or RNAi targeting various NF-κB isoforms and then challenged or not with 100 ng/ml LPS for 3 h before protein (B) or RNA (C) extraction. B, Western blot analysis was performed on various NF-κB isoforms and on α-tubulin, used as a loading control. Representative images are shown. C, real-time PCR from RNA extracts of cells transfected with scrambled RNAi or RNAi against p65 (left panel), p50 (middle panel), or p52 (right panel) was performed using primers specific for NF-κB isoforms or AQP2. Results are expressed relative to control values determined in cells transfected with scrambled RNAi and subjected to isotonic medium. Bars are the mean ± S.E. from six independent experiments. *, p < 0.05.
We next analyzed the effects of RNAi against p65, p50, and p52 on AQP2 mRNA abundance (Figs. 5, B and C). RNAi against either p65, p50, or p52 (Fig. 5B) specifically reduced target gene expression with the exception of RNAi against p52, which also decreased RelB protein expression. Interestingly, p50 knockdown was associated with increased p52 abundance. Conversely, p52 knockdown increased p50 abundance. Moreover, silencing of either p50 or p52 decreased the abundance of p100 (NF-κB2), the precursor of p52, which functionally behaves like an IκB. RNAi against p65 slightly decreased AQP2 transcription in the absence of LPS and did not affect the LPS-induced decrease of AQP2 transcription (Fig. 5C). Although RNAi against RelB strongly reduced RelB protein expression, it did not influence AQP2 expression in either the absence or presence of LPS (p > 0.05, data not shown). On the other hand, reduced expression of either p50 or p52 by RNAi decreased AQP2 mRNA levels both in the absence and presence of LPS (Fig. 5C). These results indicate that p65, p50, and p52 influence AQP2 transcription under both basal and stimulated states.
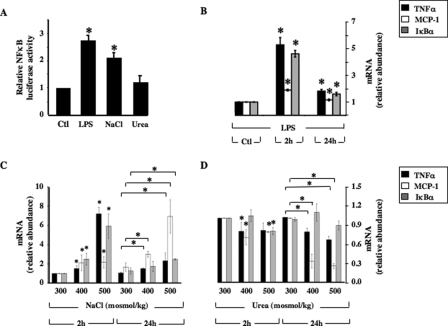
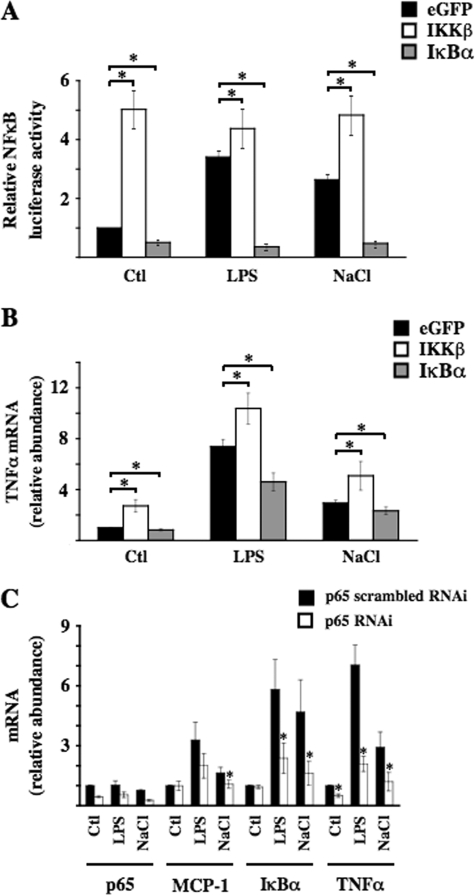
Hypertonicity Increases NF-κB Activity in mpkCCDcl4 Cells in an IKKβ- and IκBα-dependent Manner—We have previously demonstrated in mpkCCDcl4 cells the occurrence of an AVP-independent reduction of AQP2 transcriptional activity during the first hours of hypertonic stimulation (11). AQP2 transcriptional activity increased in a TonEBP-dependent manner after longer periods of hypertonic stimulation (26). Because hypertonicity might be a physiological activator of NF-κB in renal epithelial cells, we investigated the influence of hypertonicity on NF-κB activity in mpkCCDcl4 cells by exposing cells transfected with NF-κB-driven luciferase reporter plasmid to either isotonic (300 mosmol/kg) or hypertonic (500 mosmol/kg) medium for 24 h (Fig. 6A). Hypertonic NaCl, but not hyperosmotic urea, increased luciferase activity to levels comparable with that produced by 100 ng/ml LPS. In addition, in non-transfected cells both LPS (Fig. 6B) and hypertonicity (Fig. 6C) increased mRNA expression levels of three different genes under the control of NF-κB transcriptional activity, TNFα, MCP-1, and IκBα, to similar extents in a time-dependent manner. On the other hand, mRNA expression of these genes was not increased by hyperosmotic urea (Fig. 6D). Hypertonic sucrose produced results similar to those obtained by hypertonic NaCl (not shown). Because TNFα, MCP-1, and IκBα are all under the control of canonical p65-dependent NF-κB transcriptional activity (34), we tested the effects of constitutively active IKKβ and superrepressor IκBα mutants (see “Materials and Methods”) on the hypertonicity-induced increase of NF-κB activity (Fig. 7). Under isotonic conditions (control), cells transfected with mutant IKKβ or IκBα displayed a 5-fold increase and 2-fold decrease, respectively, of NF-κB-driven luciferase activity (Fig. 7A) and displayed respectively increased and decreased expression of TNFα (Fig. 7B) and MCP-1 mRNA (data not shown). Luciferase activity of cells transfected with constitutively active IKKβ mutant was not further stimulated by either LPS or hypertonicity. The relative increase of TNFα and MCP-1 mRNA expression in response to LPS and hypertonicity was greater in eGFP-transfected cells than in cells transfected with IKKβ mutant, reflecting the higher levels of basal NF-κB activation induced by the IKKβ mutant. Inversely, super-repressor IκBα mutant abolished both LPS and hypertonicity-induced luciferase activity and decreased LPS- and hypertonicity-induced TNFα and MCP-1 mRNA expression to similar extents. The observation that both LPS and hypertonicity increased NF-κB-inducible gene expression even in the presence of IκBα mutant may reflect enhanced transcriptional activities of other trans elements that bind to TNFα and MCP-1 promoters but not to the luciferase plasmid construct. It is worthwhile pointing out that NF-κB activity was decreased in non-stimulated cells expressing super-repressor IκBα mutant, suggesting that NF-κB is partially active under basal conditions in mpkCCDcl4 cells. We next analyzed the effect of reduced p65 activity on the hypertonicity-induced increase of TNFα, MCP-1, and IκBα mRNA abundance by transfecting cells with RNAi targeting p65 mRNA. Increased mRNA abundance of all three genes by either LPS or hypertonic stimulation was significantly reduced in cells transfected with RNAi targeting p65 (Fig. 7C). Collectively, these data indicate that hypertonicity stimulates NF-κB activity in mpkCCDcl4 cells via the canonical IKKβ-IκBα-p65-dependent NF-κB signaling pathway.
FIGURE 6.
NF-κB activity is increased by hypertonicity in mpkCCDcl4 cells. A, transfected cells expressing NF-κB-driven luciferase were challenged with either isotonic (Ctl, 300 mosmol/kg), hypertonic (NaCl, 500 mosmol/kg), or urea-supplemented hyperosmotic (urea, 500 mosmol/kg) medium for 12 h or were stimulated for 12 h with 100 μg/ml LPS in isotonic medium before measuring luciferase activity. Bars are the mean ± S.E. from four independent experiments. *, p < 0.05. B–D, cells were challenged with isotonic medium (Ctl, 300 mosmol/kg) or with either 100 ng/ml LPS (B), NaCl-supplemented hypertonic (400 or 500 mosmol/kg) (B) or urea-supplemented hyperosmotic (400 or 500 mosmol/kg) (C) medium for 2 or 24 h before RNA extraction. Real-time PCR was performed using primers specific for TNFα, MCP-1, or IκBα. Results are expressed relative to control values determined for each gene after 2 h of incubation in isotonic medium. Bars are the mean ± S.E. from five independent experiments. *, p < 0.05.
FIGURE 7.
Hypertonicity-induced NF-κB activation is dependent on IKKβ activity, IκBα degradation, and p65 activation. A and B, cells were transfected with an expression vector that contained either eGFP or constitutively active IKKβ or IκBα mutants together with (A) or without (B) NF-κB-driven luciferase plasmid. Cells were challenged or not with either 100 μg/ml LPS or hypertonic (NaCl, 500 mosmol/kg) medium for 12 (A) or 3 (B) h before measurement of luciferase activity (A) or to RNA extraction (B). Real-time PCR was performed using primers specific for TNFα (B). Results are expressed relative to control values determined in cells transfected with eGFP-containing cDNA and subjected to isotonic medium. Bars are the mean ± S.E. from four independent experiments. *, p < 0.05. C, cells were transfected with either scrambled RNAi or RNAi targeting p65 NF-κB and then challenged or not with either 100 ng/ml LPS or hypertonic (NaCl, 500 mosmol/kg) medium for 3 h before RNA extraction. Real-time PCR was performed using primers specific for MCP-1, IκBα, or TNFα. Results are expressed relative to control values determined in cells transfected with scrambled RNAi and subjected to isotonic medium. Bars are the mean ± S.E. from four independent experiments. *, p < 0.05. Ctl, control.
DISCUSSION
The results of the present study reveal reduced levels of AQP2 expression occurring in both cultured renal cells and native renal tissue challenged with LPS. This effect occurred both in the presence of V2 receptor antagonists and 8-bromo-cAMP. Results show that decreased AQP2 transcription results from decreased p65 and increased p50 and p52 binding to κB binding sites of the AQP2 promoter. Similar to LPS, increased NF-κB activity in response to hypertonicity was found to depend on activation of the canonical IKKβ-IκBα-p65 NF-κB pathway. Together, our findings indicate that NF-κB negatively regulates AQP2 transcription and most likely antagonizes the stimulatory effects of cAMP-responsive element-binding protein (CREB) and TonEBP under both physiological and pathophysiological conditions. This regulatory pathway might be involved in the pathogenesis of polyuria associated with renal tubulo-interstitial inflammation.
Previous studies performed on animals have documented decreased AQP2 expression after intraperitoneal LPS injection as a consequence of down-regulated V2 receptor activity (35). In addition to the identification of κB elements present in the AQP2 promoter, two other findings of the present study provide direct evidence that down-regulated AQP2 expression in response to LPS additionally arises from decreased AQP2 gene transcription. First, LPS further reduced AQP2 mRNA expression in tissue slice preparations treated with the V2 receptor antagonist SR121463B (Fig. 3). Conversely, in tissue slice preparations LPS reduced enhanced AQP2 expression induced by 8-bromo-cAMP, i.e. an effector acting downstream of V2 receptor activation. Second, LPS decreased the AVP- and hypertonicity-induced increase of AQP2 mRNA expression to similar extents in cultured CD principal cells (Fig. 2). Importantly, we have recently shown that hypertonicity increases AQP2 transcription in the absence of an increase of cAMP concentration and independently of protein kinase A activity (11, 36).
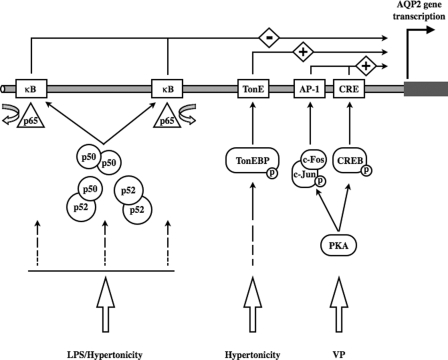
Several studies have identified regulatory motifs in the AQP2 gene that promote AQP2 expression. The influence of transcription factors that mediate the effects of vasopressin and extracellular tonicity, revealed by past studies and the present study, are schematically depicted in Fig. 8. A rise of intracellular cAMP by vasopressin induces activation of c-Fos/c-Jun and CREB that together promote AQP2 transcription by, respectively, binding to AP1 and cAMP-responsive element sites present in the AQP2 promoter (37–39). We have recently shown that binding of TonEBP to a TonE enhancer element located near the cAMP-responsive element is a major stimulus of AQP2 transcription both under isotonic and hypertonic conditions (26). In addition to these stimulatory factors, binding of negatively acting trans elements was suggested to selectively repress AQP2 transcription (40). Here we identify NF-κBas one such negatively acting transcription factor that physiologically controls AQP2 transcription and may antagonize both CREB and TonEBP stimulatory effects.
FIGURE 8.
Schematic illustration of controlled AQP2 transcription in collecting duct principal cells. By binding to cis elements of the AQP2 promoter cAMP CREB and AP-1 (c-Fos/c-Jun) enhance AQP2 transcriptional activity in response to protein kinase A-mediated vasopressin (VP) stimulation. By binding to the TonE element(s) of the AQP2 promoter, TonEBP positively regulates AQP2 transcription under both base-line conditions and in response to hypertonicity. Positive AQP2 transcriptional regulation mediated by both CREB/AP-1 and TonEBP is repressed after activation of the NF-κB pathway in response to either LPS stimulation, characteristic of inflammatory diseases, or hypertonicity. This event is mediated by p65 release from and increased p50 and p52 binding to κB elements of the AQP2 promoter. Although AQP2 transcriptional activity is maintained at low levels under iso-osmotic conditions in the absence or presence of VP, the repressive effect of NF-κB is superseded by increased TonEBP activity after longer periods of hypertonic stimulation. PKA, protein kinase A.
NF-κB stimulates or represses target gene transcription differently depending on the nature of the bound NF-κB homo- or heterodimer and on bound NF-κB co-factors. NF-κB complexes containing p65 or c-Rel generally enhance gene transcription, whereas p50 and p52 homo- and heterodimers act as repressors (41). In the present study both κB binding sites located in the first 2 kilobases of the AQP2 promoter were found to be required for LPS-induced repression of transcriptional activity. Similar functional requirement for two intact κB sites was previously described for other genes, such as for the MCP-1 and glutamate transporter genes (42–44). Both κB sites may function together to regulate AQP2 transcriptional activity, possibly via binding of specific cofactors that mediate NF-κB activity (42). Our data indicate that down-regulated AQP2 transcriptional activity after an increase of NF-κB activity is at least partly mediated by p65 release from the AQP2 promoter and an increased ability of p50 and p52 homo- and/or heterodimers to bind to the AQP2 promoter. Cells transfected with RNAi against either p65, p50, or p52 displayed reduced levels of AQP2 mRNA under both base-line conditions and after LPS challenge. Reduced AQP2 mRNA expression in cells transfected with p65 RNAi is consistent with the idea that p65 enhances constitutive AQP2 transcriptional activity. On the other hand, binding of both p50 and p52 to the AQP2 promoter is increased in response to LPS stimulation. The observation that p50 and p52 RNAi silencing, respectively, increased p52 and p50 protein expression (Fig. 5B) may indicate that reduced basal AQP2 transcription in response to RNAi silencing of either of these NF-κB isoforms occurs via increased binding of p52 (for p50 silencing) and p50 (for p52 silencing) homodimers to the AQP2 promoter. It should be noted that RNAi silencing of either p50 or p52, but not p65, decreased p100 expression. Because p100 functions as an IκB and can bind to p50 or p52 subunits, decreased p100 abundance should lead to increased amounts of biologically active p50 and p52 homo- or heterodimers. These results suggest that a balance between p100 and processed p50 and p52 subunits most likely plays a central role in regulating AQP2 transcription.
Depressed tubular function is characteristic of inflammatory diseases of the kidney such as pyelonephritis and ureteral obstruction (45, 46). Cytokines secreted by leukocytes may strongly activate NF-κB in renal epithelial cells (16, 17). The results of the present study suggest that decreased AQP2 expression after NF-κB stimulation participates in the urinary concentration defects observed in tubulo-interstitial inflammatory diseases. This interpretation is supported by observations from animal models showing that endotoxemia induced by intraperitoneal LPS injection or ureteral obstruction were associated with polyuria and decreased levels of AQP2 expression (35, 47). Down-regulated levels of AQP2 were additionally observed in animal models of puromycin aminonucleoside- and adriamycin-induced nephrotic syndrome (18, 20). In this case, excessive water reabsorption is thought to depend on increased NaCl absorption. The results of the present study suggest that increased NF-κB activity associated with nephrotic syndrome may at least be partly responsible for reduced AQP2 expression, an event that may reflect a generalized mechanism that counteracts extracellular volume expansion. It is interesting that the extent of decreased AQP2 expression in the medulla was 2-fold greater than that observed in the cortex of nephrotic animals (20). This suggests that up-regulated NF-κB activity by hypertonicity may compliment the effects mediated by cytokine release.
Together with its role in altered AQP2 expression exerted by pro-inflammatory stimuli, NF-κB may additionally mediate altered AQP2 expression in response to aldosterone. Cross-talk between aldosterone signaling and the NF-κB pathway has been demonstrated in part via an association between serum and glucocorticoid-regulated kinase 1 (SGK1) and IKKβ (48). We have previously shown that aldosterone decreases AQP2 mRNA expression in cultured CD principal cells in a mineralocorticoid receptor-dependent fashion, possibly by reducing its transcriptional activity (9). We have additionally demonstrated mineralocorticoid receptor-dependent NF-κB activation by aldosterone in CD principal cells (49), suggesting a possible role of NF-κB in down-regulated AQP2 expression induced by aldosterone.
Consistent with the findings of the present study, observations made in a previous study (50) revealed the occurrence of increased NF-κB activity induced by hypertonicity in the renal medulla. Results of that study together with those of the present study indicate that NF-κB activity is increased by hypertonicity in both kidney renal medullary interstitial cells and collecting duct principal cells. Increased NF-κB activation in renal medullary interstitial cells is illustrative of the role played by NF-κB in the kidney, which is to protect cells from the damaging effects of hypertonicity via COX2-dependent suppression of apoptosis (50). These observations highlight the potential role of NF-κB in renal cell survival under hypertonic conditions.
TonEBP activity is enhanced by hypertonicity through combined enhanced TonEBP nuclear localization, transactivation, and increased whole cell abundance (51–54). Our findings indicate that hypertonicity activates both NF-κB and TonEBP and that both factors bind to specific cis elements located in the AQP2 promoter. Because TonEBP activity is more slowly up-regulated than NF-κB activity in mpkCCDcl4 cells (Ref. 26 and this study), we propose that decreased AQP2 transcription occurring shortly after (<3 h) hypertonic challenge is associated with increased NF-κB activity. After longer periods of hypertonic challenge, when TonEBP activity is high and NF-κB activity is low, the stimulatory effect of TonEBP on AQP2 transcription would supersede the repressive effect of NF-κB and result in increased AQP2 transcription. These considerations indicate that sustained hypertonicity prevailing in the renal medulla override reduced AQP2 transcription under conditions of enhanced NF-κB activity. TonEBP abundance increases along the CD as a consequence of increased interstitial tonicity (30). This suggests that reduced AQP2 expression in tubulo-interstitial inflammatory diseases should be predominant in isotonic cortical regions, where the bulk of CD water reabsorption occurs, as opposed to TonEBP-rich hypertonic medullary regions.
In conclusion, the findings of the present study reveal that increased NF-κB activity is associated with decreased AQP2 transcriptional activity. This in turn was found to depend on altered p65, p50, and p52 NF-κB subunit binding to κB elements of the AQP2 promoter. These findings strongly indicate that NF-κB activation participates in the negative physiological control of AQP2 transcription and may antagonize increased AQP2 gene transcription by vasopressin and hypertonicity mediated by CREB and TonEBP, respectively. Moreover, strong NF-κB activation observed in renal inflammatory diseases most likely accounts for the observed decrease of AQP2 expression and accompanying urinary concentration defects.
Acknowledgments
We thank Prof. Bernard Rossier, Department of Pharmacology and Toxicology, University of Lausanne, Lausanne, Switzerland, for providing the mCCDcl1 cell line.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-DK-42479 (to H. M. K.) and DK38452 (to D. B). This work was also supported in part by Swiss National Foundation Grants 31-67878.02 and 31-109473, a Carlos and Elsie de Reuter Foundation grant, a Ernest Boninchi Foundation grant (to E. F.), and a Fondation pour la Recherche Médicale grant (to U. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: AQP, aquaporin 1; CD, collecting duct; AVP, [8-arginine]vasopressin; IKK, IκB kinases; LPS, lipopolysaccharide; CREB, cAMP-responsive element-binding protein; RNAi, RNA-mediated interference; TNF, tumor necrosis factor; TonEBP, tonicity-responsive enhancer binding protein.
References
- 1.Chou, C. L., Knepper, M. A., Hoek, A. N., Brown, D., Yang, B., Ma, T., and Verkman, A. S. (1999) J. Clin. Investig. 103 491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schnermann, J., Chou, C. L., Ma, T., Traynor, T., Knepper, M. A., and Verkman, A. S. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 9660–9664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terris, J., Ecelbarger, C. A., Nielsen, S., and Knepper, M. A. (1996) Am. J. Physiol. 271 F414–F422 [DOI] [PubMed] [Google Scholar]
- 4.Hasler, U., Mordasini, D., Bens, M., Bianchi, M., Cluzeaud, F., Rousselot, M., Vandewalle, A., Feraille, E., and Martin, P.-Y. (2002) J. Biol. Chem. 277 10379–10386 [DOI] [PubMed] [Google Scholar]
- 5.Hasler, U., Nielsen, S., Feraille, E., and Martin, P. Y. (2006) Am. J. Physiol. Renal Physiol. 290 177–187 [DOI] [PubMed] [Google Scholar]
- 6.Nielsen, S., Chou, C. L., Marples, D., Christensen, E. I., Kishore, B. K., and Knepper, M. A. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouley, R., Hasler, U., Lu, H. A., Nunes, P., and Brown, D. (2008) Semin. Nephrol. 28 266–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bustamante, M., Hasler, U., Kotova, O., Chibalin, A. V., Mordasini, D., Rousselot, M., Vandewalle, A., Martin, P. Y., and Feraille, E. (2005) Am. J. Physiol. Renal Physiol. 288 334–344 [DOI] [PubMed] [Google Scholar]
- 9.Hasler, U., Mordasini, D., Bianchi, M., Vandewalle, A., Feraille, E., and Martin, P.-Y. (2003) J. Biol. Chem. 278 21639–21648 [DOI] [PubMed] [Google Scholar]
- 10.Bustamante, M., Hasler, U., Leroy, V., de Seigneux, S., Dimitrov, M., Mordasini, D., Rousselot, M., Martin, P. Y., and Feraille, E. (2008) J. Am. Soc. Nephrol. 19 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasler, U., Vinciguerra, M., Vandewalle, A., Martin, P.-Y., and Feraille, E. (2005) J. Am. Soc. Nephrol. 16 1571–1582 [DOI] [PubMed] [Google Scholar]
- 12.Nejsum, L. N. (2005) Cell. Mol. Life Sci. 62 1692–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storm, R., Klussmann, E., Geelhaar, A., Rosenthal, W., and Maric, K. (2003) Am. J. Physiol. Renal Physiol. 284 189–198 [DOI] [PubMed] [Google Scholar]
- 14.Gerondakis, S., Grossmann, M., Nakamura, Y., Pohl, T., and Grumont, R. (1999) Oncogene 18 6888–6895 [DOI] [PubMed] [Google Scholar]
- 15.Ghosh, S., and Karin, M. (2002) Cell 109 (suppl. 1) 81–96 [DOI] [PubMed] [Google Scholar]
- 16.Husted, R. F., Zhang, C., and Stokes, J. B. (1998) Am. J. Physiol. 275 F946–F954 [DOI] [PubMed] [Google Scholar]
- 17.Kohan, D. E. (1994) J. Lab. Clin. Med. 123 668–675 [PubMed] [Google Scholar]
- 18.Apostol, E., Ecelbarger, C. A., Terris, J., Bradford, A. D., Andrews, P., and Knepper, M. A. (1997) J. Am. Soc. Nephrol. 8 15–24 [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Llama, P., Andrews, P., Ecelbarger, C. A., Nielsen, S., and Knepper, M. (1998) Kidney Int. 54 170–179 [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Llama, P., Andrews, P., Nielsen, S., Ecelbarger, C. A., and Knepper, M. A. (1998) Kidney Int. 53 1244–1253 [DOI] [PubMed] [Google Scholar]
- 21.Li, C., Wang, W., Kwon, T. H., Isikay, L., Wen, J. G., Marples, D., Djurhuus, J. C., Stockwell, A., Knepper, M. A., Nielsen, S., and Frokiaer, J. (2001) Am. J. Physiol. Renal Physiol. 281 163–171 [DOI] [PubMed] [Google Scholar]
- 22.Gaeggeler, H. P., Gonzalez-Rodriguez, E., Jaeger, N. F., Loffing-Cueni, D., Norregaard, R., Loffing, J., Horisberger, J. D., and Rossier, B. C. (2005) J. Am. Soc. Nephrol. 16 878–891 [DOI] [PubMed] [Google Scholar]
- 23.Mordasini, D., Bustamante, M., Rousselot, M., Martin, P. Y., Hasler, U., and Feraille, E. (2005) Am. J. Physiol. Renal Physiol. 289 1031–1039 [DOI] [PubMed] [Google Scholar]
- 24.Mercurio, F., Zhu, H., Murray, B. W., Shevchenko, A., Bennett, B. L., Li, J., Young, D. B., Barbosa, M., Mann, M., Manning, A., and Rao, A. (1997) Science 278 860–866 [DOI] [PubMed] [Google Scholar]
- 25.Oyama, T., Ran, S., Ishida, T., Nadaf, S., Kerr, L., Carbone, D. P., and Gabrilovich, D. I. (1998) J. Immunol. 160 1224–1232 [PubMed] [Google Scholar]
- 26.Hasler, U., Jeon, U. S., Kim, J. A., Mordasini, D., Kwon, H. M., Feraille, E., and Martin, P. Y. (2006) J. Am. Soc. Nephrol. 17 1521–1531 [DOI] [PubMed] [Google Scholar]
- 27.Breton, S., and Brown, D. (1998) J. Am. Soc. Nephrol. 9 155–166 [DOI] [PubMed] [Google Scholar]
- 28.Carranza, M. L., Feraille, E., and Favre, H. (1996) Am. J. Physiol. 271 C136–C143 [DOI] [PubMed] [Google Scholar]
- 29.Xu, D. L., Martin, P.-Y., Ohara, M., St. John, J., Pattison, T., Meng, X., Morris, K., Kim, J. K., and Schrier, R. W. (1997) J. Clin. Investig. 99 1500–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han, K. H., Woo, S. K., Kim, W. Y., Park, S. H., Cha, J. H., Kim, J., and Kwon, H. M. (2004) Am. J. Physiol. Renal Physiol. 287 878–885 [DOI] [PubMed] [Google Scholar]
- 31.Fujita, T., Nolan, G. P., Liou, H. C., Scott, M. L., and Baltimore, D. (1993) Genes Dev. 7 1354–1363 [DOI] [PubMed] [Google Scholar]
- 32.Ryser, S., Fujita, T., Tortola, S., Piuz, I., and Schlegel, W. (2007) J. Biol. Chem. 282 5075–5084 [DOI] [PubMed] [Google Scholar]
- 33.Ghosh, S., May, M. J., and Kopp, E. B. (1998) Annu. Rev. Immunol. 16 225–260 [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann, A., Leung, T. H., and Baltimore, D. (2003) EMBO J. 22 5530–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grinevich, V., Knepper, M. A., Verbalis, J., Reyes, I., and Aguilera, G. (2004) Kidney Int. 65 54–62 [DOI] [PubMed] [Google Scholar]
- 36.Hasler, U., Nunes, P., Bouley, R., Lu, H. A. J., Matsuzaki, T., and Brown, D. (2008)J. Biol. Chem 283 26643–26661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yasui, M., Zelenin, S. M., Celsi, G., and Aperia, A. (1997) Am. J. Physiol. 272 F443–F450 [DOI] [PubMed] [Google Scholar]
- 38.Hozawa, S., Holtzman, E. J., and Ausiello, D. A. (1996) Am. J. Physiol. 270 C1695–C1702 [DOI] [PubMed] [Google Scholar]
- 39.Frokiaer, J., Marples, D., Valtin, H., Morris, J. F., Knepper, M. A., and Nielsen, S. (1999) Am. J. Physiol. 276 F179–F190 [DOI] [PubMed] [Google Scholar]
- 40.Furuno, M., Uchida, S., Marumo, F., and Sasaki, S. (1996) Am. J. Physiol. 271 F854–F860 [DOI] [PubMed] [Google Scholar]
- 41.Hayden, M. S., and Ghosh, S. (2004) Genes Dev. 18 2195–2224 [DOI] [PubMed] [Google Scholar]
- 42.Leung, T. H., Hoffmann, A., and Baltimore, D. (2004) Cell 118 453–464 [DOI] [PubMed] [Google Scholar]
- 43.Ping, D., Boekhoudt, G. H., Rogers, E. M., and Boss, J. M. (1999) J. Immunol. 162 727–734 [PubMed] [Google Scholar]
- 44.Shi, X.-Z., Pazdrak, K., Saada, N., Dai, B., Palade, P., and Sarna, S. K. (2005) Gastroenterology 129 1518–1532 [DOI] [PubMed] [Google Scholar]
- 45.Kumar, A., Turney, J. H., Brownjohn, A. M., and McMahon, M. J. (2001) Nephrol. Dial. Transplant. 16 1062–1065 [DOI] [PubMed] [Google Scholar]
- 46.Klahr, S., Harris, K., and Purkerson, M. L. (1988) Pediatr. Nephrol. 2 34–42 [DOI] [PubMed] [Google Scholar]
- 47.Frokiaer, J., Christensen, B. M., Marples, D., Djurhuus, J. C., Jensen, U. B., Knepper, M. A., and Nielsen, S. (1997) Am. J. Physiol. 273 F213–F223 [DOI] [PubMed] [Google Scholar]
- 48.Zhang, L., Cui, R., Cheng, X., and Du, J. (2005) Cancer Res. 65 457–464 [PubMed] [Google Scholar]
- 49.Leroy, V., de Seigneux, Agassiz, V., Hasler, U., Rafestin-Oblin, M.-E., Vinciguerra, M., Martin, P.-Y., and Féraille, E. J. Am. Soc. Nephrol., in press [DOI] [PMC free article] [PubMed]
- 50.Hao, C. M., Yull, F., Blackwell, T., Komhoff, M., Davis, L. S., and Breyer, M. D. (2000) J. Clin. Investig. 106 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyakawa, H., Woo, S. K., Dahl, S. C., Handler, J. S., and Kwon, H. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 2538–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez-Rodriguez, C., Aramburu, J., Jin, L., Rakeman, A. S., Michino, M., and Rao, A. (2001) Immunity 15 47–58 [DOI] [PubMed] [Google Scholar]
- 53.Ferraris, J. D., Williams, C. K., Persaud, P., Zhang, Z., Chen, Y., and Burg, M. B. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woo, S. K., Dahl, S. C., Handler, J. S., and Kwon, H. M. (2000) Am. J. Physiol. Renal Physiol. 278 1006–1012 [DOI] [PubMed] [Google Scholar]