Cytoplasmic lipid droplets were considered until recently to be in the same category as glycogen granules, simple storage sites for energy, waxing and waning as metabolic energy needs dictated, but otherwise inert particles. It has become clear, however, that droplets are much more than isolated storage depots in the cell and that they can skate around on the cytoskeleton, physically interact with several organelles over short or long durations, and be beasts of burden, storing important molecules unrelated to lipids for later use. Although important clues to the panoply of droplet functions have come to light as a result of several proteomics studies, some behavioral qualities of this organelle were apparent from older morphological studies. Droplets are indeed gregarious. In this Minireview, after first considering the basic biochemical properties of lipid droplets, I shall focus on their interactions with other organelles, as manifest by morphological and dynamic studies and hinted at by proteomics. The important functions of droplets in storing and chaperoning proteins are well covered in a recent review (1) and will not be discussed here.
The Biochemistry of Lipid Droplets
Droplets store energy in the form of neutral lipids, predominantly triglycerides and steryl esters. Although best known for their storage role in mammalian adipose tissue, droplets exist in virtually all eukaryotic cells and even in some prokaryotes (2). Energy from droplets is used both in the cells of origin and at distant sites, in response to hormone stimulation of lipases, for example. In worms, fat is stored predominantly in cells lining the gastrointestinal tract (3); in insects and higher animals, there are specialized tissues for fat storage (4). In plants, droplets within oil seeds are required for germination, whereas in the anthers of flowers, they provide the lipids and proteins that coat and stabilize pollen (5).
The neutral lipid core of droplets is surrounded by a phospholipid monolayer, acyl chains pointing inward (6). Whereas triacylglycerols and steryl esters generally account for most of the mass of the core, large amounts of ether-linked glycerolipids are found in several cultured cell lines and tissues (7). Smaller amounts of diacylglycerols and fatty acids also are in the core. The phospholipids are generally similar to those in the endoplasmic reticulum (ER)2 (7, 8) with some differences in fatty acid composition (6).
Proteins can bind on the surface of droplets or extend long helical hairpins deeply into the core (9). Hydrophilic proteins have even been reported within the core (10), although it is not clear how this topology is allowed energetically.
Abundant proteins on the surface of droplets confer stability to the organelle and prevent their coalescing. In plants, oleosins play this role and provide the pollen coat (5). In mammalian adipose tissue, stability is conferred by perilipin (11). Perilipin is also essential for lipolysis, becoming highly phosphorylated upon adrenergic stimulation, which attracts hormone-sensitive lipase to the droplet. Other lipases associate with droplets in a perilipin-independent manner (12). Perilipin is a member of a larger family of animal droplet-binding proteins that share a common region, the PAT domain, named for the three original family members: perilipin, adipophilin/ADRP, and TIP47. The expression and droplet binding of individual PAT proteins vary with tissue and maturity of droplets. Although generally stabilizing droplets, unique roles of specific PAT proteins remain to be discovered (12).
Several lipid biosynthetic enzymes can associate with droplets, indicating a capacity of the organelles to grow. This is particularly well documented for Saccharomyces cerevisiae, in which several of the sterol and acylglycerol biosynthetic enzymes are localized to droplets (7, 13).
Lipid droplets have been subjected to over a dozen proteomics studies in recent years (14–17). Surprisingly, many have found proteins that regulate trafficking pathways (Table 1), such as Rab proteins, Arf1, and other proteins thought to be specific for vesicle docking events in the exocytic and endocytic pathways. Caveolins, which traffic lipids from the Golgi and plasma membrane, are frequently found in droplets (18–20). Resident proteins of the ER, mitochondria, and peroxisomes are also found to “contaminate” droplet preparations. There is growing evidence that these associations point to novel functions of droplets in organellar communication, some of which are reminders of physical associations seen decades ago (21). These proteomics data coupled with morphological evidence suggest that droplets can form stable functional interorganellar synapses for communication and transfer of molecules, as well as interact more transiently. Establishing some of these interactions may require droplet translocation through the cytoplasm, and rapid droplet movement on microtubules has been observed (22). Both kinesin and dynein can exist on single droplets, and factors have been identified in Drosophila that provide and regulate a clutch to control the direction and velocity of movement (23).
TABLE 1.
| Function | Protein or protein class |
|---|---|
| Lipid metabolism | 17β-Hydroxysteroid dehydrogenases, acyl-CoA synthetases, acyl-CoA synthetases, acyltransferases, adipocyte triglyceride lipase, CGI-58 (lipase modulator), lanosterol synthase, lipoprotein lipase |
| Droplet stability | PAT family proteins (perilipin, ADRP, TIP47, S2-12) |
| Trafficking | Arf1, caveolin-1, Rab proteins (especially Rab5s, Rab7, Rab14, Rab18), Sec22 |
| ER resident proteins | BiP, calnexin, cytochrome b5 reductase, protein-disulfide isomerase |
| Mitochondrial proteins | ADP synthase subunits |
| Cytoskeletal proteins | Actin, tubulin, spectrin, vimentin |
| Other/unknown | Alcohol dehydrogenase, CGI-49, FSP27, HSP70 |
Droplet Self-association and Fusion
Following exposure of NIH 3T3-L1 cells to oleic acid, tiny droplets are first observed near the periphery of the cell. Over time, larger ones appear that are more centrally localized, suggesting growth and movement of droplets during maturation. PAT proteins are also exchanged during this process (24). Similarly, droplets in adipose tissue-derived rat stromal vascular cells form in lamellipodia and move into the perinuclear region as they increase in size (25). Using a metabolic inhibitor, the increase in size of droplets in 3T3 cells was shown to be independent of triglyceride synthesis (26), indicating that the large droplets are fusion products of the smaller ones.
Before fusion, lipid droplets must self-associate. Droplets frequently cluster, and in yeast, they can form linear and branching chains, open to one other through nipple-like junctions (16). In 3T3 cells grown with oleic acid or in extracts derived from these cells, the clustering is nocodazole- and calcium-sensitive (26). During cluster formation, dynein becomes associated with droplets, confirming a role for microtubules in this process. The phosphorylation status of perilipin is also important because an increase in phosphorylation in response to activation by protein kinase A causes droplet dispersion, even in the absence of lipolysis. A particular site of phosphorylation, Ser492, is necessary for this effect (27).
Fusion between droplets requires SNARE coupling, similar to vesicle-targeting events or lysosomal homotypic fusion. Purified droplets from 3T3 cells contain NSF (NEM-sensitive factor), α-SNAP, and the SNAREs SNAP23, syntaxin-5, and VAMP4 (28). None of these factors are transmembrane proteins, which would be incompatible with the droplet phospholipid monolayer. Knockdown of these SNAREs results in smaller, more numerous droplets consistent with a decrease in fusion. Interestingly and of potential clinical significance, incubation of heart muscle-derived HL-1 cells with oleic acid causes the redistribution of SNAP23 from the plasma membrane to droplets and a decrease in GLUT4 transport to the cell surface. Insulin sensitivity is restored by overexpression of SNAP23 (28).
Organellar Associations
Are Droplets Simply Specialized Regions of the ER?—The weight of the evidence indicates that droplets are derived from the ER. The terminal enzymes in the synthesis of the droplet core lipids localize to the ER and often to droplets themselves. Lipid droplets in plant anther cells appear to emanate from the ER (5). Similarly, a dominant-negative form of caveolin that coats droplets traffics from the ER (29). Continuities have been frequently observed in ultrastructural studies between the cytosolic leaflet of the ER and the boundary leaflet of droplets (30). All but one green fluorescent protein-tagged droplet protein in yeast colocalized in the ER (31). The current model is that the droplet core originates as a lens of neutral lipids between the leaflets of the ER. This aggregate expands and finally buds into the cytosol with a surrounding phospholipid leaflet derived from the ER outer monolayer (32). Exchangeable PAT proteins have been proposed to curve the membrane to facilitate this reaction (24). A recent RNA interference screen in Drosophila S2 cells found evidence that COP1 coatomers play a role in droplet formation (33), but droplet budding from the ER is not a universally accepted mechanism. The absence of observed ER-droplet continuities in human monocytes has led to an alternative hypothesis that droplets grow in apposition to, not within, the ER membrane (34).
Droplets can be formed in vitro from washed microsomes upon the addition of acyl-CoAs and diacylglycerol, the substrates of the terminal enzyme for triglyceride synthesis; the resulting droplets are isolated by centrifugation (35). The reaction requires microsomal phospholipase D, dynein, and ERK2; dynein may be the target of ERK2 phosphorylation (35, 36).
Some data suggest that droplets never leave the ER. Lipid droplets isolated in the in vitro assay might have resulted from scission from the ER by centrifugation. The effects of the absence of COP1 proteins on droplet morphology might be an indirect result of defects in the retrograde pathway rather than indicating a direct role in droplet budding. Continuities between the ER and droplets are common in many cell types, suggesting permanent associations. In one study, such continuities were seen in 3T3-L1 adipocytes, epididymal adipocytes, lactating mammary gland, liver, and intestine, prompting the authors to suggest that droplets “accumulate or regress within the confines of endoplasmic reticulum membranes” (30). In S. cerevisiae, 96% of droplets were scored as associating with the ER (37), and time-lapse microscopy showed lateral motion of droplets on the surface of the ER but no dissociation.3 Most droplets in HuH-7 hepatoma cells and Vero kidney epithelial cells remain attached to the ER (38). Although droplets can migrate rapidly on microtubules, this movement may occur on the ER surface. Several proteomics studies have shown the presence of ER proteins, both luminal and membrane-associated, in purified lipid droplets. It is difficult to reconcile ER luminal proteins such as BiP as representing anything other than tightly attached ER (39). Ribosomal subunits and translation factors have also been seen; these likely represent ER-bound polysomes.
The notion that lipid droplets are specialized regions of the ER is attractive because this arrangement would provide a facile mechanism for the two compartments to exchange lipids and proteins. For example, during metabolic stress that requires rapid hydrolysis of esterified fatty acids and release of free fatty acids, there must be rapid shrinkage of the leaflet. Continuity with the ER would allow this compartment to be a sink for these leaflet components. In addition, rapid movement by microtubule motors of droplets attached to the ER would require less energy if the droplet core simply slid between the ER membrane leaflets (providing no integral ER proteins blocked the path) than if movement involved breaking and making bonds between droplet and ER proteins.
Mitochondrial Associations—Mitochondria are frequently seen densely packed close to or against lipid droplets in adipocytes (18, 21, 30). In an early study, the association of mitochondria with milk globules in lactating cells was observed, prompting the authors to suggest the presence of a “chemotactic principle” that draws mitochondria to droplets in unmilked animals (40). As a response to luteinizing hormone, droplets, mitochondria, and smooth ER in bovine oocytes form complexes, termed “metabolic units,” presumably to allow coupling of lipolysis and oxidation, which is followed by a burst of protein synthesis (41). In the following hour, the cores shrink as energy is utilized. Fluorescence resonance energy transfer analysis in porcine oocytes has been used to show that droplet-mitochondrial contacts are within 6–10 nm of each other, suggesting functional coupling (42).
Muscles need an efficient mechanism to mobilize energy substrates. In exercise-trained dogs and goats, every lipid droplet in muscle tissue was observed to be in contact with at least one mitochondrion (43). Presumably, this association allows the coupled release and oxidation of free fatty acids. Similarly, the extent of droplet-mitochondrial associations was shown recently to increase following exercise, as the serum levels of free fatty acids fell (44). In type I (highly oxidating) fibers of human skeletal muscle, mitochondria and droplets form chains of alternating organelles (45).
Association of mitochondria with lipid droplets is not limited to providing fuel. In yeast, fatty acid oxidation occurs in peroxisomes. Nevertheless, there are extensive physical couplings of droplets with mitochondria in oleic acid-cultured S. cerevisiae, and the yeast droplet proteome contains several mitochondrial proteins arising from these putative synapses (16).4 The function of these interactions is unknown; they might involve transfer of phospholipids or cations between the compartments, as occurs between mitochondria and the ER (46).
Peroxisomal Interactions—Peroxisomes are frequently seen concentrating close to droplets (21). These are often dumbbell-shaped, suggesting a dependence on droplets for membrane lipids during peroxisomal division (30). There is precedence for this relationship in plants, where glyoxysomal (peroxisomal) membrane lipids are derived directly from lipid droplets and not from the ER in germinating cotton seeds (47). Both triglycerides and fatty acids were shown to traffic from droplets to glyoxysomes in this study. Whether this pathway also exists in animal cells is not known. Thus, although ER membranes are the source of pre-peroxisomal vesicles (48), maturation or fission of peroxisomes might require droplets.
Physical associations of peroxisomes and droplets have been observed. In plants, these associations can be extensive. In cotyledons of a ped1 strain (lacking peroxisomal fatty acid β-oxidation), a marked degree of interdigitations of droplets and glyoxysomes is seen, with tubular structures within the glyoxysomes that appear to be derived from droplets; perhaps these structures are transporting triglycerides for glyoxysomal oxidation (49). In animal cells, extensive association of peroxisomes with lipid droplets in cultured COS-7 cells was observed (50). Fungi commonly exhibit peroxisome-droplet intimacies. In the oleaginous yeast Yarrowia lipolytica grown in oleic acid medium, some peroxisomes in a pex3 mutant strain (partially deficient in pre-peroxisome budding from the ER) wrap around droplets, as if attempting to access core lipids for membrane assembly (51). Extensive long-term contacts are easily found in S. cerevisiae when cultured on oleic acid; on glucose medium, the interactions are transient (16). Peroxisomes can even invade droplets with “pexopodia.” Interestingly, all but one peroxisomal protein identified in purified droplet preparations are involved in fatty acid oxidation, as if the pexopodia are specialized fatty acid oxidation machines.
Communication between droplets and peroxisomes may facilitate bidirectional lipid movement because ether lipids, which are synthesized in peroxisomes, are abundant in the droplet cores of several cell types (7). It is easy to imagine transport of these molecules across peroxisome-droplet synapses.
Lipid Droplets and Endosomal Trafficking—Evidence is growing for extensive communication between droplets and the cellular endomembrane system. Many endosomes tagged with fluorescent Rab5, Rab7, or Rab11, which label different subpopulations of endosomes, appear in close contact with droplets (52). The movement of SNARE proteins from the plasma membrane and endosomes to droplets in response to fatty acids was described earlier. The most widely documented example of droplet trafficking with endosomes involves caveolins, cholesterol- and fatty acid-binding proteins, which normally reside in caveolae on the cell surface and in the Golgi apparatus. Exogenous fatty acids cause depletion of caveolin-1 and caveolin-3 from the plasma membrane and caveolin-2 from the Golgi and accumulation of these molecules on lipid droplets, presumably transporting the added lipid (20, 53). Removal of the fatty acid restores the original distribution, demonstrating reversibility. Exogenous cholesterol also stimulates movement of caveolin-1 to droplets (19). Besides this behavior in cultured cells, caveolins also redistribute to droplets after partial hepatectomy in rats (20). Anterograde transport of caveolin from lipid droplets to the plasma membrane may also be a normal pathway because a truncation mutant of caveolin-3 (cav-3DGV) accumulates in droplets, presumably lacking the signal for transport to the cell surface (29).
Classical trafficking within the cells' exocytic and endocytic compartments requires Rab proteins for specificity. Interestingly, proteomics studies of mammalian lipid droplets invariably identify multiple Rab proteins, as many as 17 in one study (14)! The most compelling evidence for a physiological role of a Rab protein in droplet trafficking involves Rab18 (52, 54). In cultured cell models, the non-cytosolic fraction of Rab18 was found nearly exclusively on droplets. This protein is not involved in the initial formation of the droplet because it appears late in adipogenesis in 3T3 cells, 6 h after ADRP associates with droplets. Localization depends on the GTP-bound form; a constitutive GDP-bound form (S22N) does not target. Notably, hormone stimulation of lipolysis leads to an increase in Rab18 association with droplets, suggesting a function in mediating this process. Rab18 does not label all droplets and tends to decorate only smaller ones, illustrating droplet heterogeneity. When overexpressed, Rab18 causes an increase in the association of droplets with the ER; however, because overexpression also causes the release of ADRP from droplets, it is not clear if the ER association is a direct or indirect result of Rab18 expression.
Several Rab proteins have been shown to associate and dissociate in vitro with isolated droplets in a guanine nucleotide-dependent manner. More striking, binding of droplets to endosomal membranes in vitro depends on prior Rab binding (55). Although these experiments suggest an important function for Rab proteins in droplet trafficking, these proteins cannot function as they normally do in vesicle targeting because classical fusion events involving the single leaflet-enclosed droplet are impossible. The authors suggested a “kiss and run” mechanism of transient binding for Rab-dependent transfer of membrane components between droplets and other membranes (55).
As a final example of droplet interactions with organelles, droplets were shown very recently to cross the parasitophorous vacuole of HeLa cells infected with Chlamydia trachomatis. The parasite first tags the cytoplasmic droplets with a bacterial protein, which leads to their binding to and sequestration in the vacuole. The core lipids are then utilized for chlamydial metabolism (56).
Future Directions: Molecular Mechanisms and Significance of Droplet Interactions
There are several fundamental questions remaining about the biology of droplets. Few will argue that droplets are not derived from the ER, but the process of droplet formation is still obscure. Are PAT proteins involved in formation of a lens of neutral lipid between the ER leaflets? What controls the shape and directionality of cytoplasmic droplets as they bulge from the ER? Does release of droplets from the ER occur, and if so, how? Are COPI elements involved? If droplets can exist as lone organelles, how do phospholipids bind and dissociate as the core grows or shrinks in response to metabolic demands?
Droplets clearly associate with several types of organelles, including themselves (Fig. 1). They often self-aggregate, which can lead to fusion. How is this process regulated? Do droplets fuse simply to decrease their surface area-to-volume ratio, perhaps protecting them from adventitious lipolysis, or are important molecules transferred in the process? Some droplets in a cell move rapidly on cytoskeletal elements. Are droplets following a chemical gradient? Are they always moving in parallel to the ER membrane, thereby maintaining the connections? What purpose is served by these translocations?
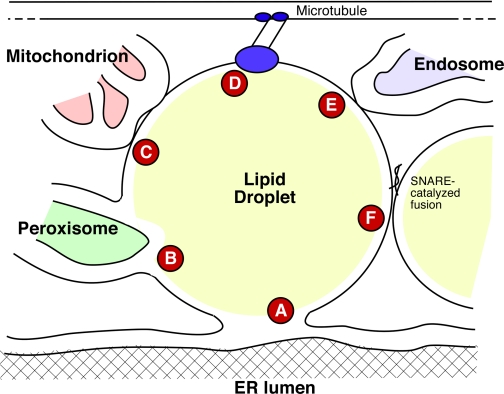
FIGURE 1.
Lipid droplet-organellar associations. A, most droplets remain attached to the ER. It is not clear whether the phospholipid monolayer is continuous with the cytoplasmic monolayer of the ER, as shown, or whether the leaflets are separate. B, peroxisomes frequently associate with droplets and can insert pexopodia into the core of the droplet. C, mitochondrial associations allow flux of fatty acyl-CoAs for oxidation; other communication also exists. D, microtubule motors associate with droplets, allowing bidirectional transport (one motor is shown). E, endosomes probably associate transiently with droplets, transferring proteins and lipids. F, droplets can undergo SNARE-mediated homotypic fusion.
How do droplets form tight synapses with mitochondria and peroxisomes? What are the proteins on each of these organelles that catalyze the binding? Are they similar to PACS-2, the protein involved in mitochondrial-ER associations (57)? Do the long-term associations involve semifusion of membranes, as has been proposed for yeast peroxisomes and droplets (16)? Presumably the transfer of lipids into peroxisomes and mitochondria for oxidation is an important function. Is this transfer passive or active? Is it regulated? Is there frequent endosome-droplet communication in vivo, and if so, are there proteins other than caveolins and SNAREs that are transported? Do the bulk of the Rab proteins that associate with droplets function in membrane-mediated trafficking to and from droplets, or are many of them simply refugee proteins (1) en route to serve elsewhere?
Rather than being wallflowers waiting to be asked to dance, droplets appear to be the life of the party, organellar butterflies. It will be fascinating to figure out the source of the droplets' charm in making new friends and to what ends this talent is used.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM084210. This work was also supported by Welch Foundation Grant I-1085. This minireview will be reprinted in the 2008 Minireview Compendium, which will be available in January, 2009.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; ADRP, adipose differentiation-related protein; SNARE, soluble NSF attachment protein receptor; SNAP, soluble NSF attachment protein.
D. Binns and J. M. Goodman, unpublished data.
J. M. Goodman, unpublished data.
References
- 1.Welte, M. A. (2007) Trends Cell Biol. 17 363–369 [DOI] [PubMed] [Google Scholar]
- 2.Packter, N. M., and Olukoshi, E. R. (1995) Arch. Microbiol. 164 420–427 [DOI] [PubMed] [Google Scholar]
- 3.Ashrafi, K. (2007) WormBook, 1–20 [DOI] [PMC free article] [PubMed]
- 4.Law, J. H., and Wells, M. A. (1989) J. Biol. Chem. 264 16335–16338 [PubMed] [Google Scholar]
- 5.Hsieh, K., and Huang, A. H. (2005) Plant J. 43 889–899 [DOI] [PubMed] [Google Scholar]
- 6.Tauchi-Sato, K., Ozeki, S., Houjou, T., Taguchi, R., and Fujimoto, T. (2002) J. Biol. Chem. 277 44507–44512 [DOI] [PubMed] [Google Scholar]
- 7.Bartz, R., Li, W. H., Venables, B., Zehmer, J. K., Roth, M. R., Welti, R., Anderson, R. G., Liu, P., and Chapman, K. D. (2007) J. Lipid Res. 48 837–847 [DOI] [PubMed] [Google Scholar]
- 8.Gennis, R. B. (1989) Biomembranes: Molecular Structure and Function, p. 21, Springer-Verlag New York Inc., New York
- 9.Capuano, F., Beaudoin, F., Napier, J. A., and Shewry, P. R. (2007) Biotechnol. Adv. 25 203–206 [DOI] [PubMed] [Google Scholar]
- 10.Robenek, H., Robenek, M. J., and Troyer, D. (2005) J. Lipid Res. 46 1331–1338 [DOI] [PubMed] [Google Scholar]
- 11.Greenberg, A. S., Egan, J. J., Wek, S. A., Garty, N. B., Blanchette-Mackie, E. J., and Londos, C. (1991) J. Biol. Chem. 266 11341–11346 [PubMed] [Google Scholar]
- 12.Brasaemle, D. L. (2007) J. Lipid Res. 48 2547–2559 [DOI] [PubMed] [Google Scholar]
- 13.Rajakumari, S., Grillitsch, K., and Daum, G. (2008) Prog. Lipid Res. 47 157–171 [DOI] [PubMed] [Google Scholar]
- 14.Bartz, R., Zehmer, J. K., Zhu, M., Chen, Y., Serrero, G., Zhao, Y., and Liu, P. (2007) J. Proteome Res. 6 3256–3265 [DOI] [PubMed] [Google Scholar]
- 15.Beller, M., Riedel, D., Jansch, L., Dieterich, G., Wehland, J., Jackle, H., and Kuhnlein, R. P. (2006) Mol. Cell. Proteomics 5 1082–1094 [DOI] [PubMed] [Google Scholar]
- 16.Binns, D., Januszewski, T., Chen, Y., Hill, J., Markin, V. S., Zhao, Y., Gilpin, C., Chapman, K. D., Anderson, R. G., and Goodman, J. M. (2006) J. Cell Biol. 173 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dugail, I., and Hajduch, E. (2007) CMLS Cell. Mol. Life Sci. 64 2452–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen, A. W., Razani, B., Schubert, W., Williams, T. M., Wang, X. B., Iyengar, P., Brasaemle, D. L., Scherer, P. E., and Lisanti, M. P. (2004) Diabetes 53 1261–1270 [DOI] [PubMed] [Google Scholar]
- 19.Le Lay, S., Hajduch, E., Lindsay, M. R., Le Liepvre, X., Thiele, C., Ferre, P., Parton, R. G., Kurzchalia, T., Simons, K., and Dugail, I. (2006) Traffic 7 549–561 [DOI] [PubMed] [Google Scholar]
- 20.Pol, A., Martin, S., Fernandez, M. A., Ferguson, C., Carozzi, A., Luetterforst, R., Enrich, C., and Parton, R. G. (2004) Mol. Biol. Cell 15 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novikoff, A. B., Novikoff, P. M., Rosen, O. M., and Rubin, C. S. (1980) J. Cell Biol. 87 180–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valetti, C., Wetzel, D. M., Schrader, M., Hasbani, M. J., Gill, S. R., Kreis, T. E., and Schroer, T. A. (1999) Mol. Biol. Cell 10 4107–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welte, M. A., Cermelli, S., Griner, J., Viera, A., Guo, Y., Kim, D. H., Gindhart, J. G., and Gross, S. P. (2005) Curr. Biol. 15 1266–1275 [DOI] [PubMed] [Google Scholar]
- 24.Wolins, N. E., Brasaemle, D. L., and Bickel, P. E. (2006) FEBS Lett. 580 5484–5491 [DOI] [PubMed] [Google Scholar]
- 25.Nagayama, M., Uchida, T., and Gohara, K. (2007) J. Lipid Res. 48 9–18 [DOI] [PubMed] [Google Scholar]
- 26.Bostrom, P., Rutberg, M., Ericsson, J., Holmdahl, P., Andersson, L., Frohman, M. A., Boren, J., and Olofsson, S. O. (2005) Arterioscler. Thromb. Vasc. Biol. 25 1945–1951 [DOI] [PubMed] [Google Scholar]
- 27.Marcinkiewicz, A., Gauthier, D., Garcia, A., and Brasaemle, D. L. (2006) J. Biol. Chem. 281 11901–11909 [DOI] [PubMed] [Google Scholar]
- 28.Bostrom, P., Andersson, L., Rutberg, M., Perman, J., Lidberg, U., Johansson, B. R., Fernandez-Rodriguez, J., Ericson, J., Nilsson, T., Boren, J., and Olofsson, S. O. (2007) Nat. Cell Biol. 9 1286–1293 [DOI] [PubMed] [Google Scholar]
- 29.Pol, A., Luetterforst, R., Lindsay, M., Heino, S., Ikonen, E., and Parton, R. G. (2001) J. Cell Biol. 152 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanchette-Mackie, E. J., Dwyer, N. K., Barber, T., Coxey, R. A., Takeda, T., Rondinone, C. M., Theodorakis, J. L., Greenberg, A. S., and Londos, C. (1995) J. Lipid Res. 36 1211–1226 [PubMed] [Google Scholar]
- 31.Habeler, G., Natter, K., Thallinger, G. G., Crawford, M. E., Kohlwein, S. D., and Trajanoski, Z. (2002) Nucleic Acids Res. 30 80–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy, D. J., and Vance, J. (1999) Trends Biochem. Sci. 24 109–115 [DOI] [PubMed] [Google Scholar]
- 33.Guo, Y., Walther, T. C., Rao, M., Stuurman, N., Goshima, G., Terayama, K., Wong, J. S., Vale, R. D., Walter, P., and Farese, R. V. (2008) Nature 453 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robenek, H., Hofnagel, O., Buers, I., Robenek, M. J., Troyer, D., and Severs, N. J. (2006) J. Cell Sci. 119 4215–4224 [DOI] [PubMed] [Google Scholar]
- 35.Marchesan, D., Rutberg, M., Andersson, L., Asp, L., Larsson, T., Boren, J., Johansson, B. R., and Olofsson, S. O. (2003) J. Biol. Chem. 278 27293–27300 [DOI] [PubMed] [Google Scholar]
- 36.Andersson, L., Bostrom, P., Ericson, J., Rutberg, M., Magnusson, B., Marchesan, D., Ruiz, M., Asp, L., Huang, P., Frohman, M. A., Boren, J., and Olofsson, S. O. (2006) J. Cell Sci. 119 2246–2257 [DOI] [PubMed] [Google Scholar]
- 37.Szymanski, K. M., Binns, D., Bartz, R., Grishin, N. V., Li, W. P., Agarwal, A. K., Garg, A., Anderson, R. G., and Goodman, J. M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 20890–20895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Targett-Adams, P., Chambers, D., Gledhill, S., Hope, R. G., Coy, J. F., Girod, A., and McLauchlan, J. (2003) J. Biol. Chem. 278 15998–16007 [DOI] [PubMed] [Google Scholar]
- 39.Prattes, S., Horl, G., Hammer, A., Blaschitz, A., Graier, W. F., Sattler, W., Zechner, R., and Steyrer, E. (2000) J. Cell Sci. 113 2977–2989 [DOI] [PubMed] [Google Scholar]
- 40.Stemberger, B. H., Walsh, R. M., and Patton, S. (1984) Cell Tissue Res. 236 471–475 [DOI] [PubMed] [Google Scholar]
- 41.Kruip, T. A. M., Cran, D. G., van Beneden, T. H., and Dieleman, S. J. (1983) Gamete Res. 8 29–47 [Google Scholar]
- 42.Sturmey, R. G., O'Toole, P. J., and Leese, H. J. (2006) Reproduction (Camb.) 132 829–837 [DOI] [PubMed] [Google Scholar]
- 43.Vock, R., Hoppeler, H., Claassen, H., Wu, D. X., Billeter, R., Weber, J. M., Taylor, C. R., and Weibel, E. R. (1996) J. Exp. Biol. 199 1689–1697 [DOI] [PubMed] [Google Scholar]
- 44.Tarnopolsky, M. A., Rennie, C. D., Robertshaw, H. A., Fedak-Tarnopolsky, S. N., Devries, M. C., and Hamadeh, M. J. (2007) Am. J. Physiol. 292 R1271–R1278 [DOI] [PubMed] [Google Scholar]
- 45.Shaw, C. S., Jones, D. A., and Wagenmakers, A. J. (2008) Histochem. Cell Biol. 129 65–72 [DOI] [PubMed] [Google Scholar]
- 46.Rizzuto, R., Pinton, P., Carrington, W., Fay, F. S., Fogarty, K. E., Lifshitz, L. M., Tuft, R. A., and Pozzan, T. (1998) Science 280 1763–1766 [DOI] [PubMed] [Google Scholar]
- 47.Chapman, K. D., and Trelease, R. N. (1991) J. Cell Biol. 115 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoepfner, D., Schildknegt, D., Braakman, I., Philippsen, P., and Tabak, H. F. (2005) Cell 122 85–95 [DOI] [PubMed] [Google Scholar]
- 49.Hayashi, Y., Hayashi, M., Hayashi, H., Hara-Nishimura, I., and Nishimura, M. (2001) Protoplasma 218 83–94 [DOI] [PubMed] [Google Scholar]
- 50.Schrader, M. (2001) J. Histochem. Cytochem. 49 1421–1429 [DOI] [PubMed] [Google Scholar]
- 51.Bascom, R. A., Chan, H., and Rachubinski, R. A. (2003) Mol. Biol. Cell 14 939–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin, S., Driessen, K., Nixon, S. J., Zerial, M., and Parton, R. G. (2005) J. Biol. Chem. 280 42325–42335 [DOI] [PubMed] [Google Scholar]
- 53.Pol, A., Martin, S., Fernandez, M. A., Ingelmo-Torres, M., Ferguson, C., Enrich, C., and Parton, R. G. (2005) Mol. Biol. Cell 16 2091–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ozeki, S., Cheng, J., Tauchi-Sato, K., Hatano, N., Taniguchi, H., and Fujimoto, T. (2005) J. Cell Sci. 118 2601–2611 [DOI] [PubMed] [Google Scholar]
- 55.Liu, P., Bartz, R., Zehmer, J. K., Ying, Y. S., Zhu, M., Serrero, G., and Anderson, R. G. (2007) Biochim. Biophys. Acta 1773 784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cocchiaro, J. L., Kumar, Y., Fischer, E. R., Hackstadt, T., and Valdivia, R. H. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 9379–9384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simmen, T., Aslan, J. E., Blagoveshchenskaya, A. D., Thomas, L., Wan, L., Xiang, Y., Feliciangeli, S. F., Hung, C. H., Crump, C. M., and Thomas, G. (2005) EMBO J. 24 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.