Abstract
Polycomb group proteins are transcriptional repressors recruited to many developmental control genes. The specificity of polycomb group protein targeting is incompletely understood. Subunits of polycomb repressive complexes (PRC) are encoded by multigene families in vertebrates. Five chromodomain-containing CBX family proteins are thought to mediate chromatin association by PRC1 complexes. We visualized the recruitment of CBX proteins to chromatin using bimolecular fluorescence complementation (BiFC) analysis, wherein fragments of fluorescent proteins fused to CBX family members and histone H3 form a fluorescent complex when the CBX proteins bind to nucleosomes. Different CBX family proteins associated with nucleosomes in different subnuclear regions in both ES cells and fibroblasts. The total populations of most CBX proteins had distributions distinct from those of the chromatin-associated complexes, indicating that most of these CBX proteins were not bound to nucleosomes. The conserved chromodomain and chromobox regions of CBX proteins were dispensable for chromatin association. The absence of H3 K27 trimethylation in EED null ES cells had minimal effects on chromatin association by CBX proteins. The BiFC complexes did not colocalize with anti-trimethyl-K27 immunofluorescence, with the exception of inactive X. Metaphase spreads derived from stable cell lines with inducible CBX fusion expression revealed reciprocal patterns of chromosome association by CBX2 and CBX6 BiFC complexes. H3.2 purified from CBX2–H3.2 BiFC complexes was enriched in trimethyl-K27, dimethyl-K4, and acetyl-K9 modifications. We conclude that different CBX proteins are recruited to distinct chromatin regions through nonconserved interactions, expanding the regulatory diversity of polycomb group proteins.
Keywords: bimolecular fluorescence complementation, embryonic stem cells, histone modification, polycomb group transcription factors, subnuclear localization
Polycomb group (PcG) proteins mediate the stable inheritance of cell states. They regulate the expression of numerous genes that control the maintenance and differentiation of vertebrate stem cells (1). Deletion of genes encoding PcG proteins in mouse results in the inability to establish ES cells (2) or impairment of embryonic or adult stem cell functions (3–6). The roles of PcG proteins in the maintenance of pluripotency suggest that they constitute a cellular memory.
Biochemical characterization of PcG proteins has identified polycomb repressive complexes 1 and 2 (PRC1 and PRC2) (7–9). PRC2 complexes have methyltransferase activity that trimethylates lysine-27 (K27) of histone H3 (10, 11). PRC1 complexes contain CBX proteins that can bind H3 trimethylated on K27 (9, 12). These complementary activities have engendered a model according to which H3 K27 trimethylation by PRC2 mediates PRC1 recruitment.
Analysis of the binding targets of several PcG proteins has identified hundreds of genes that are co-occupied by PRC1 and PRC2 subunits (13, 14). A large proportion of these genes is trimethylated on H3 K27. A null mutation in any one of the genes encoding PRC2 components (EZH2, EED, Suz12) eliminates H3 K27 trimethylation, but these mutations produce different phenotypes and have distinct effects on ES cells (2, 3, 6). Despite the absence of detectable H3 K27 trimethylation in Suz12-deficient ES cells, similar levels of CBX8 and Bmi-1 binding to most genes is observed (3). Several PRC1 components are recruited to the inactive X in EED null cells, and CBX2 is recruited to heterochromatin in male pronuclei lacking EZH2 (15, 16). Thus, in several cases PRC1 recruitment does not require PRC2 or H3 K27 trimethylation.
PRC1 binding to nucleosomes is thought to be mediated at least in part by the conserved chromodomain of CBX family proteins (CBX2, CBX4, CBX6, CBX7, and CBX8) (17–21). The isolated chromodomains of CBX family proteins can specifically recognize H3 trimethylated on K27, but some of them can also bind peptides containing other modifications in vitro (21). It is not known whether different CBX proteins target PRC1 complexes to the same or different genes or whether the regions conserved among these proteins are required for chromatin association.
Live cell imaging enables visualization of molecular processes in their native environments. Bimolecular fluorescence complementation (BiFC) is based on formation of a fluorescent complex when fragments of a fluorescent protein are brought together by an interaction between proteins fused to the fragments (22). This approach has been used to visualize histone H4 binding by bromodomain proteins (23). We have adapted the BiFC assay to compare the subnuclear distributions of chromatin-associated CBX proteins and to investigate the roles of conserved protein domains and H3 K27 trimethylation in nucleosome binding by CBX proteins in living cells.
Results
Patterns of Chromatin Association by Different CBX Proteins.
The subnuclear locations of nucleosome binding by different CBX proteins can provide information about differences in their binding specificities in the normal cellular environment. We visualized the chromatin-associated subpopulation of each CBX protein using BiFC analysis. Each CBX family member was fused to a fluorescent protein fragment, and histone H3 isoforms were fused to the complementary fragment. Nucleosome binding by the CBX fusions was predicted to result in formation of fluorescent complexes. We compared the distributions of the chromatin-associated CBX proteins with the total population of each CBX protein fused to YFP to determine whether the entire population of these proteins was associated with chromatin. All fusions were made at identical positions in the CBX proteins and at positions in H3 variants that were not predicted to alter their functions [see supporting information (SI) Results]. We initially focused on CBX protein interactions with H3.2 because it has the highest level of K27 trimethylation (24). The distributions were compared in mouse ES cells (PGK12.1) and in mouse embryo fibroblasts (MEFs).
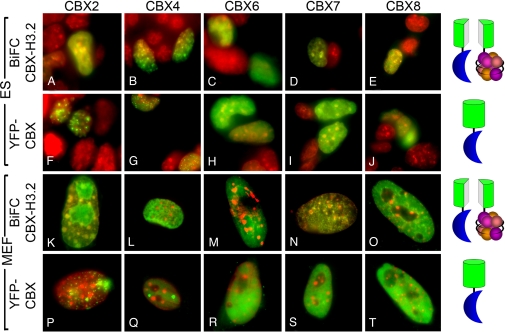
CBX2–H3.2 BiFC complexes were enriched in Hoechst-staining chromocenters, whereas the total population of CBX2 fused to YFP was localized to foci that were not associated with chromocenters (Fig. 1A vs. F and K vs. P). CBX6–H3.2 BiFC complexes as well as the total population of CBX6 fused to YFP were nearly uniformly distributed in the nucleoplasm (Fig. 1 C vs. H and M vs. R). BiFC complexes formed by CBX4, CBX7, and CBX8 with H3.2 had distinct distributions in different cell types. In most ES cells CBX4–, CBX7–, and CBX8–H3.2 BiFC complexes were enriched in chromocenters (Fig. 1 B, D, and E). In MEFs the same complexes were enriched in small foci and were excluded from regions encompassing chromocenters (Fig. 1 L, N, and O). The total populations of CBX4, CBX7, and CBX8 had similar distributions in ES cells and MEFs that were generally distinct from the distributions of BiFC complexes. CBX4 fused to YFP was localized to small foci (Fig. 1 G and Q), whereas the total populations of CBX7 and CBX8 fused to YFP were nearly uniformly distributed in the nucleoplasm, with some exclusion from chromocenters (Fig. 1 I, J, S, and T). The distributions of CBX proteins and complexes they formed on chromatin varied among individual cells (Table S1). Nevertheless, the distributions of BiFC complexes formed by each CBX protein with H3.2 in a majority of cells were distinct both from one another as well as from the distributions of the same proteins fused to YFP.
Fig. 1.
Comparison of the distributions of chromatin-associated and total CBX proteins. (A–E) Distributions of chromatin-associated CBX proteins in ES cells visualized by BiFC analysis. BiFC complexes formed by the CBX proteins indicated above the images with H3.2 fused to complementary fluorescent protein fragments (green) and Hoechst staining of DNA (red) were imaged in live cells. (F–J) Distributions of the total populations of CBX proteins fused to YFP (green) and Hoechst staining of DNA (red) imaged in ES cells. (K–O) Distributions of chromatin-associated CBX proteins in MEFs visualized by BiFC analysis as described for panels A–E. (P–T) Distributions of CBX proteins fused to YFP in MEFs imaged as described for panels F–J. The images shown represent the most frequently observed distributions in each population. The diagrams (Right) represent the BiFC complexes and YFP fusions visualized in each row.
We tested whether BiFC complex formation required specific recognition of H3 by each CBX protein by deletion of the N-terminal tail of H3. Deletion of the N-terminal tail prevents replication-dependent assembly of H3.1 and H3.2 into chromatin (25) and eliminates most sites of posttranslational modification. BiFC complex formation by all CBX proteins was reduced by 40%–80% according to quantification by flow cytometry (Fig. 2). The BiFC complexes formed by CBX proteins with tailless H3.2 accumulated in regions that excluded Hoechst staining (Fig. S1). Similar regions were occasionally observed in cells that expressed high levels of CBX proteins with wild-type H3.2 (i.e., Fig. 1K). This nonspecific BiFC complex formation did not interfere with determination of the distributions of BiFC complexes formed by CBX protein binding to H3 assembled in nucleosomes.
Fig. 2.
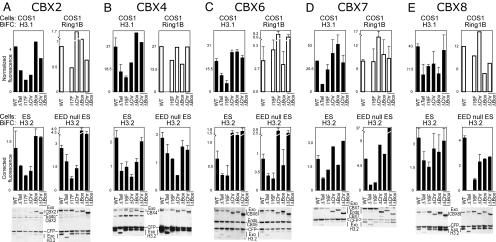
Quantitative BiFC analysis of the effects of conserved CBX protein domains and H3 K27 trimethylation on interactions with H3 and Ring1B. (A–E) Flow cytometry quantitation of the fluorescence intensities of BiFC complexes formed by the CBX proteins indicated at the top of each panel in COS1, ES, and EED null ES cells. The histograms show the fluorescence intensities of cells containing BiFC complexes formed by different CBX proteins with H3 (solid bars) or Ring1B (open bars). The mutations in CBX and H3 are indicated below each bar in the histograms. WT, wild type; ΔTail, deletion of N-terminal tail of H3.1 or H3.2; I16F, I17F, point mutation in the CBX chromodomain; ΔChr, deletion of the CBX chromodomain; ΔBox, deletion of the CBX chromobox; ΔChr ΔBox, combined deletion of both CBX domains. Normalized fluorescence = BiFC fluorescence/CFP fluorescence. Corrected fluorescence = Normalized fluorescence/Relative expression level. The relative expression levels were calculated separately for mutations in each CBX protein. The levels of exogenous (Exo) and endogenous (Endo) protein expression in ES cells and EED null ES cells were determined by Western blotting using mixtures of antibodies directed against GFP and the respective CBX proteins. CBX4 and CBX8 antibodies were not used because the endogenous proteins were not detected (Fig. S2).
We examined the levels of CBX fusion protein expression and their target gene preferences and functions to determine whether they were valid models for the endogenous CBX proteins. The CBX2, CBX6, and CBX7 fusions were expressed at levels comparable to or lower than those of the corresponding endogenous CBX proteins in ES cells (Fig. 2 and Fig. S2). The CBX4 and CBX8 fusions were expressed at similar levels, but endogenous CBX4 and CBX8 were not detected in ES cells. In ES cells that stably produced CBX2–H3.2 BiFC complexes (see below), the CBX2 fusions bound to genes recognized by endogenous CBX2 (Fig. S3) and CBX proteins fused to Venus bind selectively to genes that are recognized by endogenous PcG proteins in ES cells (26). CBX4 and CBX6 fused to the GAL4 DNA binding domain repressed an integrated reporter gene carrying GAL4 binding sites (Fig. S4). Taken together, these results suggest that the fusion proteins were valid probes for visualization of CBX protein interactions in cells.
Effects of Regions Conserved Among CBX Proteins on Association with H3 and Ring1B.
The distinct distributions of chromatin-associated CBX proteins were seemingly at odds with the proposed conserved roles of the chromodomain and chromobox regions in chromatin association by CBX proteins. We quantified the effects of mutations in these regions on BiFC complex formation by each CBX protein using flow cytometry. We initially examined BiFC complex formation with H3.1 and the Ring1B subunit of PRC1 in COS1 cells (Fig. 2 Top) where the CBX mutants were expressed at comparable levels (Fig. S5 A–E). Most CBX proteins formed BiFC complexes with all H3 variants with similar efficiencies (Fig. S5F).
Deletion of the chromodomains of CBX2, CBX4, and CBX8 reduced BiFC complex formation with H3.1 but had either no effect or increased their association with Ring1B (Fig. 2 A, B, and E; ΔChr). In contrast, deletion of the chromodomains of CBX6 and CBX7 had little effect on BiFC complex formation with H3.1 but increased their association with Ring1B (Fig. 2 C and D). More efficient complementation with Ring1B could be due to elimination of competing interactions by these deletions. The divergent effects of chromodomain deletions on BiFC complex formation with H3.1 by different CBX proteins suggest that the chromodomain did not have a conserved role in chromatin association by CBX proteins.
A single amino acid substitution in the chromodomains of all CBX proteins reduced BiFC complex formation with H3.1 and either reduced or had no effect on their association with Ring1B (Fig. 2 I16F and I17F). The general decrease in BiFC complex formation is consistent with the dominant effect of the corresponding mutation in Drosophila Pc on polycomb target gene expression (27) and suggests that this mutation affected interactions with many partners.
Chromobox deletions in CBX2, CBX4, CBX6, and CBX7 caused either no change or a small increase in BiFC complex formation with H3.1 (Fig. 2; ΔBox). Deletion of the chromobox of CBX4 reduced BiFC complex formation with Ring1B, but the corresponding deletions in CBX2, CBX6, and CBX7 had no significant effect on this interaction. Deletion of the CBX8 chromobox reduced BiFC complex formation with both H3.1 and Ring1B, possibly reflecting its lower level of expression (Fig. S5). The chromobox was therefore not necessary for interactions with either H3.1 or Ring1B by the majority of CBX proteins.
To determine whether the chromodomain and chromobox had redundant roles, we examined the effects of deleting both regions. The combined deletion caused a small reduction in BiFC complex formation by CBX2 and CBX8 but had either no effect or increased BiFC complex formation by CBX4, CBX6, and CBX7 with either H3.1 or Ring1B (Fig. 2; ΔChr ΔBox). Consequently, neither the chromodomain nor the chromobox was essential for chromatin association or Ring1B binding by CBX proteins in COS1 cells.
Determinants of Chromatin Association by CBX Proteins in ES Cells.
We quantified the effects of chromodomain and chromobox mutations in ES cells where the CBX fusions were expressed at levels comparable to those of endogenous CBX2, CBX6, and CBX7 (Fig. 2 Bottom). Almost all mutations had the same effects on BiFC complex formation with H3.2 in ES cells as with H3.1 in COS1 cells. The only cell-type- or H3-variant-specific effects were caused by chromobox deletions in CBX4 and CBX7 as well as deletion of the chromodomain or chromobox of CBX6.
The dispensability of the chromodomain in chromatin association by CBX proteins raised the question of whether K27 trimethylation affected H3 binding by these proteins. We quantified BiFC complex formation in EED null ES cells that have no detectable H3 K27 trimethylation (6). The intensities of BiFC signals in EED null ES cells were comparable to those observed in wild-type ES cells for both wild-type and mutant CBX proteins. The only difference was an increase in BiFC complex formation caused by combined chromodomain and chromobox deletions in CBX7.
Comparison of BiFC Complex Distributions with Anti-trimethyl-K27 Immunofluorescence.
To examine whether BiFC complexes formed by CBX proteins were associated with regions of high H3 K27 trimethylation, we compared the distributions of CBX–H3.2 BiFC complexes with anti-trimethyl-K27 immunofluorescence. CBX2–H3.2 and CBX7–H3.2 but not CBX4–H3.2 BiFC complexes colocalized with a bright spot of anti-trimethyl-K27 immunoreactivity corresponding to the inactive X (Fig. S6). Previously, CBX2, CBX6, CBX7, and CBX8 but not CBX4 fusions were shown to be localized to the inactive X (21). Thus, BiFC analysis reported faithfully on CBX protein recruitment to the inactive X. Apart from the inactive X and aggregates associated with overexpression, BiFC complexes did not colocalize with anti-trimethyl-K27 immunoreactivity. Thus, CBX protein binding on autosomes did not coincide with regions of high anti-trimethyl-K27 immunoreactivity in MEFs or ES cells.
Distributions of Stably Expressed CBX–H3.2 BiFC Complexes in Interphase and on Metaphase Chromosomes.
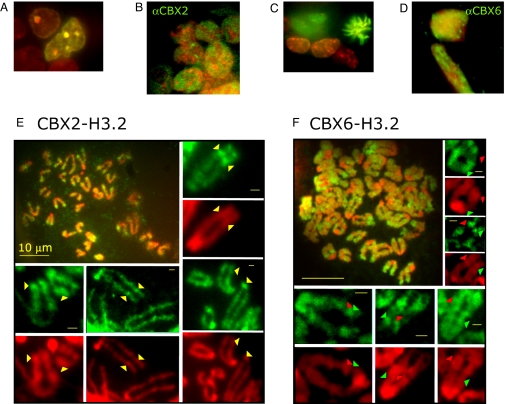
To control the timing and levels of BiFC complex formation, we developed stable cell lines that expressed the CBX2 and CBX6 fusions under the control of a doxycycline-inducible promoter and the H3.2 fusion constitutively at <1% of the level of total cellular H3. The time course of BiFC complex formation and disappearance closely followed changes in the level of CBX2 fusion protein expression (Fig. S7). The distributions of the CBX2–H3.2 and CBX6–H3.2 BiFC complexes in the stable cell lines were similar to those observed in transiently expressing cells and remained stable over several weeks of expression (compare Fig. 3 A and C with Fig. 1 A and C; Movie S1). Endogenous CBX2 was distributed in a granular pattern that was excluded from regions resembling nucleoli, whereas endogenous CBX6 was uniformly distributed in the nucleus (Fig. 3 B and D).
Fig. 3.
Distributions of CBX–H3.2 BiFC complexes during interphase and on metaphase chromosomes in stable ES cell lines. (A and C) Distributions of stably expressed CBX2–H3.2 and CBX6–H3.2 BiFC complexes, respectively (green), and Hoechst staining (red) during interphase. (B and D) Immunofluorescence detection of endogenous CBX2 and CBX6, respectively, in fixed ES cells. The spots in the αCBX6 image represent background signal that was not competed by antigen. (E and F) Metaphase spreads from cell lines stably expressing CBX2–H3.2 and CBX6–H3.2 BiFC complexes (green) and Hoechst staining (red). The montage of individual chromosomes shows regions with exclusive BiFC (green arrowheads) and Hoechst (red arrowheads) signals, as well as overlapping (yellow arrowheads) signals. (Scale bars, 10 μm in whole spreads, 1 μm in individual chromosomes.)
To determine whether BiFC complexes formed by different CBX proteins were associated with different chromosomal regions, we examined their distributions on metaphase spreads. Both CBX2–H3.2 and CBX6–H3.2 BiFC complexes formed discrete banding patterns that were symmetrically disposed on sister chromatids. CBX2–H3.2 BiFC complexes were enriched in distinct foci on the chromosomes, overlapping regions of bright Hoechst staining (Fig. 3E). In contrast, CBX6–H3.2 BiFC complexes were distributed over a larger proportion of most chromosomes and were excluded from regions of bright Hoechst staining (Fig. 3F). The reciprocal patterns of CBX2 and CBX6 BiFC complexes on metaphase chromosomes were concordant with their distinct distributions in interphase nuclei.
Histone Modifications Associated with CBX2–H3.2 BiFC Complexes.
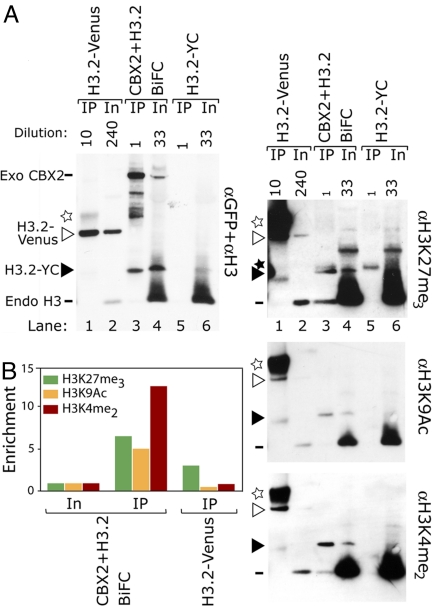
To identify histone modifications associated with CBX proteins bound to chromatin, we developed a new method based on the isolation of BiFC stabilized complexes (iBiSC). BiFC complexes were immunopurified using anti-FLAG antibodies from the stable cell lines that produced CBX2–H3.2 complexes. As a negative control, chromatin from the parental cell line was processed in parallel. The immunopurified complexes and aliquots of input chromatin were analyzed by immunoblotting using antibodies recognizing GFP, H3, and specific H3 modifications (Fig. 4A). The H3.2 fusion was coprecipitated by anti-FLAG antibodies from cells that expressed CBX2 fused to the complementary fragment but not from control cells that expressed the H3.2 fusion alone (Fig. 4A, lanes 3 and 5). On the basis of the relative efficiencies of precipitation, we estimate that the BiFC complexes were purified by >100-fold (see SI Results). We determined the ratio of the H3.2 bands between the immunopurified fraction and input chromatin on each blot. The enrichment was calculated by dividing the ratio observed on blots probes with antibodies directed against specific modifications by the ratio observed on the blot probed with anti-GFP and anti-H3 antibodies (Fig. 4A, lanes 3 and 4). The immunopurified H3.2 fusion was enriched in trimethyl-K27, acetyl-K9, and dimethyl-K4 compared with input chromatin (Fig. 4B). As a control, we measured the enrichment of histone modifications in H3.2 fused to Venus immunopurified with anti-GFP antibodies (Fig. 4A, lane 1). The H3.2 fusion immunopurified with anti-GFP antibodies was slightly enriched in trimethyl-K27 but was not enriched in acetyl-K9 or dimethyl-K4 (Fig. 4B). The same histone modifications were enriched in independent experiments, but the level of enrichment varied, possibly because of changes in H3.2 modifications caused by CBX fusion expression or BiFC complex formation (Fig. S8). The K4A and K9A but not K27A substitutions in H3.1 reduced BiFC complex formation with CBX2 (Fig. S9), suggesting that K4 and K9 but not K27 contributed to CBX2 recruitment to chromatin.
Fig. 4.
Enrichment of modifications in H3.2 associated with CBX2 identified using iBiSC. (A) Chromatin derived from cells that expressed the fusion proteins indicated above each pair of lanes was immunopurified using anti-GFP (lane 1) or anti-FLAG (lanes 3 and 5) antibodies. The total amounts of H3.2 fusions (αGFP+αH3) and H3.2 fusions containing the modifications indicated to the right of each blot were compared in the input (In) and immunopurified (IP) chromatin by immunoblotting, as described in Materials and Methods. The mobilities of the H3.2–YC and H3.2–Venus bands that were used for quantitation are indicated by open and solid triangles, respectively. The solid star marks a cross-reactive band, and the open star denotes the heavy chain. (B) Enrichment of H3.2 modifications in CBX2–H3.2 BiFC complexes and in H3.2–Venus immunopurified without BiFC complex formation.
Discussion
PcG proteins in general and the CBX family in particular have expanded in number during vertebrate evolution. The strict conservation of the chromodomain and chromobox sequences raised the question of whether different CBX proteins bound to the same or overlapping targets in chromatin. Using imaging methods based on the BiFC assay, we found that different CBX proteins bound to distinct chromatin regions. Quantitative BiFC analysis demonstrated that chromatin association by different CBX proteins was mediated by nonconserved protein regions. The nearly mutually exclusive distributions of CBX2 and CBX6 BiFC complexes indicate that these proteins were associated with largely nonoverlapping sets of target genes.
The difference between the distributions of the total population of each CBX protein fused to YFP and the chromatin-associated subpopulation visualized by BiFC complex formation with H3.2 suggested that a large proportion of each CBX protein was not associated with H3.2. This interpretation is consistent with the observation that a large fraction of CBX proteins is highly mobile in undifferentiated ES cells and in Drosophila embryos (26, 28). The CBX proteins that are not chromatin associated could be recruited to new target genes during differentiation.
The unique distributions of BiFC complexes formed by different CBX proteins demonstrate that their localization was neither a generic consequence of BiFC complex formation nor determined by the localization of the free CBX protein. CBX2–H3.2 and CBX6–H3.2 BiFC complexes exhibited contrasting distributions both in interphase and metaphase, suggesting that these patterns reflected interactions with target loci that had distinct subnuclear distributions independent of BiFC complex formation.
The role of H3 K27 trimethylation in PRC1 recruitment is widely assumed despite accumulating genetic data demonstrating PRC1 functions independent of PRC2 activity (3, 15, 16). Our experiments demonstrated that neither the chromodomains nor H3 K27 trimethylation were required for chromatin association by CBX proteins in cells, and chromatin-associated CBX proteins did not colocalize with H3 K27 trimethylation outside the inactive X. The BiFC data do not exclude the possibility that K27 methylation stabilizes CBX protein association with chromatin. These results suggest that PRC1 recruitment in the absence of K27 trimethylation is not limited to special cases, such as the inactive X and the male pronucleus (3, 15, 16).
The visualization of CBX–H3.2 BiFC complexes on metaphase chromosomes provides a unique genome-wide view of CBX protein binding. Metaphase chromosomes are more condensed than polytene chromosomes in Drosophila salivary glands, but they can be analyzed in any dividing cell type, and changes in complex distributions in response to stimuli and differentiation can be examined. Cytogenetic imaging of BiFC complexes provides high-content information that enables comparison of the genome-wide occupancy of chromatin binding proteins in single cells.
The identification of histone modifications that are associated with a particular regulatory protein complex in cells has been difficult. The iBiSC approach enabled detection of H3.2 modifications that were enriched in CBX2–H3.2 BiFC complexes. The roles of these modifications in CBX2 recruitment or functions are not clear, but “bivalent domain” H3 trimethyl-K4 and H3 trimethyl-K27 modifications correlate with PcG complex binding, and H3 acetyl-K9 modifications are altered in embryos lacking PRC1 components (13, 29). Further studies of the temporal and causal relationships between these modifications and CBX2 binding are important.
The unique patterns of chromatin association by different CBX proteins both in interphase nuclei and on metaphase chromosomes suggest that these proteins bind to nonredundant sets of target genes. Although the biologic functions of several CBX proteins have been studied in different experimental systems, no comparative studies of their functions have been performed in the same cell type. Our results provide new motivation for investigation of the mechanisms of selective CBX protein recruitment.
Materials and Methods
Plasmids, Cell Lines, and Imaging.
The construction of plasmids encoding CBX, H3, and Ring1B fusions, the cell lines, transfection protocols, fluorescence microscopy, flow cytometry, and immunologic methods are described in detail in SI Materials and Methods and Tables S2 and S3.
Derivation of Stable ES Cell Lines and Preparation of Metaphase Spreads.
ES cell lines with integrated expression constructs encoding CBX and H3.2 fused to complementary fluorescent protein fragments were established by two successive rounds of transfection. First, a cell line was established that expressed the H3.2 fusion constitutively. Second, CBX2 or CBX6 fused to the complementary fluorescent protein fragment under the control of a doxycycline-regulated promoter was introduced together with the doxycycline-responsive transactivator. Positive clones were identified by Western blotting. Metaphase spreads were prepared according to classic protocols (30) but without methanol acetic acid fixation to preserve BiFC complex fluorescence, as described in detail in SI Materials and Methods.
iBiSC and Determination of the Enrichment of Histone Modifications.
iBiSC was performed under the same conditions as ChIP analysis but without cross-linking (13). The immunoprecipitated and input chromatin were analyzed by Western blotting using antibodies recognizing specific modifications (αMod) and total H3.2 (αTotal). The enrichment of histone modifications was calculated using Eq. 1.
 |
Supplementary Material
Acknowledgments.
We thank Caroline Becker, Zunair Mahmood, and J'aime Manion for capable assistance; Terry Magnuson for providing EED null ES cells; and members of the Kerppola laboratory, in particular Xiaojun Ren, for constructive criticisms and valuable suggestions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805317105/DCSupplemental.
References
- 1.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 2.O'Carroll D, et al. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voncken JW, et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci USA. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park IK, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 6.Montgomery ND, et al. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 7.Shao Z, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 8.Levine SS, et al. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 11.Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 12.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 13.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 14.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoeftner S, et al. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puschendorf M, et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet. 2008;40:411–420. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- 17.Pearce JJ, Singh PB, Gaunt SJ. The mouse has a Polycomb-like chromobox gene. Development. 1992;114:921–929. doi: 10.1242/dev.114.4.921. [DOI] [PubMed] [Google Scholar]
- 18.Alkema MJ, et al. MPc2, a new murine homolog of the Drosophila polycomb protein is a member of the mouse polycomb transcriptional repressor complex. J Mol Biol. 1997;273:993–1003. doi: 10.1006/jmbi.1997.1372. [DOI] [PubMed] [Google Scholar]
- 19.Bardos JI, Saurin AJ, Tissot C, Duprez E, Freemont PS. HPC3 is a new human polycomb orthologue that interacts and associates with RING1 and Bmi1 and has transcriptional repression properties. J Biol Chem. 2000;275:28785–28792. doi: 10.1074/jbc.M001835200. [DOI] [PubMed] [Google Scholar]
- 20.Gil J, Bernard D, Martinez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 21.Bernstein E, et al. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 23.Kanno T, et al. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- 24.Hake SB, et al. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- 25.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 26.Ren X, Vincenz C, Kerppola TK. Changes in the distributions and dynamics of polycomb repressive complexes during embryonic stem cell differentiation. Mol Cell Biol. 2008;28:2884–2895. doi: 10.1128/MCB.00949-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagstrom K, Muller M, Schedl P. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics. 1997;146:1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ficz G, Heintzmann R, Arndt-Jovin DJ. Polycomb group protein complexes exchange rapidly in living Drosophila. Development. 2005;132:3963–3976. doi: 10.1242/dev.01950. [DOI] [PubMed] [Google Scholar]
- 29.Fujimura Y, et al. Distinct roles of Polycomb group gene products in transcriptionally repressed and active domains of Hoxb8. Development. 2006;133:2371–2381. doi: 10.1242/dev.02405. [DOI] [PubMed] [Google Scholar]
- 30.Tjio JH, Levan A. The chromosome number of man. Hereditas. 1956;42:1–6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.