Abstract
Through division into segments, animal bodies can reach higher degrees of complexity and functionality during development and evolution. The segmentation mechanisms of insects and vertebrates have been seen as fundamentally different at the anatomical and molecular levels, and consequently, independently evolved. However, this conclusion was mostly based on observations of derived insects such as Drosophila. We have cloned the Delta, Notch, and hairy genes in the cockroach Periplaneta americana, a basal insect with short germ-band development, and carried out functional assays of Notch activity during its segmentation. Our results show that, in more basal insects, segmentation involves a similar developmental mechanism to that in vertebrates, including induction of segment formation by cyclic segmental stripes of hairy and Delta expression. This result indicates that Notch-mediated segmentation is the ancestral segmentation mechanism of insects, and together with previous results in the literature [Stollewerk A, Schoppmeier M, Damen WGM (2003) Nature 423:863–865], of arthropods as well. The similarity with vertebrate segmentation might suggest that Notch-mediated segmentation is an ancient developmental mechanism inherited from a common ancestor of insects and vertebrates.
Keywords: arthropod, hairy gene, insect, segmental clock, evolution
Segmentation is a basic developmental process that divides the body into units which can then follow independent developmental programs. In this way, animals can repeatedly use their developmental genes to extend and diversify their bodies. The segmentation of vertebrates and insects, giving rise to somites and segments, respectively, has been regarded as different based on observations of embryonic morphology and developmental genetics. Vertebrate somites are formed by wavefronts of Notch signaling activity emanating from the posterior end of the embryo and leading to the formation of somite borders within a proliferating anlage (1–3) [supporting information (SI) Fig. S1A]. Notch signaling proceeds by binding of the transmembrane protein Delta to the receptor Notch and activation of downstream genes of the hairy/HES family (2, 4). A similar mechanism has been found in an arthropod, the spider Cupiennus salei (5). However, it is not known whether this similarity is the result of shared ancestry or independent parallel evolution in this spider, as functional evidence supporting a role for Notch during segmentation has not been found in any other arthropod and, in particular, seems absent in Drosophila and Tribolium species (6, 7). However, these two insect model systems have derived modes of development, including a full metamorphosis, that do not reflect the ancestral insect condition and may not be representative of insects as a whole. Particularly, Drosophila has a long germ-band development in which the embryo forms all its segments at the blastoderm stage through a mechanism involving a cascade of diffusible transcription factors (i.e., gap-segmentation) (8) (Fig. S1 B and C). More primitive insects have a short germ-band development in which most segments are formed by cell proliferation and bud off from a growth zone at the posterior end of the animal (Fig. S1 D–F). Typically, head segments and the first thoracic segment (T1) form in the blastoderm, whereas the rest of the thoracic and abdominal segments are added sequentially at the growth zone (9, 10), a process that resembles somite addition at the posterior end of vertebrate embryos. After segment border formation, the mesoderm segregates from the ectoderm and forms separated cell blocks with an internal cavity, called “somites” because of their similarity to vertebrate somites (9, 10) (Fig. S1G). In addition, RNAi works better in short-germ band insect classes such as Periplaneta and Blatella species (Dictyoptera) (12, 17) and Oncopeltus (Hemiptera) (6) than in Drosophila (Diptera), in which co-expression of Dicer is often required. These facts led us to study the role of Notch in the segmentation of Periplaneta americana.
Results and Discussion
Notch Pathway Expression in Segmental Stripes.
We have cloned several members of the Notch signaling pathway from Periplaneta: the ligand Delta (Pa-Dl), the receptor Notch (Pa-N), and the segmentation transcriptional target hairy (Pa-h; see Methods). As in other insects, a single copy of these genes seems to exist in Periplaneta, containing protein domains similar to other metazoan homologues. Amino acid sequence analyses confirm that they are homologous to the Notch, Delta, and hairy genes in other metazoan phyla. The P. americana sequences are recovered in the expected phylogenetic positions, and the overall structure of the trees is broadly consistent with the expected phylogenetic trees (SI Text and Fig. S2).
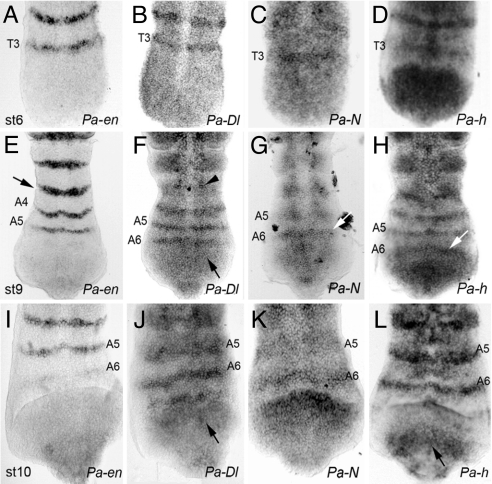
Expression of the engrailed gene (Pa-en), a segmental marker (11, 12), in Periplaneta is found in stripes at the posterior side of the segments as in other insects, and precedes the appearance of the segmental furrow, which appears abutting the posterior edge of the Pa-en stripe (Fig. 1 A, E, and I).The Pa-en stripes appear sequentially at regular intervals (≈6 h at 29°C) at the posterior end of the embryo, just anterior to the growth zone, and remain until the end of development (Fig. 3E) (12).
Fig. 1.
Expression of Pa-en, Pa-Dl, Pa-N, and Pa-h during cockroach segmentation; posterior is down. (A) Pa-en expression at stage 6 (≈96 h), before morphological signs of segmentation, in stripes at the developing segments at the growth zone up to the third thoracic segment (T3). (B) Pa-Dl expression at stage 6 is also present in stripes. (C) Pa-N expression at stage 6 in a single segmental stripe in T3. (D) Pa-h expression at stage 6. In addition to the segmental stripes, expression is observed in a domain at the posterior end of the growth zone. (E) Pa-en expression at stage 9 (≈120 h). Embryos have laid down the fifth abdominal stripe (A5). Segment furrow is forming in A3 (arrow). (F) Pa-Dl expression at stage 9 is ahead of Pa-en and is present in the sixth abdominal stripe (A6; compare with E). Anterior to A4, a diffuse pattern is observed in the epidermis, newly formed mesoderm, and specific neuroblasts (arrowhead). Posterior to A6, faint diffuse expression is seen in the posterior growth zone (arrow). (G) Pa-N expression at stage 9 has a single segmental stripe in A6. A5 expression is diffusing. (H) Pa-h expression at stage 9 is similar to that of Pa-Dl (see F) with the A6 stripe forming at the top of a stronger posterior growth zone domain (arrow). (I) Pa-en expression a stage 10 (≈128 h) is starting in a single row of cells in A6. (J) Pa-Dl expression at stage 10 is already established in A6. Expression at the posterior growth zone is easier to discriminate (arrow). (K) Pa-N segmental expression in A6. (L) Pa-h expression at stage 10 is similar to that of Pa-Dl in J.
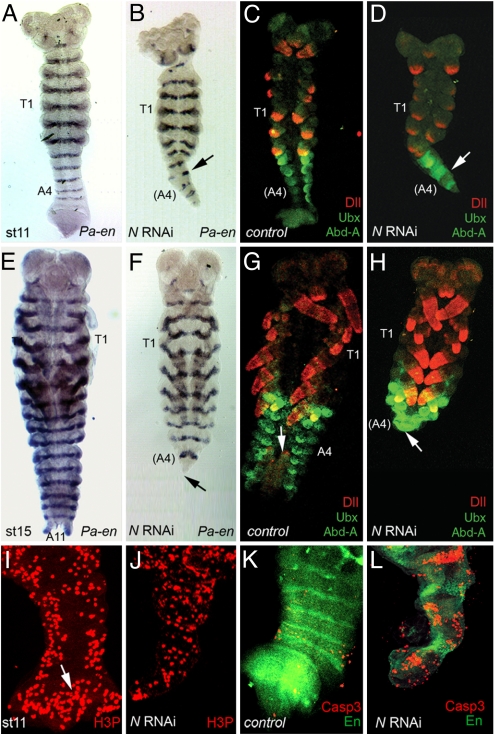
Fig. 3.
N-RNAi precludes segmentation specifically. (A) Control embryos at stage 11 (about 144 h.) have laid down the seventh abdominal stripe of Pa-en (A7). Limbs buds, somites and segment borders in more anterior segments (e.g., T1) are apparent. (B) N-RNAi-treated embryos by stage 12 develop with gaps in Pa-en stripes posterior to T1 (arrow), and the last vestigial stripe is that of A4. The growth zone is reduced and more posterior segments are absent. (C) Control embryo showing wild-type expression of Dll (red), AbdA and Ubx (green). (D) N-RNAi does not eliminate the expression of Dll (red), AbdA or Ubx (green). (E) By stage 15 control embryos have finished segmentation and contain eleven abdominal segments (A11) with stripes of Pa-en expression. (F) Stage 15 N-RNAi embryo showing normal development of the anterior segments although the embryo is truncated, and the segments posterior to A4 are absent (arrow). (G) Stage 15 control embryo showing the Dll, and AbdA and Ubx patterns of expression. The tail of the embryo is folded and the tip (the cerci) are highlighted by an arrow. (H) N-RNAi stage 15 embryo stained as in C. The posterior end of the truncated abdomen is denoted by an arrow. Dll (red), AbdA and Ubx (green) are still expressed. (I) Posterior part of a stage 11 embryo showing cells undergoing mitosis (red). The posterior growth zone contains a high number of mitotic cells (arrow). Proliferation continues in the newly formed segments. (J) Posterior part of a stage 11 N-RNAi embryo stained as in I. The posterior growth zone is much reduced but there are still cells undergoing mitosis with a lower density than the WT. (K) Posterior part of a stage 11 control embryo labeled as in L. Only a few cells scattered throughout segments show apoptosis. (L) Cell death in the posterior part of a N-RNAi stage 11 embryo. Cells undergoing apoptosis are detected by an anti-Caspase 3 antibody (red). Segments are labeled by anti-engrailed antibody (green). Cells undergoing apoptosis appear in clusters mostly after Pa-En expression, and thus segmentation, is established.
Interestingly, we also find expression of Pa-Dl in stripes at the growth zone where segments form (Fig. 1 B, F, and J). These stripes form just earlier than, and anterior to, the stripes of Pa-en expression in the developing segments. Thus, the expression of Pa-Dl appears several hours before the segregation of the mesoderm and the formation of segmental furrows. Expression of Pa-N is seen transitorily in a single segmental stripe, which appears at the same time as the earliest Pa-Dl stripe and disappears after Pa-en expression is established (Fig. 1 C, G, and K). Thus, in older, more anterior segments, the striped expression of Pa-Dl and Pa-N fades and new patterns form in mesodermal and neurogenic regions (Fig. 1F), as in other insects (7). The transitory segmental striped expression of Pa-Dl and Pa-N is not present in Drosophila, but is found in chelicerates and vertebrates, in which it underlies the role of Notch signaling in segmentation. Finally, we also observe expression of Pa-h in a pattern of stripes in the growth zone of the roach embryo. This expression has single-segmental periodicity, with one stripe of hairy per segment (Fig. 1 D, H, and L), as opposed to the double-segmental periodicity in Drosophila (13). The stripes of Pa-h emerge from the posterior growth zone at the same time as those of Pa-Dl and Pa-N, and fade similarly to those of Pa-Dl after segmental furrows start to form (Fig. 1). The timing of Pa-Dl, Pa-N, and Pa-h stripes resemble those found during vertebrate segmentation (1, 3) in that they precede both the morphological and molecular signs of segmentation and are thus compatible with a role for Notch signaling in cockroach segmentation.
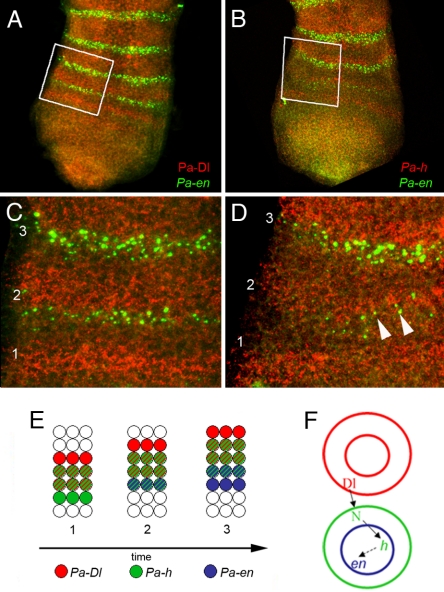
We next ascertained the relative positions of these stripes of gene expression by using double FISH. Both Pa-Dl and Pa-h stripes are anteriorly adjacent to those of Pa-en (Fig. 2 A–D). However, Pa-h and Pa-en expression overlap in the row of cells between these stripes, especially at the time of Pa-en activation (Fig. 2 B and D). These results, summarized in Fig. 2E, suggest a model of genetic regulation similar to that of vertebrates (2). In our case, anteriorly expressed Delta would signal through Notch, directly resulting in regulation of hairy expression and indirectly resulting in activation of engrailed, and hence formation of segment borders (Fig. 2F).
Fig. 2.
Spatial relationship between segmental patterns of members of the Notch pathway. (A) Growth zone of a stage 9 embryo showing Pa-Dl (red) and Pa-en (green) expression. (B) Growth zone of a stage 9 embryo showing Pa-h (red) and Pa-en (green) expression. (C) Detail of the anterior growth zone of a stage 9 embryo showing the activation of Pa-en (green) in cells posterior (Bottom) and adjacent to those expressing Pa-Dl (red). 1, no Pa-en; 2, initial activation of Pa-en in a single row of cells; 3, established expression in two to three rows. (D) Pa-h expression (red) is also anterior to Pa-en but overlaps it during initial activation (arrowheads; 2). (E) Summary of Notch pathway expression. (1) Before Pa-en activation, overlapping stripes of Pa-Dl- and Pa-h-expressing cells appear. (2) Pa-en is then activated in a single row of cells adjacent to the Pa-Dl expressing cells and overlapping with Pa-h. (3) Later, self-maintained Pa-en expression (19) is present in cells without Pa-h expression. (F) Regulatory model for the control of segmentation. Pa-Dl-expressing cells signal and activate Notch signaling in adjacent posterior cells. Notch signaling transduction directly activates the expression of Pa-h and indirectly activates the expression of Pa-en.
Notch Activity in Periplaneta Is Required for Segmentation.
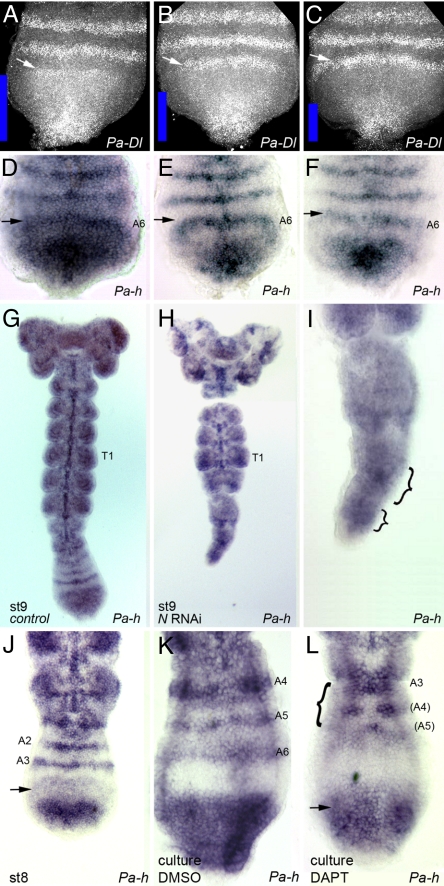
Because striped expression of Notch pathway members has occasionally been found in other insects without being apparently required for their segmentation (14, 15), we studied the functional requirements for Notch signaling in Periplaneta. First, we inhibited Notch expression by using RNAi. Following procedures in other insects (16, 17), cockroach mothers were injected with double-stranded Pa-N RNA (N-RNAi) and the offspring were studied for segmental defects. Embryos exposed to N-RNAi show no abnormalities immediately after the blastoderm stage (data not shown). Older embryos show abnormal and lost Pa-en stripes, with a gradient of severity from posterior to anterior. Pa-en stripes are eliminated in abdominal segments, show large gaps in thoracic segments, and occasionally show minor gaps in cephalic ones. In addition, segmental furrows are disrupted in thoracic and abdominal segments, and the abdominal segments and the growth zone are reduced (Fig. 3 A and B; Table S1). In the most extreme cases, the embryo is truncated and the posterior abdominal segments are missing. Remarkably, leaving N-RNAi-treated embryos to develop longer produces a more dramatic phenotype, as the addition of new posterior segments is abolished while more anterior segments continue developing (Fig. 3 E and F).
Three independent lines of evidence indicate that these N-RNAi phenotypes result from specific effects of the RNAi treatment rather than nonspecific effects. First, we observed loss of Pa-N RNA expression in N-RNAi embryos (data not shown); second, injection of double-stranded RNA for other genes does not produce this phenotype (A. Popadic, J.I.P., and J.P.C., unpublished work); and third, no alterations were observed in the expression of genes not directly involved in segmentation (Fig. 3 C, D, G, and H). Distal-less (Dll) gene expression prefigures the development of appendages in a variety of organisms including arthropods and insects (18). Expression of the Hox genes Ultrabithorax (Ubx) and abdominalA (AbdA) is required in insects to specify the correct identity of T3 and abdominal segments A1 to A8, but not for development of segmental primordia or segmental furrows (8, 19). Dll, Ubx, and Abd-A expression are still present in N-RNAi-treated embryos, even in segments showing clear morphological disruptions and loss of Pa-en (Fig. 3 C, D, G, and H). Finally, N-RNAi-treated embryos show enlargement of the cephalic nervous system (data not shown), which is similar to the neurogenic phenotype in Drosophila Notch mutant embryos (7).
To ascertain the developmental basis of segment loss in N-RNAi embryos, we studied changes in cell proliferation and death patterns. Cell proliferation is found in many cells in WT embryos, specifically in two bands running the length of the segmented part of the embryo and the growth zone (Fig. 3I). In N-RNAi embryos, the density of mitosis is lower within the very much reduced growth zone, and the mitotic pattern in the segmented region is also disorganized (Fig. 3J). Cell death is prominent in the cephalic regions of WT embryos (data not shown) but is rare toward their posterior region (Fig. 3K). In N-RNAi embryos, there is an increase in cell death in the posterior region, but mostly after segmentation has occurred (Fig. 3L). Thus, cell death is unlikely to account on its own for the phenotypes of segment loss observed, and the most likely cause involves a reduction of growth that is either a direct requirement for Pa-N in cell proliferation or an indirect consequence of the reduction in the number and size of segment primordia.
We conclude that the N-RNAi results highlight a specific requirement for Notch signaling in Pa-en expression, and in segments posterior to T1, establishment of segmental borders, segment primordia, and segment growth.
Periodic Generation of Segments and Stripes of Gene Expression by Notch.
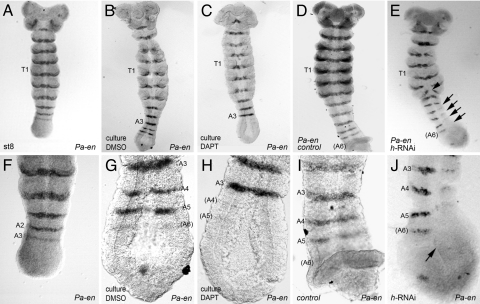
We have carried out two further independent tests of Notch signaling function during segmentation. First, we adapted a previously existing method to culture roach embryos (see Methods) to administer DAPT (2,5-bis[4-dimethylaminophenyl]-1,3,4-thiadiazole), a chemical inhibitor that interferes with Notch signaling (20). Embryos collected from a single ootheca at stage 8 are laying down the A3 stripe of Pa-en at the start of the culture (Fig. 4 A and F). Control embryos add two or three stripes of Pa-en expression (A4 to A6) at their growth zone during a 26-h culture period (Fig. 4 B and G). However, their sibling embryos cultured with DAPT do not complete any further proper Pa-en stripes (Fig. 4 C and H). Presumptive Pa-en stripes under DAPT culture consist of broken stretches at best, or at worst do not form at all. Previously laid down Pa-en stripes, such as A2 or A3, are not affected, suggesting that Pa-N is required during the activation, but not the maintenance, of Pa-en expression.
Fig. 4.
Interfering with N signaling via chemical inhibition by DAPT or via hairy RNAi disrupts segmental engrailed expression. (A) Pa-en expression in a late stage 8 embryo showing the third abdominal stripe of Pa-en (A3) being laid down before embryo culture. (B) Control embryo after 26 h of in vitro culture, new segments A4 and A5 have formed posteriorly (also see G). (C) No new Pa-en stripes are observed in a DAPT-treated embryo after 26 h of culture. Pa-en stripes that had been already formed before culture were not affected in these embryos (compare with B; details in G and H). (D) Pa-en expression in a stage 11 control embryo. The last stripe formed is A6. (E) Stage 11 h-RNAi embryo shows Pa-en expression. Pa-en is disrupted in the last abdominal segments (arrows, see detail in J). Segmentation irregularities are also observed in the third thoracic segment (T3) as the segmental furrow is truncated and a gap in Pa-en expression appears (arrowhead). (F) Posterior end of the embryo shown in A. (G) Detail of the posterior part of the embryo shown in B. (H) Detail of the DAPT-treated embryo shown in C. (I) Detail of the posterior part of the embryo shown in D. (J) Magnification of the posterior part of the h-RNAi embryo shown in E. Pa-en is present only in hemi-segments on the left. In addition, the posterior growth zone appears abnormal (arrow; compare with I). This phenotype is similar to that produced by DAPT in H.
Second, we assessed the requirement for Pa-h by h-RNAi and observed similar defects to those produced by DAPT. h-RNAi embryos display lost and abnormal Pa-en stripes and abnormal segmental furrows leading to segment fusions (Fig. 4 D, E, I, and J). Altogether, these results pinpoint the requirement for Notch signaling to the unsegmented growth zone during Pa-en activation and generation of segment primordia.
We observed the expression of Pa-Dl (Fig. 5 A–C) and Pa-h (Fig. 5 D–F) in detail in the growth zone during the formation of a segment. Roach embryos are laid sequentially in clutches inside an ootheca (9). Older embryos from one end of the ootheca do not differ in age by more than a whole segment from younger embryos from the opposite end (J.I.P., R.L., and J.P.C., unpublished data). Comparison of single-ootheca siblings thus illustrates a time series showing how segmental stripes of Pa-Dl (Fig. 5 A-C) and Pa-h (Fig. 5 D–F) are generated from the growth zone in a cyclical manner. Stripes emerge at the edge of the more diffuse expression domain at the posterior growth zone. Expression then decays behind the nascent stripe, which moves to a more anterior position. This process (i.e., decay behind the stripe and forward movement of the stripe) continues until Pa-en expression starts (Fig. 5 C and F). Pa-N expression in the growth zone also shows similar dynamic patterns, albeit fainter (data not shown). To test whether Notch signaling is required for Pa-h expression, we used N-RNAi (Fig. 5 G-I). We observe that Pa-h loses its cyclical, striped pattern and only a disorganized background remains. Similarly, in embryos cultured with DAPT cyclic Pa-h expression is also disrupted (Fig. 5 J–L). Only a few new stretches of Pa-h expression are formed, and cycling of Pa-h expression in the posterior growth zone stops.
Fig. 5.
Cyclic N signaling at the posterior growth zone controls roach segmentation. (A-C) Pa-Dl expression at the growth zone of WT embryos during the formation of the sixth abdominal stripe (arrow). Blue boxes denote Pa-Dl receding expression at the posterior growth zone. (A) The first sign of the formation of A6 starts with the up-regulation of Pa-Dl at the anterior edge of the posterior growth zone domain (arrow). Note that Pa-Dl expression covers the whole posterior growth zone (blue box). (B) Slightly later, Pa-Dl expression behind the nascent A6 stripes regresses (blue box) whereas the A6 stripe becomes stronger. (C) Finally, posterior expression of Pa-Dl remains only at the end of the growth zone, and the A6 stripe is fully formed. Note that, during this process, the distance between the A6 stripe and the posterior growth zone increases (compare with A and B). (D-F) Dynamic expression of Pa-h at the growth zone during the formation of the sixth abdominal stripe (A6; arrow) in WT embryos. (D) The presumptive sixth abdominal stripe appears by the down-regulation of Pa-h behind the anterior edge of the posterior growth zone. (E) As the A6 stripe is forming, the Pa-h pattern in the posterior growth zone regresses. (F) Finally, the Pa-h A6 stripe detaches from the posterior growth zone domain. (G-I) N-RNAi disrupts Pa-h cyclic expression. (G) Pa-h pattern of expression in a stage 10 control embryo. (H) N-RNAi stage 10 embryo shows Pa-h expression reduced posterior to T1 but remaining in more anterior segments in the somites, brain, and midline. (I) Detail of the posterior part of the N-RNAi embryo in E. No Pa-h segmental stripes appear; instead, a diffuse and irregular pattern is observed in the growth zone (brackets; compare with D-F). (J-L) DAPT treatment disrupts cyclic Pa-h expression. (J) Pa-h expression pattern in a stage 8 embryo before culture. The A3 stripe has just been laid down. In addition, dynamic Pa-h expression at the posterior growth zone is observed (arrow). (K) Control embryo after 26 h in culture has laid down three Pa-h stripes, similar to that observed with Pa-en expression (Fig. 4 A and F and B and G). (L) DAPT-treated embryo cultured for 26 h shows a few abortive segmental stretches of Pa-h expression (bracket) posterior to A3. No further posterior stripes are formed, and a longer unstained and unsegmented area develops anterior to the posterior growth zone domain, which appears irregular (arrow).
In summary, our analyses of expression patterns and DAPT and RNAi treatments reveal a requirement for Notch signaling during segmentation of Periplaneta americana. Stripes of Pa-Dl, Pa-h, and Pa-N expression appear shortly before Pa-en stripes and several hours before the morphological onset of segmentation in the region where presumptive segments form. The stripes of Pa-Dl appear anterior to the positions where Pa-en stripes will form, whereas Pa-h stripes overlap both domains. These expression patterns are compatible with the hypothesis that Pa-Dl signals through Pa-N to regulate Pa-h and Pa-en expression, and this hypothesis is corroborated by the DAPT and N-RNAi functional experiments. These functional results also confirm that the timing of Notch signaling is coupled to the sequential timing of segment development, a result reminiscent of the interplay between Notch signaling and the vertebrate segmentation clock (2–4). N-RNAi embryos at stage 15 show that, after an initial window during segment formation, Notch signaling has no major requirement in segmentation. Similarly, when we administer DAPT, we obtain defects only in the specific segments that are forming at the time. Finally, the results regarding the role of hairy in P. americana and vertebrates are also comparable. hairy expression appears in stripes that originate at the unsegmented zone in a cyclical manner, although the length of the region of the embryo where such oscillation takes place is smaller in the roach than in vertebrates. Blocking Notch signaling in both systems does not completely eliminate hairy expression, but abolishes its periodic pattern, and altering hairy expression leads to segmentation defects similar to those observed when Notch function is perturbed (1–3).
Ancestral Notch-Mediated Segmentation.
Our results support Notch-mediated segmentation as a basal developmental mechanism for insects (Fig. S3A). More derived insects such as Drosophila and Tribolium seem to have lost Notch-mediated segmentation and replaced it with gap segmentation (8, 16) in correlation with the evolution of specialized ovaries to support it (21). A gradual mechanism has been suggested for this evolutionary shift (22), and in this view, it would be possible for different insect species to have all, some, or none of their segments develop under the control of Notch, according to the insect's position in the phylogenetic tree, with closer relatives of Drosophila more likely to lack a role for Notch in segmentation. In Periplaneta N-RNAi embryos, we occasionally observed minor Pa-en expression defects in cephalic segments anterior to T1, but total loss of Pa-en and truncation of growth occur only in more posterior segments. This suggests that the requirement for Notch is less strong anterior to T1, and fits with the observation that these are the segments formed in the blastoderm (J.I.P., R.L., and J.P.C., unpublished data and refs. 9, 10) and thus presumably under the control of gap segmentation as in other short-germ band insects (6, 22). In this view, the segmental stripes of expression of Serrate in Drosophila and fringe in grasshoppers, which do not seem to have a role in segmentation (14, 15), might be seen as vestiges of an ancestral Notch segmentation mechanism. In Drosophila at least, the Notch segmentation mechanism seems to remain in a derived form having been co-opted for the formation of leg segments (23–25). The independent role of Notch in neurogenesis seems conserved as well (7).
The ancestral role of Notch segmentation is reinforced by results in the chelicerate Cupiennus salei. Notch, Delta, and other pathway members are expressed and required during segmentation in this spider (5), and homology with the Notch-mediated vertebrate segmentation clock has been debated, but not proven, as a functional requirement for Notch during segmentation had not been shown in any other arthropod (6, 16, 22). However, the striking similarity between Periplaneta and Cupiennus in the Pa-Dl, Pa-N, and Pa-h striped expression patterns in the developing segments and growth zone—and in the Notch RNAi effects on morphology and engrailed and hairy expression—suggests, as the most parsimonious explanation, that Notch segmentation is homologous in both lineages, insects and chelicerates, and therefore ancestral not only to insects but to arthropods as well (Fig. S3A). Such ancestral Notch-mediated segmentation would also explain the expression in stripes of Notch-related genes in another arthropod lineage, myriapods, represented by the centipede Strigamia maritima (26). Conservation of Notch segmentation in arthropods could be quite common.
Our findings re-ignite the question of whether Notch signaling has been independently recruited for segmentation in vertebrates and arthropods, or whether they have both inherited it from a common ancestor (Fig. S3A). Notch signaling activity is present in Cnidarians (27) and thus Notch signaling could have been recruited independently during the evolution of segmentation in separate vertebrate and arthropod ancestors. It would remain to be explained what constraints limited evolution's choice to the same signaling pathway given the ancestral presence of other pathways capable of similar developmental functions, such as Wnt and Hh (19, 27). Similarly, a putative conservation of Notch segmentation from a common ancestor begs the question of the constraints and mechanisms that have conserved this role of Notch and adapted it to changing embryonic architectures. Such a conservation would also fuel the debate about whether the last common ancestor of arthropods and vertebrates was a simple or complex animal (28, 29), and would raise the question of why, when, and how segmentation was lost in unsegmented species. In this regard, the recent findings in hemichordates supporting a common origin of dorsal-ventral patterning in vertebrates and insects are interesting (30). The conservation of Notch segmentation could also extend to lophotrochozoans. In annelids, expression of engrailed at segment borders (11) and dynamic expression of Notch pathway members during segmentation (31) has been reported. In primitive mollusks with metameric organization, recent results also reveal the presence of engrailed stripes (32).
An intermediate possibility is segmentation may have evolved independently from a simpler Notch-mediated metameric organization involving repeated organs or body boundaries in a common ancestor of protostomes and deuterostomes. Notch signaling is a good candidate to drive this hypothetical transition from metamerism to a more advanced segmental organization, as it is involved in extant animals in the segregation of repeated pattern elements from equivalence groups, in the establishment of boundaries or patterning lines, and in the segregation of specific cell lineages (2, 4, 7, 23–25). It is not difficult to imagine a Notch-mediated evolutionary transition from the development of repeated pattern elements along the body axis to their association with patterning boundaries, culminating in the association of these patterning boundaries with a lineage and fate restriction—the essence of segmentation in chordates, arthropods, and lophotrochozoans (Fig. S3B). Further studies should clarify which of these elements of segmental organization were present in the common ancestor, which evolved independently in each segmented lineage, and which have been lost in apparently non-metameric species.
Methods
Periplaneta americana Rearing.
Adult cockroaches were kept at 29°C. Freshly laid oothecae were collected and placed in a humidified chamber at 29°C. Embryos were staged according to laying date and morphology (9).
Cloning Pa-Dl Pa-N and Pa-h Genes.
We designed degenerate primers based on conserved amino acid domains in arthropod Delta exons 4 and 6. Periplaneta genomic DNA was used for PCR amplification. Low-specificity PCR reactions with outer primers (Dl3F, Dl3R; see SI Text) were followed by nested PCR reactions with inner primers (Dl3F, Dl4R). Expected-size PCR fragments were sequenced. Gene-specific primers were generated to isolate overlapping fragments of Pa-Dl cDNA using a λZAP II cDNA library [gift from B. Marie and J. Bacon (University of Sussex, Brighton, UK)] as template. PCR fragments were assembled into a full-length Pa-Dl cDNA of 3.2 kb. Degenerated primers used for Pa-N cloning are described by Stollewerk et al (5). Specific primers were designed to isolate Pa-N cDNA by RACE (Ambion). A 1.5-kb fragment of the 5′ end of the Pa-N transcript was isolated. For Pa-h degenerate primers using the second-exon, conserved sequences from other arthropods were used initially. Subsequently specific outer primers and nested primers were generated for 5′ and 3′ PCR reactions by using a λZAP II cDNA library as a template, and fragments were assembled into a full-length 1.7-kb Pa-h cDNA.
ISH and FISH.
We followed the protocol described by Marie and Bacon (12). digoxigenin antisense RNA probes were made using a 1.4-kb Pa-Dl cDNA fragment, a 1.5-kb Pa-N cDNA fragment, a 1.4-kb Pa-h cDNA fragment, and a 1.3-kb Pa-en2 cDNA fragment. Probes were hydrolyzed into smaller fragments as described by Moens (Division of Basic Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA). For FISH, we used TSA system protocols (Perkin–Elmer).
Immunocytochemistry.
We dissected oothecae in cold PBS solution and fixed embryos in cold 4% paraformaldehyde for 20 min. Afterward, we followed standard Drosophila protocols. The primary antibodies used recognize homologous proteins in a range of arthropod species. The mouse monoclonal antibodies and dilutions were: anti-Dll (18) at 1:200; FP6.87 antibody that recognizes Ubx and Abd-A proteins [Rob White (University of Cambridge, Cambridge)] at 1:10; and anti-En 4F11 at 1:5 (11). We used a rabbit anti-cleaved caspase 3 from Cell Signaling Technology at 1:50 to detect apoptosis and a rabbit anti-phosphorylated Histone 3 (Upstate) to label cells in mitosis at 1:1,000. Secondary antibodies conjugated with biotin or fluorophores were from Jackson Immunochemicals.
Embryo Culture and DAPT Treatment.
Embryos from stage 8 onward were cultured according to the method described by Wang and Denburg (33), which allows normal development for up to 48 h, with some modifications. Embryos were dissected in sterile culture medium at 29°C and transferred rapidly into a sterile low-adhesion tissue culture plate with prewarmed culture medium. Culture plates were placed in a humidified chamber at 29°C. Culture medium was replaced every 12 h. For DAPT treatment, a 10-mM stock in DMSO was diluted to 100 μM in culture medium. Control embryos were treated with the same amount of DMSO. Treatment with 50 μM of DAPT produced slightly weaker phenotypes but allowed longer culture.
RNAi.
Forward- and reverse-specific primers containing T7 polymerase promoter sequences were used to amplify the Pa-N RACE and Pa-h 3′ cDNA templates, giving rise to 700-bp and 850-bp fragments respectively. In vitro transcription of these fragments was carried out with the T7 RiboMAX kit (Promega). For Pa-N, dsRNA for injection was 0.5 μg/μl, whereas for Pa-h, dsRNA for injection was 1.5 μg/μl. Maternal injection was done as described by Cruz et al (17) by loading 4 μl dsRNA into a Hamilton 75N syringe and injecting newly molted adult virgin females in the sternites between the fourth and fifth abdominal segments. After 2 h, females were injected again to counteract loss of dsRNA resulting from wound leakage. Injected females were reared with males at 29°C, and oothecae were collected daily and cultured as described earlier. No segmental defects were observed in control embryos from mothers injected with dH20.
Supplementary Material
Acknowledgments.
We thank B. Marie for techniques and materials; R. Phillips and S. A. Bishop for technical help; and A. Popadic, J. Baguña, R. Ray, and C. Alonso for manuscript comments. This work was funded by a Wellcome Trust Senior Fellowship reference no. 057730 (J.P.C.) and a Biotechnology and Biological Sciences Research Council studentship (to R.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (Pa-Dl accession number FJ222590; Pa-N accession number FJ222591; Pa-H accession number FJ222592).
See Commentary on page 16411.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804093105/DCSupplemental.
References
- 1.Palmeirim I, Henrique D, IshHorowicz D, Pourquie O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- 2.Pourquie O. The segmentation clock: Converting embryonic time into spatial pattern. Science. 2003;301:328–330. doi: 10.1126/science.1085887. [DOI] [PubMed] [Google Scholar]
- 3.Jiang YJ, et al. Notch signalling and the synchronization of the somite segmentation clock. Nature. 2000;408:475–479. doi: 10.1038/35044091. [DOI] [PubMed] [Google Scholar]
- 4.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 5.Stollewerk A, Schoppmeier M, Damen WGM. Involvement of Notch and Delta genes in spider segmentation. Nature. 2003;423:863–865. doi: 10.1038/nature01682. [DOI] [PubMed] [Google Scholar]
- 6.Tautz D. Segmentation. Dev Cell. 2004;7:301–312. doi: 10.1016/j.devcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Campos-Ortega JA, Knust E. Genetics of early neurogenesis in Drosophila melanogaster. Ann Rev Gen. 1990;24:387–407. doi: 10.1146/annurev.ge.24.120190.002131. [DOI] [PubMed] [Google Scholar]
- 8.Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development (Cambridge, U.K.) 1987;101:1–22. [PubMed] [Google Scholar]
- 9.Lenoir-Rousseaux JJ, Lender T. Table de developpement embryonnaire de Periplaneta americana. Bull Soc Zool France. 1970;95:737–751. [Google Scholar]
- 10.Anderson DT. In: Developmental Systems: Insects. Counce SJ, Waddington CH, editors. Vol. 1. London: Acad. Press; 1972. pp. 95–163. [Google Scholar]
- 11.Patel NH, et al. Expression of engrailed proteins in arthropods, annelids, and chordates. Cell. 1989;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- 12.Marie B, Bacon JP. Two engrailed-related genes in the cockroach: cloning, phylogenetic analysis, expression and isolation of splice variants. Dev Genes Evol. 2000;210:436–448. doi: 10.1007/s004270000082. [DOI] [PubMed] [Google Scholar]
- 13.Ingham PW, Howard KR, Ish-Horowicz D. Transcription pattern of the Drosophila segmentation gene hairy. Nature. 1985;318:439–445. [Google Scholar]
- 14.Dearden P, Akam M. A role for Fringe in segment morphogenesis but not segment formation in the grasshopper, Schistocerca gregaria. Dev Genes Evol. 2000;210:329–336. doi: 10.1007/s004270000072. [DOI] [PubMed] [Google Scholar]
- 15.Bachmann A, Knust E. Dissection of cis-regulatory elements of the Drosophila gene Serrate. Dev Genes Evol. 1998;208:346–351. doi: 10.1007/s004270050190. [DOI] [PubMed] [Google Scholar]
- 16.Choe CP, Miller SC, Brown SJ. A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc Natl Acad Sci USA. 2006;103:6560–6564. doi: 10.1073/pnas.0510440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cruz J, Mane-Padros D, Belles X, Martin D. Functions of the ecdysone receptor isoform-A in the hemimetabolous insect Blattella germanica revealed by systemic RNAi in vivo. Dev Biol. 2006;297:158–171. doi: 10.1016/j.ydbio.2006.06.048. [DOI] [PubMed] [Google Scholar]
- 18.Panganiban G, Sebring A, Nagy L, Carroll S. The development of crustacean limbs and the evolution of arthropods. Science. 1995;270:1363–1366. doi: 10.1126/science.270.5240.1363. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Arias A. In: The Development of Drosophila melanogaster. Bate CM, Martinez-Arias A, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1993. pp. 517–608. [Google Scholar]
- 20.Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3:688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel NH, Condron BG, Zinn K. Pair-rule expression patterns of even-skipped are found in both short- and long-germ beetles. Nature. 1994;367:429–434. doi: 10.1038/367429a0. [DOI] [PubMed] [Google Scholar]
- 22.Peel A, Akam M. Evolution of segmentation: Rolling back the clock. Curr Biol. 2003;13:708–710. doi: 10.1016/j.cub.2003.08.045. [DOI] [PubMed] [Google Scholar]
- 23.de Celis JF, Tyler DM, de Celis J, Bray SJ. Notch signalling mediates segmentation of the Drosophila leg. Development (Cambridge, U.K.) 1998;125:4617–4626. doi: 10.1242/dev.125.23.4617. [DOI] [PubMed] [Google Scholar]
- 24.Bishop SA, Klein T, Arias AM, Couso JP. Composite signalling from Serrate and Delta establishes leg segments in Drosophila through Notch. Development (Cambridge, U.K.) 1999;126:2993–3003. doi: 10.1242/dev.126.13.2993. [DOI] [PubMed] [Google Scholar]
- 25.Rauskolb C, Irvine KD. Notch-mediated segmentation and growth control of the Drosophila leg. Dev Biol. 1999;210:339–350. doi: 10.1006/dbio.1999.9273. [DOI] [PubMed] [Google Scholar]
- 26.Chipman AD, Arthur W, Akam M. A double seament periodicity underlies segment generation in centipede development. Curr Biol. 2004;14:1250–1255. doi: 10.1016/j.cub.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 27.Technau U, et al. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Gen. 2005;21:633–639. doi: 10.1016/j.tig.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 28.De Robertis EM. Evo-devo: variations on ancestral themes. Cell. 2008;132:185–195. doi: 10.1016/j.cell.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erwin DH, Davidson EH. The last common bilaterian ancestor. Development (Cambridge, U.K.) 2002;129:3021–3032. doi: 10.1242/dev.129.13.3021. [DOI] [PubMed] [Google Scholar]
- 30.Lowe CJ, et al. Dorsoventral patterning in hemichordates: Insights into early chordate evolution. PLOS Biol. 2006;4:1603–1619. doi: 10.1371/journal.pbio.0040291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thamm K, Seaver EC. Notch signaling during larval and juvenile development in the polychaete annelid Capitella sp. Dev Biol. 2008;320:304–318. doi: 10.1016/j.ydbio.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Jacobs DK, et al. Molluscan engrailed expression, serial organization, and shell evolution. Evol Dev. 2000;2:340–347. doi: 10.1046/j.1525-142x.2000.00077.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Denburg JL. A role for proteoglycans in the guidance of a subset of pioneer axons in cultured embryos of the cockroach. Neuron. 1992;8:701–714. doi: 10.1016/0896-6273(92)90091-q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.