Abstract
Chemical cross-linking is an attractive approach to map peptide-protein and protein-protein complexes. Previously, we explored 3,4-dihydroxylphenylalanine (DOPA) as a protein cross-linking agent upon periodate oxidation.(Burdine, L.; Gillette, T. G.; Lin, H-J.; Kodadek, T. J. Am. Chem. Soc. 2004, 126, 11442–11443) We report here a study on the chemistry of DOPA-protein cross-linking. First, using a peptide nucleic acid (PNA) templated system, we identified the α-amino, ε-amino of Lys, imidazole of His, and thiol of Cys as functional groups capable of attacking DOPA ortho-quinone. Second, we demonstrated that periodate-induced DOPA-protein cross-linking could be carried out efficiently at neutral pH in the presence of excess aliphatic 1,2-diols such as ethylene glycol, lactose, and ATP. This result indicated that DOPA-protein cross-linking and 1,2-diol oxidative cleavage proceed via different mechanisms and, that carbohydrates will not interfere with this process when carried out in crude cell extracts or on intact cells.
Protein-protein interactions are of fundamental importance in almost all biological processes. In some cases, these are strong, stable contacts while in others they represent much weaker, transient interactions. In principle, chemical cross-linking followed by detailed product analysis is an attractive approach to map protein-protein contacts under native conditions. However, this is complicated by the low reactivity of typical bifunctional cross-linking reagents and their tendency to produce false positives (reviewed in references1, 2). As part of a program to develop a new generation of more efficient and useful cross-linking reagents, we have shown recently that molecules containing 3,4-dihydroxylphenylalanine (DOPA) often cross-link efficiently to their protein receptors when treated with sodium periodate.3 Under these conditions, DOPA is converted to an ortho-quinone intermediate that can be attacked by nearby nucleophiles, resulting in a stable cross-link.4 Using as a model system a high affinity complex comprised of the yeast Gal80 protein and a DOPA-containing peptide mimicking the Gal4 activation domain,5–7 we showed that this chemistry provided a high yield of cross-linked product when the purified components were mixed together in standard biochemical buffers or in crude cell extracts. The reaction was relatively rapid, reaching completion in less than one minute. Finally, control experiments demonstrated that no coupling occurred unless the DOPA-containing molecule was specifically bound to the protein receptor, suggesting that false positives will be rare using this chemistry.
As DOPA appears to be a promising cross-linking agent with which to study peptide-protein and protein-protein interactions, it is important to understand the mechanism of cross-linking in more detail. While it seems clear that the critical intermediate is an ortho-quinone, it is less clear what amino acid residues on the protein partner are capable of coupling to this species. This information would be of interest not only to better understand the periodate-mediated cross-linking process, but is also relevant to the behavior of native DOPA, an important neurotransmitter. A number of syndromes, such as Parkinson’s disease, may involve cross-linking of oxidized DOPA to receptors.8 Biochemical studies of this reaction indicated that cysteine is most likely the amino acid responsible for DOPA-protein coupling,9, 10 but it is unclear what other amino acids might participate in this process.
In this study, we report the results of experiments designed to test the reactivity of various amino acids with the DOPA-derived ortho-quinone. As mentioned above, our preliminary experiments indicated that this reaction occurs efficiently only when this intermediate and a suitable amino acid are brought into close proximity by a peptide-protein or protein-protein interaction. Thus, we decided to investigate this chemistry in the context of a templated system11–15. Specifically, we used short complementary peptide nucleic acid (PNA)16 strands to bring together tethered amino acid and DOPA residues through annealing, prior to triggering DOPA oxidation with sodium periodate. Cross-linked products were detected by high performance liquid chromatography (HPLC) and matrix assisted laser desorption / ionization – time of flight (MALDI-TOF) mass spectroscopy. Using this approach, we have identified several amino acids capable of coupling with the DOPA-derived ortho-quinone. We also studied the effect of excess aliphatic diols, including sugars, on DOPA-protein cross-linking chemistry. This is relevant to the utility of the reaction in complex mixtures containing glycoconjugates since periodate-mediated cleavage of sugars is a well-known reaction. Surprisingly however, we show that cross-linking reactions involving DOPA are unaffected by aliphatic diols.
Results
Experimental Design
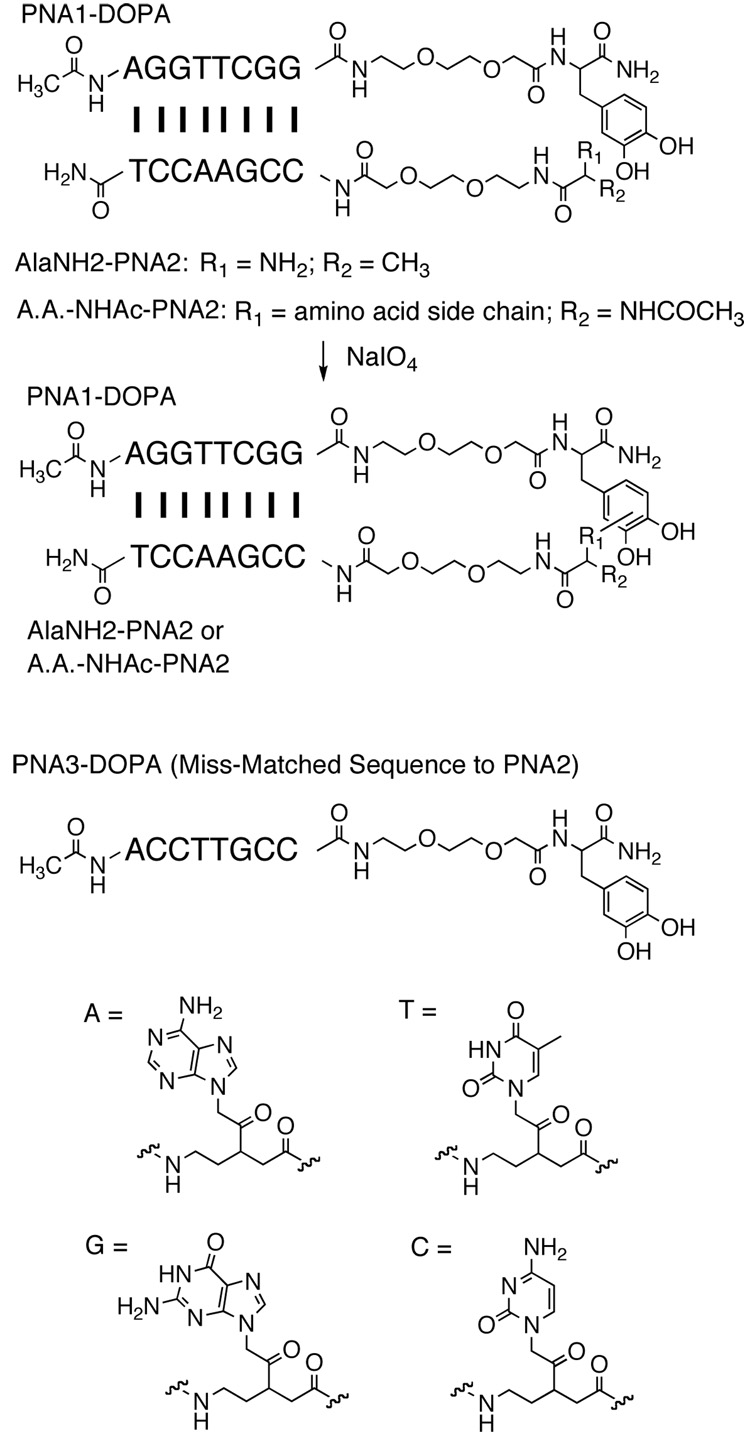
To provide a general templated system in which to explore the reactivity of oxidized DOPA with various amino acids, DOPA was conjugated to the C-terminus of an octameric PNA via an acetyl-ethyleneglycol-ethylamine (AEEA) linker and an amino acid was conjugated to the N-terminus of a complementary octameric PNA strand via the same linker (Figure 1). 1.7 µM amino acid-PNA2 and 1.0 µM PNA1- DOPA conjugates were mixed together, heated at 99 °C for 1 min and cooled slowly to room temperature to allow the PNA strands to anneal. 1 mM sodium periodate was then added to trigger the cross-linking reaction, which was allowed to proceed for 90 seconds at room temperature. The mixture was injected onto a C-18 reverse phase analytical column for analysis by HPLC. Under the chromatographic conditions employed, sodium periodate eluted from the column almost immediately, effectively quenching the reaction. As shown below, the PNA strands separated under these conditions unless they had been covalently cross-linked by a reaction between the amino acid and the ortho-quinone. Thus, the appearance of new peaks in the chromatogram was taken as evidence of products and these were characterized by mass spectrometry.
Figure 1.
Chemical structures of PNA-amino acid and PNA-DOPA conjugates employed in this study.
Coupling Of The Quinone Intermediate With Nitrogen Nucleophiles
We first examined potential coupling between oxidized DOPA and the α-amino group. Alanine was conjugated to PNA2 to form AlaNH2-PNA2 (Figure 1). In the absence of sodium periodate treatment, the annealed complex AlaNH2-PNA2 / DOPA-PNA1 yielded two well-separated peaks on a hydrophobic HPLC column (peak 1 and 2, Figure 2A). MALDI-TOF analysis of each confirmed that these corresponded to the individual strands (Figure 2B), showing that the PNA-PNA duplex was unstable and that the strands separated under the chromatographic conditions employed. When 1 mM sodium periodate was added to the complex, a new peak (peak 3, Figure 2A) emerged at longer retention time (~ 26 min), accompanied by reduced intensities of peaks 1 and 2. Peak 3 showed a strong signal in the mass spectrum at of m/e = 4952.72 ± 2.11. This value matches the sum of the measured mass values of AlaNH2-PNA2 (peak 1) and PNA1-DOPA (peak 2), arguing that it represents the cross-linked product. This product is presumably the result of α-amino attack on the ortho-quinone intermediate by Michael addition as illustrated in Figure 2C. Judging from the HPLC trace (Fig. 2A), the reaction proceeded in > 80% yield, as judged by the relative peak heights of peaks 1 (present in 1.7-fold molar excess) and 3.
Figure 2.
α-Amino group cross-linking to DOPA ortho-quinone. A. HPLC spectra of annealed AlaNH2-PNA2 / PNA1-DOPA complex in phosphate buffered saline containing 0.5 mM DTT before and after addition of 1 mM sodium periodate. B. MALDI-TOF mass spectra of the peaks indicated in the HPLC spectra. C. Proposed cross-linking mechanism. D. Same as A, but with AlaNH2-PNA2 / PNA3-DOPA as reactants. E. Same as A, but with Ala-NHAc-PNA2 / PNA1-DOPA as reactants.
To determine if PNA-PNA hybridization was required for the formation of this product, we repeated the experiment with mismatched PNAs 2 and 3 (see Fig. 1) conjugated to alanine and DOPA, respectively. In this case, when sodium periodate was added, no cross-linked product was detected (Figure 2D). This experiment demonstrated that α-amino-DOPA cross-linking requires the close proximity of the ortho-quinone intermediate and the amine nucleophile provided by PNA-PNA interaction. Note that without sodium periodate oxidation, the AlaNH2-PNA2 / PNA3-DOPA mixture has an additional peak on HPLC (peak3, Figure 2D). This peak was assigned as the DTT adduct of PNA3-DOPA base on the findings that it has a mass value 152 more than that of PNA3-DOPA and it was absent when DTT was eliminated from the solution (data not shown). Such adducts were observed periodically in the course of this study and are assumed to be the result of air oxidation of some of the DOPA-containing molecules followed by capture of the ortho-quinone by a thiol group in DTT via a Michael reaction.
To determine if the nucleophilic α-amino group is essential for cross-linking, as anticipated, we carried out a nearly identical experiment using the matched PNAs, except that the α-amino group attached to strand 2 was acetylated (Figure 2E). In this case, addition of periodate to the duplex PNA conjugates did not produce a new peak, suggesting that clean covalent coupling of the two strands did not occur. However, we note that the peak corresponding to the DOPA-conjugated PNA strand was diminished substantially, suggesting that it underwent other reactions, probably several, that did not produce a single, characterizable product. This result suggests that clean coupling requires a relatively potent nucleophile.
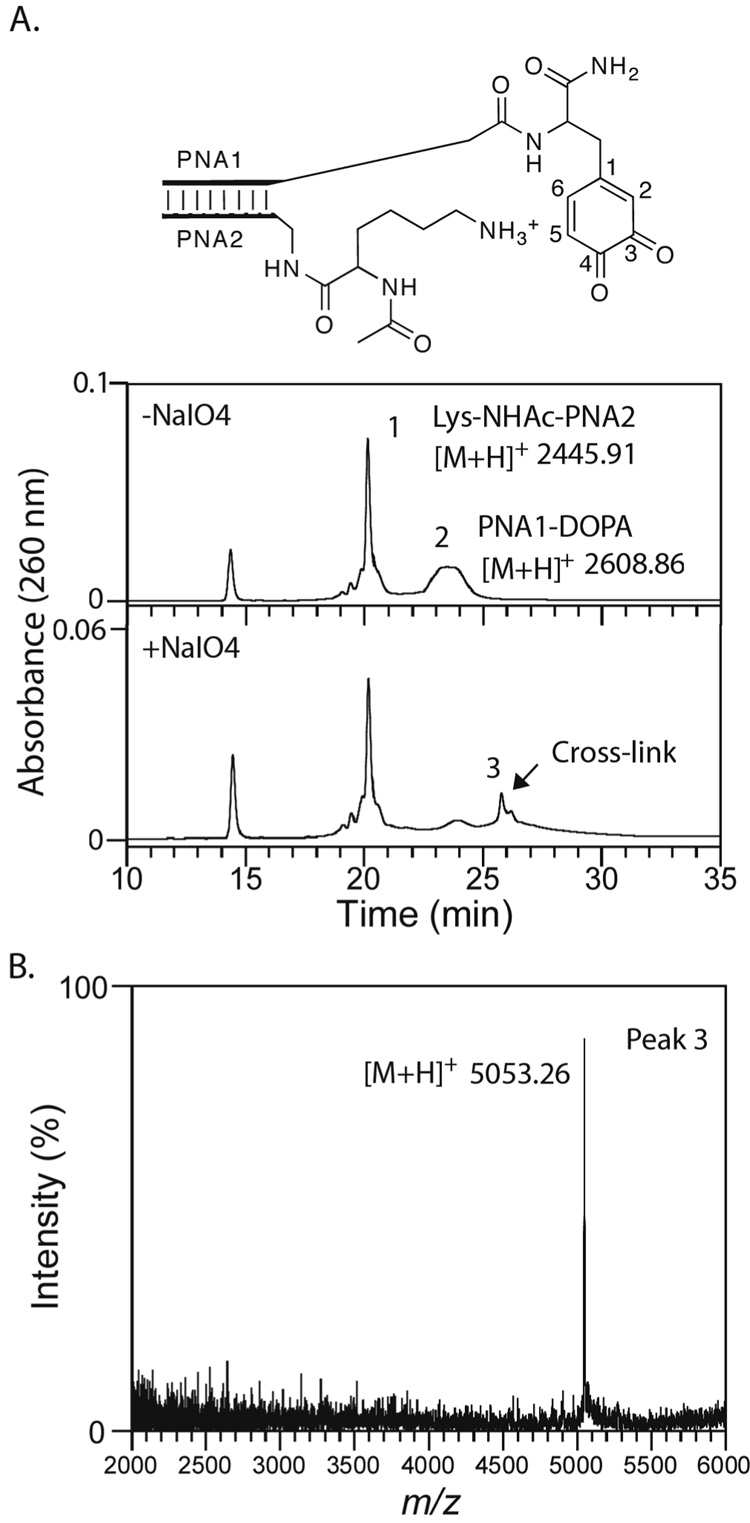
With the analytical system established, we turned to study the reactivity of other nucleophilic potential partners for the oxidized DOPA residue, specifically lysine and histidine. These amino acids, with the α-amino group blocked by acetylation, were conjugated to PNA2 and hybridized to PNA1-DOPA. Upon sodium periodate treatment, the Lys-NHAc-PNA2 / PNA1-DOPA complex yielded a distinct cross-link peak on HPLC (peak 3, Figure 3A). Analysis of this fraction by mass spectrometry revealed a dominant peak at m/e = 5053.26 ± 1.82 (Figure 3B). This mass value was consistent with a Michael addition mechanism, differing by only 1.5 mass units from that predicted product, a value within the experimental error of the measurement (0.05%). The cross-linking yield was lower than that observed for coupling of the α-amino group of alanine, possibly due to the higher pKa value of the ε-amino group.
Figure 3.
Lysine cross-linking to DOPA ortho-quinone. A. HPLC spectra of annealed Lys-NHAc-PNA2 / PNA1-DOPA complex in phosphate buffered saline containing 0.5 mM DTT before and after addition of 1 mM sodium periodate. B. MALDI-TOF mass spectrum of the cross-link peak indicated in the HPLC spectra.
Fig. 4A shows the result when the same experiment was carried out with the His-NHAc-PNA2 conjugate. Again, a new peak in the HPLC was observed whose mass was consistent with the formation of a cross-linked adduct (Figure 4B) (1.3 mass units difference from the predicted Michael adduct, which was within the experimental error).
Figure 4.
Histidine cross-linking to DOPA ortho-quinone. A. HPLC spectra of annealed His-NHAc-PNA2 / PNA1-DOPA complex in phosphate buffered saline containing 0.5 mM DTT before and after addition of 1 mM sodium periodate. B. MALDI-TOF mass spectrum of the cross-link peak indicated in the HPLC spectra.
Cysteine Side Chain Cross-linking To DOPA Ortho-Quinone
Cysteine would be expected to be perhaps the most reactive partner for nucleophilic coupling with the ortho-quinone intermediate.10 Cys-NHAc-PNA2 and PNA1-DOPA were annealed in a phosphate buffered saline solution containing 0.5 mM DTT. Even without addition of sodium periodate, a distinct product was detectable by HPLC (peak 3, Figure 5A). The major mass peak exhibited m/e = 5027.41 ± 2.03, which was 1.0 ± 2.0 less than the total found mass values of Cys-NHAc-PNA2 and PNA1-DOPA (1 mass unit difference from the predicted cross-linked product). This argues that a coupling reaction via Michael addition had occurred. In the absence of periodate, only small amounts of the ortho-quinone exist, formed presumably by air oxidation. Apparently, cysteine was able to capture these intermediates very efficiently. This periodate-independent coupling nonetheless required templating. When Cys-NHAc-PNA2 was mixed with PNA3-DOPA, a conjugate with a mismatched PNA sequence (Figure 1) in 0.5 mM DTT, no cross-linked product was detected (Figure 5C). Upon sodium periodate induction, the Cys-NHAc-PNA2 / PNA1-DOPA gave a broad peak with a retention time about that anticipated for a cross-linked product based on the experiments discussed above (Figure 5A). Unfortunately, we were unable to cleanly identify the identities of the cross-linked products. A possible reason for this complication was that under oxidative condition, the Cys-DOPA cross-linked product reacted further through subsequent oxidations and additions.
Figure 5.
Cysteine cross-linking to DOPA ortho-quinone. A. HPLC spectra of annealed Cys-NHAc-PNA2 / PNA1-DOPA complex in phosphate buffered saline containing 0.5 mM DTT before and after addition of 1 mM sodium periodate. B. MALDI-TOF mass spectrum of the cross-link peak indicated in the HPLC spectra. C. HPLC spectrum of Cys-NHAc-PNA2 / PNA3-DOPA mixture in phosphate buffered saline containing 0.5 mM DTT.
Scanning Other Amino Acids For DOPA-reactive Groups
To evaluate the reactivity of other amino acid side chains with oxidized DOPA, we made a number of amino acid-PNA2 conjugates, including Arg-NHAc-PNA2, Glu-NHAc-PNA2, Met-NHAc-PNA2, Ser-NHAc-PNA2, Thr-NHAc-PNA2, Trp-NHAc-PNA2, Tyr-NHAc-PNA2, Phe-NHAc-PNA2, and Gln-NHAc-PNA2. In these experiments, DTT was omitted from the buffer in hopes of allowing less reactive functional groups to participate in the coupling reaction without competition with this dithiol. In order to have a comprehensive data set, we also repeated the experiments described above using AlaNH2-PNA2, Ala-NHAc-PNA2, Lys-NHAc-PNA2, His-NHAc-PNA2, and Cys-NHAc-PNA2 in the absence of DTT. The results are summarized in Figure S1 (see supplementary materials). Among all the amino acids studied, only α-amino (Figure S1A), ε-amino (Lys, Figure S1F), imidazole (His, Figure S1E), and thiol (Cys, Figure S1L) groups were able to attack the DOPA ortho-quinone in such a way that an identifiable product was produced (note that cross-link peaks have retention times of ~ 26 min in all of the chromatograms under the conditions employed). Interestingly, all four groups were able to form at least low levels of cross-linked products even without sodium periodate, again presumably due to air oxidation of the DOPA moiety. Cysteine was, again, exceptionally reactive. Despite the fact that a considerable amount of the starting Cys-NHAc-PNA2 formed a dimer through disulfide bond (Figure S1L), the cross-linking yield was significant (peak 3, Figure S1L). In all four cases, the cross-linking yields were significantly increased with sodium periodate induction.
Our experimental conditions also allowed us to evaluate the modification of amino acids under sodium periodate oxidation. Most amino acids were inert to sodium periodate except cysteine and methionine. As shown in Figure S1L, Cys-NHAc-DOPA (peak 1) was modified to form peak 1’. Peak 1’ has a mass value 48 more than of Cys-NHAc-DOPA, indicating that the thiol group (-SH) was converted to sulfonic acid (-SO3H) by periodate oxidation to form CysO3H-NHAc-PNA2. In the case of methionine (Figure S1G), the peak of Met-NHAc-DOPA was shifted to a shorter retention time on the HPLC when sodium periodate was added. This peak (peak 1’, Figure S1G) gave a mass value 16 more than that of Met-NHAc-DOPA, suggesting that the sulfide group (-S-CH3) was converted to the sulfoxide group (-SO-CH3).
Aliphatic 1,2-Diols Do Not Interfere With Cross-linking
Fig. 6A shows the presumed mechanism by which sodium periodate reacts with DOPA to form the active ortho-quinone intermediate. This raises the important practical question of whether aliphatic 1,2-diols will act as inhibitors of periodate-mediated cross-linking of DOPA-containing molecules, which are well known to react with periodate to form carbonyl compounds that result from oxidative cleavage of the C-C bond joining the alcohols (Fig. 6B). Cells are rich in sugars that could compete for limiting periodate when this reaction is employed in the presence of intact cells or in cell extracts.
Figure 6.
Proposed mechanisms of periodate-induced DOPA ortho-quinone formation (A) and periodate-induced 1,2-diol oxidative cleavage (B).
To probe this point, we examined the periodate-mediated cross-linking of the Gal80 protein (Gal80p) and the Gal80-binding peptide (Gal80BP) in solutions containing a large excess of ethylene glycol, a standard substrate for periodate-induced oxidative 1,2-diol cleavage. Gal80BP (NH2-YDQDMQNNTFDDLFWKEGHR-COOH) was selected by phage display17 as a ligand for Gal80p (KD ≈300 nM).7 We have shown previously that a DOPA-substituted Gal80BP-Gal80p complex cross-linked efficiently with periodate induction.3
A six-histidine (His6)-tagged Gal80p was mixed with the Gal80-DOPA-BP with a biotin labeled at the N-terminus of the peptide (both present at 1 µM) in neutral phosphate buffered saline containing 0.5 mM DTT and ethylene glycol. The mixtures were treated with 1 mM sodium periodate for 30 seconds and the reaction was then quenched with a protein loading buffer containing 100 mM DTT. The cross-linked products were probed with a NeutrAvidin-horse radish peroxidase (HRP) conjugate. As shown in Figure 7A, at up to 100 mM ethylene glycol (100-fold higher than the sodium periodate concentration), there was no detectable reduction of the yield of Gal80p-Gal80BP cross-linked product (lane 3–8, Figure 7A).
Figure 7.
Chemical cross-linking between His6-Gal80p and biotinylated Gal80-DOPA-BP in buffers containing 1,2-diols and at different pH. A. Cross-linking reactions in the presence of ethylene glycol. Lane 1–2: cross-linked products obtained in the absence of ethylene glycol. Lane 3–8: cross-linked products in the presence of ethylene glycol at the concentrations of 100 mM, 50 mM, 10 mM, 1 mM, 0.1 mM, and 0.001 mM. B. Same as A, but in the presence of lactose. Lane 1–2: cross-linked products obtained in the absence of lactose. Lane 3–8: cross-linked products in the presence of lactose at concentrations of 100 mM, 50 mM, 10 mM, 1 mM, 0.1 mM, and 0.001 mM. C. Same as A, but in the presence of ATP. Lane 1: cross-linked product obtained in the absence of ATP. Lane 2–5: cross-linked products in the presence of ATP at concentrations of 0.1 mM, 0.5 mM, 1 mM, and 10 mM. D. Cross-linking reactions at different pH. Lane 1–2: cross-linked products obtained at pH 7.0. Lane 3–13: cross-linked products at pH 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, and 9.0. Cross-linked products were probed with a NeutrAvidin-horse radish peroxidase (HRP) conjugate. Probing the blot with anti-His6 antibody showed that equivalent amounts of protein were loaded.
We then examined the same reaction in the presence of lactose and ATP, two common 1,2-diols in cells. ATP contains a cis- axial 1,2-diol, presumably facilitating the C-C cleavage upon periodate treatment.18 The Gal80p-Gal80BP cross-link was not reduced by either a large excess of lactose (up to 100-fold, land 3–8, Figure 7B) or ATP (up to 10-fold, lane 2–5, Figure 7C).
On the surface, the insensitivity of the cross-linking process to such a large excess of aliphatic 1,2-diols was surprising. However, one potential explanation is that the oxidative cleavage of these species is optimally carried out within the pH range 3–5,19 while the peptide-protein cross-linking reaction is carried out at neutral pH. To probe this issue, the His6-Gal80p - Gal80-DOPA-BP cross-linking experiments were repeated at different pHs. As shown in Fig. 7D, cross-linking occurred efficiently at neutral or slightly alkaline pH (lane 8–13, Figure 7C). Very little cross-linked product was detected below pH 6.5 (lane 3–7, Figure 7C). The very different pH optima of the cross-linking process and aliphatic 1,2-diol cleavage lead us to propose that they must proceed by significantly different mechanisms. A model to explain these observations is presented below.
Discussion
We demonstrated previously that periodate-triggered oxidation of DOPA-containing molecules results in covalent cross-linking to their protein receptors.3 This technique is useful for the analysis of protein-protein and protein-ligand interactions in large complexes. For example, we applied this methodology recently to identify the direct binding partners of the Gal4 transactivation domain in the proteasome, a >2 MDa complex.20 Treatment of a DOPA-containing Gal4 activation domain derivative and immuno-purified yeast proteasome with sodium periodate resulted in the production of only two major cross-linked products that were shown to correspond to coupling of the Gal4 domain to the Sug1/Rpt6 and Sug2/Rpt4 proteins, thus confirming a model for Gal4-proteasome interaction that was originally proposed on the basis of genetic evidence and “pull-down” experiments utilizing in vitro translated proteins studied outside of the context of the proteasome.21, 22
These and other results obtained using this chemistry have suggested that it will be of considerable utility in probing interactions between biomolecules in complex environments. Therefore, we have been interested in better understanding its mechanism. While it seems clear that the active intermediate is an ortho-quinone produced by periodate-mediated oxidation of DOPA, other aspects of the reaction were less well understood. In particular, we were interested in two issues: 1) better understanding what nucleophilic side chains in a protein are capable of linking covalently to the ortho-quinone intermediate, and 2) asking if other 1,2-diols that are abundant in cells would interfere with this reaction.
The first point is highly relevant to the detailed characterization of cross-linked products, for example at the level of tryptic peptides in the mass spectrometer.2 Unfortunately, branched structures such as cross-linked peptides do not ionize efficiently in mass spectrometers (unpublished observation) and the peaks that represent these products are difficult to identify in complex mass spectra even for high-yield reactions. However, much higher sensitivity can be achieved by carrying out a focused search for candidate ions in the mass spectrometer based on a detailed knowledge of the reaction.23, 24 Of course, this requires one to know the precise masses of candidate products, thereby necessitating a knowledge of the reactive amino acids and how they couple to the active intermediate.
We originally attempted to address this problem by simply mixing amino acids with DOPA in the presence of periodate, but even at high concentrations of reactants, these experiments provided poor results. Indeed, in all of the biochemical experiments that we have done, it has become apparent that efficient coupling of protein side chains to the oxidized DOPA will occur only if the two reactants are held in close proximity. We thus developed the PNA templated system reported here to force the two potential reactants into close proximity. These experiments demonstrated that the α-amino group, the amino group in the side chain of lysine, and the imadazole side chain of histidine are all sufficiently nucleophilic to capture the oxidized DOPA moiety. The thiol of cysteine, as expected, was extraordinarily reactive and coupled to the ortho-quinone with the greatest efficiency. However, detailed structures of the cross-linked products remained to be determined.
Somewhat curious was the failure to observe the cross-link when tyrosine was conjugated to one of the PNAs. Tyrosine was speculated to interact with DOPA through radical coupling,25 similar to DOPA-DOPA dimer formation.26 In our assay, no distinct Tyr-DOPA cross-link was detected.
Other useful information that was obtained during this study is that most amino acids were found to be inert to sodium periodate except cysteine and methionine. Upon sodium periodate treatment, the thiol group on Cys was oxidized to the sulfonic acid group (-SO3H) and the sulfide group (-S-CH3) on methionine was modified to the sulfoxide group (-SO-CH3). With DOPA-protein cross-linking chemistry cleanly defined, identification of unknown cross-linked proteins through mass data analysis should be facilitated.
Our results also provide valuable information for studies on DOPA as a neurotransmitter and a toxic molecule contributing to neurodegenerative disorders. Cysteine-DOPA coupling is well documented as a possible mechanism for protein modification and malfunction.8 Recently, a physiological study indicated that parkin, a cysteine-rich protein involved in Parkinson’s disease was most likely modified by DOPA under oxidative stress.27 On the other hand, it has been found that DOPA ortho-quinone cross-linked to the neuronal protein α-synuclein to stabilize the α-synuclein protofibril state, which was potentially pathogenic.25, 28, 29 It is unclear how DOPA reacts with α-Synuclein since this protein does not contain cysteine. Our results revealed the high sensitivity of cysteine-DOPA reaction. While studies on other amino acid-DOPA reactions related to neurological disorders are rare,30 our results show that α-amino, Lys, and His are also capable to attack DOPA ortho-quinone with substantial yields. Lys-DOPA, His-DOPA, and Cys-DOPA cross-links have been identified by proteolytic and chemical digestions of natural proteins such as lysyl oxidase cofactor31, insect cuticle proteins32, and mussel adhesive proteins33. Our results provide direct chemical evidence to support these findings and suggest that amino acid-DOPA proximity is important for cross-linking reactions.
The second issue that we examined was the ability of 1,2-diols, such as sugars to interfere with the periodate-mediated cross-linking reaction. This is important with respect to understanding whether this process is likely to be useful for the analysis of macromolecular interactions on cell surfaces or in other venues that are rich in sugars. We find that in vitro, even a very large excess of ethylene glycol, lactose or ATP had essentially no effect on the yield of cross-linking of a DOPA-containing Gal80-binding peptide and Gal80p. Clearly, there must be a significant mechanistic difference between the mechanisms of ortho-quinone formation and C-C oxidative cleavage, despite the fact that both are thought to proceed via an intermediate periodoester (Fig. 6). We speculate that there may be two reasons. First, catechols (pKa ≈ 9.2 –.9.734, 35) are far more acidic than aliphatic alcohols including 1,2-diols (pKa > 1436). The relatively high nucleophilicity of a catechol perhaps allows it to form the periodoester more rapidly than aliphatic 1,2-diols. Second, the rate of aliphatic 1,2-diol C-C bond cleavage reaches a maximum at pH 3–5, and decreases greatly at neutral and higher pH.18 The lower efficiency of 1,2-diol C-C bonding cleavage at neutral and slightly alkaline pH may allow high yield of DOPA-protein cross-linking within this pH range. In any case, the empirical result is that sugars clearly do not interfere with this cross-linking process, suggesting that it will be useful for the analysis of glycosylated proteins, between proteins that reside on cell surfaces and other applications where sugars will be present in large excess over the DOPA residue.
Experimental Section
Synthesis
Peptide nucleic acid (PNA) monomers Fmoc-T-OH, Fmoc-C(Bhoc)-OH, Fmoc-G(Bhoc)-OH, Fmoc-A(Bhoc)-OH, carboxyl activator O-(7-azabenzo-triazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU), and 1-hydroxy-7-azabenzotriazole (HoAt) were purchased from Applied Biosystems. Diisopropylethylamine (DIPEA) and 2,6-lutidine were from Sigma-Aldrich. Fmoc-XAL-PEG-PS resin was from Applied Biosystems. Fmoc-8-amino-3,6-dioxaoctanoic acid (Fmoc-acetyl-ethyleneglycol-ethylamine, Fmoc-AEEA-OH) was from Peptides Inernational. Fmoc-DOPA(acetonid)-OH was from NOVAbiochem. All Fmoc amino acid monomers were from BACHEM and Advanced Chemtech.
PNA1-DOPA, AA-PNA2, and PNA3-DOPA were synthesized manually in a 25 mL reaction vessel (ChemGlass) at 5 µmol scale using standard fluorenylmethoxy-carbonyl (Fmoc) chemistry. Fmoc-XAL-PEG-PS resin was used as the solid phase. Fmoc deblocking was done by treating the resin in 20 % piperidine in dimethylformamide (DMF) (5 min × 2). Fmoc-DOPA(acetonid)-OH, Fmoc-AEEA-OH, and Fmoc-amino acid couplings were carried out in 2 mL of anhydrous dimethylformamide (DMF) solution containing 25 µmol of each monomer, 25 µmol of HATU, 25 µmol of HoAt, 6.4 µL of DIPEA, and 2.6 µL of 2,6-lutidine. For each PNA monomer coupling, 25 µmol of each monomer, 25 µmol of HATU, 25 µmol of HoAt, 6.4 µL of DIPEA, and 2.6 µL of 2,6-lutidine were mixed in 2 mL of anhydrous N-methyl-2-pyrolidinone (NMP) and applied to the resin. A stream of nitrogen passed through the reaction solution from the bottom of the reaction vessel to mix the resin with the reaction mixture. Each coupling was allowed at room temperature for 30 min. When a PNA synthesis was finished, the resin was treated with 1 mL of 20 % acetic anhydride in DMF containing 6.4 µL of DIPEA to acetylize the N-terminus. The resin was thoroughly washed with DMF, dichloromethane, and methanol, dried in vacuum, and cleaved in 400 µL of a cleavage mixture containing 80 % trifluoroacetic acid (TFA) and 20 % m-cresol. The resin was then filtered by an Ultrafree-MC centrifugal filter unit (Millipore). To the filtrate was added 2 mL of cold ethyl ether. The white precipitant was collected by centrifugation and purified by HPLC on a C18 semi-preparative column using a 1 % acetonitrile / min elution gradient. MALDI-TOF [M+H]+: PNA1-DOPA calculated 2608.04, found 2608.89; PNA3-DOPA calculated 2488.02, found 2489.09; Cys-NHAc-PNA2 calculated 2419.97, found 2420.92; Ala-NHAc-PNA2 calculated 2388.00, found 2388.94; Lys-NHAc-PNA2 calculated 2445.06, found 2445.82; His-NHAc-PNA2 calculated 2454.02, found2455.28; Tyr-NHAc-PNA2 calculated 2480.03, found 2481.21; Arg-NHAc-PNA2 calculated 2473.07, found 2474.29; AlaNH2-PNA2 calculated 2345.99, found 2346.95; Ser-NHAc-PNA2 calculated 2404.00, found 2404.85; Met-NHAc-PNA2 calculated 2448.00, found 2448.60; Trp-NHAc-PNA2 calculated 2503.04, found 2504.80; Thr-NHAc-PNA2 calculated 2418.01, found 2418.91; Glu-NHAc-PNA2 calculated 2446.01, found 2446.69; Phe-NHAc-PNA2 calculated 2464.03, found 2464.20; Gln-NHAc-PNA2 calculated 2445.02, found 2445.22.
Periodate Induced DOPA-Amino Acid Cross-linking Reactions
All PNA samples in water, or in 0.5 mM dithiothreitol (DTT) were prepared freshly before reactions and analyses. The concentrations of PNA1-DOPA and PNA3-DOPA stock solutions were controlled below 4 µM. In a 200 µL reaction volume was added 1 µM PNA1-DOPA (or PNA3-DOPA), 1.7 µM amino acid-PNA2, 10 mM sodium phosphate, 138 mM NaCl, 2.7 mM KCl, and 0.5 mM DTT. All reactions were carried out at pH 7.0 except that for His-NHAc-PNA2, which was carried out at pH 6.4. The solution was heated at 99 °C for 1 min and slowly cooled to room temperature in 1 h. 1 mM sodium periodate was added at room temperature for 90 s and the sample was injected in a Waters HPLC system on a C18 analytical column. The elution gradient was 1 % acetonitrile / min. Reactions were repeated in a similar buffer without DTT.
Periodate-Triggered Gal80p-Gal80BP Cross-linking In Ethylene Glycol, Lactose, ATP, And In Various pH
Biotinylated Gal80-DOPA-BP and His6-tagged Gal80p were prepared as described before.3 The peptide (1 µM) and the protein (1 µM) were incubated for 5 min at room temperature in reaction buffers containing 25 mM Tris, pH 7.4, 150 mM NaCl, 25 mM MgCl2,0.5 mM DTT, and ethylene glycol (lactose, or ATP) at various concentrations. For pH dependent cross-linking experiments, the reaction buffers contained 10 mM sodium phosphate, 137 mM NaCl, 2.7 mM KCl, and 0.5 mM DTT at various pH. Sodium periodate was added to a final concentration of 1mM. The reaction was quenched 30 s later by the addition of 6x protein-loading buffer containing 100 mM DTT. The final sample volume was 30 µL. The samples were then run on an SDS-PAGE gradient gel (4 – 20%). Peptide-protein cross-links were detected by western blotting as described before.3
Supplementary Material
HPLC spectra of cross-linking experiments on 14 amino acids in phosphate buffered saline. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgment
This work was supported by grants from the National Institute of Health (GM58175) and the Welch Foundation (I-1299).
References
- 1.Edwards AM, Kus B, Jansen R, Greenbaum D, Greenblatt J, Gerstein M. Trends Genet. 2002;18:529–536. doi: 10.1016/s0168-9525(02)02763-4. [DOI] [PubMed] [Google Scholar]
- 2.Denison C, Kodadek T. J. Proteome Res. 2004;3:417–425. doi: 10.1021/pr034071j. [DOI] [PubMed] [Google Scholar]
- 3.Burdine L, Gillette TG, Lin HJ, Kodadek T. J. Am. Chem. Soc. 2004;126:11442–11443. doi: 10.1021/ja045982c. [DOI] [PubMed] [Google Scholar]
- 4.Burzio LA, Waite JH. Biochemistry. 2000;39:11147–11153. doi: 10.1021/bi0002434. [DOI] [PubMed] [Google Scholar]
- 5.Johnston SA, Salmeron JM, Dincher SS. Cell. 1987;50:143–146. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- 6.Ma J, Ptashne M. Cell. 1987;50:137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- 7.Han Y, Kodadek T. J. Biol. Chem. 2000;275:14979–14984. doi: 10.1074/jbc.275.20.14979. [DOI] [PubMed] [Google Scholar]
- 8.Stokes AH, Hastings TG, Vrana KE. J. Neurosci. Res. 1999;55:659–665. doi: 10.1002/(SICI)1097-4547(19990315)55:6<659::AID-JNR1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 9.Rosengren E, Lindereliasson E, Carlsson A. J. Neural Transm. 1985;63:247–253. doi: 10.1007/BF01252029. [DOI] [PubMed] [Google Scholar]
- 10.Kato T, Ito S, Fujita K. Biochim. Biophys. Acta. 1986;881:415–421. doi: 10.1016/0304-4165(86)90034-6. [DOI] [PubMed] [Google Scholar]
- 11.Orgel LE. Nature. 1992;358:203–209. doi: 10.1038/358203a0. [DOI] [PubMed] [Google Scholar]
- 12.Bohler C, Nielsen PE, Orgel LE. Nature. 1995;376:578–581. doi: 10.1038/376578a0. [DOI] [PubMed] [Google Scholar]
- 13.Gartner ZJ, Liu DR. J. Am. Chem. Soc. 2001;123:6961–6963. doi: 10.1021/ja015873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li XY, Liu DR. Angew. Chem.-Int. Edit. 2004;43:4848–4870. doi: 10.1002/anie.200400656. [DOI] [PubMed] [Google Scholar]
- 15.Zhou QB, Rokita SE. Proc. Natl. Acad. Sci. USA. 2003;100:15452–15457. doi: 10.1073/pnas.2533112100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen PE, Egholm M, Berg RH, Buchardt O. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 17.Smith GP, Petrenko VA. Chem. Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 18.Bunton CA. Oxidation in organic chemistry, Part A. New York: Academic Press; 1965. pp. 189–204. [Google Scholar]
- 19.Furniss BS, Hannaford AJ, Smith PWG, Tatchell AR. Vogel's textbook of practical organic chemistry. 5 ed. London: Longman; 1989. pp. 454–455. [Google Scholar]
- 20.Archer CT, Burdine L, Kodadek T. Mol. BioSys. 2005;5–6:366–372. doi: 10.1039/b510019d. [DOI] [PubMed] [Google Scholar]
- 21.Swaffield JC, Melcher K, Johnston SA. Nature. 1995;374:88–91. doi: 10.1038/374088a0. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez F, Delahodde A, Kodadek T, Johnston SA. Science. 2002;296:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- 23.Trester-Zedlitz M, Kamada K, Burley SK, Fenyo D, Chait BT, Muir TW. J. Am. Chem. Soc. 2003;125:2416–2425. doi: 10.1021/ja026917a. [DOI] [PubMed] [Google Scholar]
- 24.McLachlin DT, Chait BT. Curr. Opin. Chem. Biol. 2001;5:591–602. doi: 10.1016/s1367-5931(00)00250-7. [DOI] [PubMed] [Google Scholar]
- 25.Conway KA, Rochet JC, Bieganski RM, Lansbury PT. Science. 2001;294:1346–1349. doi: 10.1126/science.1063522. [DOI] [PubMed] [Google Scholar]
- 26.Burzio LA, Waite JH. Protein Sci. 2001;10:735–740. doi: 10.1110/ps.44201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, Selkoe DJ. Nat. Med. 2005;11:1214–1221. doi: 10.1038/nm1314. [DOI] [PubMed] [Google Scholar]
- 28.Goldberg MS, Lansbury PT. Nat. Cell Biol. 2000;2:E115–E119. doi: 10.1038/35017124. [DOI] [PubMed] [Google Scholar]
- 29.Rochet JC, Outeiro TF, Conway KA, Ding TT, Volles MJ, Lashuel HA, Bieganski RM, Lindquist SL, Lansbury PT. J. Mol. Neurosci. 2004;23:23–33. doi: 10.1385/jmn:23:1-2:023. [DOI] [PubMed] [Google Scholar]
- 30.Montine TJ, Farris DB, Graham DG. J. Neuropathol. Exp. Neurol. 1995;54:311–319. doi: 10.1097/00005072-199505000-00004. [DOI] [PubMed] [Google Scholar]
- 31.Wang SX, Mure M, Medzihradszky KF, Burlingame AL, Brown DE, Dooley DM, Smith AJ, Kagan HM, Klinman JP. Science. 1996;273:1078–1084. doi: 10.1126/science.273.5278.1078. [DOI] [PubMed] [Google Scholar]
- 32.Kramer KJ, Kanost MR, Hopkins TL, Jiang HB, Zhu YC, Xu RD, Kerwin JL, Turecek F. Tetrahedron. 2001;57:385–392. [Google Scholar]
- 33.Zhao H, Waite JH. Biochemistry. 2005;44:15915–15923. doi: 10.1021/bi051530g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saleem MMM, Wilson MT. Biochem. J. 1982;201:433–444. doi: 10.1042/bj2010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schweigert N, Hunziker RW, Escher BI, Eggen RIL. Environ. Toxicol. Chem. 2001;20:239–247. [PubMed] [Google Scholar]
- 36.Ballinger P, Long FA. J. Am. Chem. Soc. 1960;82:795–798. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HPLC spectra of cross-linking experiments on 14 amino acids in phosphate buffered saline. This material is available free of charge via the Internet at http://pubs.acs.org.