Abstract
Sterols are major components of the plasma membrane, but their functions in this membrane are not well understood. We isolated a mutant defective in the internalization step of endocytosis in a gene (ERG2) encoding a C-8 sterol isomerase that acts in the late part of the ergosterol biosynthetic pathway. In the absence of Erg2p, yeast cells accumulate sterols structurally different from ergosterol, which is the major sterol in wild-type yeast. To investigate the structural requirements of ergosterol for endocytosis in more detail, several erg mutants (erg2Δ, erg6Δ, and erg2Δerg6Δ) were made. Analysis of fluid phase and receptor-mediated endocytosis indicates that changes in the sterol composition lead to a defect in the internalization step. Vesicle formation and fusion along the secretory pathway were not strongly affected in the ergΔ mutants. The severity of the endocytic defect correlates with changes in sterol structure and with the abundance of specific sterols in the ergΔ mutants. Desaturation of the B ring of the sterol molecules is important for the internalization step. A single desaturation at C-8,9 was not sufficient to support internalization at 37°C whereas two double bonds, either at C-5,6 and C-7,8 or at C-5,6 and C-8,9, allowed internalization.
INTRODUCTION
Eukaryotic cells are able to internalize extracellular molecules and plasma membrane components via endocytosis (Mellman, 1996; Geli and Riezman, 1998). Endocytic functions include uptake of some nutrients and adaptation to environmental signals by down-regulation of signal receptor molecules present in the plasma membrane. In receptor-mediated endocytosis, a ligand binds specifically to its receptor present in the plasma membrane, leading to the internalization of the ligand–receptor complex into small vesicles (Mellman, 1996; Riezman et al., 1996). Over the past years, genetic approaches in the yeast Saccharomyces cerevisiae have greatly aided in the identification and characterization of proteins that function at different stages along the endocytic pathway (Riezman et al., 1996; Geli and Riezman, 1998; Wendland et al., 1998). Of particular interest was the discovery that actin plays a fundamental role in the internalization step in yeast. A requirement for actin has also been reported for endocytosis in animal cells (Parton et al., 1994; Deckert et al., 1996; Lamaze et al., 1997). Various studies revealed that several of the identified yeast proteins have homologous counterparts in animals, and, contrary to previous belief, yeast and animal cells appear to use endocytic machinery that shows at least some mechanistic similarities (Geli and Riezman, 1998). In comparison with some membrane trafficking events, however, the molecular mechanisms underlying endocytosis remain poorly understood.
Over the past years, it has become apparent that in addition to proteinaceous factors, lipids play an important role in endocytosis (Anderson, 1998; Kobayashi et al., 1998). Recent attention has been given to sterols, essential components of cellular membranes in eukaryotic cells. Sterols are mainly present in the plasma membrane (Lange, 1991; Zinser et al., 1993), and this localized concentration may reflect a specific function of sterols in this membrane. In animal cells, the major sterol is cholesterol (Lange, 1991). Based on the raft hypothesis, cholesterol is proposed to interact with sphingolipids to form lipid microdomains or so-called lipid rafts that serve as platforms for many cellular events such as membrane trafficking and signal transduction (Simons and Ikonen, 1997; Brown and London, 1998). For membrane trafficking, these lipid rafts may be involved in the lateral recruitment and subsequent internalization of specific proteins (Harder and Simons, 1997; Brown and London, 1998). Studies using drugs that sequester cholesterol or block sterol biosynthesis at an early step in the biosynthetic pathway support a role of cholesterol in endocytosis in animal cells. Depletion for cholesterol leads to a loss of invaginated caveolae and caveolae-like domains (Rothberg et al., 1990; Rothberg et al., 1992; Schnitzer et al., 1994; Hailstones et al., 1998) and to a flattening of clathrin-coated pits (Rodal et al., 1999; Subtil et al., 1999). Furthermore, it inhibits internalization of proteins, including the bacterial cholera toxin (Orlandi and Fishman, 1998), the transferrin-receptor (Rodal et al., 1999) and glycosylphosphatidylinositol-anchored proteins such as the folate receptor, alkaline phosphatase, and CD59 (Chang et al., 1992; Cerneus et al., 1993; Deckert et al., 1996).
In yeast, it is unknown whether sterols serve a similar function in endocytosis. The major sterol of yeast is ergosterol, which, like cholesterol, is mainly present in the plasma membrane (Zinser et al., 1993). Mammalian cells can acquire cholesterol either by endogenous biosynthesis or by internalization of extracellular sterols via receptor-mediated endocytosis or receptor-mediated transfer (Fielding and Fielding, 1997). In contrast, yeast relies only on endogenous ergosterol biosynthesis. They are unable to take up sterols from the extracellular medium under aerobic growth conditions (Trocha and Sprinson, 1976; Keesler et al., 1992). Most ERG genes of the ergosterol biosynthetic pathway are essential, and only five proteins functioning in the final steps of the pathway are encoded by nonessential ERG genes (Lees et al., 1995; Parks and Casey, 1995; Daum et al., 1998). Thus, yeast cells containing erg mutations in the late part of the biosynthetic pathway are viable but are unable to synthesize ergosterol. Each erg mutant accumulates, however, a distinct set of sterols that differ from ergosterol in specific structural features, thus leading to changes in the membrane composition (Lees et al., 1995). In yeast, two functions of ergosterol have been examined in more detail, the so-called “sparking function” and the bulk membrane function. For the sparking function, sterols in nanomolar concentrations are required for yeast cells to complete the cell cycle. Only sterols with specific structural features are sufficient to overcome this cell cycle arrest in the G1 to S transition (Rodriguez and Parks, 1983; Lorenz et al., 1989). In contrast, a number of sterols can fulfill the bulk membrane function in yeast (Nes et al., 1993). This function is important for modulating the fluidity and permeability of the plasma membrane. Changes in the sterol composition have been reported to increase or decrease the sensitivity of the yeast cell to certain drugs (Lees et al., 1995; Parks and Casey, 1995), to decrease the activity of plasma membrane proteins (Gaber et al., 1989; Welihinda et al., 1994), and to decrease cell–cell fusion during mating (Gaber et al., 1989; Tomeo et al., 1992). Overall, however, the physiological roles of sterols in yeast remain largely unknown.
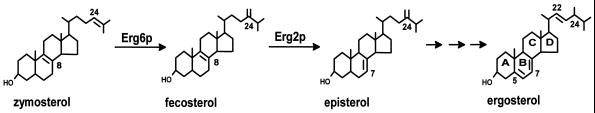
In the present studies, we show that ergosterol is required for the internalization step of endocytosis in yeast. Previously, the end11-1 mutant was isolated in a genetic screen for yeast mutants defective in endocytosis (Munn and Riezman, 1994). Analysis of fluid phase and receptor-mediated endocytosis demonstrated that end11-1 is defective in the first step of endocytosis, the internalization step (Munn and Riezman, 1994). It also exhibits a reduced growth rate at 24 and 37°C. We report here that END11 is allelic to ERG2, a gene that encodes the C-8 sterol isomerase that acts in the late part of the ergosterol biosynthetic pathway (Arthington et al., 1991). Erg2p converts fecosterol to episterol by isomerizing a C-8,9 double bond to a C-7,8 double bond in the B ring of the sterol molecule (Figure 1). Yeast strains containing mutations in the ERG2 gene lack the C-8 sterol isomerase activity and are not able to synthesize ergosterol (Arthington et al., 1991). They accumulate sterols different from ergosterol that lack the double bond at C-7,8. The identification of END11 as ERG2 indicates that sterols different from ergosterol may not be able to support endocytosis in yeast. To gain a better understanding of how the endocytic defect correlates with changes in the sterol composition, we analyzed endocytosis in erg mutants (erg2Δ, erg6Δ, and erg2Δerg6Δ) known to synthesize different sets of sterols. Erg6p is the C-24 sterol methyltransferase that acts immediately upstream of Erg2p in the ergosterol biosynthetic pathway (Gaber et al., 1989; Figure 1). In contrast to Erg2p, Erg6p modifies the side chain of the sterol molecule by methylating zymosterol at the C-24 position to produce fecosterol (Gaber et al., 1989; Figure 1). Thus, erg6Δ mutant strains have been reported to accumulate sterols lacking proper side chain modifications. The erg2Δerg6Δ double mutant strain lacks both C-8 sterol isomerase and C-24 sterol methyltransferase activities and has been reported to accumulate mainly zymosterol (Bard et al., 1977). When compared with ergosterol, zymosterol lacks both a proper B ring desaturation as well as a proper side chain modification (Figure 1). Analysis of these ergΔ mutants allowed us to correlate the endocytic defects with the sterol composition of each ergΔ mutant. Our work highlights the importance of specific structural features present in the ergosterol molecule for the internalization step of endocytosis in yeast.
Figure 1.
Late pathway of ergosterol biosynthesis in yeast with emphasis on the steps catalyzed by Erg6p (C-24 sterol methyltransferase) and Erg2p (C-8 sterol isomerase).
MATERIALS AND METHODS
Media and Strains
Yeast strains used in this study are listed with the relevant genotypes in Table 1. SD selective medium and YPUATD medium were prepared as described by Munn et al. (1995), except that 40 μg/ml tryptophan were added to YPUAD after autoclaving. Where specified, nystatin (Life Technologies, Paisley, United Kingdom) was added to SD complete medium at a final concentration of 33 U/ml. Plasmid propagation was carried out in Escherichia coli strain DH5α (Sambrook et al., 1989). Bacterial strains were grown in Luria–Bertani medium containing 100 μg/ml ampicillin where necessary to select for plasmids (Sambrook et al., 1989). All solid media for growth of yeast and bacteria contained 2% Bactoagar (Difco, Detroit, MI).
Table 1.
Strains and plasmids used in this study
| Strain/plasmid | Genotype/description | Source |
|---|---|---|
| Strains | ||
| RH448 | MATa his4 leu2 ura3 lys2 bar1 | Lab strain |
| RH1201 | MATα/MATa his4/his4 leu2/leu2 ura3/ura3 lys2/lys2 bar1/bar1 | Lab strain |
| RH1737 | MATa his4 leu2 ura3 bar1 sec18 | Lab strain |
| RH1800 (RH144-3D) | MATa his4 leu2 ura3 bar1 | Lab strain |
| RH1894 (RH296-10D) | MATα his4 ura3 erg6Δ | Lab strain |
| RH2622 (RH522-2C) | MATa his4 leu2 ura3 ade6? lys2? bar1 end11(erg2)-1 | Lab strain |
| RH2635 (RH144-3A) | MATα his4 leu2 ura3 bar1 | Lab strain |
| RH2180 (SF838-9D vps1-Δ2) | MATα his4 leu2 ura3 ade6 pep4 vps1-Δ2::LEU2 | T. Stevens |
| RH2894 [RH144-3A ERG2(END11)::URA3] | MATα his4 leu2 ura3 bar1 ERG2(END11)::URA3 | This study |
| RH2895 (RH2894 × RH2622) | MATα/MATa his4/his4 leu2/leu2 ura3/ura3 ADE6/ade6? LYS2/lys2? bar1/bar1 end11(erg2)-1/ERG2(END11)::URA3 | This study |
| RH2896 [RH1201 erg2(end11)-Δ1::URA3] | MATα/MATa his4/his4 leu2/leu2 ura3/ura3 lys2/lys2 bar1/bar1 erg2(end11-Δ1)::URA3/ERG2(END11) | This study |
| RH2897 | MATa his4 leu2 ura3 lys2 erg2(end11)-Δ1::URA3 bar1 | This study |
| RH2878 (RH407-1B) | MATa his3 leu2 ura3 ade2 lys2 bar1 | Lab strain |
| RH3610 (RH1894 × RH2897, 559) | MATα/MATa his4/his4 leu2/LEU2 ura3/ura3 lys2/LYS2 erg2(end11)-Δ1::URA3/ERG2(END11) erg6Δ/ERG6 bar1/bar1? | This study |
| RH3611 (RH559-8A) | MATα his4 ura3 erg6Δ erg2(end11)-Δ1::URA3 bar1? | This study |
| RH3612 (RH559-9B) | MATα his4 ura3 erg2(end11)-Δ1::URA3 erg6Δ bar1? | This study |
| RH3614 (RH3612 × RH2878, 562) | MATα/MATa his4/HIS4 leu2/LEU2 ura3/ura3 his3/HIS3 ade2/ADE2 lys2/LYS2 bar1/bar1? erg2(end11)-Δ1::URA3/ERG2(END11) erg6Δ/ERG6 | This study |
| RH3616 (RH562-8D) | MATa leu2 ura3 erg2(end11)-Δ1::URA3 erg6Δ bar1 | This study |
| RH3617 (RH562-1A) | MATa his (his3, his4, or his3 his4) ura3 erg2(end11)-Δ1::URA3 erg6Δ bar1 | This study |
| RH3622 | MATa his4 leu2 ura3 erg6Δ::LEU2 bar1 | This study |
| Plasmids | ||
| pBluescript KS− | ApR E. coli cloning vector | Stratagene |
| YCplac111 | ApR, LEU2-marked CEN/ARS E. coli/S. cerevisiae shuttle vector | Gietz and Sugino, 1988 |
| Ylplac211 | ApR, URA3-marked S. cerevisiae integration vector | Gietz and Sugino, 1988 |
| pEND11.2 | YCplac111 genomic library plasmid carrying ERG2(END11) on a 6.3-kb partial-Sau3A fragment | This study |
| pBKS-END11 | 2.9-kb BamHI fragment containing ERG2(END11) in pBKS | This study |
| Ylplac211-ERG2 | 2.9-kb BamHI fragment containing ERG2(END11) in Ylplac211 | This study |
| pend11-Δ1::URA3 | pBKS-END11 in which the BgIII–SphI fragment extending from nucleotides −110 to +495 relative to the start of ERG2(END11) was replaced by a 1.1-kb BamHI fragment containing URA3 | This study |
| pIU222 | YCp50 carrying a copy of ERG6 in which an internal 400bp XbaI–SaII fragment was replaced by the wild-type LEU2 gene | Gaber et al, 1989 |
Yeast Genetic Techniques and DNA Manipulations
Mating of haploid strains of yeast, sporulation of diploid strains, and tetrad dissection were performed generally as described in Sherman et al. (1974). A Leitz (Wetzlar, Germany) micromanipulator attached to a microscope with fixed stage (Wild, Heerbrugg, Switzerland) was used for tetrad dissection. The plasmids used in this study are shown in Table 1. The lithium acetate method of yeast cell transformation was used for introduction of plasmids into yeast cells (Gietz et al., 1992; Munn et al., 1995). Plasmids were recovered from yeast cells by the method of Ward (1990). Standard molecular procedures were performed according to the methods of Sambrook et al. (1989).
Cloning of the Wild-Type END11 Gene
An S. cerevisiae genomic library constructed in the LEU2-marked centromere vector YCplac111 (Gietz and Sugino, 1988; constructed and kindly provided by Fatima Cvrckova, Institute of Molecular Pathology, Vienna, Austria) was transformed into the end11-1 mutant (RH2622). Leu+ transformants were selected at 24°C. Approximately 1 × 105 transformant colonies were pooled and replated at 37°C on SD selective medium to select for temperature-resistant growth. Leu+ transformants that displayed improved growth on SD selective medium at 37°C were chosen for further analysis. Plasmids were isolated from five yeast transformants, amplified in E. coli strain DH5α, and reintroduced into the end11-1 mutant strain. One plasmid showed a reproducible ability to restore wild-type growth at 37°C to cells carrying the end11-1 mutation. This plasmid contained an insert of 6.3 kb and was named pEND11.2. The minimal DNA insert required for complementation of the end11-1 growth defect was identified as a 1.2-kb PstI–KpnI fragment by creating various deletions at the left and right ends of the insert and testing the deleted clones for complementation of the growth defect of end11-1. DNA sequence analysis of the 1.2-kb PstI–KpnI fragment revealed an open reading frame encoding a protein of 222 amino acids. Comparison with sequences in the nonredundant protein database showed that the identified gene is ERG2, which encodes C-8 sterol isomerase (Arthington et al., 1991).
To test whether ERG2 is the locus affected in end11-1 mutants or an unlinked gene that can suppress end11-1, the cloned ERG2 gene was tagged with URA3 to perform integrative mapping of the URA3-tagged ERG2 gene and end11-1. A 2.9-kb BamHI fragment carrying the complete ERG2 gene was cloned from pEND11.2 into the integration vector YIplac211 (marked with URA3; Gietz and Sugino, 1988) to create YIplac211-ERG2. The tagged ERG2 construct was integrated into the genome of a haploid wild-type strain (RH2635) at the chromosomal locus corresponding to the cloned DNA sequence to create RH2894. RH2894 was crossed to the end11-1 haploid RH2622 to create the diploid strain RH2895. Consistent with the tagged ERG2 locus being tightly linked to end11-1, all Ura+ haploids derived from RH2895 were nystatin sensitive (END11) and all Ura− haploids were nystatin resistant (end11-1).
DNA Sequence Analysis
For DNA sequence analysis, inserts were subcloned into pBluescript KS II or pBluescript SK II vectors (Stratagene, La Jolla, CA). Double-stranded DNA was prepared and denatured with sodium hydroxide treatment before primer annealing using a modification of the method of Chen and Seeburg (1985). DNA sequencing was carried out with a Sequenase II kit according to the manufacturer’s specifications (United States Biochemical, Cleveland, OH). Reactions were resolved by electrophoresis using a Sequi-Gen DNA Sequencing Cell (Bio-Rad, Hercules, CA). DNA sequences were analyzed using the University of Wisconsin (Madison, WI) Genetics Computer Group programs run on a VAX/VMS computer system at the Universität Rechnung Zentrum at the University of Basel.
Construction of erg Deletion Alleles
To disrupt the ERG2/END11 gene, the 2.9-kb BamHI fragment from pEND11.2, which contains the entire open reading frame, was subcloned into pBluescript KS− (Stratagene) to create pBKS-END11. The BglII–SphI fragment extending from nucleotides −110 to +495 relative to the start of translation of ERG2 was then replaced by a 1.1-kb BamHI fragment containing URA3 to generate pend11-Δ1::URA3. During the cloning, the BamHI sites flanking URA3 were destroyed by base filling and blunt-end ligation, leaving only the outer BamHI sites. Digestion of pend11-Δ1::URA3 with BamHI releases a URA3 fragment with ERG2/END11-flanking sequences. This fragment was used for disruption of ERG2/END11 in the homozygous wild-type diploid strain RH1201. Sporulation of the heterozygous diploid strain (RH2896) generated by the disruption event yielded Ura+ recombinant haploid spores in which the ERG2/END11 locus was deleted [erg2(end11)Δ1::URA3]. The strain RH2897 is derived from a spore of RH2896. The ERG6 gene was disrupted in RH1800 using the erg6Δ::LEU2 construct pIU222 (Gaber et al., 1989).
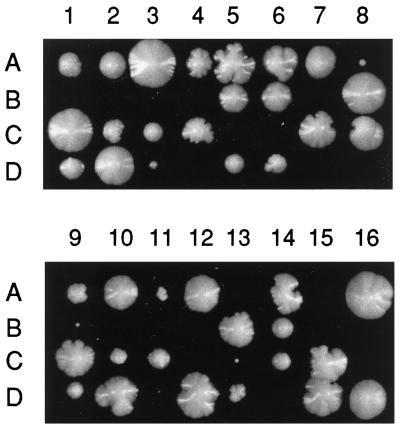
The strain RH1894 contains an unmarked disruption of the ERG6 gene (erg6Δ). It is a MATα erg6Δ haploid derived from a cross of MD59 to RH1800. To create an erg2Δerg6Δ heterozygous double mutant strain (RH3610), RH2897 was crossed to RH1894. After sporulation of RH3610, tetrads were dissected onto YPUATD solid medium and incubated at 24°C. Most presumed double mutant spores were inviable, but two MATα double mutant spores survived of 16 tetrads (64 spores) dissected (RH3611 [8A] and RH3612 [9B]; see Figure 2). These spores gave rise to extremely slow-growing colonies. RH3612 was then crossed to the wild-type strain RH2878 to create the diploid RH3614. RH3614 was sporulated, and tetrads were dissected. Two mutations conferring nystatin-resistance segregated in this cross, consistent with RH3612 being a true erg2Δerg6Δ double mutant. Two viable MATa haploids arising from the RH3614 diploid, RH3616 and RH3617, were of presumed genotype erg2Δerg6Δ and were retained. Like RH3611 and RH3612, RH3616 and RH3617 also grew extremely slowly. In crosses of RH3616 and RH3617 to the wild-type MATα strain RH2635, two mutations conferring nystatin resistance were segregating. This is consistent with RH3616 and RH3617 having the genotype erg2Δerg6Δ. For reasons we do not understand, in these crosses most of the erg2Δerg6Δ double mutant spores were viable (but formed tiny colonies).
Figure 2.
Dissection plate of the diploid strain erg2Δ/ERG2 erg6Δ/ERG6 (RH3610). Spores (A–D) from each tetrad (1–14) were allowed to germinate and grow on YPUATD at 24°C. Inviable spores were those predicted to have the genotype erg2Δerg6Δ. A few spores of this genotype were viable (8A and 9B) but grew extremely slowly.
Endocytosis Assays
For fluid phase endocytosis assays, cells were incubated with Lucifer yellow carbohydrazide (LY, dilithium salt; Fluka, Buchs, Switzerland) in YPUATD medium for 1 h at 24°C, washed, examined microscopically, and photographed as described by Munn and Riezman (1994). The [35S]α-factor was prepared as described by Munn and Riezmen (1994). α-Factor uptake and degradation assays were performed on cells grown at 24°C to a final density of 0.7–1.0 × 107 cells/ml in YPUATD medium. The internalization assays were carried out at 24 or 37°C using the continuous presence protocol with a 15-min preshift to the respective temperatures before adding the [35S]α-factor (Dulic et al., 1991). Internalization (in percentage) was calculated by dividing internalized counts (pH1-resistant counts) by the total cell-associated counts (pH6-resistant counts) for each time point. Values correspond to the means of three or four experiments. For α-factor degradation assays, [35S]α-factor was allowed to prebind to cells on ice for 50 min. Subsequent incubation was at 37°C. Samples were taken at times indicated and diluted in pH1 (internalized counts) or pH6 (total cell-associated counts) buffer. Subsequent cell extractions and separation of intact from degraded radiolabeled α-factor were done as described by Dulic et al. (1991).
Carboxypeptidase Y Delivery to the Vacuole
Cells were grown in SD containing 0.2% yeast extract at 24°C to an A600 of 0.5–1.0. Then 20 A600 units of cells were harvested, washed in SD, and resuspended in 2.5 ml of fresh SD (preheated to 37°C). After 15 min of incubation at 37°C, the cells were metabolically labeled with [35S]methionine and [35S]cysteine (Tran35S-label or EASYTAG EXPRESS protein labeling mix 35S; New England Nuclear, Boston, MA) at 37°C for 5 min and chased with unlabeled methionine, cysteine, and sulfate as described by Munn et al. (1995). At 0, 5, 10, and 30 min after addition of the chase, samples were removed to microfuge tubes on ice containing sodium azide and sodium fluoride (20 mM final concentration each). The cells were collected by centrifugation and directly lysed by agitation in 500 μl of 2% SDS with 0.5 g of 0.5-mm-diameter glass beads. After heating the lysates immediately to 90°C, the debris were sedimented by centrifugation. The cleared supernatants were subjected to immunoprecipitation with carboxypeptidase Y (CPY)-specific antibodies followed by protein A-Sepharose (Pharmacia, Uppsala, Sweden). After washing several times in TNET (100 mM Tris-HCl, pH 8, 100 mM NaCl, 5 mM EDTA, 1% Triton X-100) and once in 20 mM Tris-HCl, pH 7.5, the immunoprecipitates were dissolved in 2× Laemmli sample buffer (Laemmli, 1970) and heated to 90°C for 3 min. Immunoprecipitates were resolved on 7.5% SDS-polyacrylamide gels (Laemmli, 1970). The gels were dried, and the radiolabeled CPY was visualized with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Secretion of p2 CPY was examined using the colony immunoblotting assay described by Rothman et al. (1986). The extremely slow-growing erg2Δerg6Δ strain was patched onto a YPUATD plate and incubated for 1 d at 24°C. Then the other strains to be tested were patched onto the YPUATD plate and grown for an additional 1 d. The plate was then overlaid with a nitrocellulose filter (BA85, 0.45 μm; Schleicher & Schuell, Dassel, Germany) and incubated for an additional 1 d at 24°C to allow the yeast patches to grow to the same density. The nitrocellulose filter was removed, and cells were eluted from the filter by several distilled water washes. The filter was blocked in 2% skim milk, 1× PBS, and 0.1% Tween 20 for 1–2 h and probed with a rabbit polyclonal antiserum against CPY or calmodulin followed by goat anti-rabbit immunoglobulin G coupled to horseradish peroxidase (Bio-Rad). After several washes with PBS, the presence of p2 CPY antigen on the filter was visualized by using an enhanced chemiluminescence kit (Amersham, Arlington Heights, IL) and XAR x-ray film (Kodak, Rochester, NY).
Invertase Secretion Assays
The invertase secretion assays were performed according to the methods of Novick and Schekman (1979) with some modifications. Yeast cells were grown at 24°C to an A600 of 0.2–0.5 in YPUATD medium. Ten A600 units of cells were harvested, washed in distilled water, and induced for invertase expression by resuspension in YPUAT(low D)S medium (containing 0.05% glucose and 2% sucrose) prewarmed to 37°C. Cell samples were taken after 0, 15, 30, 45, and 60 min of induction at 37°C. Membrane transport was blocked by addition of sodium azide to 10 mM final concentration and transfer to ice. Cells of each sample were washed twice in ice-cold 10 mM sodium azide and resuspended in 2 ml of ice-cold 10 mM sodium azide. The final A600 was then adjusted to 0.5 with ice-cold 10 mM sodium azide. Two 0.5-ml aliquots of each sample were transferred to fresh microfuge tubes. To one aliquot of cells, 50 μl of distilled water were added, and the cells were left on ice (whole cells). To the other aliquot of cells, 50 μl of 10% Triton X-100 were added. Cells of this aliquot were permeabilized by freezing in liquid nitrogen and thawing at room temperature (lysates) and placed on ice. Aliquots of whole cells and lysates were assayed for invertase enzyme activity using the method of Goldstein and Lampen (1975). Ten-microliter samples were added in duplicate to 25 μl of 0.2 M sodium acetate, pH 4.9, and 12.5 μl of 0.5 M sucrose on ice. For the standard curve, known amounts of glucose (0, 12.5, 25, 50, 75, and 112.5 nmol) were used in place of the sample and sucrose. The tubes were incubated at 37°C for 10 min and then placed on ice. The reaction was terminated, and the invertase was inactivated by addition of 50 μl of 100 mM potassium phosphate buffer, pH 7, and heating to 90°C for 3 min followed by chilling on ice. After addition of 500 μl of solution C (50 μg/ml glucose oxidase [Aspergillus niger; Fluka], 10 μg/ml horseradish peroxidase [Fluka], 10 mM potassium phosphate buffer, pH 7, 300 μg/ml o-dianisidine [Sigma, St. Louis, MO], and 38% wt/vol glycerol), the samples were incubated at 30°C for 20 min. The assay was terminated, and the color was developed by addition of 750 μl of 6 M HCl. Absorbance at 540 nm was determined spectrophotometrically.
The readings for the duplicate samples were averaged. The internal invertase activity was calculated from the difference between the total invertase activity (from lysates) and the surface invertase activity (from whole cells) at each time point. The A540 values for the duplicate glucose standards were also averaged and plotted to give a glucose standard curve. The A540 values for the total, external, and internal invertase in the cell samples were then converted into nanomoles of glucose formed using the calculated slope of the glucose standard curve as a conversion factor (usually ∼1 A540 unit represents 100 nmol of glucose). The internal and external invertase activity was then expressed as micromoles of glucose formed per A600 unit of cells per minute and plotted as a function of time after invertase induction.
Sterol Analysis
Cells were grown to early logarithmic phase (0.7–1.0 × 107 cells/ml) in YPUATD medium at 24°C. Total sterols were extracted from whole cells based on a procedure by Folch et al. (1957). Alkaline hydrolysis was carried out as described by Lewis et al. (1987). Briefly, ∼1 × 109 cells were harvested by centrifugation, resupended in prewarmed YPUATD medium, and incubated for 30 min at 24 or 37°C. Harvested cells were then washed twice in distilled water to remove traces of the YPUATD medium. Cells were resuspended in a lysis solution comprising 1.5 ml of methanol (100%), 1 ml of 0.5% Pyrogallol (wt/vol in 100% methanol), and 1 ml of 60% KOH and heated for 2 h at 85°C. Total sterols were then extracted (three times) with 3 ml of petroleum ether. Upper phases were combined, dried under constant nitrogen gas, and stored at −20°C. Before subjection to gas–liquid chromatography (GLC) and GLC-mass spectrometry (GLC-MS) analysis, dried sterols were resuspended in 0.5 ml of cyclohexane (Fluka). Individual sterols were analyzed by GLC (HP 5 column) and GLC-MS (HP 5-MS column; Hewlett Packard) as described by van den Hazel et al. (1999). Relative retention times of sterols were in agreement with previous reports (Patterson, 1971; Xu et al., 1988; Nes et al., 1989). The abundance estimate of each sterol was based on two independent experiments analyzed in duplicate by GLC.
Sterol Structure Drawings
The sterol structures shown in Figures 1 and 8 were drawn using the ChemSketch 3.5 software purchased from Advanced Chemistry Development (Toronto, Ontario, Canada).
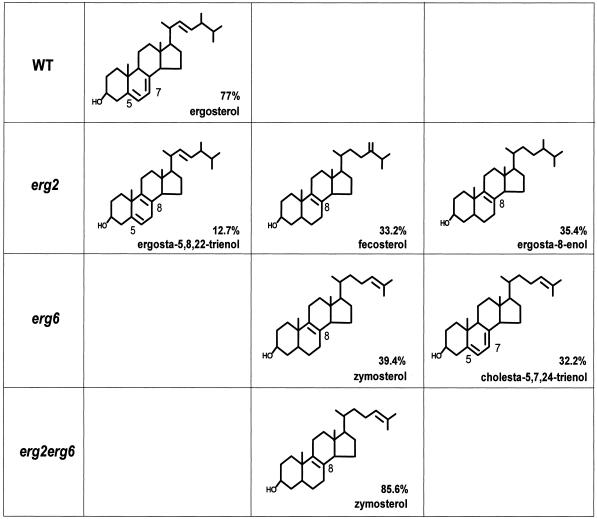
Figure 8.
Chemical structures of the most abundant sterols (>10%) present in wild-type (RH448), erg2Δ (RH2897), erg6Δ (RH3622), and erg2Δerg6Δ (RH3616) cells. The abundance of each sterol within a strain is given as a percentage of total sterol.
RESULTS
END11 Is Allelic to ERG2, the Gene Encoding C-8 Sterol Isomerase
The wild-type END11 gene was isolated by complementation cloning based on the growth defect associated with end11-1. For this purpose, the end11-1 mutant strain was transformed with a yeast genomic library carried on a centromeric vector. Sequence analysis of a complementing plasmid, pEND11.2, revealed that the plasmid carried the ERG2 gene (Arthington et al., 1991). Transformation of end11-1 cells with the ERG2-containing plasmid restored cell growth to wild-type rates (our unpublished results). In addition, mutant cells transformed with this plasmid were able to accumulate the fluorescent dye LY in vacuoles (our unpublished results), indicating that the plasmid-borne ERG2 gene was able to complement the fluid phase endocytic defect of end11-1 (Munn and Riezman, 1994). An integrative mapping strategy further demonstrated that the ERG2 gene and the end11-1 mutation are tightly linked (see MATERIALS AND METHODS).
The ERG2 gene encodes C-8 sterol isomerase, an enzyme that functions in a late step of the ergosterol biosynthetic pathway (Arthington et al., 1991). Absence of ergosterol is known to confer nystatin-resistant growth. Nystatin is an antifungal drug that interacts selectively with membrane ergosterol but not with sterols different from ergosterol (Lees et al., 1995). As reported for erg2 mutant strains (Arthington et al., 1991), end11-1 is able to grow on nystatin. Taken together, these results confirm that END11 is allelic to ERG2.
erg2Δ, erg6Δ, and erg2Δerg6Δ Mutants Exhibit Defects in the Internalization Step of Endocytosis
The identification of END11 as ERG2 was the first indication that ergosterol is required for endocytosis in yeast. To examine the in vivo requirement for ergosterol in endocytosis in more detail, we made use of ergΔ mutants affected in the late ergosterol biosynthetic pathway. Each of these mutants accumulates a distinct set of sterols with structural differences specific to the ergosterol molecule (Lees et al., 1995; Parks et al., 1995). For this purpose, the ERG2 and ERG6 genes were disrupted by integration replacements (see MATERIALS AND METHODS). Erg2p, the C-8 sterol isomerase, is involved in changes of the B ring desaturation, whereas Erg6p, the C-24 sterol methyltransferase, modifies the sidechain of the sterol molecule (Figure 1). An erg2Δerg6Δ double mutant strain lacks both Erg2p and Erg6p activities and has been reported to accumulate mainly zymosterol (Bard et al., 1977; Figure 1; see below).
Consistent with previous reports, deletions of the ERG2 and ERG6 genes did not cause lethality (Gaber et al., 1989; Arthington et al., 1991; Hardwick and Pelham, 1994; Welihinda et al., 1994). The erg2Δ strains showed a slow-growth phenotype at 24 and 37°C similar to that observed for end11-1 (our unpublished results). The erg6Δ strain grew at wild-type rates at 24°C, but its growth was reduced at 37°C (our unpublished results). Double mutant strains (erg2Δerg6Δ) were isolated after mating erg2Δ and erg6Δ haploid strains, sporulation, and dissection of the diploid. Most double mutant spores were inviable, but occasionally they gave rise to extremely slow-growing colonies (Figure 2; see spores 8A and 9B). Spores that were wild type, erg2Δ, or erg6Δ were nearly all viable and grew relatively well. When surviving double mutant haploids were crossed back to a wild-type strain, each resulting diploid gave rise to wild-type, erg2Δ, erg6Δ, as well as erg2Δerg6Δ mutant spores. In each case, the double mutant spores were mostly inviable, demonstrating that the surviving double mutant haploids from the first cross used as one parent did not possess an extragenic suppressor mutation. Two of these slow-growing erg2Δerg6Δ double mutants were retained for further analysis.
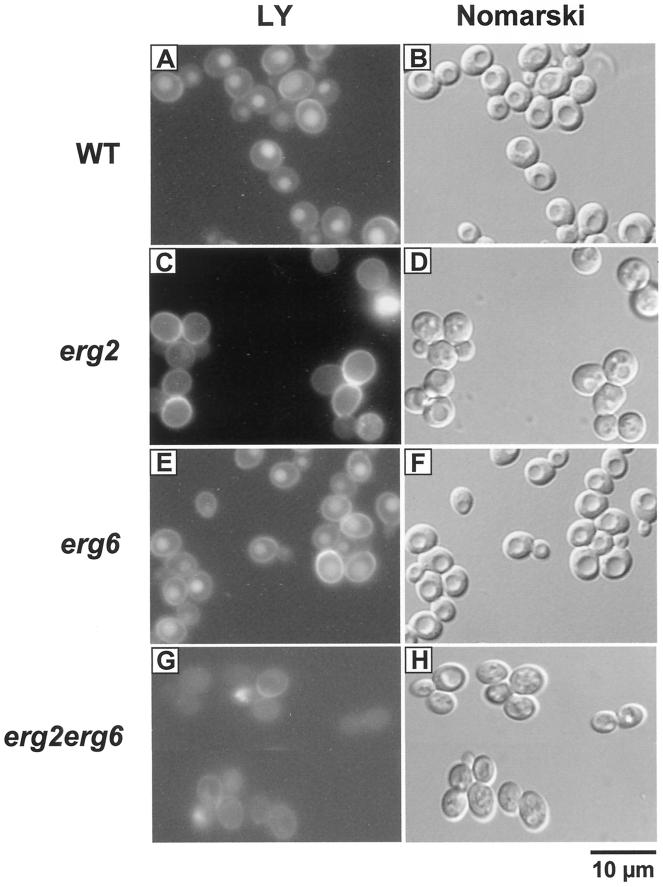
Fluid phase and receptor-mediated endocytosis were examined in erg2Δ, erg6Δ, and erg2Δerg6Δ mutant strains and compared with endocytosis in the wild-type strain. In assays for fluid phase endocytosis, yeast cells were incubated with the fluorescent dye LY at 24°C for 1 h and observed by fluorescence microscopy. As shown for the wild-type strain (Figure 3A; Riezman, 1985), endocytic uptake of the dye resulted in LY accumulation in yeast vacuoles that were visible as indentations in the Nomarski image (Figure 3B). The erg6Δ cells accumulated LY in vacuoles to similar levels as wild-type cells (Figure 3E), indicating no apparent block in fluid phase endocytosis. In contrast, erg2Δ and erg2Δerg6Δ cells were clearly defective in fluid phase endocytosis, because little or no fluorescent dye was present in vacuoles (Figure 3, C and G, respectively). The block in fluid phase endocytosis of erg2Δ cells was consistent with the results obtained for end11-1 (Munn and Riezman, 1994). It should be noted that some erg2Δ and erg2Δerg6Δ cells did not contain a recognizable vacuole in Nomarski images; however, those containing an obvious vacuole did not accumulate the fluorescent dye (Figure 3, D and H). The strong cytoplasmic staining in some erg2Δerg6Δ cells (Figure 3G) may be due to loss of viability that was also observed in subsequent experiments. We also observed that many erg2Δerg6Δ cells exhibited stronger cytoplasmic staining than wild-type or ergΔ single mutant cells indicating that the ergΔ double mutant cells may be more permeable to the dye.
Figure 3.
Analysis of fluid phase endocytosis in ergΔ mutant cells. Wild-type (RH448; A and B), erg2Δ (RH2897; C and D), erg6Δ (RH3622; E and F) and erg2Δerg6Δ (RH3616; G and H) cells were incubated with LY at 24°C. To visualize the localization of LY, cells were viewed by FITC-fluorescence optics (A, C, E, and G). The same fields of cells were examined by Nomarski optics to visualize the vacuoles (which are apparent as indentations in the cell profiles; B, D, F, and H). Bar, 10 μm.
To assay the first step of receptor-mediated endocytosis, radiolabeled α-factor was added to ergΔ mutant and wild-type cells that had been preincubated for 15 min at 24 or 37°C. Internalization of the α-factor receptor–ligand complex at the given temperature was then examined by determining the percentage of internal radiolabeled α-factor at specific time points. Consistent with previous reports (Munn and Riezman, 1994; Munn et al., 1995), internalization of α-factor in wild-type cells was similar at 24 and 37°C (Figure 4). The ergΔ single and double mutant cells exhibited reduced internalization at 24°C, but the defect was stronger at 37 than at 24°C (Figure 4). While wild-type cells internalized most of the α-factor within 30 min (Figure 4A), erg2Δ cells exhibited a defect in α-factor internalization similar to that reported previously for end11-1 cells (Figure 4A; Munn and Riezman, 1994). These results indicate that cells lacking the C-8 sterol isomerase activity have a defect in the internalization step, the first step of endocytosis. Internalization was also reduced in erg6Δ cells (Figure 4B), but to a lesser extent than in erg2Δ cells (Figure 4A). In contrast to LY uptake, α-factor internalization is a quantitative assay (Dulic et al., 1991), and it is possible that a mild endocytic defect in erg6Δ cells was not apparent in the LY accumulation experiment (Figure 3). In erg2Δerg6Δ double mutant cells, α-factor internalization was completely abolished at 37°C (Figure 4C). Thus at 37°C, cells lacking both C-24 sterol methyltransferase and C-8 sterol isomerase activities show endocytic defects as severe as those observed in the tightest end mutants blocked in the internalization step of endocytosis, such as end3-1, end4-1 (Raths et al., 1993), and act1-1 (Kübler and Riezman, 1993).
Figure 4.
Analysis of the internalization step of receptor-mediated endocytosis in ergΔ mutant cells. Internalization assays were performed at 24°C (open symbols) and 37°C (closed symbols) on erg2Δ (RH2897; A), erg6Δ (RH3622; B), and erg2Δerg6Δ (RH3616; C) mutant and wild-type cells (RH1800; A–C). Internalization of radiolabeled α-factor was expressed as a percentage by dividing internalized counts by the total cell-associated counts for each time point.
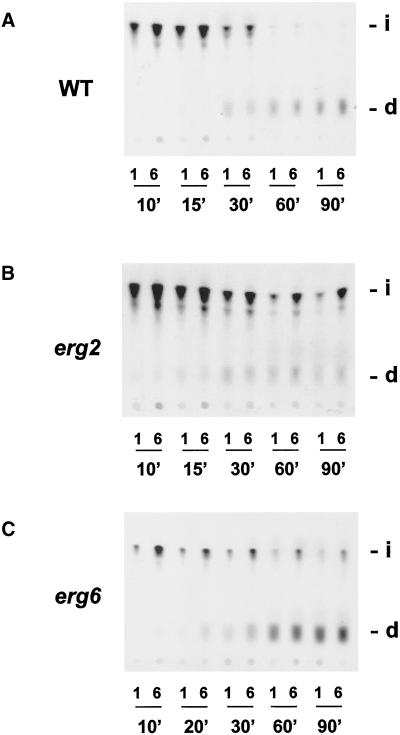
After internalization of the α-factor receptor–ligand complex, the complex moves through early and late endosomal compartments to the vacuole where it is degraded. To determine whether later steps in the endocytic pathway were also affected in ergΔ mutant cells, α-factor degradation assays were performed at 37°C (Dulic et al., 1991). In wild-type cells, most of the α-factor was internalized within 30 min (Figure 5A; pH1 resistant) and then delivered to the vacuole as evident by the accumulation of degraded α-factor at later timepoints (Figure 5A, d). Consistent with a defect in the internalization step, some intact α-factor remained at the cell surface of erg2Δ and erg6Δ mutant cells even after 90 min (Figure 5, B and C, i; pH 6 resistant). The erg2Δ and erg6Δ mutant cells did not, however, exhibit strong defects at a postinternalization step, because degraded α-factor was present at later time points (Figure 5, B and C, d). We were unable to perform the degradation assay on the erg2Δerg6Δ double mutant because insufficient amounts of α-factor were internalized in this ergΔ mutant because of the severity of the defect. Based on these endocytic assays, we conclude that specific sterols are required for the internalization step of endocytosis in yeast.
Figure 5.
Analysis of α-factor degradation in ergΔ mutant cells. α-Factor degradation assays were performed at 37°C on wild-type (RH1800; A), erg2Δ (RH2897; B), and erg6Δ (RH3622; C) cells. At times indicated, samples were taken and diluted in pH1 buffer (1; internalized counts) or pH6 buffer (6; total cell-associated counts). Degradation of α-factor was assessed by extracting cell-associated radioactivity and separating degraded α-factor (d; indicating delivery to the vacuole) from intact α-factor (i) on TLC plates. d, degraded α-factor; i, intact α-factor; n′, time in minutes.
erg2Δ, erg6Δ, and erg2Δerg6Δ Mutations Do Not Affect Maturation of CPY or Secretion of Invertase to the Plasma Membrane
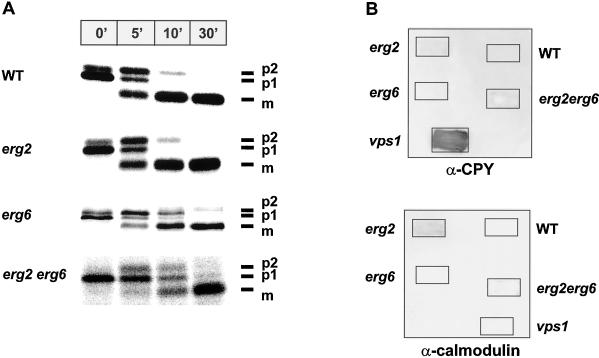
Ergosterol is predominately present in the plasma membrane, but it is also found in significant amounts in secretory vesicles (Zinser et al., 1993). We therefore investigated whether the sterol requirement is specific for the formation of endocytic vesicles or whether vesicular trafficking through the secretory pathway also requires ergosterol. By monitoring maturation of the newly synthesized CPY precursor, we followed the vesicular transport of CPY from the endoplasmic reticulum (ER) to the Golgi and then to the vacuole in the ergΔ mutant and wild-type strains (Figure 6A). It has been previously shown for wild-type cells (Stevens et al., 1982; Klionsky et al., 1990) that upon translocation into the ER, CPY is core glycosylated to generate a form with an apparent molecular mass of 67 kDa (p1). This precursor protein is further glycosylated to the 69-kDa form (p2) in an early Golgi compartment, and upon arrival in the vacuole, the p2 form is cleaved to the mature and active CPY of 61 kDa (m). Pulse–chase labeling experiments followed by immunoprecipitation of CPY showed that CPY matures in ergΔ single and double mutant strains with kinetics similar to those observed in the wild-type strain (Figure 6A). The p1 form of CPY seems to be converted to the p2 form slightly more slowly than in wild-type cells, but this defect is mild when compared with the defects in growth rate and endocytosis exhibited in the erg2Δerg6Δ mutant cells by the double mutant. These results are in agreement with previous reports that end11-1 (Munn and Riezman, 1994) and erg6Δ (Hardwick and Pelham, 1994; isolated as sed6) do not have a defect in CPY maturation.
Figure 6.
Sorting of newly synthesized CPY to the vacuole in ergΔ mutant cells. (A) Maturation of newly synthesized CPY in wild-type (RH1800), erg2Δ (RH2897), erg6Δ (RH3622), and erg2Δerg6Δ (RH3617) cells. Cells were grown at 24°C, shifted to 37°C for 15 min, and then pulse labeled at 37°C for 5 min with [35S]methionine and [35S]cysteine followed by a chase with nonradioactive sulfate, methionine, and cysteine at 37°C. CPY was immunoprecipitated from samples taken at specified time points of chase (minutes). The immunoprecipitates were resolved on 7.5% SDS-polyacrylamide gels and visualized on a PhosphorImager. p1, core glycosylated precursor form of CPY; p2, fully glycosylated precursor form of CPY; m, proteolytically processed, mature CPY; n′, time in minutes. (B) Sorting of CPY to the vacuole in wild-type (RH1800), erg2Δ (RH2897), erg6Δ (RH3622), erg2Δerg6Δ (RH3617), and vps1Δ (RH2180) cells. Cells were grown as patches on YPUATD solid medium in contact with nitrocellulose filters at 24°C. Washed filters were probed with either rabbit polyclonal antibodies against CPY (α-CPY) or calmodulin (α-calmodulin) followed by goat anti-rabbit immunoglobulin G coupled to horseradish peroxidase. Any CPY secreted or any calmodulin released from the cells was visualized by enhanced chemiluminescence.
Some mutants defective in transport of CPY to the vacuole (vacuole protein-sorting [vps] mutants) do not exhibit defects in maturation of intracellular CPY but still missort significant amounts of p2 CPY to the cell surface (Robinson et al., 1988). Therefore, we also assayed secretion of CPY in the ergΔ mutants by colony immunoblotting using antibodies against CPY. As expected, a vps1 control strain showed high levels of CPY secretion (Figure 6B, α-CPY; Robinson et al., 1988). However, none of the ergΔ mutants or the wild-type strain showed significant secretion of CPY into the medium (Figure 6B, α-CPY). The release of trace amounts of CPY antigen in erg2Δ and erg2Δerg6Δ mutant strains were likely caused by occasional cell lysis, because these strains also released low levels of calmodulin, a cytoplasmic protein (Figure 6B, α-calmodulin).
To determine whether ergΔ mutations affect protein secretion to the plasma membrane, we analyzed secretion of invertase. In wild-type cells, invertase is secreted into the periplasm between the plasma membrane and the cell wall. Initial attempts to measure invertase secretion by pulse–chase radiolabeling of spheroplasts were unsuccessful, because ergΔ mutant strains had a tendency to lyse upon removal of the cell wall, even in the presence of osmotic support. As an alternative approach, internal and external invertase activities were measured using enzyme latency assays that do not require the removal of the cell wall (Rothman et al., 1986). In these assays, it is possible to differentiate between internal and external invertase activities, because the substrate (sucrose) can diffuse easily across the cell wall but not the plasma membrane. Internal invertase activity was then calculated from the difference between external invertase activity in whole cells and total invertase activity in cell lysates at each time point. Even though invertase activity was induced to different levels within each strain, erg2Δ [Figure 7B, (e)], erg6Δ [Figure 7C, (e)], and erg2Δerg6Δ [Figure 7D(e)] mutant strains secreted invertase with wild-type kinetics [Figure 7A, WT (e)]. Similar amounts of internal invertase were detected in wild-type cells and any of the ergΔ mutant cells. In contrast, a sec18 mutant strain (defective in ER-to-Golgi transport of secreted proteins) did not secrete invertase into the periplasm but accumulated invertase internally (Figure 7A; Novick et al., 1981). Taken together, these data indicate that sterols present in erg2Δ, erg6Δ, and erg2Δerg6Δ mutant strains are capable of supporting vesicle formation and fusion throughout the secretory pathway, including vesicle fusion to the plasma membrane, but do not allow the formation of endocytic vesicles at the plasma membrane.
Figure 7.
Secretion of invertase in ergΔ mutant cells. Invertase activities were assayed in wild-type (RH1800; A), sec18 (RH1737; A), erg2Δ (RH2897; B), erg6Δ (RH3622; C), and erg2Δerg6Δ (RH3617; D) cells. Internal invertase activity (i) was calculated from the difference between the total invertase activity (cell lysate) and the surface invertase activity (whole cell; e) at each time point. Internal and external invertase activities were expressed as micromoles of glucose formed per A600 units of cells per min and plotted as a function of time after invertase induction. Open symbols, extracellular invertase activities (e); closed symbols, intracellular invertase activities (i); O.D., optical density.
Determining the Sterol Composition of ergΔ Single and Double Mutants
The sterol composition of erg2Δ, erg6Δ, and erg2Δerg6Δ mutant strains has been described previously for cells in stationary growth phase (Bard et al., 1977; Gaber et al., 1989; Arthington et al., 1991). It appears, however, that the sterol composition can vary depending on growth conditions and the stage of cellular growth (Leber et al., 1995). All of our previously described experiments were performed on cells in early log phase, a growth phase at which the exact sterol composition of the ergΔ mutants and the wild-type strain is unknown. To correlate the internalization defects with the sterol compositions of the ergΔ mutants, it was therefore necessary to determine the sterol composition of each ergΔ mutant and wild-type strain under the same growth conditions used for the experiments described above. As mentioned previously, ergΔ single and double mutant cells exhibited defects in α-factor internalization after incubation at 24°C; however, these defects were more severe after incubation at 37°C (Figure 4). To investigate whether these effects on internalization were due to changes in the overall sterol composition, total sterols were isolated from whole yeast cells that were grown to early log phase at 24°C and then incubated for 30 min at either 24 or 37°C. Individual sterols were separated by GLC and GLC-MS (van den Hazel et al., 1999). Based on the retention time and the mass spectrum of each sterol, we were able to identify nearly all sterols present in the ergΔ mutant and wild-type strains (Table 2). In addition, we calculated the abundance of each sterol within a given strain (Table 2). Preincubation of cells at 24 or 37°C did not lead to significant changes in the overall sterol composition in any of the analyzed strains (Table 2; our unpublished results). This result indicates that the more severe internalization defect observed in the ergΔ mutant cells when incubated at 37°C is not due to an altered sterol composition.
Table 2.
Analysis of the sterol composition and the relative abundance of sterols within wild-type (RH448), erg2 (RH2897), erg6 (RH3622), and erg2erg6 (RH3616) cells
| Sterol | % sterol | Mass |
|---|---|---|
| WT | ||
| Zymosterol | 6.9 | 384 |
| Ergosterol | 77 | 396 |
| Fecosterol | 3 | 398 |
| A | 5.1 | 398 |
| Episterol | 3.2 | 398 |
| Lanosterol | 3.8 | 426 |
| erg2 | ||
| Squalene | 1.4 | 410 |
| Zymosterol | 5 | 384 |
| Ergosta-5,8,22-trienol | 12.7 | 396 |
| Ergosta-7,22-dienol | 4.3 | 398 |
| Fecosterol | 33.2 | 398 |
| Ergosta-8-enol | 35.4 | 400 |
| B | 1.6 | 396 |
| 4-m-Cholesta-8,24-dienol | 2.5 | 398 |
| Lanosterol | 2.9 | 426 |
| 4,4-dm-Cholesta-8,24-dienol | 1.1 | 412 |
| erg6 | ||
| Squalene | 1.3 | 410 |
| Cholesta-5,8,24-trienol | 2.8 | 382 |
| Zymosterol | 39.4 | 384 |
| Cholesta-5,7,24-trienol | 32.2 | 382 |
| Cholesta-7,24-dienol | 7.3 | 384 |
| Cholesta-5,7,22,24-tetraenol | 7.9 | 380 |
| C | 2.7 | 398 |
| Lanosterol | 3.1 | 426 |
| 4,4-dm-Cholesta-8,24-dienol | 2.3 | 412 |
| erg2erg6 | ||
| Squalene | 2.7 | 410 |
| Zymosterol | 85.6 | 384 |
| D | 3.7 | 382 |
| E | 1.5 | 398 |
| Lanosterol | 3.3 | 426 |
| 4,4-dm-Cholesta-8,24-dienol | 2.1 | 412 |
After growth at 24°C to midlog phase, cells were incubated for 30 min at 37°C. Total sterols were isolated from whole cells and subjected to GLC and GLC-MS to identify individual sterols and to determine the abundance of each sterol within a strain. WT, wild type; 4,4-dm-cholesta-8,24-dienol, 4,4-dimethyl-cholesta-8,24-dienol; 4-m-cholesta-8,24-dienol, 4-methyl-cholesta-8,24-dienol; A–E, identification of sterols uncertain.
Many of the ergosterol biosynthetic enzymes are able to act on a range of sterol substrates, indicating that the ergosterol biosynthetic pathway is not strictly linear (reviewed in Lees et al., 1995; Parks and Casey, 1995). An erg mutant can therefore accumulate a variety of sterol analogues rather than only one ergosterol precursor. Overall, the sterol compositions of the ergΔ mutant and wild-type strains were in agreement with previous reports (Bard et al., 1977; Gaber et al., 1989; Arthington et al., 1991). The predominant sterol in the wild-type strain was ergosterol (77%; Table 2 and Figure 8). As expected, none of the ergΔ strains accumulated any detectable amount of ergosterol. These results are in agreement with previous studies, which reported that under aerobic growth conditions yeast cells are unable to internalize sterols from the extracellular medium (Trocha and Sprinson, 1976; Keesler et al., 1992). Consistent with the absence of the C-8 sterol isomerase activity, the erg2Δ mutant strain accumulated sterols that lack a C-7,8 double bond. The major sterols in this mutant were ergosta-8-enol (35.4%), fecosterol (33.3%), and ergosta-5,8,22-trienol (12.7%; Table 2 and Figure 8). Consistent with a lack of the C-24 sterol methyltransferase activity, all sterols in the erg6Δ mutant strain were missing the C-24 methylation, and zymosterol (39.4%) and cholesta-5,7,24-trienol (32.2%) were the most abundant sterols (Table 2 and Figure 8). The erg2Δerg6Δ double mutant strain accumulated nearly exclusively zymosterol (85.6%), an intermediate in the late part of the ergosterol biosynthetic pathway. Based on these sterol data, we were able to correlate the endocytic phenotypes of the ergΔ mutant and wild-type strains to their sterol composition and abundance of the major sterols.
DISCUSSION
In the present study, we identified a novel function of sterols in yeast, that is, facilitating endocytosis. The first evidence for this endocytic function came from the identification of END11, whose gene product is required for the internalization step of endocytosis (Munn and Riezman, 1994), as ERG2. This gene encodes the C-8 sterol isomerase, which functions in a late step of ergosterol biosynthesis (Arthington et al., 1991; Figure 1). Cells with a deleted ERG2 gene show internalization defects similar to those observed for end11-1. These endocytic defects are due to the altered sterol composition rather than to a novel function of Erg2p in endocytosis. An internalization defect, although a weaker one, was also observed in an erg6Δ strain that lacks the C-24 sterol methyltransferase activity of Erg6p. This transferase has no structural nor functional similarities to Erg2p, an isomerase. Furthermore, studies using animal cells indicate that depletion of cholesterol leads to internalization defects of certain proteins (Chang et al., 1992; Cerneus et al., 1993; Deckert et al., 1996; Orlandi and Fishman, 1998; Rodal et al., 1999).
In animal cells, the sterol requirement has been mainly investigated by using drugs such as filipin, nystatin, and β-cyclodextrin that bind and sequester cholesterol (Chang et al., 1992; Cerneus et al., 1993; Deckert et al., 1996; Orlandi and Fishman, 1998; Rodal et al., 1999; Subtil et al., 1999). In contrast, the yeast ergΔ mutants allowed us to examine the in vivo requirement for sterols without the use of drugs and sterol depletion. Moreover, we were able to assess the importance of particular structural features of the ergosterol molecule for the internalization step, because the ergΔ mutants accumulate sterols that are different from ergosterol. After the identification of End11p as Erg2p, the C-8 sterol isomerase involved in the B ring modification of ergosterol, we chose to examine endocytosis in erg mutants reported to accumulate sterols with changes in the side chain modification and in both B ring desaturation and side chain modification. The obvious choice was to construct an erg6Δ strain lacking the C-24 sterol methyltransferase activity (Gaber et al., 1989) and an erg2Δerg6Δ strain lacking both C-8 sterol isomerase and C-24 sterol methyltransferase activities. Interestingly, internalization of the α-factor receptor–ligand complex was inhibited to different extents in each ergΔ strain. By taking the sterol composition and sterol abundance of each ergΔ mutant and the wild type into account, we were able to correlate defects in internalization to the altered sterol composition. More importantly, it enabled us to correlate the internalization defect to a specific portion of the ergosterol molecule, namely to the B ring and its desaturation state.
Ergosterol, the main sterol in the wild-type strain, can fully support the internalization step of endocytosis at 24 and 37°C. In contrast, erg2Δerg6Δ cells with zymosterol as their predominant sterol showed the strongest defect in internalization. Zymosterol has a double bond at C-8,9 but lacks both of the double bonds (C-5,6 and C-7,8) present in the B ring of ergosterol (Table 2 and Figure 8). In addition, zymosterol does not have the C-24 methylation present in ergosterol. The C-24 methylation did not appear to play a major role in the internalization step, because all sterols present in the erg6Δ strains lack this modification (Table 2 and Figure 8), and the erg6Δ strain did not exhibit a strong internalization defect. It seems more likely that the state of the B ring desaturation is mainly responsible for the endocytic defects. The rate of internalization correlated well with the abundance of sterols with two double bonds at C-5,6 and C-7,8 or at C-5,6 and C-8,9, and it correlated inversely with the abundance of sterols with a single desaturation at C-8,9. The erg2Δerg6Δ double mutant cells had the highest amount of sterols with a single C-8,9 desaturation in the B ring (>85%) and also showed the strongest internalization defect. In addition, >75% of sterols in erg2Δ cells had a single C-8,9 desaturation in the B ring. Its internalization defect was less severe than that observed in the erg2Δerg6Δ double mutant strain, but it was more severe than that in erg6Δ cells. Even though the erg2Δ strain did not accumulate any C-7,8 sterols, it showed some internalization of α-factor. It is reasonable to assume that the relatively low amounts of ergosta-5,8,24-trienol (12.7%), a sterol containing two desaturations in the B ring at positions C-5,6 and C-8,9, is responsible for this α-factor internalization. These results indicate that even though a single desaturation in the B ring at C-8,9 is not able to support internalization, an additional C-5,6 desaturation (leading to a B ring desaturation at both C-5,6 and C-8,9) allows internalization of α-factor at 37°C. In addition to the C-8,9 sterols (∼40%), the erg6Δ strain accumulated >30% of cholesta-5,7,24-trienol, a sterol containing the exact B ring desaturation (C-5,6 and C-7,8) present in ergosterol. It is likely that cholesta-5,7,24-trienol is able to support endocytosis, but there is simply not enough of this sterol present in the erg6Δ strain to support endocytosis with wild-type kinetics.
Taking these arguments together, we conclude that the state of the B ring desaturation is critical for the internalization step of endocytosis. While a single desaturation at C-8,9 was not sufficient to support internalization of α-factor at 37°C, the presence of two double bonds in the B ring, either a combination of C-5,6 and C-7,8 or C-5,6 and C-8,9, allowed internalization to occur at this temperature. The methylation at C-24 in the side chain did not appear to be required for the internalization step of endocytosis, because its absence did not have a strong effect on α-factor uptake. At this point, the exact location of each sterol remains unknown in the ergΔ strains. It is reasonable to assume that the sterols with altered structural features were present in the plasma membrane, because the majority of sterols are usually found in this membrane (Zinser et al., 1993).
Clearly, sterols play an important role in the internalization step of endocytosis in yeast perhaps similar to the role of cholesterol in endocytosis in animal cells. Even though the overall structures of ergosterol and cholesterol are similar, three structural differences are evident in the B ring and the side chain. While cholesterol has only a single desaturation at C-5,6, ergosterol possesses two double bonds at C-5,6 and C-7,8. The side chain of ergosterol contains a desaturation at C-22,23 and a methyl group at C-24 that are both absent in cholesterol (for review, see Parks and Casey, 1995). Interestingly, the C-8 sterol isomerase (Erg2p) and the C-24 sterol methyltransferase (Erg6p) represent two of the enzyme activities that confer the structural differences between ergosterol and cholesterol. The absence of these two enzyme activities leads to the accumulation of zymosterol. As evident from the strong endocytic defect in the erg2Δerg6Δ double mutant, zymosterol, containing a single C-8,9 desaturation in the B ring was unable to support internalization in yeast at 37°C. It seems likely that in general, ergosterol and cholesterol serve similar functions, but the structure of the sterols may be adjusted to suit the specific needs of the organism the sterols are present in. In a single-cell organism, such as yeast, ergosterol may satisfy a variety of functions, including some that may not be critical in multicellular organisms. In support of this hypothesis, other yeast lipids such as phopholipids and sphingolipids also differ from the respective animal lipid in certain structural features (Daum et al., 1998; Dickson, 1998).
Animal cells are able to obtain cholesterol by two ways: endogenously, by producing the sterol de novo in the ER, and exogenously, by internalizing cholesterol from the extracellular medium (Fielding and Fielding, 1997). In contrast, yeast has to rely on ergosterol produced endogenously, because under normal, aerobic growth conditions yeast is unable to take up sterols from the extracellular medium (Trocha and Sprinson, 1976; Keesler et al., 1992). Consistent with these reports, we showed in the present studies that none of the ergΔ mutants was able to internalize ergosterol from the growth medium despite the presence of yeast extract, because no ergosterol was detected by GLC and GLC-MS analysis (Table 2). Furthermore, the α-factor internalization defect in erg2Δ cells was similar whether the yeast cells were grown in synthetic medium lacking any lipids or in YPUATD containing yeast extract (our unpublished results). Although desirable, we were not able to perform ergosterol feedback experiments in these ergΔ mutants because of the sterol exclusion under growth conditions that are required for the endocytic assays.
Even though a role for sterols in endocytosis is evident, the question remains how sterols participate in the internalization step at the plasma membrane. Ergosterol may serve as a site for the interaction of the endocytic machinery with the plasma membrane. Similar to what has been observed for animal cells (for review, see Harder and Simons, 1997; Anderson, 1998; Brown and London, 1998), ergosterol may associate with sphingolipids to form lipid rafts that serve as platforms for the recruitment and subsequent internalization of specific proteins. Microdomains with some features in common with animal cell microdomains are present in yeast membranes (Kübler et al., 1996). These microdomains have been proposed to function in signaling (Kübler et al., 1996); however, neither their lipid composition nor their role in internalization has been determined. Little evidence exists for sterol-rich and -poor regions in yeast plasma membranes (Bottema et al., 1983), nor is it known whether ergosterol can associate with sphingolipids to form lipid rafts (Leber et al., 1997). In animal cells, depletion of cholesterol prevents clustering of glycosylphosphatidylinositol-anchored proteins into lipid rafts and, thus, blocks their subsequent internalization (Chang et al., 1992; Rothberg et al., 1990; Rothberg et al., 1992). If the main role of ergosterol lies in the recruitment and clustering of proteins into sterol-rich domains, one might expect that only receptor-mediated but not fluid phase endocytosis would be defective in ergΔ mutants. This is not true for erg2Δ and erg2Δerg6Δ, because in addition to defects in α-factor receptor internalization, these ergΔ mutants exhibit strong defects in fluid phase endocytosis. These results point toward a more general defect in endocytosis in ergΔ mutants.
Ergosterol may also provide a membrane environment required for the correct conformation of plasma membrane proteins that function in endocytosis. In support of this hypothesis, several plasma membrane proteins (Gaber et al., 1989; Welihinda et al., 1994) have been reported to function at reduced levels in erg6Δ mutants. We do not believe, however, that the endocytic defects observed in the ergΔ mutants are due to an inactivation of Ste2p, the α-factor receptor. The α-factor internalization and α-factor degradation assays show that Ste2p is able to bind α-factor, indicating that Ste2p is functional in plasma membranes with an altered sterol composition. In addition, the internalization defect in the ergΔ mutants is not specific for ligand-induced endocytosis of Ste2p, because we observed a severe block in fluid phase endocytosis in erg2Δ and erg2Δerg6Δ mutants.
Finally, ergosterol may play a crucial role in the physical formation of endocytic vesicles. Sterols are rigid and compact molecules consisting of a planar, nonpolar ring system and a single polar functional group, a hydroxyl group at C-3 (Brown, 1998). The orientation and conformation of sterols are important for their interaction with other lipids and influence membrane permeability and membrane fluidity (Brown, 1998; Daum et al., 1998). As previously reported, alterations in the sterol composition change the fluidity of membranes, and the membrane fluidity decreases from wild type to erg6Δ to erg2Δ to erg2Δerg6Δ, with the double mutant showing the highest membrane rigidity (Lees et al., 1979). Interestingly, an increase in membrane rigidity correlates well with the severity of the internalization defect for the ergΔ mutants reported in the present studies. It is interesting to note that even though the sterol compositions were similar at 24 and 37°C, each ergΔ mutant exhibited stronger internalization defects at higher temperatures. In particular, α-factor internalization was completely blocked in the erg2Δerg6Δ mutant strain at 37°C. It is therefore possible that the internalization defect is merely due to a physical restriction in the formation of endocytic vesicles imposed by a change in sterol composition in the ergΔ mutant membranes. In support of this hypothesis, the removal of cholesterol from animal cells directly correlates with the flattening of clathrin-coated pits (Rodal et al., 1999; Subtil et al., 1999) and leads to a loss of the characteristic flask-shape morphology of caveolae (Rothberg et al., 1990, 1992; Chang et al., 1992; Hailstones et al., 1998). Thus, cholesterol appears to affect the morphology and curvature of the plasma membrane in animal cells, and ergosterol may function in a similar manner in the yeast plasma membrane. At this point, however, we cannot exclude the possibility that ergosterol serves a combination of the proposed functions.
The present study identified sterols as a novel requirement for endocytosis in yeast. These results are consistent with the role of cholesterol in endocytosis in animal cells. The genetics of yeast, however, provide a powerful tool to understand the in vivo functions of specific portions of the ergosterol molecule in endocytosis. As shown here, the analysis of certain ergΔ mutants demonstrated that the desaturation state of the sterol B ring is crucial for the ability of sterols to function in the internalization step. In the future, a systematic analysis of other viable ergΔ single and double mutants that accumulate sterols with altered structural features should give a more defined view on how sterols function in endocytosis.
ACKNOWLEDGMENTS
We thank the Riezman laboratory and in particular Dr. M. Isabel Geli, for stimulating discussions throughout the progression of this work and also thank Drs. Janet Lee and Mohan Balasubramanian for many comments and suggestions on the manuscript. We are grateful to Drs. F. Cvrckova and R. Gaber for sending strains and plasmids. We also thank Drs. Günther Daum and Dagmar Zweytick for help with the sterol identification and Dr. Erich Leitner for help with the GLC-MS. This work was funded by the Kanton Baselstadt and by grants to H.R. from the Swiss National Science Foundation and to A.H.-P. from the Human Frontier Science Program (LT-194/98) and the European Molecular Biology Organization (ASTF9273). During the completion of this work, A.L.M. was supported by the National Science and Technology Board of Singapore. H.P. was supported by the Fonds zur Förderung der wissenschaftlichen Forschung in Oesterreich (project 12076).
REFERENCES
- Anderson RG. The caveolae membrane system. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- Arthington BA, Bennett LG, Skatrud PL, Guynn CJ, Barbuch RJ, Ulbright CE, Bard M. Cloning, disruption and sequence of the gene encoding yeast C-5 sterol desaturase. Gene. 1991;102:39–44. doi: 10.1016/0378-1119(91)90535-j. [DOI] [PubMed] [Google Scholar]
- Bard M, Woods RA, Barton DH, Corrie JE, Widdowson DA. Sterol mutants of Saccharomyces cerevisiae: chromatographic analyses. Lipids. 1977;12:645–654. doi: 10.1007/BF02533759. [DOI] [PubMed] [Google Scholar]
- Bottema CDK, McLean-Bowen CA, Parks LW. Role of sterol structure in the thermotropic behavior of plasma membranes in Saccharomyces cerevisiae. Biochim Biophys Acta. 1983;734:235–248. [Google Scholar]
- Brown DA, London E. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- Brown RE. Sphingolipid organization in biomembranes: what physical studies of model membranes reveal. J Cell Sci. 1998;111:1–9. doi: 10.1242/jcs.111.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerneus DP, Ueffing E, Posthuma G, Strous G J, van der Ende A. Detergent insolubility of alkaline phosphatase during biosynthetic transport and endocytosis. Role of cholesterol. J Biol Chem. 1993;268:3150–3155. [PubMed] [Google Scholar]
- Chang WJ, Rothberg KG, Kamen BA, Anderson RG. Lowering the cholesterol content of MA104 cells inhibits receptor-mediated transport of folate. J Cell Biol. 1992;118:63–69. doi: 10.1083/jcb.118.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Seeburg PH. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985;4:165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Deckert M, Ticchioni M, Bernard A. Endocytosis of GPI-anchored proteins in human lymphocytes: role of glycolipid-based domains, actin cytoskeleton, and protein kinases. J Cell Biol. 1996;133:791–799. doi: 10.1083/jcb.133.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC. Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu Rev Biochem. 1998;67:27–48. doi: 10.1146/annurev.biochem.67.1.27. [DOI] [PubMed] [Google Scholar]
- Dulic V, Egerton M, Elguindi I, Raths S, Singer B, Riezman H. Yeast endocytosis assays. Methods Enzymol. 1991;194:697–710. doi: 10.1016/0076-6879(91)94051-d. [DOI] [PubMed] [Google Scholar]
- Fielding CJ, Fielding PE. Intracellular cholesterol transport. J Lipid Res. 1997;38:1503–1521. [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Gaber RF, Copple DM, Kennedy BK, Vidal M, Bard M. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol Cell Biol. 1989;9:3447–3456. doi: 10.1128/mcb.9.8.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli MI, Riezman H. Endocytic internalization in yeast and animal cells: similar and different. J Cell Sci. 1998;111:1031–1037. doi: 10.1242/jcs.111.8.1031. [DOI] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-bp restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Goldstein A, Lampen JO. Beta-d-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- Hailstones D, Sleer LS, Parton RG, Stanley KK. Regulation of caveolin and caveolae by cholesterol in MDCK cells. J Lipid Res. 1998;39:369–379. [PubMed] [Google Scholar]
- Harder T, Simons K. Caveolae, DIGs, and the dynamics of sphingolipid-cholesterol microdomains. Curr Opin Cell Biol. 1997;9:534–542. doi: 10.1016/s0955-0674(97)80030-0. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Pelham HR. SED6 is identical to ERG6, and encodes a putative methyltransferase required for ergosterol synthesis. Yeast. 1994;10:265–269. doi: 10.1002/yea.320100213. [DOI] [PubMed] [Google Scholar]
- Keesler GA, Casey WM, Parks LW. Stimulation by heme of steryl ester synthase and aerobic sterol exclusion in the yeast Saccharomyces cerevisiae. Arch Biochem Biophys. 1992;296:474–481. doi: 10.1016/0003-9861(92)90600-2. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Herman PK, Emr SD. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev. 1990;54:266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Gu F, Gruenberg J. Lipids, lipid domains and lipid-protein interactions in endocytic membrane traffic. Semin Cell Dev Biol. 1998;9:517–526. doi: 10.1006/scdb.1998.0257. [DOI] [PubMed] [Google Scholar]
- Kübler E, Dohlman HG, Lisanti MP. Identification of Triton X-100 insoluble membrane domains in the yeast Saccharomyces cerevisiae. Lipid requirements for targeting of heterotrimeric G-protein subunits. J Biol Chem. 1996;271:32975–32980. doi: 10.1074/jbc.271.51.32975. [DOI] [PubMed] [Google Scholar]
- Kübler E, Riezman H. Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 1993;12:2855–2862. doi: 10.1002/j.1460-2075.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamaze C, Fujimoto LM, Yin HL, Schmid SL. The actin cytoskeleton is required for receptor-mediated endocytosis in mammalian cells. J Biol Chem. 1997;272:20332–20335. doi: 10.1074/jbc.272.33.20332. [DOI] [PubMed] [Google Scholar]
- Lange Y. Disposition of intracellular cholesterol in human fibroblasts. J Lipid Res. 1991;32:329–339. [PubMed] [Google Scholar]
- Leber A, Fischer P, Schneiter R, Kohlwein SD, Daum G. The yeast mic2 mutant is defective in the formation of mannosyl-diinositolphosphorylceramide. FEBS Lett. 1997;411:211–214. doi: 10.1016/s0014-5793(97)00692-3. [DOI] [PubMed] [Google Scholar]
- Leber R, Zinser E, Hrastnik C, Paltauf F, Daum G. Export of steryl esters from lipid particles and release of free sterols in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1995;1234:119–126. doi: 10.1016/0005-2736(94)00270-y. [DOI] [PubMed] [Google Scholar]
- Lees ND, Bard M, Kemple MD, Haak RA, Kleinhans FW. ESR determination of membrane order parameter in yeast sterol mutants. Biochim Biophys Acta. 1979;553:469–475. doi: 10.1016/0005-2736(79)90302-x. [DOI] [PubMed] [Google Scholar]
- Lees ND, Skaggs B, Kirsch DR, Bard M. Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae—a review. Lipids. 1995;30:221–226. doi: 10.1007/BF02537824. [DOI] [PubMed] [Google Scholar]
- Lewis TA, Rodriguez RJ, Parks LW. Relationship between intracellular sterol content and sterol esterification and hydrolysis in Saccharomyces cerevisiae. Biochim Biophys Acta. 1987;921:205–212. doi: 10.1016/0005-2760(87)90020-8. [DOI] [PubMed] [Google Scholar]
- Lorenz RT, Casey WM, Parks LW. Structural discrimination in the sparking function of sterols in the yeast Saccharomyces cerevisiae. J Bacteriol. 1989;171:6169–6173. doi: 10.1128/jb.171.11.6169-6173.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- Munn AL, Riezman H. Endocytosis is required for the growth of vacuolar H(+)-ATPase-defective yeast: identification of six new END genes. J Cell Biol. 1994;127:373–386. doi: 10.1083/jcb.127.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn AL, Stevenson BJ, Geli MI, Riezman H. end5, end6, and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1721–1742. doi: 10.1091/mbc.6.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes WD, Janssen GG, Crumley FG, Kalinowska M, Akihisa T. The structural requirements of sterols for membrane function in Saccharomyces cerevisiae. Arch Biochem Biophys. 1993;300:724–733. doi: 10.1006/abbi.1993.1100. [DOI] [PubMed] [Google Scholar]
- Nes WD, Xu SH, Haddon WF. Evidence for similarities and differences in the biosynthesis of fungal sterols. Steroids. 1989;53:533–558. doi: 10.1016/0039-128x(89)90030-5. [DOI] [PubMed] [Google Scholar]
- Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi PA, Fishman PH. Filipin-dependent inhibition of cholera toxin: evidence for toxin internalization and activation through caveolae-like domains. J Cell Biol. 1998;141:905–915. doi: 10.1083/jcb.141.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks LW, Casey WM. Physiological implications of sterol biosynthesis in yeast. Annu Rev Microbiol. 1995;49:95–116. doi: 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- Parton RG, Joggerst B, Simons K. Regulated internalization of caveolae. J Cell Biol. 1994;127:1199–1215. doi: 10.1083/jcb.127.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GW. Relation between structure and retention time of sterols in gas chromatography. Anal Chem. 1971;43:1165–1170. [Google Scholar]
- Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H. Endocytosis in yeast: several of the yeast secretory mutants are defective in endocytosis. Cell. 1985;40:1001–1009. doi: 10.1016/0092-8674(85)90360-5. [DOI] [PubMed] [Google Scholar]
- Riezman H, Munn A, Geli MI, Hicke L. Actin-, myosin- and ubiquitin-dependent endocytosis. Experientia. 1996;52:1033–1041. doi: 10.1007/BF01952099. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal SK, Skretting G, Garred O, Vilhardt F, van Deurs B, Sandvig K. Extraction of cholesterol with methyl-beta-cyclodextrin perturbs formation of clathrin-coated endocytic vesicles. Mol Biol Cell. 1999;10:961–974. doi: 10.1091/mbc.10.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RJ, Parks LW. Structural and physiological features of sterols necessary to satisfy bulk membrane and sparking requirements in yeast sterol auxotrophs. Arch Biochem Biophys. 1983;225:861–871. doi: 10.1016/0003-9861(83)90099-1. [DOI] [PubMed] [Google Scholar]
- Rothberg KG, Heuser JE, Donzell WC, Ying YS, Glenney JR, Anderson RG. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- Rothberg KG, Ying YS, Kamen BA, Anderson RG. Cholesterol controls the clustering of the glycophospholipid-anchored membrane receptor for 5-methyltetrahydrofolate. J Cell Biol. 1990;111:2931–2938. doi: 10.1083/jcb.111.6.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Hunter CP, Valls LA, Stevens TH. Overproduction-induced mislocalization of a yeast vacuolar protein allows isolation of its structural gene. Proc Natl Acad Sci USA. 1986;83:3248–3252. doi: 10.1073/pnas.83.10.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schnitzer JE, Oh P, Pinney E, Allard J. Filipin-sensitive caveolae-mediated transport in endothelium: reduced transcytosis, scavenger endocytosis, and capillary permeability of select macromolecules. J Cell Biol. 1994;127:1217–1232. doi: 10.1083/jcb.127.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink G, Lawrence C. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1974. [Google Scholar]
- Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Stevens T, Esmon B, Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Subtil A, Gaidarov I, Kobylarz K, Lampson MA, Keen JH, McGraw TE. Acute cholesterol depletion inhibits clathrin-coated pit budding. Proc Natl Acad Sci USA. 1999;96:6775–6780. doi: 10.1073/pnas.96.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomeo ME, Fenner G, Tove SR, Parks LW. Effect of sterol alterations on conjugation in Saccharomyces cerevisiae. Yeast. 1992;8:1015–1024. doi: 10.1002/yea.320081204. [DOI] [PubMed] [Google Scholar]
- Trocha PJ, Sprinson DB. Location and regulation of early enzymes of sterol biosynthesis in yeast. Arch Biochem Biophys. 1976;174:45–51. doi: 10.1016/0003-9861(76)90322-2. [DOI] [PubMed] [Google Scholar]
- van den Hazel HB, Pichler H, do Valle Matta MA, Leitner E, Goffeau A, Daum G. PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J Biol Chem. 1999;274:1934–1941. doi: 10.1074/jbc.274.4.1934. [DOI] [PubMed] [Google Scholar]
- Ward AC. Single-step purification of shuttle vectors from yeast to high frequency back-transformation into E. coli. Nucleic Acids Res. 1990;18:5319. doi: 10.1093/nar/18.17.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welihinda AA, Beavis AD, Trumbly RJ. Mutations in LIS1 (ERG6) gene confer increased sodium and lithium uptake in Saccharomyces cerevisiae. Biochim Biophys Acta. 1994;1193:107–117. doi: 10.1016/0005-2736(94)90339-5. [DOI] [PubMed] [Google Scholar]
- Wendland B, Emr SD, Riezman H. Protein traffic in the yeast endocytic and vacuolar protein sorting pathways. Curr Opin Cell Biol. 1998;10:513–522. doi: 10.1016/s0955-0674(98)80067-7. [DOI] [PubMed] [Google Scholar]
- Xu SH, Norton RA, Crumley FG, Nes WD. Comparison of the chromatographic properties of sterols, select additional steroids and triterpenoids: gravity-flow column liquid chromatography, TLC, gas-liquid chromatography and high-performance liquid chromatography. J Chromatogr. 1988;452:377–398. doi: 10.1016/s0021-9673(01)81462-x. [DOI] [PubMed] [Google Scholar]
- Zinser E, Paltauf F, Daum G. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J Bacteriol. 1993;175:2853–2858. doi: 10.1128/jb.175.10.2853-2858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]