Summary
The mechanism of recombination-activating gene (RAG)-mediated rearrangement exists in all jawed vertebrates, but the organization and structure of immunoglobulin (Ig) genes, as they differ in fish and among fish species, reveal their capability for rapid evolution. In systems where there can exist 100 Ig loci, exon restructuring and sequence changes of the constant regions led to divergence of effector functions. Recombination among these loci created hybrid genes, the strangest of which encode variable (V) regions that function as part of secreted molecules and, as the result of an ancient translocation, are also grafted onto the T-cell receptor. Genomic changes in V-gene structure, created by RAG recombinase acting on germline recombination signal sequences, led variously to the generation of fixed receptor specificities, pseudogene templates for gene conversion, and ultimately to Ig sequences that evolved away from Ig function. The presence of so many Ig loci in fishes raises interesting questions not only as to how their regulation is achieved but also how successive whole-locus duplications are accommodated by a system whose function in other vertebrates is based on clonal antigen receptor expression.
Introduction
Adaptive immunity is based on the unique recombination mechanism that generates the antigen receptor repertoire of lymphocytes. The structure, organization, and expression patterns of the immunoglobulin (Ig) genes are not only well conserved among mammals but remain largely the same among tetrapods (Fig. 1); detailed descriptions and maps of the human and mouse heavy (H)- and light (L)-chain loci are available (1). In this article, we begin with a brief review of the mammalian Ig model before comparing salient features –in organization, expression, and diversity of effector function – with Ig systems from non-mammals, focusing primarily on those in fish, which provide the greatest apparent differences.
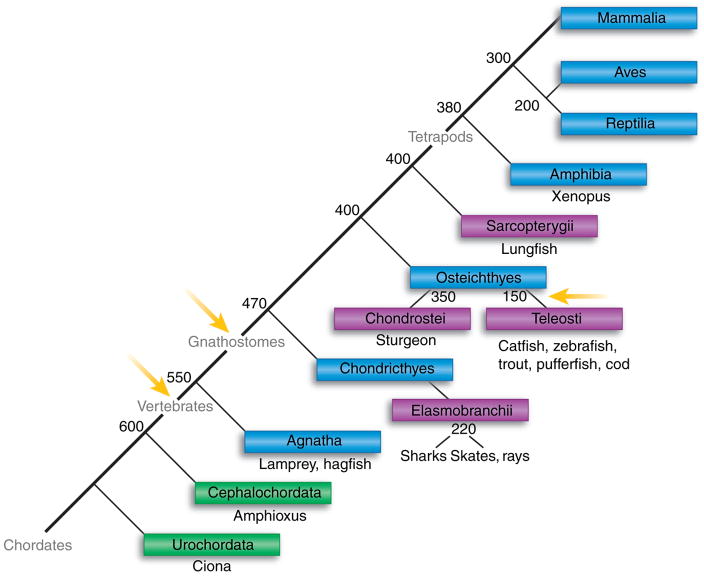
Fig. 1. . Evolution of the chordates.
The phylogenetic relationships among chordates is shown (boxes) with notations of the major animal models in each taxon beneath the boxes. The adaptive immune system, as defined by rearranging antigen receptor genes in the immunoglobulin superfamily and by the major histocompatibility complex, has been found only in the jawed vertebrates (gnathostomes). Blue boxes denote vertebrate classes, green subphyla, and purple lower taxa. Yellow arrows indicate the timing of proposed genome-wide duplications. Numbers denote when the taxa emerged in evolution (millions of years ago).
The emergence of adaptive immunity and rearranging genes followed the period of great genomic re-structuring in ancestral chordates. We examine genomic changes affecting the processes that promote repertoire diversification and enable modifications in effector function in antibodies. The Ig of tetrapod animals are encoded by two to four loci that contain many tandemly duplicated gene segments, whereas the earliest jawed vertebrates, sharks and skates, possess 100–200 Ig loci through whole-locus duplications. The bony fishes, a later diverging branch, underwent yet another whole-genome duplication, and the Ig gene system of each teleost species is different, as if they were snapshot demonstrations of the rapid evolution of Ig genes and organization. Cartilaginous fishes evolved the alternative organization for genes, and such systems took a different evolutionary strategy from those in tetrapods to create unique and variant antigen receptors that can be secreted like Ig. Finally, we introduce novel shark genes encoding ligand-binding sites that are expressed with T-cell receptors (TCRs) as well as with secreted molecules; it is the plasticity of their Ig system that also enabled the evolution of genes bridging B- and T-cell function.
Ig genes in mouse and other tetrapods
Ig in the mouse is encoded by one H-chain locus (IgH) and two L-chain loci (IgL) called κ and λ. A single species of Ig receptor is present on the cell surface of any one B lymphocyte, and it consists of two H chains and two L chains, the result of monoallelic expression of the IgH locus and of the κ or the λ locus (isotype exclusion). Estimations of the Ig-combining site repertoire range from 107 to 1010 different receptors, each one somatically generated by recombination of different gene components in differentiating B lymphocytes.
The variable (V) region at the N-terminus of H- and L-chain polypeptides together form the antigen-combining site. The rest of the H-chain constant (C) region contains effector functions that enable the antibody – antigen complex to be processed or be disposed of. The Ig genes are not functional until the V regions have been successfully assembled, a process that entails the joining together of various gene segments that may be distantly located. This cut-and-paste process involves double-strand DNA breaks and removal of the intervening DNA.
The mouse IgH locus is about three megabases, consisting of >170 V-gene segments spread over two megabases, followed after an interval of 80 kb by 13 diversity (D)-gene segments, followed by four joining (J)-gene segments, and the C-region exons. The C exons following J encode the first C region (μ) that is expressed during the life of a B lymphocyte. There are seven other C region isotypes, and these are expressed as a result of alternative splicing (∂) or through additional recombination events occurring after antigen stimulation (γ3, γ1, γ2b, γ2a, ε, α). Although the Ig cell-surface receptor is a monomer with two combining sites, the secreted antibody can be a pentamer (IgM), dimer (sIgA), or monomer (IgA, IgD, IgE, and IgG).
Expression of the recombinase recombination-activating gene (RAG) signals the first of the pro-B-cell events involving H-chain rearrangement (reviewed in 1–3). By an as yet uncharacterized process of activation, one chromatin domain encompassing the region D downstream to the Cμ exons becomes available to the RAG action. The RAG recognizes recombination signal (RS) sequences flanking the 3′ end of D and the 5′ end of the J-gene segments and promotes double-strand DNA breaks at the border of the RS and the gene segment, while generating hairpins at the D- and J-coding ends. The intervening DNA, bearing the RS ends, is eliminated. Although the recognition and nicking by RAG is precise, the subsequent asymmetric opening of the two hairpinned ends produces overhangs that are variously trimmed back or added to by terminal deoxynucleotidyl transferase (TdT) before ligation to each other by components of the non-homologous end-joining pathway. The result of the RAG action, V(D)J recombination, is not only a rearrangement of a D-gene segment to a J-gene segment but also the generation of a DJ fusion that is highly heterogeneous in the junctional sequence.
The second part of the 2-step recombination process involves the activation of the chromatin domain(s) encompassing the DNA upstream of the DJ fusion. When the V-gene segments become recombinogenic, RAG joins one of the V-gene segments to the fused DJ. Because of the imprecision of the joint formations, there is one chance in three that the VDJ sequence, spliced to Cμ, contains an open-reading frame whose transcript encodes a protein. In the event of a non-functional or non-productive VDJ, the rearrangement process continues on the IgH locus of the homologous chromosome.
With a successful H-chain rearrangement, the B cell moves on to another stage in differentiation and commences L-chain rearrangement. After the B-cell receptor (BCR) is expressed on the cell surface, the B cell exits the bone marrow and enters the circulation. When its receptor has recognized and bound antigen, the activated B cell proliferates and undergoes additional differentiation. The expression of activation-induced cytidine deaminase (AID) leads to DNA lesions in the Ig loci. Within the VDJ and VJ regions, repair of the lesions by various pathways, including error-prone polymerases and mismatch repair, results in single nucleotide changes that can change the coding sequence. This process, somatic hyper-mutation, generates mutant V sequences that are selected for improved binding capacity. Other AID-mediated DNA lesions occur in the highly repetitive region 5′ of the Cμ (switchbox) and that of one of the other CH regions, causing deletion of the intervening DNA that contains Cμ; in this way, the γ or α or ε C genes replace Cμ in its position downstream of the rearranged VDJ. This second process is called H-chain class or isotype switch. The V region previously part of μH chain is now expressed with an H chain of different effector function.
Among tetrapods, V(D)J recombination occurs in the same way; it is the timing and nature of postrearrangement V region diversification that differs. In chicken and rabbit, the primary repertoire is principally generated through gene conversion events on the V region, utilizing templates from upstream pseudo-V genes. These events, including the hypermutation that occurs after activation by antigen, are AID dependent.
The primary difference in Ig between humans and frogs or even humans and mouse is the number and nature of the CH region isotypes (4). For example, Xenopus B cells can switch from IgM to an IgG analog (IgY) that is present only as a single copy; Xenopus do not have an IgE but do express a multimeric IgA equivalent. The Xenopus or chicken IgY and IgA are identifiable by function, and unlike their mammalian counterparts, they lack a hinge. The independent development of the hinge in IgG and in IgA after mammalian radiation illustrates the structural flexibility of the C-region exons in accommodating to functional needs of the species.
The organization of the tetrapod IgH locus is intrinsic to its operation and to its function. The many tandemly duplicated gene segments provide a combinatorial selection process that enhances the junctional diversity. The segregation of chromatin domains encompassing V and D/J/C allows for differential activation of the rearranging components of the locus, and the two-step recombination process contributes to the establishment and maintenance of monoallelic expression of the H-chain gene. The arrangement of tandem C-region exons allows for substitution of one C region for another. All these features contribute to generating an antigen receptor that is highly diversified at the ligand-binding site, clonally expressed, and that, as a secreted antibody, directs the removal of its bound target.
How they got that way
The Ig of sharks and skates is encoded by the multiple cluster organization, wherein an H- or L-chain isotype is encoded by miniloci present in several copies, each containing one V-gene segment and one set of C-region exons (5). In cartilaginous fishes, all the Ig and Ig-related molecules are encoded by such miniloci. In teleost fishes, the IgH is translocon, but the IgL are multiclustered (6). Whether in shark or in human, the V genes rearrange and they hypermutate (reviewed in 7); these phenomena are conserved, although the gene organizations differ. In this review, we compare and contrast the fish and tetrapod Ig, their Ig gene organizations, and in the process speculate as to how they may have evolved.
Only jawed vertebrates carry those components of the adaptive immune system (RAG, rearranging genes) that enable expression of lymphocyte antigen receptors. A recent comprehensive search for immune-related molecules in the genome of the protochordate Ciona intestinalis (Fig. 1) concluded that there was no evidence for Ig, TCR, RAG1/2, major histocompatibility complex (MHC) class I and II, or AID (8), although a sequence that was equally related to TdT and polymerase μ was found (8, 9, and unpublished observations). Ig superfamily (IgSF) V-like sequences have existed since sponges (10), and even V sequence fused to recognizable J sequence (G-strand) existed prior to jawed vertebrates (11). Two kinds of C domains (C2 and I-set) are widespread, but it is only the C1-type that is found in MHC, Ig, TCR, and tapasin (12), and the first C1 domain that can be identified in non-vertebrates is part of a nectin-like sequence in Ciona. This finding suggests that the ancestral structural forms related to vertebrate lymphocyte antigen receptors were present in protochordates.
Some time after the divergence of the jawless fishes, the most primitive vertebrates, there evolved those elements that are essential to the adaptive immune system, which, in terms of genes and pathways, are conserved from sharks to humans. The event believed to have transformed the early vertebrate immune system was the introduction by horizontal transfer of sequences encoding components of a nuclease that evolved into the V(D)J recombinase RAG.
Origin of rearranging genes
The RAG transposon hypothesis
When the RS sequences flanking the 3′ end of the V-gene segments and the 5′ end of the J-gene segment of the λ L-chain locus were discovered, it was clear that these were recognition motifs promoting the joining of the VJ and the deletion of the DNA in between. The RS of the V-gene segment consisted of a heptamer and a nonamer separated by a 12-bp spacer sequence (12-RS), whereas the RS of the J consisted of the reverse complement of the heptamer and nonamer but with a 23-bp spacer (23-RS). Only a 12-RS and 23-RS gene combination rearranges together (12/23 rule). The inverted repeats were reminiscent of the motifs found at the termini of integrated transposable elements. Sakano and coworkers (13) speculated that these shared features of the vertebrate V(D)J recombination system and a prokaryotic insertion system could indicate a common enzymatic pathway or evolutionary origin.
The isolation of the recombinase components (RAG1 and RAG2) and their lack of introns suggested a bacterial or viral origin (14, 15). The current hypothesis is that a DNA transposon, probably encoding RAG, was introduced into the genome of a primitive vertebrate ancestor (13, 16–18). Either this event or a subsequent integration resulted in the splitting of an ancestral V-like gene into two components (Fig. 2). The first evidence for such a scenario was the demonstration of latent transposase activity in RAG (19, 20). Of the excision and transposition phases of transposase action, the latter has been superseded and suppressed by the excision/V(D)J recombinase activity in RAG.
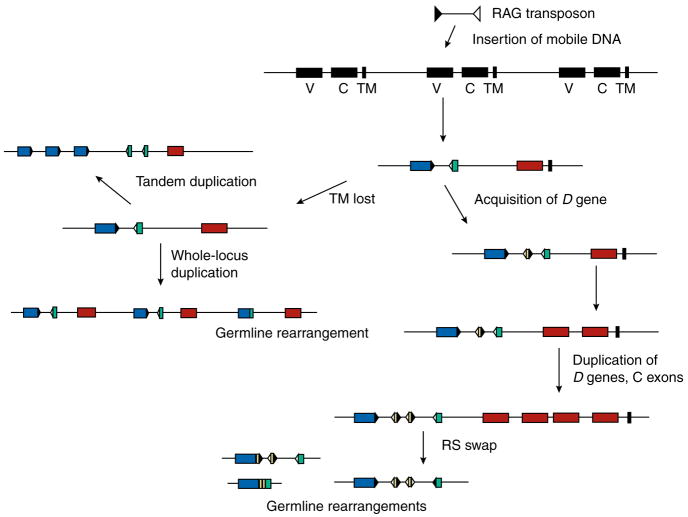
Fig. 2. Hypothetical scheme for the evolution of immunoglobulin gene systems.
In the ancestral jawed vertebrate, a mobile DNA sequence inserted in an archaic gene, splitting it into V (blue) and J (green) segments with recombination signal (RS) sequences at their flanks (black and white triangles representing RS with 12 and 23-bp spacers). We suggested that the ancient gene was part of a family of receptors (TM is transmembrane, black), and the L-chain genes evolved from those that lost the TM. Expansion of the genes could have occurred by tandem duplications of the gene segments, resulting in the translocon organization in tetrapods, or by duplication of the whole locus, which generated the multicluster organization in sharks and skates. The V evolved in structure; D sequences (yellow) were acquired. The number of C region domains (brown) could be increased with tandem gene duplication and divergence. The recombination-activating gene (RAG)-mediated events (germline-rearrangement in VJ; and VD, VDDJ, and RS swap at bottom) are discussed in the text.
Mechanistic parallels have been drawn between RAG and the insect Hermes transposase: a nick on the top (non-transferred) strand initiates, respectively, the first break at the RS for V(D)J recombination and the first step in transposition of the hAT element (21). In both cases, a double-strand break is the result, with a hairpinned intermediate at one end of the break. But, the objectives are complementary: for Hermes, the host DNA is left to repair the hairpins, while the excised intervening DNA with open ends is the target and object for transposition; for RAG, it is resolving the sealed hairpins at either end of the two breaks that unites the formerly separated Ig gene segments, while the excised DNA is discarded.
The RAG1 core, which contains the catalytic activity in combination with RAG2, shows sequence similarities with the Transib superfamily of transposases that are present in nematodes, insects, and sea urchin. Although the action of Transib transposases is not known, analysis of the terminal inverted repeats of the Transib transposon revealed striking identity to the V(D)J RS signals, not only in the heptamer and nonamer-like sequences present but also that these motifs are separated by sequence lengths corresponding to the RS 12/23 spacers (22). Thus, the biochemical action and the recognition motifs of RAG have their parallels in transposon systems and argue strongly for the nature of RAG’s origins.
The proposed origin of RAG by horizontal transfer in an ancestral vertebrate explains the presence of the recombinase genes (RAG1 and RAG2) and the coevolved rearranging Ig and TCR genes in every vertebrate from cartilaginous fishes to mammals and their absence in the earlier jawless vertebrates, hagfish and lamprey, or in protochordates (Fig. 1). The presence of RAG1-like sequences in the sea urchin suggests that mobile elements carrying RAG had entered the genomes of various organisms but that its presence in vertebrates was the result from a chance integration into a context (the V gene) leading to its selection.
Speculations on the ancestral antigen receptor
What was the disrupted V gene selected for? One has to keep in mind that the end result of this breakage and re-joining is sequence diversification at the junction. The nature of the double-strand break initiated by RAG recombinase led to hairpinned DNA ends that, when repaired, generated heterogeneous joints. Although DNA sequence change can be introduced by mutation or gene conversion, sequence length changes are unique. It is length variation that generates the varied topography of the combining site: depressions, protrusions, and deep pockets. The primary action of RAG is to introduce a spectrum of sequence lengths, routinely and in one place – a loop structure at the surface – tolerated in the protein (23). The acquisition of D genes to create two joints in H chain, further enhancing junctional diversity, supports this notion (24).
If sequence diversification was the characteristic selected for, probably it was also essential to the original ancestral V gene, and combined with the consideration that the RAG transposon insertion must initially destroy function, we propose in Fig. 2 that ur-V gene must have been part of a multi-gene system. Going further, we have speculated that hypermutation as a diversification mechanism might have already existed and thus preceded rearrangement (7, 25) and that rearrangement was selected for because it enhanced hypermutation. The ancestral unsplit genes were most probably involved in an immune function, and perhaps these genes were eventually superseded by the more versatile products of the rearranging genes.
Expansion of the rearranging genes
In the 80–100 million year interval between divergence of jawless fish and emergence of Chondrichthyes (Fig. 1), the ancestral jawed vertebrate acquired RAG and RAG-mediated rearranging genes. There are no living representatives from the early jawed fishes (Placoderms) that preceded sharks and skates. With an immunological fait accompli in terms of Ig, TCR, MHC components, and functional pathways already established in cartilaginous fishes, the evolutionary events leading to the present antigen receptor function and gene organization must remain highly speculative.
What has been deduced from comparative genome studies is that before the emergence of jawed vertebrates, there occurred possibly either two whole-genome duplications or one whole-genome duplication plus multiple segmental duplications (26, 27). The extensive duplication events, whatever their nature, occurred before and after divergence of jawless fishes (28, 29) (Fig. 1, arrows). Thus, the incipience and evolution of the rearranging genes probably overlapped in the time of widespread duplications and the genome revisions that followed in the diploidization process. Kasahara (30) has suggested that the increase in gene numbers provided an opportunity for the creation of novel molecules for the adaptive immune system. Possibly, it was in the time of genetic sorting out that cartilaginous fishes diverged, survived, and acquired their alternative Ig-gene organization. Another lineage with the Ig in translocon organization gave rise to all other vertebrates. The relative paucity of gene duplication events thereafter (31) is perhaps reflected in the stability of their Ig-gene organization.
There was an additional fish-specific genome-wide duplication in Teleostei, proposed to have facilitated the burst of speciation among bony fishes (32). Like the conserved tetrapod Ig organization, those of bony fish are also translocon in nature, but those IgH so far analyzed in catfish (33, 34), zebrafish (35), and trout (36) differ from each other and contain species-unique variations in terms of duplicated regions, pseudogenes, and position of V-gene segments, a very complex situation perhaps acquired in the course of genome restructuring. These events may also underlie the unusual heterogeneity in L-chain gene organization among teleosts that is discussed later. Findings of re-organization in other bony fish genes, such as the MHC (37) and homeobox (38) genes, support this idea.
Independent tandem gene duplication events are frequent and are proposed to arise at 0.01 per gene per million years, a rate on the same order as mutation per nucleotide (39). The split V-gene multiplied, and duplications were retained increasing diversity. Duplications of the individual V(D)J-gene segments and of C-region genes led to the translocon organization that is today present in all tetrapods; in all systems so far studied, there is one IgH and one to three IgL loci. At the other extreme, duplication of the entire Ig locus, initially observed in sharks, resulted in the multiple cluster organization (40). The Ig (IgM) and IgSF immune molecules that are secreted in serum (IgW, NAR, and IgM1gj) are all encoded by independent loci in cartilaginous fishes; in the nurse shark, there are 100 such loci per genome (Hsu and Flajnik, unpublished results).
The advantage of the translocon organization is plain to see: multiple tandem duplications of gene segments within one locus ensure that all components remain under the control of one set of regulatory elements. From this standpoint, it is not clear how the multicluster organization avoided the classical gene-dosage problem; that is, in any instance involving a duplication of a full complement of exons, there is the potential for disturbing the intracellular stoichiometric balance. With Ig genes, the requirement must be that the newly duplicated cluster, like the other clusters, is not expressed in every B cell. Thus, the qualification, if there is one, is that whole-Ig-locus duplications can be retained as long as the overall regulatory control of Ig expression is not disturbed. Although there is evidence for the expression of a single H-chain species per B cell in skates (41), the regulation of the multiple Ig loci is largely unknown, and it is not clear how the expression pattern is achieved (see Antigen receptor exclusion).
Because the DNA breakpoints in unequal crossing-over occur at random, duplications may not include the entire gene and its cis-regulatory components. We speculate that complete sets of regulatory elements might exist in only few places, and these exert control over several Ig clusters. Thereafter, the ‘incomplete’ loci are preferentially retained if duplicated. This hypothesis is supported by a study comparing regulatory elements among several multicluster genes. Although the authors felt that perhaps not all the experimental subclones contained intact regulatory regions, they concluded that only two loci contained good and comparable enhancer activity of six salmon IgL loci from two linkage groups (42).
Evolution of V-gene segments
The ligand-binding possibilities of V regions were expanded by increasing the gene and gene segment number. Junctional diversity was further improved upon by the generation of D-gene segments at the IgH and by the involvement of TdT, both of which extended the length variation spectrum. The tandem array of V, D, and J in a translocon organization allowed for a combinatorial selection of gene segments, in addition to the junctional diversity. In contrast, for multiclustered elasmobranch Ig genes, recombination takes place only within a locus; no intercluster recombination has been found. The V regions at the various loci diverged in sequence, but the primary Ig repertoire is essentially created by junctional diversity.
All vertebrate Ig V regions hypermutate and perhaps can undergo gene conversion. We have shown that shark NAR and IgL chains mutate extensively (25, 43–45), so that this diversification mechanism is an important and ancient one. We have found similar patterns of hypermutation (point mutations along with the unique shark tandem mutations of two to five) at two IgH loci and at seven loci of two L-chain isotypes in nurse shark (46). The cis-regulatory elements targeting somatic hypermutation must have existed in the ancestral Ig gene, before H- and L-chain divergence.
Germline-joined V genes
RAG is hypothesized to have the central role in creating rearranged genes (24). We have suggested that RAG further acted on germline substrates with RS sequences, modifying the organization and structure of these genes in evolution. RAG, originally a DNA transposase, would have been expressed in germ cells; it could have continued to be expressed in the germ cells of some vertebrates to generate structures that appear to be rearrangements occurring in the germline.
Some of the clustered loci in cartilaginous fishes carry inheritable rearranged V regions. L-chain loci can contain rearranged VJ, and some H-chain loci partially (VD-D-J and VDD-J) and fully rearranged VDDJ in the germline. The function of these genes is not known; possibly the V region specificity occupies a special niche in the antibody repertoire (45). For example, there are three functional loci encoding one nurse shark L-chain isotype (NS5), one of which is germline-joined and encodes an inframe VJ sequence with a CDR3 of six codons. The normal spectrum of CDR3 from the two rearranging genes is 9–14 codons, and no CDR3 of less than nine codons has been isolated, even from perinatal nurse sharks. This germline-joined V region undoubtedly contributes to forming a combining site with a topology very different from those with somatically recombined V regions.
The question of the relationship of the rearranging and germline-joined Ig genes was intriguing: do the joined VJ in this early vertebrate represent some form of the ancestral V gene before it was able to rearrange (5)? Unexpectedly, we discovered a way of solving this mystery (24). One nurse shark L-chain isotype (NS4) consisted of 60 loci: six are rearranged and the rest are split genes. Because they were very closely related, a phylogenetic tree could be constructed (Fig. 4). All the nurse shark NS4 sequences (Fig. 4, top pink block), germline-rearranged (R) or split (S), clustered together and away from the homologous horn shark type III genes. Because all horn shark type III genes are split, the nurse shark germline-joined genes arose after species divergence and are therefore not ‘ancestral’ genes. Furthermore, the rearranged (R) genes, instead of clustering together, appeared to be more related to split (S) genes, suggesting that not only did the rearranged and split genes share common split ancestors but that VJ-joining in germ cells occurred several times, independently, over millions of years. A molecular clock showed that the divergence of the most closely related pair, R4/S31, occurred 7 million years ago. Thus, the events leading to the germline-joining in R4 occurred any time between then and now, that is, fairly recently.
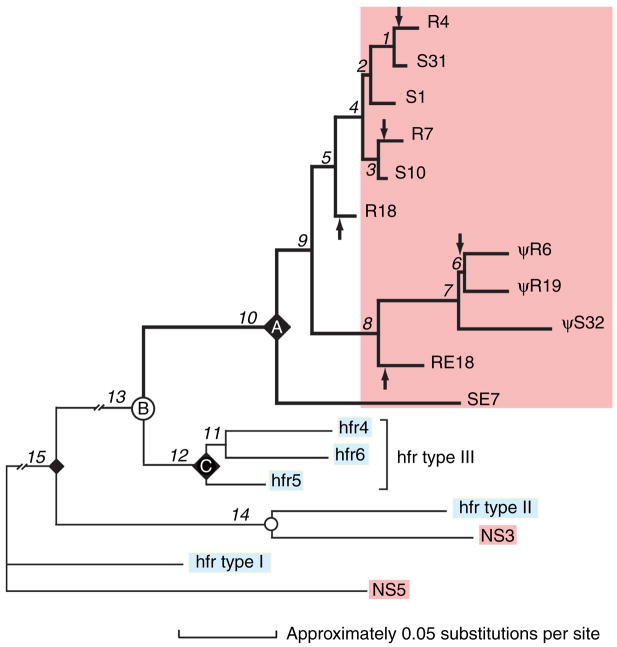
Fig. 4. Phylogenetic tree of nurse shark light (L)-chain loci.
The three L-chain isotypes of nurse shark (pink) and of horn shark homologs (blue) were analyzed as to their phylogenetic relationship. The various L-chain isotypes grouped together as NS4/type III, NS3/type II, and NS5/type I, suggesting respective orthology in the two species. The gene expansion and diversification of NS4 genes (pink block) occurred independently of the horn shark type III homolog (nodes A and C representing common ancestor of respective genes, node B representing speciation event). Three clades (clade I R4, S31, S1, R7, S10, R18; clade II R6, R19, S32, RE18; clade III SE7) contain mixed germline-rearranged NS4 (R) and non-rearranged or split NS4 (S) genes. Assuming that L-chain genes joined in the germline cannot revert to split genes, it is estimated there are at least five independent rearrangement events in the evolutionary lineage. Arrows designate branches in which germline rearrangements in NS4 gene occurred. Because their closest relative is almost always a split gene, it is most parsimonious to infer that the ancestor at node A must have been split. Reproduced from The Journal of Experimental Medicine, 2000;191:1637–147, by copyright permission of The Rockefeller University Press.
Because all the germline-coding flanks were very uniform, it was possible to compare the germline-joined VJ with somatically derived rearrangements from lymphocyte cDNA. The fine-structure analyses of their joints are presented in Table 1. In two genes (R7 and R4), cleavage apparently occurred at the RS to generate hairpins, and processing the hairpins resulted in a telling P region at the V flank and trimming at the J flank. This precisely describes steps in V(D)J recombination and suggests that the germline-joined Ig genes are formed by the same process that takes place in lymphocytes.
Table 1.
Derivation of joints in shark light (L) chain germline-rearranged genes
| Clone
|
V-gene segment
|
N
|
J-gene segment
|
|---|---|---|---|
| R7 | |||
| S10 | TGTCAGAGTGCATATTACAGCTCCTCCGA | GTTTGCGTTC | |
| R7 | TGTCAGAGTGCATATTACAGCTCCTCCGATC | TTGCGTTC | |
| R4 | |||
| S1 | TGTCAGGGTGCATATGGCAGCTACTCCGA | GTTTGCGTTC | |
| S31 | TGTCTGGGTGCATATAGCAGCTACTCCGA | GTTTGCGTTC | |
| R4 | TGTCTGGGTGCAGATGGCAGCTACCCCGATC | TTGCGTTC | |
| R6, R19, R18, RE18 | |||
| SE32 | TGTCAGAGTGCATATGGCAGCTACTCCGA | GTTTGCGTTC | |
| S11 | TGTCAGCAAAGTAGGACCTACCCTTGG | GTTTACGTTC | |
| *S32 | TGTCAGCAAGGTTCCAGCTTCCCCTA | ACATACGTTC | |
| RE18 | TGTCAGCAGTATAGCAGCTTCCCGTA | TGCGTTC | |
| *R6 | TGTCAGCAAGGTCACAGTTCCCCTT | TACGTTC | |
| *R19 | TGTCAGCAAGGTCACAGCTTCTCTT | ACGTTC | |
| R18 | TGTCAGCAGTATAACAGCTCCCCCTA | TGCGTTC | |
| CDNA | |||
| Clade I | |||
| S1 | TGTCAGGGTGCATATGGCAGCTACTCCGA | GTTAGCGTTC | |
| S10 | TGTCAGAGTGCATATTACAGCTCCTCCGA | GTTTGCGTTC | |
| S37 | TGTCTGGGTGCATATAGCAGCTACTCCGA | GTTTGCGTTC | |
| S31 | TGTCTGGGTGCATATAGCAGCTACTCCGA | GTTTGCGTTC | |
| SE32 | TGTCAGAGTGCATATGGCAGCTACTCCGA | GTTTGCGTTC | |
| *S5 | TGTCAACAAGGTTACAGCTGGTCTAA | GCATACGTTC | |
| cDNA6 | TGTCAGAGTGCATATTACAGCTCCTCCGATCGG | CCTAG | GTTC |
| cDNA14 | TGTCAGAGTGCATATTACATGTCCTCCGATC | CAAAA | GCGTTC |
| cDNA18 | TGTCAGAATACATATAGCAGCAGGTCCGATC | A | GTTTACGTTC |
| cDNA19 | TGTCAGAATGCATATAGCAGCTACTCCGATCGG | TC | TTC |
| cDNA20 | TGTCAGAGTGCATATGGCAGCTACTCCG | GA | GCGTTC |
| cDNA21 | TGTCTGGGTGCATATAGCAGCTCCTC | AGG | GTTTGCGTTC |
| cDNA23 | TGTCAGAGTGCATATTACAGCTCCTCCGATC | ATAG | GTTC |
| cDNA24 | TGTCAGAGTGCATATGGCAGCTACTCCGA | C | TTTGCGTTC |
| cDNA25 | TGTCAGAATAGATATGGCAGCTACCCCG | G | TGCGTTC |
| cDNA26 | TGTCAGGGTGCAGATAGCAGCTACTCC | CTCC | TTGCGTTC |
| cDNA27 | TGTCAGGGTGCATATGGCAGCTACTCCG | CC | TTAGCGTTC |
| cDNA28 | TGTCAGAGTGCATATGGCAGCTACTCCG | G | AGCGTTC |
| cDNA30 | TGTCAGAGTGCATATGGCAGCTACTCCG | GG | GCGTTC |
| cDNA31 | TGTCTGGGTGCATATAGCAGCTACTCC | CCG | GCGTTC |
| cDNA32 | TGTCAGAGTGCATATGGCAGGTACTC | GGG | TGCGTTC |
| Clade II | |||
| *S32 | TGTCAGCAAGGTTCCAGCTTCCCCTA | ACATACGTTC | |
| Clade III | |||
| S3 | TGTCAGCAAAGTAGGACCTACCCTGGG | GTTTACGTTC | |
| S11 | TGTCAGCAAAGTAGGACCTACCCTTGG | GTTTACGTTC | |
| *S15 | TGTCAGCAAAGTGAGAGCTTCCCTCGA | GTTTACGTTC | |
| SE7 | TGTCAGCAAAGTGAGAGCTTGCCTTGG | GCTTACGTTC | |
| cDNA22 | TGTCAGCAGTATAGCAGCTACCC | CA | TTACGTTC |
The junctions of the germline-joined (R) NS4 genes (highlighted) are compared with the V and J flanks from the most closely related split genes (S), here shown without the RS sequences. Assuming that the split ancestor of R7 (top) resembled S10, the R7 joint could have been produced by cleavage at the RS and formation of hairpins at the V and J flanks, followed by nicking and processing of the hairpins to generate the P region (underlined TC) at the V flank and trimming of the J flank by two nucleotides before re-ligation. R4 also carries a possible P region, indicating a hairpinned intermediate. The other R genes show only trimming at both flanks. cDNA junctions (as labeled) are similar in occasionally carrying P regions (underlined); the main difference is that N region is present in almost all NS4 somatic rearrangements. Asterisk indicates pseudogene.
We concluded that recombination events in germ cells involving Ig/RS sequences are mediated by RAG. Both RAG1 and RAG2 mRNA are found in large quantities in zebrafish oocytes, at levels comparable with the lymphopoietic kidney, although it is not yet known whether the protein is present (Changchien and Hsu, unpublished results). Germline-joined genes have not so far been observed in zebrafish, but an in-frame VDJ exists within the IgH locus of another teleost fish, the catfish (47). Because the same NS4 and NS5 germline-joined genes have been found in several unrelated nurse sharks that had demonstrated polymorphism for MHC class I genes, we concluded that these germline rearrangement events are rare but that once established in the population the genes became widespread (24, 48, 49) (Figs 3 and 4, Table 1).
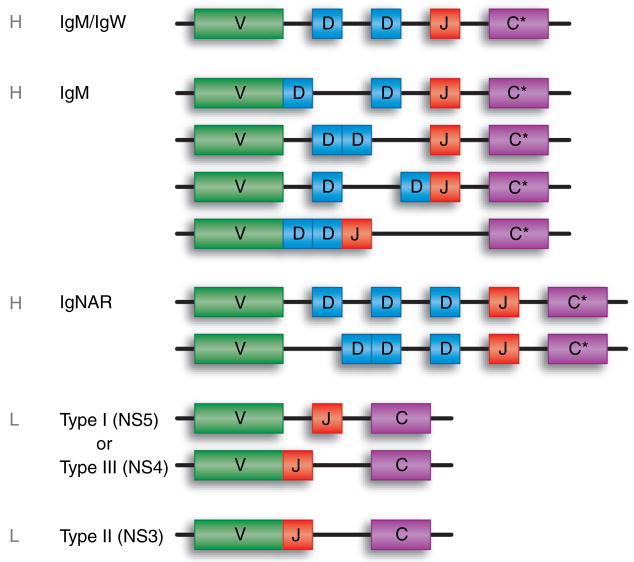
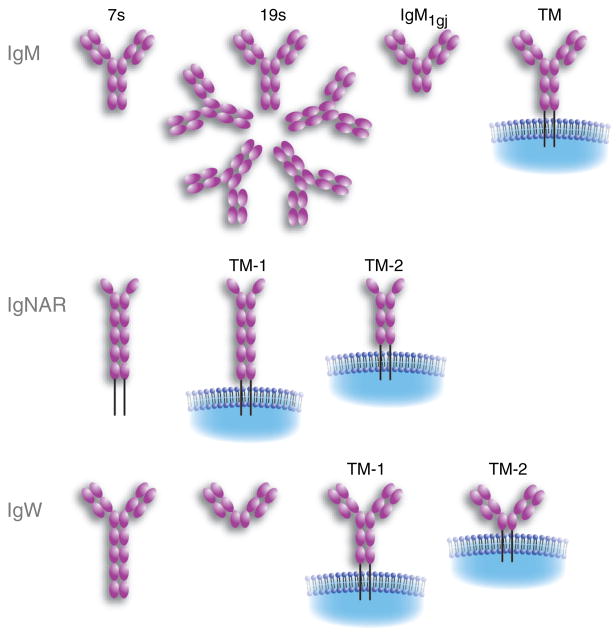
Fig. 3. Cartilaginous fish immunoglobulin (Ig) gene organization.
Contiguous boxes making contact indicate a germline-joined form of the gene. V, variable region exon; D, diversity segment; J, joining segment; C, constant gene or exon (C* signifies that multiple exons encode the C regions of the H-chain isotypes) found only in jawed vertebrates.
De novo creation of gene segments: VD
In chickens, there is a single functional V-gene segment available for rearrangement at both IgH and IgL loci (50). After Hand L-chain genes recombine productively, a gene conversion mechanism is initiated to diversify the rearrangement, utilizing one or more of the many upstream V pseudogenes as templates. At the IgH locus, these pseudo-Vs do not have RS at the 3′ flank, but rather in place of the RS are stretches that resemble D or D and J sequences. Thus, the template V genes appear to be fused VD and perhaps VDJ. In an avian ancestor, there was an initial segmental duplication including V and D or V-, D-, and J-gene segments. Germline-joining of some of these elements occurred at some point, followed by expansion of the VD/VDJ as the chicken system channeled its diversification pathway into a gene conversion mechanism.
Other genomic changes by RAG
Certain configurations of gene segments and RS sequences have been observed variously in different vertebrate genomes. Fig. 2 shows some observed and hypothesized Ig (and TCR) reorganizations based on RS recognition. Aside from the partial (VD, VDD, etc.) and complete rearrangements of V-gene segments, another kind of RAG-mediated change has been observed in shark H-chain multiclusters. Of four IgH germline genes isolated from horn shark (51), three carried the following RS arrangement, VH(23-RS) - (12-RS)D1(23-RS) - (12-RS)D2(12-RS) - (23-RS)JH, whereas the fourth one was VH(23-RS) - (12-RS)D1(12-RS) - (23-RS)D2(12-RS) - (23-RS)JH. In other words, it looked as if the 12/23 RS pairs between D1 and D2 were ‘swapped’. Susanna Lewis (52) suggested that this strongly resembled ‘hybrid joint’ formations mediated by RAG, where the DNA between the 12-RS and 23-RS is not eliminated but re-inserted, either in the orientation as before (open and shut) or inverted so that the re-ligated RS sequences are joined to different coding flanks. The coding flank that originally possessed a 12-RS now is linked to the 23-RS and vice versa. There did appear, in fact, to have been an inversion event, as borne out by comparing the intervening sequence between D1 and D2 among the four clones (51).
This potential for separating coding flank (V or J) from its RS can explain certain observed RS associations that are not compatible with the evolutionary relationships among the coding sequences (52). For instance, the κ and λ L-chain V-gene segments are more closely related to each other than either one is to the H-chain V-gene segments, but the Vκ flanks carry 12-RS (and Jκ 23-RS), whereas the Vλ and VH have 23-RS and the Jλ 12-RS. Because the RS sequences are simply extensions of the flanks, the switching around of RS motifs is otherwise difficult to explain with respect to the closer relationship between L-chain genes.
In summary, although V sequences have variously duplicated and mutated during vertebrate evolution, their structure and gene organization are largely conserved. There are many instances, however, where the V genes (that is, sequences flanked by RS) appear to have been manipulated by RAG in the germline. How might such sequences evolve? We speculate that a germline VDJ gene might evolve away from conventional antibody function, because germline-joining makes a gene with a preassembled V region functionally independent of RAG (23, 24). This independence means that expression does not have to be linked to and limited to the programming of B cells. Perhaps the germline-rearranged IgM1gj gene (48), described below, is such an example. IgM1gj encodes a polypeptide that interacts with L chain, but it does not seem to have transmembrane exons and is only secreted. This Ig-like molecule is not a cell-surface receptor, although it retains an antibody-combining site. Lacking the means for signal transduction and being of invariant sequence, IgM1gj is no longer an Ig.
Evolution of C regions
We speculate that the ancestral V gene may have encoded a cell-surface receptor (Fig. 2, top, drawn with membrane exons) like many other immune-related genes. Before or perhaps after it acquired the ability to rearrange, it functioned as a dimer. After segmental (or whole-genome) duplications, events that would have occurred before the appearance of Chondrichthyes, the divergence of rearranging loci into H- and L-chain genes perhaps began with the loss of the membrane-spanning sequence in the latter. Because L-chain molecules by themselves have no signal transduction property, they coupled with molecules that did; the H-chain genes could have been generated by duplication events that expanded it to four C region domains.
All vertebrates possess IgM, the isotype expressed earliest in development, and at least one other kind of Ig or Ig-like H-chain isotype. In the IgH translocon situation, tandemly duplicated C regions diverged by similar tactics. Extensive modification or deletion of exons also occurred. An example of the former is the generation of the hinge exon, a truncated sequence created through the mutation and shifting of the splice sites; in the murine γ2b C-region gene, the remains of a severed exon are discernible in the intervening sequence (53). Where the hinge was identified as the 5′ extension of the CH exon, as in the human or mouse-α genes, it was hypothesized that the shifting of the 5′ acceptor splice site into the upstream pyrimidine-rich intervening sequence created a proline-rich sequence that increased the flexibility of the antibody arms (54). The formation of the α C region involved the concomitant deletion of the former CH2 exon, although elimination of a C-region exon can occur without obvious compensations elsewhere. At the RNA level, alternative splicing plays a similar role in truncating the H chain (5.7S and 7.8S Ig in birds and turtles), creating smaller Ig molecules with probably a subset of effector functions.
Tetrapods have an Ig class that is primarily involved in mucosal immunity: IgA in endotherms and IgX in Xenopus. The H chain of the single ‘IgY’ class in amphibian, birds, and reptiles is related by sequence to both ε and γ (55), although as far as known, it functions as an IgG analog. The expansion of IgG subclasses is a mammalian invention. The IgH in bony fish is in translocon organization, and until 1997, bony fish were believed to have only IgM, found as a tetramer. The discovery of an IgD homolog in this group was entirely unexpected (discussed below) (34). Thus, tetrapods possess IgM, IgA, and IgG equivalents and share with bony fish, by function or by sequence homology, IgM and IgD (56).
However, a third novel bony fish isotype called IgZ in zebrafish (35) and IgT in trout (36) was uncovered in screens of the EST and genomic databases. Its genomic organization parallels the TCR α-δ locus in that the IgZ/T D, J, and C elements are found between the VH and Cμ/Cδ exons. IgZ/T is a 5-domain H chain predicted to associate with L chains. The authors propose that lymphocytes bearing IgZ may be B1-cell equivalents, but a preliminary VH repertoire analysis does not suggest that unique sets of V regions are expressed on IgZ compared with IgM. IgT is not preferentially expressed over IgM early in trout development, and there is high expression in the spleen throughout ontogeny into adulthood. The effector function of IgZ/T is not known, but there may be species-specific differences in expression during ontogeny and in adult life among bony fishes.
The nurse shark possesses four H-chain Ig isotypes, brought about by the divergence of the C sequences among the many loci into IgM, IgW, IgM1gj, and NAR H-chain sequences (Fig. 5). Not only does the sequence but also the number of C-region exons (3–6) differ considerably among the H-chain isotypes. In addition, for IgW loci, alternative splicing generates different forms using the same set of C-region genes. The plethora of Ig loci in shark is very much worthwhile investigating in detail, if only as an example of evolutionary experimentation on duplicated immune function genes. Among the criteria (57) postulated to contribute to determine whether a duplicated gene is retained in the genome-subfunctionalization, dominant-negative phenotypes, dosage effects, and partitioning of function, none precisely describes the relationship among the shark H chain isotypes. The multiplicity of IgM loci might be described as subfunctionalization, in the sense that one cluster by itself, even hypermutating extensively, does not produce a large-enough repertoire sufficient for the animal’s protection. It also seems that many varied IgM loci are not sufficient by themselves. The duplicated loci with rearranging genes allowed for expansion of effector function, but the roles and relationships of IgM to IgW or to any of the others have yet to be elucidated. What is known of these unique H-chain isotypes is described below, as are the gaps in our knowledge of IgM.
Fig. 5. Cartilaginous fish immunoglobulin heavy chain (IgH) isotypes in all of their described forms.
Refer to the text for descriptions of each isotype (76, 97). TM, transmembrane form.
Organization and expression of Ig and IgSF genes in multiple clusters
The cluster organization was originally discovered in the horn shark (40), where the IgM H-chain genes were estimated to be present at about 100–200 copies. Our model, the nurse shark, has a genome size less than half of the horn shark, and we have defined the Ig and Ig-related loci more precisely: there are about 15 IgM loci, 4–7 IgW loci, one IgM1gj locus, four NAR loci, and 70 IgL loci consisting of three isotypes, six NS3, about 60 NS4, and four NS5 (45).
IgM
In horn shark, nurse shark, little skate, and ratfish (Fig. 1), an IgM H-chain minilocus is arranged as one V, two D, and one J-gene segments encoding the V region, that when productively recombined and transcribed, are spliced to four Cμ exons; the fourth exon also carries the tailpiece for the secreted form. The cell-surface receptor, as in mammals, is generated by alternative splicing to two transmembrane exons located downstream of Cμ4 (5) (Fig. 5).
Because recombination of the V-gene segments only takes place within its own cluster, the cluster arrangement lacks the additional diversity derived from the combinatorial mixing of V(D)J elements in a translocon organization. This, however, is offset by more diversification opportunities at the junctions (51). First, there are two D segments. The configuration of their RS signals is such that not only can joining occur, V to D1 to D2 to J, but combinations including V/D2/J, V/inverted D2/J, V/D1/inverted D2/J, and V/inverted D2 + D1/J are possible, some of which have been found (46). N-region addition occurs at every junction.
The heterogeneity of the V-gene segments themselves are limited, by traditional definition, to being all members of one VH family, sharing 70% or more identity at the nucleotide level. However, in the course of characterizing cDNA sequences from the neonatal nurse shark, we found that, like in the sandbar shark (58), the V regions were distinct and could be classified into five groups that differed by 74–80% identity (59). Also a phylogram including all known VH sequences from IgM of sandbar shark and horn shark showed that the VH clustered within the species. It is probable that, after species divergence, selection operated to cull and expand Ig, a process that involved entire loci in the course of duplications and eliminations.
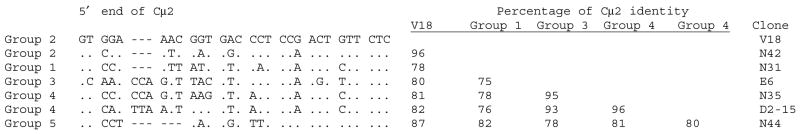
As expected, the group classifications can be extended to the C regions to which the VH are linked (46). The Cμ2 and Cμ3 domains are the most variable in IgM evolution, and in the case of the nurse shark Cμ2, the five groups are also distinguished from each other by insertion/deletion at the 5′ end of the Cμ2 exon (Fig. 6). This is reminiscent of the kind of changes that occurred at the 5′ region of the α exon, which generated the hinge. The volatility of the CH2 exon is not only manifested in its transformation into a hinge region in mammalian IgG but also its disappearance from the shark IgM1gj, as discussed in the next section.
Fig. 6. Comparison of the 5′ end of the second domain in nurse shark μ sequences.
Various cDNA sequences classified as groups 1–5 are compared with reference Cμ 2 from a genomic phage clone. They differ from V18 by 13–22% over 212 bp. Dots indicate identity, dashes gaps.
IgM is present in all jawed vertebrates and has been assumed to be the primordial Ig isotype. It is also the isotype expressed earliest in development in all tetrapods; until recently, it was believed to be the case in fish as well, but this view has changed recently (see later). It has been known for a long time that in all elasmobranchs, IgM is present at very high amounts in the plasma and that it is found in two forms: multimeric (19S) and monomeric (7S) (60, 61). It is unlikely that the two forms are encoded by different gene clusters: (i) peptide maps are identical (Flajnik, unpublished); (ii) early work by Clem (62) found the sequences of the cysteine-containing tail of 19S and 7S H chains to be identical; and (iii) all identified germline VH families are represented for the 19S form (59, Hsu, unpublished data). Although most studies (but not all) reported that 19S and 7S are not differentially regulated during an immune response, in our hands and in one previous study, the 19S response wanes over time, and a stable 7S titer is maintained for periods of up to 2 years after immunization (63, 64). In addition, antigen-specific 7S antibodies observed late in the response have a higher binding strength than those found early, suggesting a maturation of the response, also generally at odds with the previous literature. Finally, when specific antibody titers were allowed to drop, a memory response was observed that was exclusively of the 7S class. This work has shown that a ‘switch’ indeed occurs in the course of an immune response; whether the switch is due to an induction of the 19S-producing cells to become 7S producers or whether there are lineages of 19S-and 7S-producing B cells is an open question. Our working hypothesis is that expression of J chain is important for regulating whether a B cell makes 19S or 7S Ig, but of course that could be at the lineage level or the switch level (64).
IgM1gj
While examining the expression of Ig during ontogeny, we discovered an isotype related to IgM that diverged from the typical IgM class over 100 million years ago, based on phylogenetic analyses (48). Like other four-domain H-chain isotypes in mammals, as mentioned above, there has been a deletion of the CH2 domain, resulting in a four-domain molecule. Like all other cartilaginous fish Ig genes, the IgM1gj H chain is encoded in a single cluster. The V domain is encoded by a complete germline-joined gene, apparently rare among functional H chains in nurse sharks (Hsu, unpublished observations). The CDR3 is very short, and a putative D segment was identified based upon horned shark Ig germline sequences in the databases. The IgM1gj H-chain associates covalently with an L chain encoded from the NS5 germline-joined cluster (probably not exclusively), in which the V region is also encoded by a germline-joined gene with a short CDR3 (45, Flajnik et al., unpublished observations). Because the canonical cysteine in the CH1 domain that bonds to L chains is not present in the IgM1gj H chain, we believe that the H–L association will be unusual, and we hope to resolve this association with a crystallographic analysis. Both the H and L chains (as well as the germline-joined NS4 L chain described earlier) are expressed in relatively high amounts early in ontogeny and are almost entirely supplanted by expression of split genes at approximately 5 months after birth. We have not detected a transmembrane (TM) form of this H chain, and preliminary analysis with a monoclonal antibody specific for IgM1gj shows it to be highly expressed throughout the red pulp of young animals. In adults, we detect a few single secretory cells scattered over the red pulp (Haines and Flajnik, unpublished observations). Expression of this isotype, like all of the germline-joined genes found, is perpetuated at relatively high levels in the epigonal organ, a bone marrow equivalent in the elasmobranchs (48, 65). Such expression is mirrored for the zebrafish IgZ H chain, which is the first Ig class to be expressed and is preferentially expressed in primary lymphoid tissues (kidneys) in adult life (35).
IgNAR
While making monoclonal antibodies specific for nurse shark IgM, we detected a novel secreted molecule composed of two covalently associated H chains, each having six domains, with no associated L chains (43). We named this isotype new antigen receptor (NAR), and subsequently renamed it IgNAR, because functionally it shares more features with Ig isotypes than with TCRs. The V-gene sequence of this isotype is no more closely related to VH than it is to VL or TCRV, suggesting that it is either related to the ancestral antigen receptor V or evolved and diverged to such a degree that its true phylogenetic affiliation can no longer be perceived (66). The C domains are most similar to another isotype, IgW (66–68), and we hypothesize that an IgNAR VDJ cluster recombined with an IgW cluster early in cartilaginous fish history to create this new isotype (see later). The free and flexible V domains, which are tethered to the C domains via a hinge-like region, bind to antigens as single domains (66, 69). The V is encoded by a paucicopy gene family, of which only two genes are expressed in adult life. These type I and type II genes are very similar in sequence and are distinguished primarily by a different number of non-canonical cysteine residues that regulate the formation of very different tertiary structures (see below). Each IgNAR V cluster contains three D segments, which are almost always included in the CDR3, implicating four rearrangement events (order of rearrangement not determined). The rearrangements and accompanying N-region additions result in long and heterogeneous CDR3, but the constraints imposed by the non-canonical disulfide bonds result in longer CDR3 in type I (mean of 16) than type II (mean of 10) (69).
In type I IgNAR V, two cysteines are almost always found in CDR3, usually encoded by preferred reading frames of the D segments (66). These cysteines bond to non-canonical cysteines in FR2 and FR4, presumably to stabilize the non-associated V domain (69). Type I CDR3 is thus ‘pulled down’ to the body of the IgNAR V domain. In type II IgNAR V, there is only one cysteine in CDR3 that bonds to a non-canonical cysteine in CDR1. In this case, the CDR3 is ‘raised up’ and makes intimate interactions with CDR1 (70). The very different structures of type I and type II IgNAR serve to explain the differential selection on CDR1 and CDR2, as revealed via somatic hypermutation (71).
Because we were able to identify all of the IgNAR V germ-line genes, we could study somatic hypermutation with unambiguous template genes. The mutations do not generate the primary repertoire, as we originally proposed, because they are detected in secreted but not TM forms of IgNAR (72). Thus, the data suggest that the mutations occur after antigenic stimulation. We were initially surprised at the very high levels of mutation in this gene, and we proposed that IgNAR was responsible for secondary immune responses and IgM for a ‘first line of defense’ (43). However, with the discoveries of high mutation levels in L-chain genes (25, 45) and the increased binding strength of 7S IgM over the course of an immunization (64), our original hypothesis was clearly naïve (see below). Nevertheless, the high mutation levels in IgNAR suggest, contrary to earlier studies humoral immunity in sharks, that antibodies are capable of augmenting their affinities.
One of the four IgNAR clusters, called type III, is expressed early in development, like IgM1gj (71). This NAR cluster contains only two D regions (apparently D1 and D2 are fused). It is interesting that a cluster that is only partially germline-joined nevertheless has a similar expression pattern (preferential, early) as those genes that are entirely preassembled. The CDR3 of this IgNAR type is remarkably constrained in its size, despite undergoing three rearrangement events, suggesting that it is under strong selection for binding to a particular epitope.
IgW
All elasmobranchs studied to date have another isotype called IgW. It was probably discovered long ago in skates as a non-IgM-secreted isotype called IgR, but no protein sequence of this molecule has been obtained for confirmation (73). Subsequently, an Ig gene was discovered in skates encoding a three-domain molecule with an unusual secretory tail that was named IgX (which was unfortunate, considering there was already another isotype with that name in amphibians) (74). A high-molecular weight species detected using Northern blotting with an IgW probe suggested that there might be a longer form of this isotype, subsequently shown to be true in the sandbar shark (IgW) (75), nurse shark (IgNAR C – it was so-named because the C domains had highest similarity to IgNAR C domains) (67), and skate (68). It was originally believed that sharks only expressed the long (seven-domain) IgW form, but they were later shown to have both secretory forms (76). The reason for the discrepancy was shark-to-shark variation in expression of the short form, for unknown reasons. The major IgW TM form, such as IgM, is composed of five domains, but variants with three domains, like the secretory forms, were also detected (76). Very little is known about the function of this isotype, as IgW-specific monoclonal antibodies have not been generated as they have for IgM and IgNAR.
IgW was thought to be a dead-end isotype in the cartilaginous fish, but a homolog was found recently in the lungfish (77). It also is present in two secreted forms, one with eight domains and the other, like in the elasmobranchs, with three domains (unfortunately, the secretory tail was not sequenced and the TM form was not studied). Recently, we found an Ig isotype in Xenopus tropicalis that is most related to the lungfish IgW. Computer searches of databases for the X. tropicalis genome project uncovered a new isotype flanked by the IgM and IgX genes at the IgH locus (Ohta and Flajnik, manuscript in preparation). The deduced amino acid sequence obtained from the exons on the genomic scaffold suggests a nine-domain molecule. The N-terminal C domains and the TM regions are most similar to mouse and human IgD regions, and its genomic location also suggests that it is an IgD equivalent. Thus, these new data reveal that like IgM, IgW/D is an isotype that was present at the emergence of all extant vertebrate taxa.
An interesting feature of this IgD/W locus is that it is highly plastic in evolution, both in terms of the number of domains in different fish species (76–78) and the plethora of splice variants found, at least in cartilaginous fish (76). In cartilaginous fish, two of the C domains were derived approximately 250 million years ago by a tandem duplication event (67), and there was a Xenopus-specific, two-domain tandem duplication event as well. Within teleost fish, the number of C exons for this isotype is different in various species (78), and the secreted and TM forms seem to be encoded by different loci in the catfish (34). In addition to the splice variants described in the cartilaginous fish and lungfish above, in teleosts the IgM C1 domain exon is spliced into the IgD transcript (56). Even in mammals, there are different numbers of C domains in different species and even exons that have emerged quite recently in evolution (79). It is our impression that this is the Ig locus that evolution ‘plays with’, perhaps using it for different functions in vertebrate taxa. We are obviously at the beginning of our analysis of this system, but perhaps an evolutionary perspective of IgD/W will eventually lead to an understanding of the function of this slippery isotype.
NAR-TCR
The four types of TCR were described in cartilaginous fish, initially in skates, by Litman and colleagues (80), disproving the hypothesis that T cells had not emerged at this phylogenetic level (81). We decided to study the genetics and expression of elasmobranch TCR genes in more detail in the nurse shark. While analyzing the TCR Vδ repertoire, we uncovered an entirely new form of this chain, which encodes three domains, V-V-C (82). The C is encoded by the single-copy Cδ gene, and the membrane-proximal V is encoded by a Vδ gene that rearranges to the DJδ elements. The membrane distal V domain is encoded by a gene in the NAR family, found in a rearranging VDJ cluster typical of all cartilaginous fish Ig clusters. The NAR-TCR V genes, unlike IgNAR V genes, only have a single D region in each cluster. The particular Vδ loci linked to each NAR-TCR gene – called NAR-TCR-supporting Vδ – encode a cysteine in CDR1 that probably makes a disulfide bridge with the NAR-TCR V domain. The J segment of the rearranged NAR-TCR V-gene splices at the RNA level directly to the supporting Vδ segment, which has lost its leader exon.
This organization probably arose from an IgNAR V cluster that translocated to the TCRαδ load locus upstream of a Vδ-gene segment. After modifications of the supporting Vδ genes, this entire V-V gene set duplicated and diverged several times in different species of sharks. We estimate that about 25% of the expressed nurse shark TCRαδ repertoire is composed of this new type of TCR (encoded by 15–20 V-V genes in this species) and have proposed that the typical γ/δ TCR acts as a scaffold upon which sits the single-chain NAR V, like old King Midas on his throne.
Our interpretation is that true to the proposal that γ/δ TCRs interact with free antigen (83), the NAR V is providing a binding site that can interact with antigen in a different way than conventional heterodimeric Vs. Thus, this is the first case in which a particular V region family has been shown to be associated with a BCR and TCR. In the case of the BCR, the function probably resides within the Fc portion of IgNAR, and for the TCR, the function (cytokine secretion, killing) lies within the T cell itself.
Ontogeny
Ig and TCR gene expression early in development have been studied in two species to date, the skate (84) and the nurse shark (24, 25, 45, 48, 59, 65, 71). In both species, antigen receptor CDR3 junctions are diversified with N-region additions early in ontogeny, unlike what has been found in several other vertebrates. Nevertheless, the junctions are generally smaller than those of adults (46, 59). The 19S form of IgM is almost exclusively found (at about equal levels with IgM1gj, both at low levels) at birth, with 7S reaching adult levels at about 6 months of age (65, 85, Rumfelt and Flajnik, unpublished). IgNAR type I and type II rise in concentration concurrently with 7S IgM. This expression pattern during ontogeny mirrors the appearance-specific Ig after immunization (64). IgW has been a tougher nut to crack: both the long and short secretory forms can be expressed at birth in some sharks, and in others, there is a delay in the expression of the short form (76). Expression of IgW early in development breaks the mold that IgM is the isotype expressed earliest in development.
Regarding development of the lymphoid tissues, the spleen has been studied in some detail (65). White pulp present at birth consists entirely of IgM-positive B cells, with secretory cells (19S and IgM1gj producing) found in the red pulp. The B cells at this stage are class II-negative. By 5 months after birth, there are additionally large Ig− white pulp zones, presumably containing T cells, that are infiltrated by class II+ dendritic cells and a few IgM and IgNAR secretory cells.
The primary lymphoid tissues’ epigonal (and Leydig in skates) organ and thymus are the only tissues that express RAG genes at high levels throughout life (48, 65, 84). Expression of the germline-joined genes, including the NS4 and NS5 genes described above and IgM1gj, are expressed during adult life in relatively high amounts in the epigonal organ (24, 45, 48). This expression suggests that either there is a repository of secretory cells maintained that express these specificities or developing B cells may express such genes before extinguishing expression as the cells mature.
Translocon versus cluster in evolution
We have previously suggested that the cluster organization provides freedom for evolution of the system that the translocon does not (24, 48, 49). In the cartilaginous fish, clusters can evolve away to develop new functions, as described above. Some of these germline-joined genes expressed early in development may have unique binding specificities, similar to those proposed for certain V regions in mammalian B and T cells. In mammals, to ensure that the binding sites of certain genes are evolutionarily selected, ‘tricks’ have been used, such as rearrangement before TdT expression and homology-based joins (86). In contrast, the germline-joined clusters, in some cases, have reverted to become innate receptors, expressed early in ontogeny, and no further tricks are necessary to ensure the integrity of their binding site. The NAR-TCR described above is a perfect example of how the cluster organization has been used to create an entirely different type of TCR; such a novel rearranging gene could not have been easily assembled in a pure translocon system. Finally, new isotypes can be generated by fundamentally changing the nature of the V region, as is seen in the evolution of IgNAR and IgW; the precedent of a NAR VDJ cluster translocating to the TCRαδ locus suggests that a similar gene chimera between NAR and IgW occurred early in cartilaginous fish evolution (66, Criscitiello and Flajnik, manuscript submitted).
One disadvantage of the cluster organization is that rearrangement only occurs within a cluster, limiting combinatorial diversity. However, it is not clear how important this feature is, considering that the CDR3 are extremely diverse. Another disadvantage is the apparent lack of extensive H-chain switching, i.e. selecting for a particular VDJ rearrangement and then shuffling it onto a new isotype to modify function. Instead, in the cartilaginous fish, there are likely to be lineages of B cells that produce either IgM, IgNAR, or IgW, more similar to T cells that express either α/β or α/δ TCR from their earliest developmental stages. This expression would suggest that B cells with different types of receptors would compete for antigen in a humoral immune response, which seems not to be the most efficient way to regulate a humoral response. If true, having IgM as a first line of defense and Ig NAR as a second line of defense is not a viable framework. More likely, both types of B cell become stimulated early in the response, and the very different types of antibodies provide different conformations for binding to antigens and perhaps different effector functions.
Translocon plus cluster in bony fishes
We have been discussing the merits of translocon and clustered organization as if they were independent systems that could not coexist, whereas in fact, in teleost fishes such as catfish, trout, and cod, it has been established that the IgH is in a translocon arrangement but their IgL are clustered (reviewed in 5, 87). Although this finding (6) was initially surprising because H- and L-chain gene organization were supposed to have coevolved, there is in fact little direct mechanistic connection between H- and L-chain expression. H- and L-chain rearrangement generally do not occur simultaneously, and their activation occurs at separate stages of mammalian B-cell development. That B-cell differentiation in bony fish is similar in this respect may be inferred by the presence of N region in zebrafish H chain CDR3 (88), in contrast to an absence of TdT activity during L-chain rearrangement (Pulham and Hsu, unpublished results).
The term ‘translocon’ was originally coined by Gally and Edelman (89) and referred to any Ig locus that rearranges and contained all the genetic elements encoding an isotype. Thus, in humans, there are three translocons encoding the H and L chains. However, in the recent literature, translocon organization has taken to mean a locus with multiple V and few or one C genes, in contrast to clusters, which are multiple loci each containing one to three V and one C genes. Although it is implicit that there is one translocon-type locus per isotype, this aspect has not been strictly adhered to.
Except for recent cases of polyploidization in amphibians and in the Salmonid family, the IgH in vertebrates exists either as a single locus with translocon organization or as multiple clusters. This either/or situation appeared to be paralleled in L chains, applicable to three L-chain isotypes in fishes and Xenopus (87). The teleost IgL analyzed in the literature are all clustered, although they are considered not directly related to the multiple IgL loci in cartilaginous fishes (90). There is a tetrapod-like IgL organization in the Siberian sturgeon, which is evolutionarily placed between cartilaginous fishes and teleosts (Fig. 1). The sturgeon IgL contains multiple V and J and few C regions, but there are probably two loci per genome equivalent. Lundqvist et al. (90) referred to this situation as translocon and suggested that the ancestral IgL of Osteichthyes and all tetrapods evolved independently from Chondrichthyes and carried multiple V genes and one C. A separate development in teleost fish led to reduction of V-gene number and duplication of the minilocus, recreating a multicluster-style IgL organization that exists in catfish, trout, cod, Fugu, and salmon.
However, we have recently found evidence of continuing flexibility in IgL organization in bony fishes. The zebrafish genome is currently not yet complete, but using what is available, we have been able to locate sequences encoding its three L-chain isotypes. Using the cDNA V and C sequences from the studies that defined the L-chain isotypes (91), we identified and mapped the germline V- and J-gene segments and their C exons. To our surprise, although the zebrafish type 1 L chain is largely of the cluster type as described in other teleosts, its type 2 and type 3 L-chain loci contain many V-gene segments and few C genes (Fig. 7). This kind of IgL arrangement has not been observed before. Preliminary genomic Southern blotting detected two type 3 C sequences and multiple V in zebrafish genomic DNA (Pulham and Hsu, unpublished results), supporting the conclusions drawn from the database.
Fig. 7. Comparison of gene organizations of the three zebrafish light (L) chain isotypes.
The V and C regions of the three zebrafish L-chain isotypes (90) were used separately to search the database www.ensembl.org/Danio_rerio/index.html. The V-gene segments were identified not only by homology with published V sequence but also by the adjoining RS. Isotype 1: A chunk size of 156 kb contained two C regions; the published genomic Southern revealed multiple bands for C sequences, so isotype 1 in part encoded by genes in a multicluster organization. Isotype 2: 12 V, one J, and one C genes were located within 27 kb on a chunk size of 109 kb. Isotype 3: eight V, one J, and one C genes were found on chromosome 5; description in text. Blue boxes indicate V-gene segments, black boxes J-gene segments, and pink boxes C-region exons.
A taxonomically close relative of zebrafish is catfish, whose L chains have been classified into two isotypes, F and G, which are encoded by 70 and 15 clusters, respectively (92, 93). The F L chain is phylogenetically related to zebrafish type 3 (Criscitiello and Hsu, unpublished results). If the ancestral F/type 3 gene were cluster-type, then, with the expansion of V-gene segment number and paring down of the number of loci to two per genome, the zebrafish type 3 L chain appears to be evolving toward a translocon situation. Moreover, in zebrafish, mixed L-chain organizations have been observed for the first time to coexist in one animal.
Another interesting conclusion is that it appears the regulatory processes for activation and expression of the bony fish L-chain genes can accommodate these varied situations. It may be that L-chain regulation differs from H chain, but the nature of this accommodation is an issue worthy of pursuit in the future.
Antigen receptor exclusion
Allelic exclusion of the translocon-encoded BCR exists in mammals and birds as well as in amphibians (94, 95). We have been investigating Ig expression in nurse shark and believe that more than one locus is activated at a time, because of the large proportion of genomic non-productive rearrangements detected (45). Moreover, we have suggested that H- and L-chain rearrangement occurs concurrently in developing B cells in sharks, based on the finding that N-region additions are inserted as frequently in L chains as in H chains CDR3 in nurse shark. This is the most parsimonious explanation. Without sequential rearrangement there would be no requirement for a surrogate L chain; the H-chain dimer IgNAR also obviously does not require such an L chain, either. Thus, we speculate that although several loci are activated at once, the formation of a functional receptor would be all that is required to allow a shark B cell to proceed in its development.
Such a mechanism predicts that there would be many failed/incomplete rearrangements, as well as a higher level of multiple producers, compared with the translocon arrangement. A first analysis of this system in skates suggested that only a single productive μH chain is expressed in most B cells (41), and the authors suggest that the regulation is akin to what governs the multiple loci of the olfactory receptor system. At this time, no one knows the basis for the monoallelic expression of olfactory receptors, and there is some speculation that the olfaction gene expression is based on limiting activation factors. While this is also a hypothesis for L-chain κ gene allelic exclusion (96), it is probably not the explanation for the fish B cells, where several lgL clusters might be under the regulation of one enhancer (42).
In any model, the presence of the germline-joined genes must be taken into account. The type II (NS3) L chain loci do not need recombination, and they would be expressed sooner than a competing L-chain locus that must rearrange. Whether one locus or many loci are activated at one time, the germline joined would be expected to take over the repertoire, and they do not. Only in the nurse shark have the expression of all three L-chain isotypes been studied, and we found that the relative expression levels of the different genes roughly correlate with the number of genes for each Ig subtype. Furthermore, in the cases where the gene organizations are mixed (split and germline-joined such as NS4 and NS5, Fig. 4) within a family, the germline-joined loci appear early in development and are extinguished later. We have preliminary evidence for multiple L-chain RNA transcripts in single cells (Hsu and Flajnik, unpublished observations and 41), and we suggest tentatively that perhaps not all potentially functional rearrangements are used as part of the BCR. Indeed in the skate study several cells cantained more than one productive lgW H chain sequence(41). Perhaps all are downregulated except the ones that are actually expressed as the BCR. The regulatory situation is not fully explored, but it is certainly of interest to reconcile the fish situation with the current chromatin-domain activation model of the translocon IgH locus.
Some questions among many
We have described how Ig genes may have evolved, from the first V-like gene disrupted by the RAG transposon to the complex multiloci systems in extant vertebrates. The wealth of different C- and V-gene sequences and the varied gene organizations demonstrate how much is still unknown regarding Ig function and regulatory processes. For instance, although IgM is universally conserved, novel isotypes found in non-mammals do not always have a homolog. The functional niche occupied by IgW in sharks and the ever-changing forms of IgD in other animals remain to be discovered. The single-chain V-region-combining site shared by both Ig (as IgNAR) and TCR (as NAR-TCR) suggests that a category of antigens exists that can be recognized only by this unusual conformation and raises questions as to how this specialized array of specificities is compensated for in higher vertebrates. The plasticity of Ig gene configurations is well demonstrated in sturgeon and zebrafish compared with other teleost fishes; exploration of IgL in older extant fishes will give insight into the evolution of Ig gene organization.
Acknowledgments
In memory of Jan Moor-Jankowski. We thank Louis Du Pasquier for reading this manuscript, Jürgen Brosius for his timely input, and Karolina Malecek and Julie Brandman for their invaluable assistance. This work was supported by National Institutes of Health Grants GM068095 and RR06603.
References
- 1.Honjo T, Alt FW, Neuberger M, editors. Molecular Biology of B Cells. Elsevier Academic Press; 2004. [Google Scholar]
- 2.Schlissel MS. Regulating antigen-receptor gene assembly. Nat Rev Immunol. 2003;3:890–899. doi: 10.1038/nri1225. [DOI] [PubMed] [Google Scholar]
- 3.Chowdhury D, Sen R. Regulation of immunoglobulin heavy-chain gene rearrangements. Immunol Rev. 2004;200:182–196. doi: 10.1111/j.0105-2896.2004.00177.x. [DOI] [PubMed] [Google Scholar]
- 4.Hsu E. The variation in immunoglobulin constant regions in evolution. Semin Immunol. 1994;6:383–391. doi: 10.1006/smim.1994.1048. [DOI] [PubMed] [Google Scholar]
- 5.Litman GW, Anderson MK, Rast JP. Evolution of antigen binding receptors. Annu Rev Immunol. 1999;17:109–147. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]
- 6.Daggfeldt A, Bengtén E, Pilstrom L. A cluster type organization of the loci of the immunoglobulin light chain in Atlantic cod (Gadus morhua L) and rainbow trout (Oncorhynchus mykiss Walbaum) indicated by nucleotide sequences of cDNAs and hybridization analysis. Immunogenetics. 1993;38:199–209. doi: 10.1007/BF00211520. [DOI] [PubMed] [Google Scholar]
- 7.Du Pasquier L, Wilson M, Greenberg AS, Flajnik MF. Somatic mutation in ectothermic vertebrates: musings on selection and origins. Curr Top Microbiol Immunol. 1998;229:199–216. doi: 10.1007/978-3-642-71984-4_14. [DOI] [PubMed] [Google Scholar]
- 8.Azumi K, et al. Genomic analysis of immunity in a Urochordate and the emergence of the vertebrate immune system: ‘Waiting for Godot’. Immunogenetics. 2003;55:570–581. doi: 10.1007/s00251-003-0606-5. [DOI] [PubMed] [Google Scholar]
- 9.Bartl S, Baish MA, Flajnik MF, Ohta Y. Identification of class I genes in cartilaginous fish, the most ancient group of vertebrates displaying an adaptive immune response. J Immunol. 1997;159:6097–6104. [PubMed] [Google Scholar]
- 10.Du Pasquier L. The phylogenetic origin of antigen-specific receptors. Curr Top Microbiol Immunol. 2000;248:160–185. [PubMed] [Google Scholar]
- 11.Du Pasquier L, Zucchetti W, De Santis R. Immunoglobulin superfamily receptors in protochordates: before RAG time. Immunol Rev. 2004;198:233–248. doi: 10.1111/j.0105-2896.2004.00122.x. [DOI] [PubMed] [Google Scholar]
- 12.Hood L, Kronenberg M, Hunkapiller T. T cell antigen receptors and the immunoglobulin supergene family. Cell. 1985;40:225–229. doi: 10.1016/0092-8674(85)90133-3. [DOI] [PubMed] [Google Scholar]
- 13.Sakano H, Huppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- 14.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 15.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 16.Thompson CB. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity. 1995;3:531–3539. doi: 10.1016/1074-7613(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 17.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 18.Lewis SM, Wu GE. The old and the restless. J Exp Med. 2000;191:1631–1636. doi: 10.1084/jem.191.10.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 20.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- 22.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis SM, Wu GE, Hsu E. The origin of V(D)J diversification. In: Honjo T, Alt FW, Neuberger MS, editors. Molecular Biology of B Cells. Amsterdam: Elsevier Academic Press; 2004. pp. 473–489. [Google Scholar]
- 24.Lee SS, Fitch D, Flajnik MF, Hsu E. Rearrangement of immunoglobulin genes in shark germ cells. J Exp Med. 2000;191:1637–1648. doi: 10.1084/jem.191.10.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SS, Tranchina D, Ohta Y, Flajnik MF, Hsu E. Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity. 2002;16:571–582. doi: 10.1016/s1074-7613(02)00300-x. [DOI] [PubMed] [Google Scholar]
- 26.Spring J. Genome duplication strikes back. Nat Genet. 2002;31:128–129. doi: 10.1038/ng0602-128. [DOI] [PubMed] [Google Scholar]
- 27.Furlong RF, Holland PWH. Were vertebrates octoploid? Philos Trans R Soc Lond B Biol Sci. 2002;357:531–544. doi: 10.1098/rstb.2001.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holland PWH, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994:125–133. [PubMed] [Google Scholar]
- 29.Escriva H, Manzon L, Youson J, Laudet V. Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol Biol Evol. 2002;19:1440–1450. doi: 10.1093/oxfordjournals.molbev.a004207. [DOI] [PubMed] [Google Scholar]
- 30.Kasahara M, Nakaya J, Takahata N. Chromosomal duplication and the emergence of the adaptive immune system. Trends Genet. 1997;13:90–92. doi: 10.1016/s0168-9525(97)01065-2. [DOI] [PubMed] [Google Scholar]
- 31.Robinson-Rechavi M, Boussau B, Laudet V. Phylogenetic dating and characterization of gene duplications in vertebrates: the cartilaginous fish reference. Mol Biol Evol. 2004;21:580–586. doi: 10.1093/molbev/msh046. [DOI] [PubMed] [Google Scholar]
- 32.Hoegg S, Brinkmann H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol. 2004;59:190–203. doi: 10.1007/s00239-004-2613-z. [DOI] [PubMed] [Google Scholar]
- 33.Ghaffari SH, Lobb CJ. Organization of immunoglobulin heavy chain constant and joining region genes in the channel catfish. Mol Immunol. 1992;29:151–159. doi: 10.1016/0161-5890(92)90096-g. [DOI] [PubMed] [Google Scholar]
- 34.Bengtén E, et al. The IgH locus of the channel catfish, Ictalurus punctatus, contains multiple constant region gene sequences: different genes encode heavy chains of the membrane and secreted IgD. J Immunol. 2002;169:2488–2497. doi: 10.4049/jimmunol.169.5.2488. [DOI] [PubMed] [Google Scholar]
- 35.Danilova N, Bussmann J, Jekosch K, Steiner LA. The immunoglobulin heavy-chain locus in zebrafish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nat Immunol. 2005;6:295–302. doi: 10.1038/ni1166. [DOI] [PubMed] [Google Scholar]
- 36.Hansen JD, Landis ED, Phillips RB. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish. Proc Natl Acad Sci USA. 2005;102:6919–6924. doi: 10.1073/pnas.0500027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelley J, Walter L, Trowsdale J. Comparative genomics of major histocompatibility complexes. Immunogenetics. 2005;56:683–695. doi: 10.1007/s00251-004-0717-7. [DOI] [PubMed] [Google Scholar]
- 38.Amores A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 39.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 40.Hinds KR, Litman GW. Major reorganization of immunoglobulin VH segmental elements during vertebrate evolution. Nature. 1986;320:546–549. doi: 10.1038/320546a0. [DOI] [PubMed] [Google Scholar]
- 41.Eason DD, Litman RT, Luer CA, Kerr W, Litman GW. Expression of individual immunoglobulin genes occurs in an unusual system consisting of multiple independent loci. Eur J Immunol. 2004;34:2551–2558. doi: 10.1002/eji.200425224. [DOI] [PubMed] [Google Scholar]
- 42.Bengtén E, Stromberg S, Daggfeldt A, Bradley GM, Pilstrom L. Transcriptional enhancers of immunoglobulin light chain genes in Atlantic cod (Gadus morhua) Immunogenetics. 2000;51:647–658. doi: 10.1007/s002510000176. [DOI] [PubMed] [Google Scholar]
- 43.Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 44.Diaz M, Velez J, Singh M, Cerny J, Flajnik MF. Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: the translesion synthesis model of somatic hypermutation. Int Immunol. 1999;11:825–833. doi: 10.1093/intimm/11.5.825. [DOI] [PubMed] [Google Scholar]
- 45.Fleurant M, Changchien L, Chen CT, Flajnik MF, Hsu E. Shark Ig light chain junctions are as diverse as in heavy chains. J Immunol. 2004;173:5574–5582. doi: 10.4049/jimmunol.173.9.5574. [DOI] [PubMed] [Google Scholar]
- 46.Malecek K, Brandman J, Brodsky JE, Ohta Y, Flajnik MF, Hsu E. Somatic hypermutation and junctional diversification at immunoglobulin heavy chain loci in the nurse shark. J Immunol. 2005;175:8105–8115. doi: 10.4049/jimmunol.175.12.8105. [DOI] [PubMed] [Google Scholar]
- 47.Ghaffari SH, Lobb CJ. Structure and genomic organization of a second cluster of immunoglobulin heavy chain gene segments in the channel catfish. J Immunol. 1999;162:1519–1529. [PubMed] [Google Scholar]
- 48.Rumfelt LL, Avila D, Diaz M, Bartl S, McKinney EC, Flajnik MF. A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc Natl Acad Sci USA. 2001;98:1775–1780. doi: 10.1073/pnas.98.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flajnik MF. Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol. 2002;2:688–698. doi: 10.1038/nri889. [DOI] [PubMed] [Google Scholar]
- 50.Reynaud CA, Dahan A, Anquez V, Weill JC. Somatic hyperconversion diversifies the single Vh gene of the chicken with a high incidence in the D region. Cell. 1989;59:171–183. doi: 10.1016/0092-8674(89)90879-9. [DOI] [PubMed] [Google Scholar]
- 51.Kokubu F, Litman R, Shamblott MJ, Hinds K, Litman GW. Diverse organization of immunoglobulin VH gene loci in a primitive vertebrate. EMBO J. 1988;7:3413–3422. doi: 10.1002/j.1460-2075.1988.tb03215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis SM. Evolution of immunoglobulin and T-cell receptor gene assembly. Ann NY Acad Sci. 1999;870:58–67. doi: 10.1111/j.1749-6632.1999.tb08865.x. [DOI] [PubMed] [Google Scholar]
- 53.Tucker PW, Marcu KB, Newell N, Richards J, Blattner FR. Sequence of the cloned gene for the constant region of murine gamma 2b immunoglobulin heavy chain. Science. 1979;206:1303–1306. doi: 10.1126/science.117549. [DOI] [PubMed] [Google Scholar]
- 54.Tucker PW, Slightom JL, Blattner FR. Mouse IgA heavy chain gene sequence: Implications for evolution of immunoglobulin hinge exons. Proc Natl Acad Sci USA. 1981;78:7684–7688. doi: 10.1073/pnas.78.12.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warr GW, Magor KE, Higgins DA. IgY: clues to the origins of modern antibodies. Immunol Today. 16:392–398. doi: 10.1016/0167-5699(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 56.Wilson M, Bengtén E, Miller NW, Clem LW, Du Pasquier L, Warr GW. A novel chimeric Ig heavy chain from a teleost fish shares similarities with IgD. Proc Natl Acad Sci USA. 1997;94:4593–4598. doi: 10.1073/pnas.94.9.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolfe KH. Yesterday’s polyploids and the mystery of diploidization. Nat Rev Genet. 2001;2:333–3341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- 58.Shen SX, Bernstein RM, Schluter SF, Marchalonis JJ. Heavy-chain variable regions in carcharhine sharks development of a comprehensive model for the evolution of VH domains among the gnathanstomes. Immunol Cell Biol. 1996;74:357–364. doi: 10.1038/icb.1996.63. [DOI] [PubMed] [Google Scholar]
- 59.Rumfelt LL, Lohr RL, Dooley H, Flajnik MF. Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol. 2004;5:8. doi: 10.1186/1471-2172-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marchalonis J, Edelman GM. Polypeptide chains of immunoglobulins from the smooth dogfish (Mustelus canis) Science. 1966;154:1567–1568. doi: 10.1126/science.154.3756.1567. [DOI] [PubMed] [Google Scholar]
- 61.Clem LW, De Boutaud F, Sigel MM. Phylogeny of immunoglobulin structure and function. II Immunoglobulins of the nurse shark. J Immunol. 1967;99:1226–1235. [PubMed] [Google Scholar]
- 62.Klapper DG, Clem LW. Phylogeny of immunoglobulin structure and function: characterization of the cysteine-containing peptide involved in the polymerization of shark IgM. Dev Comp Immunol. 1977;1:81–92. doi: 10.1016/s0145-305x(77)80002-5. [DOI] [PubMed] [Google Scholar]
- 63.Voss EW, Sigel MM. Valence and temporal change in affinity of purified 7S and 18S nurse shark anti-24 dinitrophenyl antibodies. J Immunol. 1972;109:665–673. [PubMed] [Google Scholar]
- 64.Dooley H, Flajnik MF. Shark immunity bites back: affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur J Immunol. 2005;35:936–945. doi: 10.1002/eji.200425760. [DOI] [PubMed] [Google Scholar]
- 65.Rumfelt LL, McKinney EC, Taylor E, Flajnik MF. The development of primary and secondary lymphoid tissues in the nurse shark Ginglymostoma cirratum. B-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny in the spleen. Scand J Immunol. 2002;56:130–148. doi: 10.1046/j.1365-3083.2002.01116.x. [DOI] [PubMed] [Google Scholar]
- 66.Roux KH, et al. Structural analysis of the nurse shark (new) antigen receptor (NAR): molecular convergence of NAR and unusual immunoglobulins. Proc Natl Acad Sci. 1998;95:11804–11809. doi: 10.1073/pnas.95.20.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greenberg AS, Hughes AL, Guo J, Avila D, McKinney EC, Flajnik MF. A novel ‘chimeric’ antibody class in cartilaginous fish: IgM may not be the primordial immunoglobulin. Eur J Immunol. 1996;26:1123–1129. doi: 10.1002/eji.1830260525. [DOI] [PubMed] [Google Scholar]
- 68.Anderson MK, et al. A long form of the skate IgX gene exhibits a striking resemblance to the new shark antigen IgW and IgNARC genes. Immunogenetics. 1999;49:56–67. doi: 10.1007/s002510050463. [DOI] [PubMed] [Google Scholar]
- 69.Stanfield RL, Dooley H, Flajnik MF, Wilson IA. Crystal structure of a shark single domain antibody V region in complex with lysozyme. Science. 2005;305:1770–1773. doi: 10.1126/science.1101148. [DOI] [PubMed] [Google Scholar]
- 70.Streltsov VA, Varghese JN, Carmichael JA, Irving RA, Hudson PJ, Nuttall SD. Structural evidence for evolution of shark Ig new antigen receptor variable domain antibodies from a cell-surface receptor. Proc Natl Acad Sci USA. 2004;101:12444–12449. doi: 10.1073/pnas.0403509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Diaz M, Stanfield RL, Greenberg AS, Flajnik MF. Structural analysis, selection, and ontogeny of the nurse shark new antigen receptor (IgNAR): Identification of a new locus preferentially expressed early in development. Immunogenetics. 2002;54:501–512. doi: 10.1007/s00251-002-0479-z. [DOI] [PubMed] [Google Scholar]
- 72.Diaz M, Greenberg AS, Flajnik MF. Somatic hypermutation of the new antigen receptor gene (NAR) in the nurse shark does not generate the repertoire: possible role in antigen-driven reactions in the absence of germinal centers. Proc Natl Acad Sci USA. 1998;95:14343–14348. doi: 10.1073/pnas.95.24.14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kobayashi K, Tomonaga S, Kajii T. A second class of immunoglobulin other than IgM present in the serum of a cartilaginous fish, the skate Raja kenoji: isolation and characterization. Mol Immunol. 1984;21:397–404. doi: 10.1016/0161-5890(84)90037-3. [DOI] [PubMed] [Google Scholar]
- 74.Harding FA, Amemiya CT, Litman RT, Cohen N, Litman GW. Two distinct immunoglobulin heavy chain isotypes in a primitive cartilaginous fish, Raja erinacea. Nucleic Acids Res. 1990;18:6369–6376. doi: 10.1093/nar/18.21.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bernstein RM, Schluter SF, Shen S, Marchalonis JJ. A new high molecular weight immunoglobulin class from the carcharhine shark implications for the properties of the primordial immunoglobulin. Proc Natl Acad Sci. 1996;93:3289–3293. doi: 10.1073/pnas.93.8.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rumfelt LL, Diaz M, Lohr RL, Mochon E, Flajnik MF. Unprecedented multiplicity of Ig transmembrane and secretory mRNA forms in the cartilaginous fish. J Immunol. 2004;173:1129–1139. doi: 10.4049/jimmunol.173.2.1129. [DOI] [PubMed] [Google Scholar]
- 77.Ota T, Rast JP, Litman GW, Amemiya CT. Lineage-restricted retention of a primitive immunoglobulin heavy chain isotype within the Dipnoi reveals an evolutionary paradox. Proc Natl Acad Sci USA. 2003;100:2501–2506. doi: 10.1073/pnas.0538029100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saha NR, Suetake H, Kikuchi K, Suzuki Y. Fugu immunoglobulin D a highly unusual gene with unprecedented duplications in its constant region. Immunogenetics. 2004;56:438–447. doi: 10.1007/s00251-004-0693-y. [DOI] [PubMed] [Google Scholar]
- 79.Zhao Y, et al. Artiodactyl IgD: The missing link. J Immunol. 2002;169:4408–4416. doi: 10.4049/jimmunol.169.8.4408. [DOI] [PubMed] [Google Scholar]
- 80.Rast JP, Anderson MK, Strong SJ, Luer C, Litman RT, Litman GW. α, β, γ, and δ T cell antigen receptor genes arose early in vertebrate phylogeny. Immunity. 1997;6:1–11. doi: 10.1016/s1074-7613(00)80237-x. [DOI] [PubMed] [Google Scholar]
- 81.Smith LC, Davidson EH. The echinoid immune system and the phylogenetic occurrence of immune mechanisms in deuterostomes. Immunol Today. 1992;13:356–362. doi: 10.1016/0167-5699(92)90172-4. [DOI] [PubMed] [Google Scholar]
- 82.Criscitiello M, Saltis M, Flajnik M. An evolutionary mobile antigen receptor variable region gene: doubly rearranging NAR-TCR genes in sharks. Proc Natl Acad Sci USA. doi: 10.1073/pnas.0507074103. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chien YH, Jores R, Crowley MP. Recognition by γ/δ T cells. Annu Rev Immunol. 1996;14:511–532. doi: 10.1146/annurev.immunol.14.1.511. [DOI] [PubMed] [Google Scholar]
- 84.Miracle AL, et al. Complex expression patterns of lymphocyte-specific genes during the development of cartilaginous fish implicate unique lymphoid tissues in generating an immune repertoire. Int Immunol. 2001;13:567–580. doi: 10.1093/intimm/13.4.567. [DOI] [PubMed] [Google Scholar]
- 85.Fidler JE, Clem LW, Small PA., Jr Immunoglobulin synthesis in neonatal nurse sharks (Ginglymostoma cirratum) Comp Biochem Physiol. 1969;31:365–371. doi: 10.1016/0010-406x(69)91660-0. [DOI] [PubMed] [Google Scholar]
- 86.Feeney AJ. Predominance of the prototypic T15 anti-phosphorylcholine junctional sequence in neonatal pre-B cells. J Immunol. 1991;147:4343–4350. [PubMed] [Google Scholar]
- 87.Bengtén E, Wilson M, Miller N, Clem LW, Pilstrom L, Warr GW. Immunoglobulin isotypes: structure, function, and genetics. Curr Top Microbiol Immunol. 2000;248:189–219. doi: 10.1007/978-3-642-59674-2_9. [DOI] [PubMed] [Google Scholar]
- 88.Danilova N, Hohman VS, Kim EH, Steiner LA. Immunoglobulin variable-region diversity in the zebrafish. Immunogenetics. 2000;52:81–91. doi: 10.1007/s002510000255. [DOI] [PubMed] [Google Scholar]
- 89.Gally JA, Edelman GM. The genetic control of immunoglobulin synthesis. Annu Rev Genet. 1972;6:1–46. doi: 10.1146/annurev.ge.06.120172.000245. [DOI] [PubMed] [Google Scholar]
- 90.Lundqvist M, Bengtén E, Stromberg S, Pilstrom L. Ig light chain gene in the Siberian sturgeon (Acipenser baeri) J Immunol. 1996;157:2031–2038. [PubMed] [Google Scholar]
- 91.Haire RN, Rast JP, Litman RT, Litman GW. Characterization of three isotypes of immunoglobulin light chains and T-cell antigen receptor alpha in zebrafish. Immunogenetics. 2000;51:915–923. doi: 10.1007/s002510000229. [DOI] [PubMed] [Google Scholar]
- 92.Ghaffari SH, Lobb CJ. Structure and genomic organization of immunoglobulin light chain in the channel catfish. An unusual genomic organizational pattern of segmental genes. J Immunol. 1993;151:6900–6912. [PubMed] [Google Scholar]
- 93.Ghaffari SH, Lobb CJ. Structure and genomic organization of a second class of immunoglobulin light chain genes in the channel catfish. J Immunol. 1997;159:250–258. [PubMed] [Google Scholar]
- 94.Lee SS, Hsu E. Allelic exclusion in anamniotic vertebrates. Semin Immunol. 1999;11:329–336. doi: 10.1006/smim.1999.0189. [DOI] [PubMed] [Google Scholar]
- 95.Du Pasquier L, Hsu E. Immunoglobulin expression in diploid and polyploid interspecies hybrid of Xenopus: evidence for allelic exclusion. Eur J Immunol. 1983;13:585–890. doi: 10.1002/eji.1830130714. [DOI] [PubMed] [Google Scholar]
- 96.Liang HE, Hsu LY, Cado D, Schlissel MS. Variegated transcriptional activation of the immunoglobulin κ locus in pre-B cells contributes to the allelic exclusion of light-chain expression. Cell. 2004;118:19–29. doi: 10.1016/j.cell.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 97.Dooley H, Flajnik MF. Antibody repertoire development in cartilaginous fish. Dev Comp Immunol. 2006;30:43–56. doi: 10.1016/j.dci.2005.06.022. [DOI] [PubMed] [Google Scholar]