Abstract
A critical step in the degradation of many eukaryotic mRNAs is a decapping reaction that exposes the transcript to 5′ to 3′ exonucleolytic degradation. The dual role of the cap structure as a target of mRNA degradation and as the site of assembly of translation initiation factors has led to the hypothesis that the rate of decapping would be specified by the status of the cap binding complex. This model makes the prediction that signals that promote mRNA decapping should also alter translation. To test this hypothesis, we examined the decapping triggered by premature termination codons to determine whether there is a down-regulation of translation when mRNAs were recognized as “nonsense containing.” We constructed an mRNA containing a premature stop codon in which we could measure the levels of both the mRNA and the polypeptide encoded upstream of the premature stop codon. Using this system, we analyzed the effects of premature stop codons on the levels of protein being produced per mRNA. In addition, by using alterations either in cis or in trans that inactivate different steps in the recognition and degradation of nonsense-containing mRNAs, we demonstrated that the recognition of a nonsense codon led to a decrease in the translational efficiency of the mRNA. These observations argue that the signal from a premature termination codon impinges on the translation machinery and suggest that decapping is a consequence of the change in translational status of the mRNA.
INTRODUCTION
Many eukaryotic mRNAs decay through a pathway of turnover that is initiated by shortening of the poly(A) tail followed by degradation of the body of the transcript (for review, see Beelman and Parker, 1995; Jacobson and Peltz, 1996). In Saccharomyces cerevisiae, Chlamydomonas reinhardtii (Gera and Baker, 1998), and possibly other eukaryotes (Lim and Maquat, 1992; Higgs and Colbert, 1994; Couttet et al., 1997), mRNA deadenylation leads to decapping of the mRNA, which is then followed by rapid 5′ to 3′ exonucleolytic degradation (Decker and Parker, 1993; Hsu and Stevens, 1993; Muhlrad et al., 1994, 1995). Decapping is an important step in the turnover of yeast mRNAs, because it precedes the decay of the body of the transcript and is also a site of control, as individual mRNAs are decapped at different rates (Muhlrad et al., 1994, 1995; Caponigro and Parker, 1996).
Decapping also occurs in a conserved process termed “mRNA surveillance” or “nonsense-mediated decay,” whereby aberrant mRNAs, including those containing a premature translation stop codon, are rapidly decapped independent of deadenylation (Losson and Lacroute, 1979; Leeds et al., 1991; Peltz et al., 1993; Pulak and Anderson, 1993; Muhlrad and Parker, 1994). How a nonsense-containing transcript is recognized as aberrant and subsequent mRNA decapping is triggered is unclear. It has been hypothesized that the recognition of an mRNA as aberrant would trigger an alteration in the complex of translation initiation factors assembled on the 5′ cap structure and thereby allow decapping (Muhlrad and Parker, 1994; Hagan et al., 1995; Peltz and Jacobson, 1996). A prediction of this model is that the recognition of an mRNA as aberrant would also alter translation of the mRNA, and that such an alteration would precede decapping. To examine the relationship between the triggering of decapping by premature translation termination and the translation efficiency of the mRNA, we devised a reporter system that enabled us to measure both the levels and decay rates of mRNAs containing premature termination codons, as well as to monitor the levels of the proteins being produced from the upstream open reading frame. Analysis of the decay and translation of these reporter mRNAs indicated that when an mRNA was recognized as “nonsense containing,” the amount of protein produced per transcript was reduced. This observation suggests that the signal from a premature termination codon impinges on the translation machinery, and that decapping may be a consequence of the change in translational status of the mRNA.
MATERIALS AND METHODS
Yeast Strains
Yeast strains used in this study are as follows: yRP1209, MATa, leu2, lys2, his4, trp1, ura3, cup1Δ∷URA3; yRP1277, MATa, leu2, lys2, his4, trp1, ura3, cup1Δ∷URA3, dcp1Δ∷URA3; yRP1212, MATa, leu2, lys2, his4, trp1, ura3, cup1Δ∷URA3, upf1Δ∷URA3; and yRP1235, MATa, leu2, lys2, his4, trp1, ura3, cup1Δ∷URA3 dcp1Δ∷URA3, upf1Δ∷URA3.
Plasmids
The DCP1 disruption plasmid pRP716 was used to remove DCP1 and replace it with URA3 (Beelman et al., 1996). A UPF1 gene disruption plasmid (URA3) was obtained from Audrey Atkins (University of Nebraska). A URA3 disruption of CUP1 was made using plasmid pBX-1 originally from Dennis Thiele (University of Michigan).
The control plasmid B55PGK1pG (pRP871) contains the GAL1 upstream activating sequence followed by PGK1 containing a Bg1II site 55 bases into the mRNA and also a poly(G) tract inserted in the 3′ untranslated region (UTR) as previously described (Decker and Parker, 1993). This plasmid is a 2μ TRP1 plasmid.
The construct expressing the PGK1(cup1)UAApG (pRP869) consists of the 144-bp GAL1 upstream activating sequence, the 5′ portion of PGK1 through amino acid 7, a Flag peptide inserted in frame into a Bg1II site (B55), followed by two CYS codons, amino acid 2 through the stop codon of the CUP1 gene, and then the rest of the PGK1 mRNA sequence containing a poly(G) insert in the 3′ UTR.
The construct expressing the PGK1(cup1)UAApG(ΔDSE) (pRP870) is identical to pRP869 until after the stop codon of CUP1. In this case, there are 27 more bases of PGK1, a polylinker region containing several restriction sites, and then the PGK1 3′ UTR sequence from 24 bases before the poly(G) insertion through the 3′ UTR.
The control plasmid PGK1(cup1)pG (pRP960) is identical to plasmid pRP869, except there is a precise deletion of the stop codon after the CUP1 open reading frame.
RNA
RNA was made from midlog cultures grown in selective media according to Caponigro et al. (1993). Standard 1.25% formaldehyde Northern blots were run with 10 μg of RNA per lane and probed with an oligo complementary to the poly(G) (oRP121).
Protein Analysis
Protein extracts were made by first harvesting half of a 20-ml culture grown in selective media to midlog. The other half of the same culture was used for RNA preparation. Pellets used for protein preparation were resuspended in 100 μl of sample buffer (final concentration, 125 mM Tris, pH 6.8, 1% SDS, 2% glycerol, 10% β-mercapto-ethanol). Cells were lysed by adding glass beads, boiling for 3 min, vortexing for 1 min, boiling for 3 min, and vortexing for 2 min. The extract was removed from the glass beads by puncturing the tube with a needle and spinning into another tube. The supernatant was the collected, and aliquots were run on denaturing 15% acrylamide-SDS gels. The gels were transferred to 0.2μ nitrocellulose. Western blots were performed using Flag primary antibodies (Eastman Kodak, Rochester, NY) at a 1∶10,000 dilution and goat anti-mouse HRPAb (Boehringer Mannheim, Indianapolis, IN) secondary antibodies also at a 1∶10,000 dilution. Blocking was done in PBS and Tween 20 containing 10% milk and 5% BSA, followed by 10% milk blocking in the presence of antibodies. Pierce (Rockford, IL) Ultra developer was used to visualize protein bands.
Protein gels were stained with GelCode Blue (Pierce) to visualize and compare protein loading between extract samples.
Copper plate assays were done at 30°C by patching cells onto selective media, then to YEP galactose plates, and finally replica plating onto galactose-selective minimal plates containing copper concentrations from 0 to 1.5 mM.
RESULTS
Reporter System
To assay the rates of translation and turnover in response to recognition of an early nonsense codon, we constructed a nonsense-containing mRNA in which the polypeptide encoded ahead of the stop codon would have biological activity. In such a construct we would be able to assess the amount of protein produced from a transcript without a requirement for stop codon readthrough or translation reinitiation for protein production. The particular mRNA constructed was a variant of the PGK1 mRNA wherein we introduced the open reading frame from the CUP1 gene early in the PGK1 open reading frame, followed immediately by a termination codon (see MATERIALS AND METHODS). This termination codon was in a region of the PGK1 transcript where nonsense codons are known to trigger rapid mRNA degradation (Peltz et al., 1993; Muhlrad and Parker, 1994). We used the CUP1 open reading frame for two reasons. First, the CUP1 protein is a Cu++-binding protein whose cellular levels are easily measured by growth on different concentrations of copper. Second, previous work has shown that the ability of nonsense codons to trigger mRNA decay diminishes as the ribosome translates further into open reading frames (Losson and Lacroute, 1979; Peltz et al., 1993; Pulak and Anderson, 1993). Because the CUP1 open reading frame consists of only 61 amino acids, we hypothesized that it would not be of sufficient length to prevent recognition of the introduced nonsense codon as a premature termination site. In addition, to allow direct detection of the CUP1 polypeptide, we inserted the Flag epitope in the 5′ end of the construct. The resulting mRNA, termed PGK1(cup1)UAApG, is shown in the second schematic on the left in Figure 1.
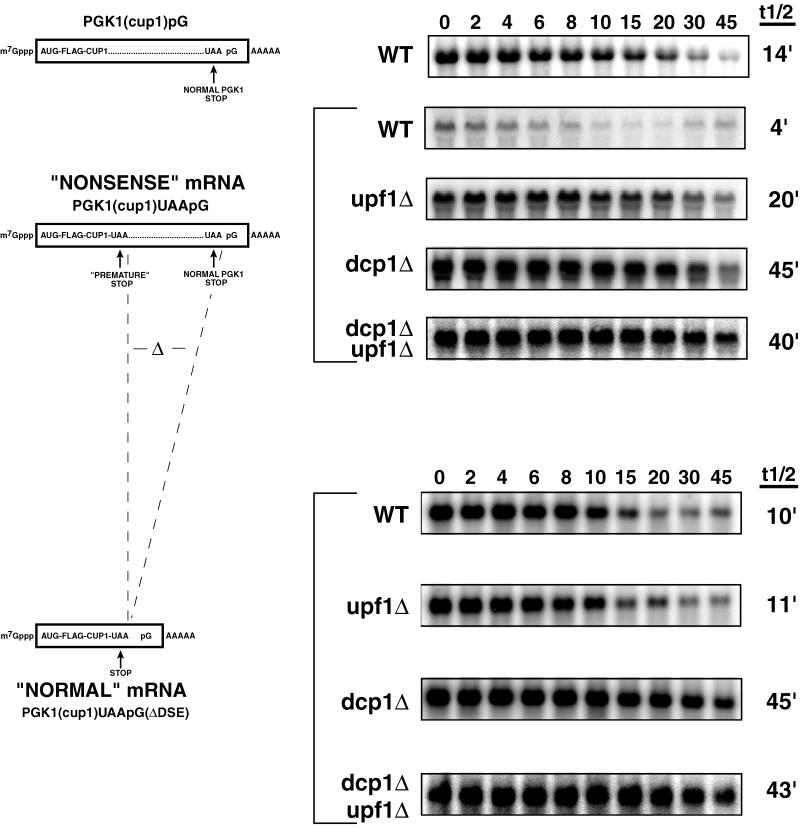
Figure 1.
Northern analysis of the PGK1(cup1) containing transcripts. Diagrams on the left represent the mRNAs: PGK1(cup1)pG, PGK1(cup1)UAApG, and PGK1(cup1)UAApG(ΔDSE). They show the location of the insertions of the Flag tag, the CUP1 open reading frame, both the premature and normal stop codons, and a poly(G) tag sequence. The region of the DSE deletion is designated with two dashed lines between the diagrams and is depicted as Δ. Strain names are labeled to the right of the construct cartoons; WT, wild-type. Northern blots showing decay after transcriptional repression are shown for each construct in the respective strains. Above each panel is the time in minutes after repression of transcription. Average half-life values from at least two experiments are given to the right of each Northern blot.
Three observations indicated that the PGK1(cup1)UAApG mRNA was subject to nonsense-mediated decay. First, comparison of the decay rates of the PGK1(cup1)UAApG mRNA (t1/2 = 4 min) to a control mRNA wherein the premature stop codon was precisely deleted (t1/2 = 14 min) showed that the nonsense codon destabilized the mRNA (Figure 1). Second, the rapid decay of the PGK1(cup1)UAApG mRNA (t1/2 = 4 min) was prevented in a upf1Δ strain (t1/2 = 20 min) (Figure 1), which is known to specifically affect nonsense-mediated decay (Leeds et al., 1991). The mRNA was also stabilized in a dcp1Δ strain (t1/2 = 45 min) (Figure 1), which removes the decapping enzyme required for both normal and nonsense-mediated decay. Third, the decay of the PGK1(cup1)UAApG mRNA was slowed and made independent of the Upf1p by deletion of the PGK1 coding region 3′ of the stop codon in a construct termed PGK1(cup1)UAApG(ΔDSE) (Figure 1). This deletion removed sequences (referred to as the downstream sequence element [DSE]) known to be required 3′ of the termination codon for nonsense-mediated decay (Peltz et al., 1993). In summation, these observations indicated that the PGK1(cup1)UAApG transcript was a substrate for nonsense-mediated decay.
Nonsense-containing mRNAs Produce Less Protein per mRNA than Normal Transcripts
To test the hypothesis that recognition of an mRNA as nonsense containing repressed translation and activated decapping, we measured the steady-state levels of mRNA and protein for both the PGK1(cup1)UAApG and the PGK1(cup1)UAApG(ΔDSE) mRNAs in wild-type, upf1Δ, and dcp1Δ strains. The ratio of the mRNA levels to the corresponding protein levels then gives a measure of the overall translational efficiency of the mRNA. In addition, as an important control, we performed a similar experiment in a dcp1Δupf1Δ double mutant strain (see below).
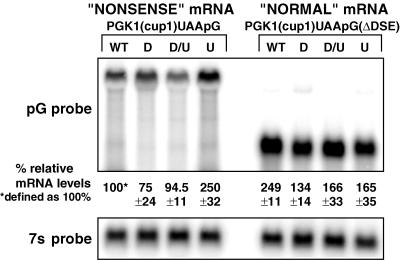
The steady-state levels of the PGK1(cup1)UAApG or PGK1(cup1)UAApG(ΔDSE) transcripts in each strain were determined on Northern blots for multiple cultures (more than five in each case). Representative gels are shown in Figure 2, in which the signal was standardized to the levels of the 7S RNA (Caponigro et al., 1993). The important observations to note are as follows. First, in wild-type strains, the PGK1(cup1)UAApG(ΔDSE) mRNA was present at two to three times the level of the PGK1(cup1)UAApG transcript. This is consistent with the differences in mRNA decay rates for these mRNAs (Figure 1). Second, in upf1Δ strains the amount of the PGK1(cup1)UAApG transcript increased two- to threefold compared with wild type, consistent with the requirement of the UPF1 gene product for nonsense-mediated decay (Figure 1). Surprisingly, despite the almost 10-fold increase in PGK1(cup1)UAApG transcript half-life in the dcp1Δ and the dcp1Δupf1Δ strains, the levels of this mRNA were essentially unchanged compared with the wild-type strain. This observation suggests that there are other important features of mRNA metabolism that play a role in determining the steady-state levels of transcripts in addition to the cytoplasmic mRNA decay rate (see DISCUSSION).
Figure 2.
Relative mRNA levels of the PGK1(cup1)UAApG and the PGK1(cup1)UAApG(ΔDSE) transcripts. The upper Northern blot shows analysis of the PGK1(cup1)UAApG (left four lanes) and the PGK1(cup1)UAApG(ΔDSE) (right four lanes) transcripts. Yeast strains are designated above each lane as follows: wild type (WT), dcp1Δ (D), upf1Δ (U), and dcp1Δupf1Δ (D/U). The relative percent mRNA levels and error values for each mRNA species from five or more experiments are shown below each lane, with the PGK1(cup1)UAApG mRNA in the wild-type strain defined as 100%. The lower panel shows the same Northern blot probed for the 7S ribosomal RNA, which was used to standardize the lanes.
The levels of the Flag-Cup1 protein produced under different conditions were determined by Western blotting equal amounts of cell protein using antibodies directed against the Flag epitope. The analysis was done on the same samples that were used to measure mRNA levels. The first comparison to note is between the PGK1(cup1)UAApG mRNA, which is recognized as nonsense containing in wild-type cells, and the PGK1(cup1)UAApG(ΔDSE), which is recognized as a “normal” mRNA in wild-type cells. In wild-type cells very low levels of protein were produced from the PGK1(cup1)UAApG mRNA (Figure 3A, lane 1). In contrast, the amount of protein resulting from the PGK1(cup1)UAApG(ΔDSE) construct in the same strain, which was no longer a substrate for nonsense-mediated decay, was substantially higher (Figure 3A, lane 3). An identical stained gel (Figure 3B) shows that equal amounts of total cell protein were loaded in each lane.
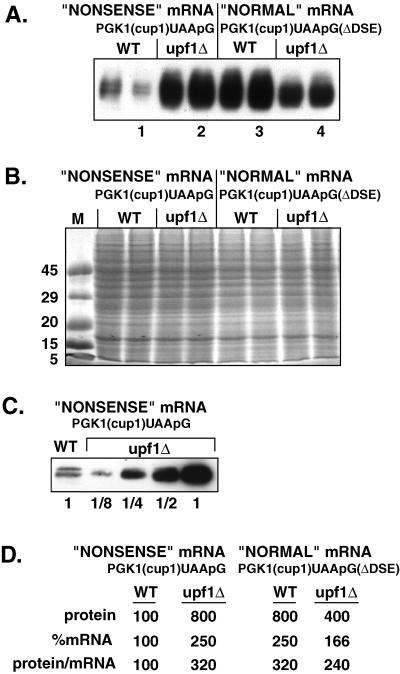
Figure 3.
Relative protein levels of the PGK1(cup1)UAApG and the PGK1(cup1)UAApG(ΔDSE) transcripts in wild-type and upf1Δ strains. (A) Western analysis using the FLAG antibody to measure protein levels produced from the PGK1(cup1)UAApG and PGK1(cup1)UAApG(ΔDSE) constructs, labeled at the top. Strain names are given above each lane set, which consist of two independent experiments run side by side. One lane of each set is numbered left to right as reference for the text. (B) Stained protein gel containing the identical samples to A. (C) Dilution comparison Western analysis of the PGK1(cup1)UAApG containing wild-type (WT) and upf1Δ strains. Dilution factors are given below each lane. (D) The percent relative mRNA and protein levels as well as protein per mRNA ratios are given for each sample type. The level of the protein produced from the nonsense mRNA PGK1(cup1)UAApG in wild-type cells is set at 100%. Values are averages based on at least five independent experiments.
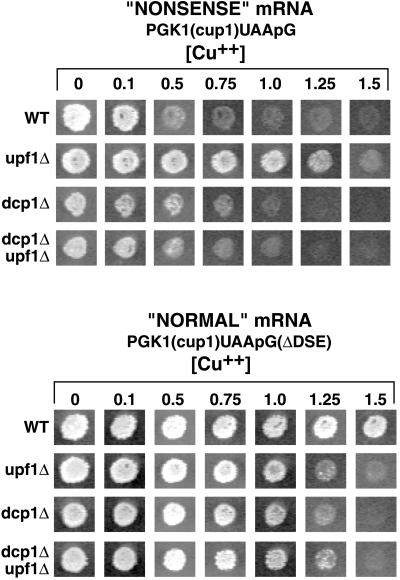
To determine the differences in protein levels, dilutions of the samples from these experiments were compared by Western analysis (Figure 3C). This comparison indicated that there was approximately eightfold more protein produced from the PGK1(cup1)UAApG(ΔDSE) mRNA compared with the PGK1(cup1)UAApG mRNA. This eightfold difference in protein levels was consistently seen in multiple experiments, indicating that this difference was highly reproducible. An important point was that this eightfold increase was greater than expected, because the steady-state levels of the PGK1(cup1)UAApG(ΔDSE) mRNA in wild-type strains were only 2.5-fold higher than the PGK1-(cup1)UAApG transcript (Figure 2). Thus, the deletion of the DSE leads to an mRNA that is not only more stable than the full-length transcript but also allows the production of approximately two to three times more protein per transcript. Consistent with the increased protein levels produced from the PGK1(cup1)UAApG(ΔDSE) mRNA, strains expressing this mRNA were able to grow on media containing higher concentrations of copper than strains expressing the PGK1(cup1)UAApG mRNA (Figure 4).
Figure 4.
Copper phenotypes of the PGK1(cup1)UAApG and the PGK1(cup1)UAApG(ΔDSE) transcripts in wild-type and deletion strains. The top portion shows yeast colonies containing the PGK1(cup1)UAApG construct from four different strains designated on the left. From top to bottom: wild-type (WT), upf1Δ, dcp1Δ, and the double deletion dcp1Δupf1Δ. Colonies were grown for 3 d on selective media containing galactose and increasing millimolar levels of copper designated above each column. The lower panels are identical to above, except that these strains contain the PGK1(cup1)UAApG(ΔDSE) construct.
Similar results were seen when nonsense-mediated decay was blocked in trans by the deletion of the UPF1 gene. In upf1Δ strains, the PGK1(cup1)UAApG mRNA was increased 2.5-fold (Figure 2), yet the levels of the encoded proteins increased eightfold compared with the wild type (Figure 3, A, compare lanes 1 and 2, and D). Again, consistent with the Western analysis, upf1Δ strains expressing the PGK1(cup1)UAApG mRNA were more copper resistant in plate assays than wild-type strains containing the same construct (Figure 4). Moreover, to rule out the possibility that the differences in protein levels were due to changes in protein stability, we analyzed protein decay of the PGK1(cup1)UAApG construct in both wild-type and upf1Δ strains. We determined that the decay rates of the Flag-Cup1 protein were the same in both strains (our unpublished results). This indicated that the differences in protein steady-state levels that we observed were not due to differences in protein decay rate and instead were due to differences in the rate of protein production. These results suggest that preventing recognition of an mRNA as aberrant by removing the Upf1 protein increases the translation efficiency of that transcript. In contrast, the upf1Δ had little effect on the protein produced per transcript for the normal control PGK1(cup1)UAApG(ΔDSE) mRNA (Figure 3).
We also examined the effects of removing the decapping enzyme on the expression of the PGK1(cup1)UAApG and PGK1(cup1)UAApG(ΔDSE) mRNAs by using a strain deleted for the DCP1 gene, which encodes the decapping enzyme (Beelman et al., 1996; LaGrandeur and Parker, 1998). Unexpectedly, the amount of protein per PGK1(cup1)-UAApG mRNA decreased in the dcp1Δ strain compared with the wild-type strain (Figure 5, A, compare lanes 1 and 2, and B). This observation suggested that perhaps the slow growth rate of the dcp1Δ strain, or other unknown changes to cellular physiology, made the interpretation of the protein to mRNA levels difficult in the dcp1Δ strain and implied that there might be a global decrease in translation in the dcp1Δ strain. Consistent with this result, the amount of protein produced per PGK1(cup1)UAApG(ΔDSE) mRNA, which is not subjected to nonsense-mediated decay, was also reduced in the dcp1Δ strain (Figure 5A, lanes 1 and 4).
Figure 5.
Relative protein levels of the PGK1(cup1)UAApG and the PGK1(cup1)UAApG(ΔDSE) transcripts in dcp1Δ and dcp1Δupf1Δ strains. (A) Western analysis of PGK1(cup1)UAApG and PGK1(cup1)UAApG(ΔDSE) constructs in the wild-type (WT), dcp1Δ, and dcp1Δupf1Δ strains. Strain names and constructs are given above the Western blots, and lanes are labeled below as a reference for the text. FLAG antibodies were used to probe the Western blots. (B) Values for protein levels, percent mRNA levels, and ratios of protein per mRNA are shown for each lane corresponding to the Western blots in A. These values are from at least five independent experiments. To allow comparisons, we have set the levels of the protein produced from the nonsense mRNA PGK1(cup1)UAApG in dcp1Δ cells at 100%.
To test whether the underlying alterations in growth rate in the dcp1Δ strain were complicating the analysis in this strain, we reasoned that the analysis of a dcp1Δupf1Δ strain should be informative. If the conclusions drawn from comparing wild-type versus upf1Δ strains were correct, then in the dcp1Δupf1Δ double mutant we should see no change in the mRNA decay rate or the steady-state level of transcript compared with the dcp1Δ strain. However, there should be an increase in the amount of protein being produced per transcript for mRNAs that would normally be recognized as aberrant. This effect would be due to the requirement for the UPF1 protein to recognize the mRNA as aberrant.
Comparison of the PGK1(cup1)UAApG mRNA in the dcp1Δ and dcp1Δupf1Δ double mutant strains indicated that mRNA decay rates were basically unchanged in each strain (Figure 1). However, there was more protein produced per PGK1(cup1)UAApG mRNA in the dcp1Δupf1Δ double mutant strain compared with the dcp1Δ strain (Figure 5A, compare lanes 2 and 3). These results indicated that the translation of the PGK1(cup1)UAApG mRNA is repressed in a Upf1p-dependent manner that is independent of the decapping enzyme. This provides evidence that the recognition of the mRNA as nonsense containing leads to a repression of translation. In contrast, the amount of protein per transcript for the PGK1(cup1)UAApG(ΔDSE) mRNA, which is not subjected to nonsense-mediated decay, was unchanged between the dcp1Δ and dcp1Δupf1Δ strains (Figure 5A, compare lanes 4 and 5). We interpreted these observations to indicate that in the absence of the UPF1 protein the mRNA is no longer recognized as aberrant, and this leads to an increase in the translational efficiency of the mRNA.
DISCUSSION
Recognition of an mRNA as Nonsense Containing Leads to an Inhibition of Translation
Three lines of evidence suggest that when an mRNA is recognized as nonsense containing or aberrant, the translation of that transcript is repressed. First, the amount of protein produced per PGK1(cup1)UAApG transcript was increased when the DSE was deleted, thus preventing the recognition of this mRNA as aberrant (Figure 3). Second, the amount of protein produced per PGK1(cup1)UAApG transcript was increased in the upf1Δ strain compared with the wild-type strain (Figure 3). Third, comparison of dcp1Δ and dcp1Δupf1Δ strains, wherein mRNA decay and mRNA levels are the same, indicated that the dcp1Δupf1Δ strain produced more protein per PGK1(cup1)UAApG transcript than the dcp1Δ strain (Figure 5). We interpret these observations to suggest that when mRNAs are recognized as nonsense containing, they are both repressed for translation and activated for decapping.
Our data indicate that the Upf1p acts upstream of the decapping enzyme in the mechanism of mRNA decay induced by nonsense codons. The critical observation was that the upf1Δ both prevented the repression of translation and activation of decapping, whereas the dcp1Δ prevented only activation of mRNA decapping. A role for the Upf1p early in the mRNA surveillance pathway is also suggested by two prior observations. First, specific alleles of Upf1p can affect suppression of nonsense codons, thereby implying a role of this protein in the translation termination process (Weng et al., 1996a,b). Second, and consistent with a role in termination per se, the Upf1p has been shown to physically interact with the termination factors RF1 and RF2 (Czaplinski et al., 1998). Moreover, it should be noted that because translation is repressed in a dcp1Δ strain and not in a dcp1Δupf1Δ strain, the Dcp1p itself is not required for the process that leads to down-modulation of translation. This result indicates that translational repression of nonsense-containing mRNAs occurs before decapping.
An important issue is how recognition of an mRNA as nonsense containing leads to the activation of decapping and the repression of translation. The first phase of this process is the manner in which normal mRNAs are distinguished from abnormal transcripts. The available literature suggests that the difference between normal and abnormal transcripts is the relationship between the translation termination codon and both positive and negative sequence elements within the mRNA (reviewed by Hilleren and Parker, 1999; Peltz et al., 1999). Recognizing an abnormal transcript also appears to require hydrolysis of ATP by Upf1p, although the specific role of the ATP hydrolysis is unclear. In one model, the Upf1p uses ATP hydrolysis to scan 3′ of the termination codon to identify downstream sequences that trigger decay (Hentze and Kulozik, 1999; Peltz et al., 1999). In this view, recognition of downstream sequences would then trigger a signal transduction process that leads to repression of translation and mRNA decapping. An alternative possibility is that the Upf1p serves as a sort of internal clock to distinguish proper from improper termination contexts. In this view, ATP hydrolysis by Upf1p might alter the nature of translation termination in a manner that affects translation initiation rate (for more discussion, see Hilleren and Parker, 1999). An important goal of future work will be to resolve the manner in which aberrant termination is coupled to translation repression.
Regardless of the specific mechanism by which the nonsense-containing signal reaches the 5′ end to promote decapping, it should be noted that deadenylation-independent decapping is not a general response to any block to translation initiation. This conclusion is based on the observation that other blocks to translation initiation do not trigger deadenylation-independent decapping. For example, inhibition of translation initiation by loss of function mutations in initiation factors (Schwartz and Parker, 1999) or with stem-loop structures in the 5′ UTR (Muhlrad et al., 1995) does not lead to deadenylation-independent decapping. This implies that the recognition of an mRNA as nonsense containing triggers a specific alteration to the 5′ end that allows rapid decapping.
Dcp1Δ Cells Show Additional Abnormalities
In our experiments the dcp1Δ cells showed two unexpected abnormalities. First, the amount of steady-state mRNA in a dcp1Δ strain was lower than expected for the observed change in decay compared with wild-type cells. For example, although the PGK1(cup1)UAApG transcript showed an almost 10-fold increase in mRNA half-life in both the dcp1Δ and dcp1Δupf1Δ double mutant strains, the levels of the transcript were only slightly higher than in wild-type strains. The lower than expected levels of particular mRNA species in the dcp1Δ strains are not limited to our constructs and are also seen with GAL1, GAL7, and GAL10 mRNAs (our unpublished results). We do not currently understand the basis for the differences in steady-state levels, but this clearly indicates that there must be additional factors that modulate mRNA steady-state levels. There are two simple possibilities. First, there might be a feedback mechanism such that transcription is reduced in response to a decrease in mRNA turnover. Alternatively, transcription may be proceeding at a normal pace, but there is a limited number of proteins required for stable production of a messenger ribonuclear particle (mRNP). In this latter view, the extra mRNAs that would be accumulating would titrate the limiting mRNP components and therefore lead to the production of mRNAs lacking critical mRNA-binding proteins. Such unfinished mRNPs might then be rapidly degraded, perhaps in the nucleus, at a rate too fast to be transiently observed. It should be noted that the observation that steady-state mRNA levels and mRNA decay rates do not always correlate indicates that the use of steady-state levels to measure mRNA decay rates may be misleading.
The second surprising feature of the dcp1Δ strain was that the amount of protein being produced per mRNA was significantly reduced. Importantly, this was true even for mRNAs that were not recognized as nonsense containing. Specifically, the amount of protein produced from the PGK1(cup1)UAApG(ΔDSE) transcript was reduced in dcp1Δ cells. This observation suggests that dcp1Δ cells have a defect in translation attributable to any number of possibilities. First, the Dcp1p could be functioning as an important component of the translation initiation machinery (in addition to being a degradative enzyme). Second, the mRNAs that are accumulating in dcp1Δ cells will be largely lacking the poly(A) tail and therefore would be at a translational disadvantage, thereby leading to less protein per mRNA. Third, it is possible that the translation defect is due to an indirect alteration of cellular metabolism caused by the dcp1Δ. For example, it is known that in dcp1Δ strains mRNAs are degraded 3′ to 5′ (Jacobs Anderson and Parker, 1998), and it is possible that in dcp1Δ cells, the 5′ portion of the mRNA that might accumulate titrates cap-binding proteins, essentially acting as a cap competitor.
ACKNOWLEDGMENTS
We thank Audrey Atkins for the UPF1 deletion plasmid. We also thank members of the Parker lab for helpful comments. This work was supported by a grant from the Howard Hughes Medical Institute.
REFERENCES
- Beelman CA, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- Caponigro G, Muhlrad D, Parker R. A small segment of the MATα1 transcript promotes mRNA decay in yeast: a stimulatory role for rare codons. Mol Cell Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G, Parker R. mRNA turnover in yeast promoted by the MATα1 instability element. Nucleic Acids Res. 1996;24:4304–4312. doi: 10.1093/nar/24.21.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T. Messenger RNA deadenylation precedes decapping in mammalian cells. Proc Natl Acad Sci USA. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Gonzalez CI, Peltz SW. Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. Bioessays. 1999;8:685–696. doi: 10.1002/(SICI)1521-1878(199908)21:8<685::AID-BIES8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes & Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes & Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Gera JF, Baker EJ. Deadenylation-dependent and -independent decay pathways for alpha1-tubulin mRNA in Chlamydomonas reinhardtii. Mol Cell Biol. 1998;3:1498–1505. doi: 10.1128/mcb.18.3.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan KW, Ruiz-Echevarria MJ, Quan Y, Peltz SW. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol Cell Biol. 1995;15:809–823. doi: 10.1128/mcb.15.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- Higgs DC, Colbert JT. Oat phytochrome A mRNA degradation appears to occur via two distinct pathways. Plant Cell. 1994;6:1007–1019. doi: 10.1105/tpc.6.7.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, Parker R. mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA. 1999;5:711–719. doi: 10.1017/s1355838299990519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CL, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs Anderson JS, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- LaGrandeur T, Parker R. Isolation and characterization of DCP1p, the mRNA decapping enzyme. EMBO J. 1998;17:1487–1496. doi: 10.1093/emboj/17.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes & Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Lim SK, Maquat LE. Human β-globin mRNAs that harbor a nonsense codon are degraded in murine erythroid tissues to intermediates lacking regions of exon I or exons I and II that have a cap-like structure at the 5′ termini. EMBO J. 1992;11:3271–3278. doi: 10.1002/j.1460-2075.1992.tb05405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci USA. 1979;76:5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ to 3′ digestion of the transcript. Genes & Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on three mRNA sequence elements and at least one trans-acting factor. Genes & Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes & Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996a;10:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996a;10:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]