Abstract
Cold acclimation in woody plants may have special features compared to similar processes in herbaceous plants. Recent studies have shown that circadian clock behavior in the chestnut tree (Castanea sativa) is disrupted by cold temperatures and that the primary oscillator feedback loop is not functional at 4°C or in winter. In these conditions, CsTOC1 and CsLHY genes are constantly expressed. Here, we show that this alteration also affects CsPRR5, CsPRR7 and CsPRR9. These genes are homologous to the corresponding Arabidopsis PSEUDO-RESPONSE REGULATOR genes, which are also components of the circadian oscillator feedback network. The practically constant presence of mRNAs of the 5 chestnut genes at low temperature reveals an unknown aspect of clock regulation and suggests a mechanism regulating the transcription of oscillator genes as a whole.
Introduction
Circadian clocks allow organisms to adapt to periodic environmental changes in light and temperature. In plants, circadian clock performance increases growth, survival and competitive advantage [1], [2]. Although the clock components do not seem to be conserved among kingdoms, clock mechanisms in different organisms are remarkably similar [3]–[5]. In the model plant Arabidopsis, it was initially proposed that circadian rhythms are based on a feedback loop in which two partially redundant proteins, LHY (late elongated hypocotyl) and CCA1 (circadian clock associated 1), negatively control their own synthesis by inhibiting the expression of the positively regulating TOC1 (timing of cab, chlorophyll a/b binding protein, expression 1) transcription factor [6]. However, current evidence indicates that the Arabidopsis oscillator comprises several interlocking feedback loops, comparable to those identified in the circadian systems of other eukaryotes [7]–[9]. Besides having established roles for the primary clock genes LHY, CCA1 and TOC1 in the oscillator feedback network, experimental data point to the participation in this mechanism of several PSEUDO-RESPONSE REGULATOR (PRR) genes, belonging to the same family as TOC1/PRR1 [10]–[14]. This family consists of five members that are expressed after dawn in the sequential order PRR9→PRR7→PRR5→PRR3→TOC1 [15]. A feedback loop between PRR9/PRR7 and LHY/CCA1 was initially proposed whereby LHY and CCA1 trigger the expression of PRR9 and PRR7, and the corresponding PRR proteins feed back to regulate LHY and CCA1 expression [11], [13]; PRR5 was later proposed to also participate in such a loop [12]. Accordingly, computational models of the Arabidopsis circadian oscillator include this feedback loop between PRR9/PRR7 and LHY/CCA1 [16], [17].
In a recent attempt to elucidate the role played by low temperatures in the onset of winter dormancy in woody plants, we showed that circadian clock behavior in the chestnut tree is disrupted in response to cold [18]. The chestnut genes CsTOC1 and CsLHY, which are homologous to essential components of the circadian oscillator in Arabidopsis, were observed to cycle daily during vegetative growth as expected. However, during winter, the presence of high non-oscillating levels of CsTOC1 and CsLHY mRNAs indicates alteration of the circadian clock. In addition, we were able to induce a similar disruption by chilling (4°C) chestnut seedlings [18]. To determine the extent to which this chestnut clock disruption affects other elements of the oscillator feedback network, we investigated the behavior of chestnut PRR genes during winter dormancy and in response to cold. Here, we report that the interrupted circadian behavior observed previously in CsTOC1 and CsLHY expression is also true of CsPRR5, CsPRR7 and CsPRR9.
Results and Discussion
The Circadian Behavior of the Chestnut PRR5, PRR7 and PRR9 Genes
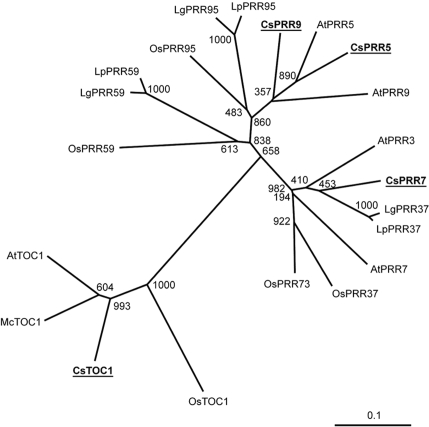
To help clarify the behavior of the chestnut circadian clock response to cold, additional members of the PRR gene family were identified by screening a chestnut stem cDNA library. Three full-length cDNA clones, homologous to Arabidopsis PRR genes were isolated. Each clone encoded a polypeptide containing the two domains characteristic of the Arabidopsis PRR protein family (PRR1, PRR3, PRR5, PRR7 and PRR9): the pseudo-receiver domain and the CCT (CONSTANS, CONSTANS-LIKE, and TOC1) motif. To explore possible genetic relationships between these chestnut polypeptides and the PRRs previously characterized in Arabidopsis and other species [15], [19]–[21], we constructed a phylogenetic tree (Figure 1). The topology of the phylogram revealed three groups of PRR proteins, as has been observed by Murakami et al. [19]. One of these groups, which includes CsTOC1/CsPRR1, comprises the PRR1 proteins of the different species. The other two groups each included representatives of two PRR proteins: PRR5 and PRR9 in one group, and PRR3 and PRR7 in the other. In these last two groups, the PRR proteins of rice, Lemna gibba and Lemna paucicostata were designated two numbers (37 or 73, and 59 or 95), since it was difficult to estimate which PRR from rice or the two Lemna species corresponded to which Arabidopsis PRR [19], [21]. Of the three chestnut pseudo-response regulators identified in the present study, two were found to group with the proteins PRR5 and PRR9. These PRRs were denoted CsPRR5 and CsPRR9 based on the relative distances shown in the phylogram to Arabidopsis PRR5 and PRR9. The third chestnut PRR we could infer from the clones isolated, grouped within the PRR3 and PRR7 protein class. Given the characteristics of the phylogram and the fact that we did not isolate the gene coding for the other PRR protein of this group in the chestnut, it is difficult to ascribe the corresponding homology to this CsPRR. We opted for tentatively designating it as CsPRR7 rather than CsPRR3 based on the similar circadian expression pattern of the gene to that shown by the Arabidopsis PRR7 gene (Figure 2; see below) [15].
Figure 1. Phylogram of PRR proteins.
A non-rooted neighbor-joining phylogenetic tree of PRR proteins was constructed using amino acid sequences of the pseudo-receiver domain (55). Species identifiers are: At, Arabidopsis thaliana; Cs, Castanea sativa; Lg, Lemna gibba; Lp, Lemna paucicostata; Mc, Mesembryanthemum crystallinum; Os, Oryza sativa. The following are the accession numbers for the proteins: AtTOC1 (AF272039); AtPRR3 (BAB13744); AtPRR5 (BAB13743); AtPRR7 (BAB13742); AtPRR9 (BAB13741); CsTOC1 (AY611028); CsPRR5 (ABV53464); CsPRR7 (ABV53463); CsPRR9 (ABV53465); LgPRR37 (BAE72700); LgPRR59 (BAE72701); LgPRR95 (BAE72702); LpPRR37 (BAE72697); LpPRR59 (BAE72698); LpPRR95 (BAE72699); McTOC1 (AAQ73525); OsTOC1 (BAD38854); OsPRR37 (BAD38855); OsPRR59 (AK120059) (KOME database); OsPRR73 (BAD38856); OsPRR95 (BAD38857). Numbers at each branch point indicate the bootstrap replicates (out of 1000) giving rise to the topology.
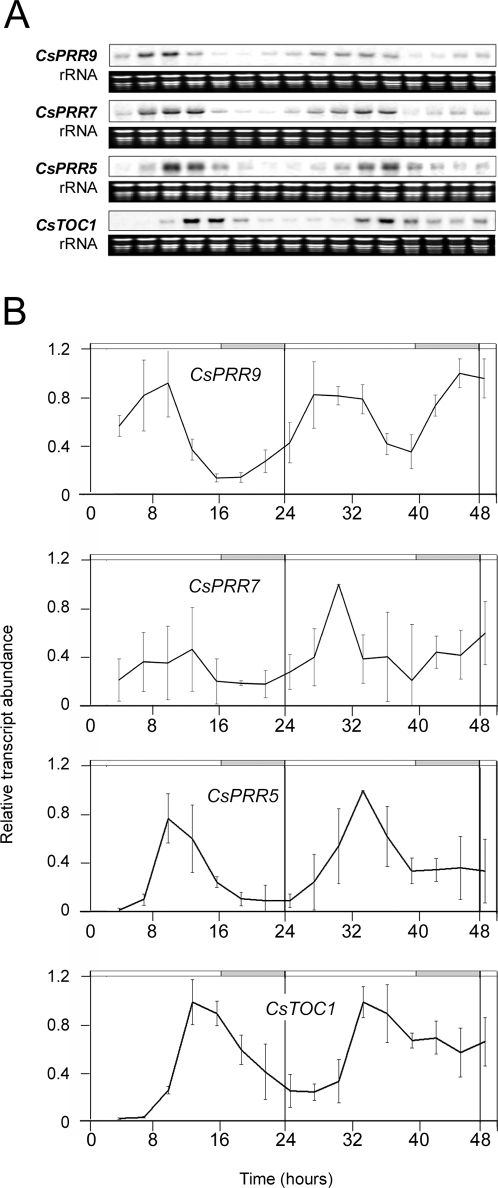
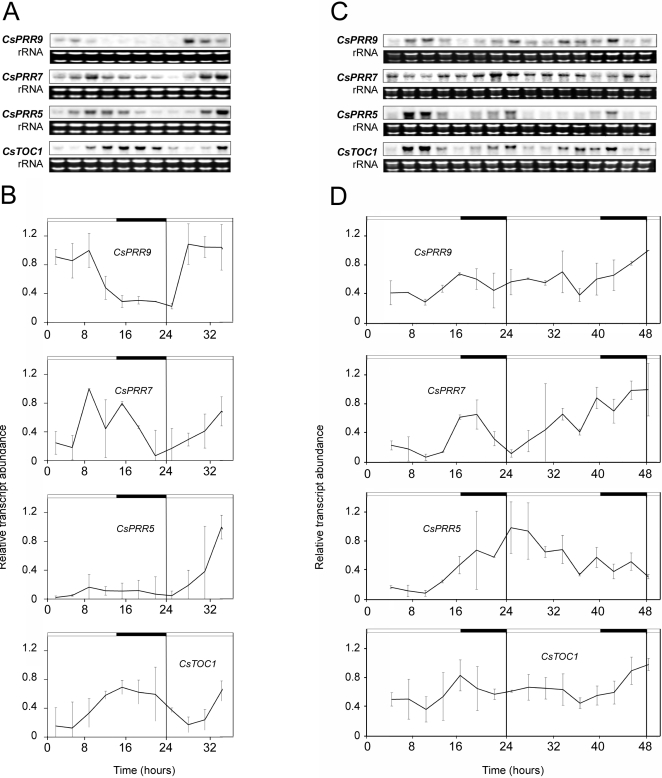
Figure 2. CsPRR gene expression levels in chestnut leaves under LL conditions.
CsPRR gene expression rhythms observed in leaves taken from 16- to 24 week-old chestnut seedlings grown under standard conditions (LD, 22°C) subsequently transferred to conditions of LL and 22°C. Samples were collected at 3-h intervals. (A) Northern blot analysis. The rRNA loading reference was detected by staining gels with ethidium bromide. (B) Quantitative RT-PCR analysis. Relative transcript abundances are shown in the graphs. Data are means from three biological replicates. The open and shaded bars above the graphs represent subjective day and night lengths, respectively.
To test the circadian behavior of the CsPRR genes, we analyzed their expression in leaves collected from 16- to 24 week-old chestnut seedlings grown under continuous light (LL) conditions in a temperature-controlled (22°C) growth chamber over a 48 h period. RNA samples were analyzed by Northern blot hybridization using probes specific for each gene CsTOC1/PRR1, CsPRR5, CsPRR7, and CsPRR9. These probes were sequences of the non-conserved region of PRR family genes, flanked by the pseudo-receiver and CCT domains. We observed the diurnal oscillation of transcript levels of the CsPRRs over an approximate 24 h period (Figure 2). These chestnut PRR transcripts started to accumulate after subjective dawn in the order PRR9 and PRR7→PRR5→TOC1. Despite the concurrent appearance of PRR9 and PRR7 mRNAs, PRR9 mRNA peaked earlier than PRR7 mRNA. This expression order of the CsPRR genes resembles that of the Arabidopsis PRR genes (PRR9→PRR7→PRR5→PRR3→TOC1) more than the order noted in the monocotyledons rice or Lemna spp. [15], [19], [21]. For example, circadian analysis of OsPRR in rice also indicates their sequential expression but in a different order: OsPRR73 and OsPRR37→OsPRR95 and OsPRR59→OsPRR1 [19]. Differences among species in the temporal order of PRR gene expression could be essential, given the direct role these genes play in the machinery of the circadian oscillator [11]–[14]. In particular, the feedback loop proposed among PRR9, PRR7, and possibly PRR5 and LHY/CCA1, which is thought to form part of a multiple-loop network [7]–[9], could be affected by these differences. The expression interval of each PRR could also be a determining factor for their participation in clock output pathways. The findings of a recent study suggest that PRR9, PRR7 and PRR5 regulate flowering time in Arabidopsis through the CONSTANS- dependent pathway [22]. Interestingly, a genetic complementation analysis in which the rice genes OsPRR1 and OsPRR37 were introduced into the corresponding Arabidopsis loss-of-function mutants (toc1 and prr7 respectively) revealed that these genes are only partially interchangeable between these species [23].
The Circadian Behavior of CsPRR Genes is Disrupted in Response to Low Temperatures
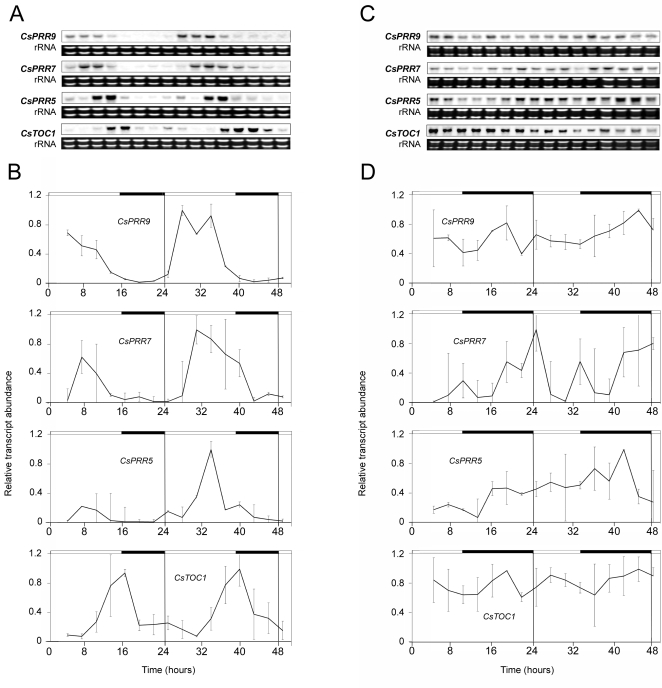
When we addressed whether the circadian behavior of CsPRR5, CsPRR7 and CsPRR9 was modified during winter dormancy or in response to cold, similar changes were observed to those described previously for the central oscillator genes, CsLHY and CsTOC1 [18]. We examined the expression of the CsPRR genes in adult chestnuts grown under natural light and temperature conditions in Zarzalejo, Madrid (4°11′W, 40°35′N): first in June, which is when vegetative growth takes place, and then in December, when temperatures are low and these trees are in a state of endodormancy. In samples collected in June, oscillatory expression patterns were observed for the four CsPRR genes. The characteristics of their fluctuations were consistent with those described above for plantlets exposed to continuous light. This behavior was noted both in stem and leaf samples (Figures 3A, 3B, and S1). In contrast, in stem samples collected in December, the circadian expression of the CsPRR genes was modified. Thus, rather than exhibiting typical daily cycles, CsPRR mRNA levels remained consistently high (Figure 3C and 3D). Hence, the previously described altered expression of CsTOC1 and CsLHY in winter was similarly shown by CsPRR5, CsPRR7 and CsPRR9.
Figure 3. CsPRR gene expression in stems of adult chestnuts under different seasonal conditions.
Stems (second-year branch internodes) collected in June (A and B) and December (C and D). Samples were collected at 3-h intervals. (A and C) CsPRR northern blot analysis. The rRNA loading reference was detected by staining gels with ethidium bromide. (B and D) Quantitative RT-PCR analysis. Relative transcript abundances are shown in the graphs. Data are means from two biological replicates. Open and filled bars above each graph represent natural day and night lengths, respectively, as provided by the National Institute of Meteorology, Madrid.
We were also able to confirm that, as for CsTOC1 and CsLHY, this alteration is not intrinsic to the state of endodormancy. Endodormancy is caused by plant endogenous factors and, once established, no growth can be achieved until a chilling requirement has been satisfied. In order for bud break to occur, plants need to be exposed to low temperatures for a cumulative number of hours. Thus, endodormant plants that had not yet satisfied the chilling requirement, were transferred to growth chambers kept at 22°C and subjected to a long day (LD) photoperiod (16 h light / 8 h dark). In these conditions, the plants remain in a state of endodormancy yet they gradually lose their characteristics exclusive to the cold response. After one week at 22°C, expression levels of the CsPRR genes in stem tissue had recovered their circadian rhythm (Figure 4).
Figure 4. Recovery of circadian CsPRR gene expression in endodormant chestnut seedlings.
CsPRR gene expression rhythms observed in stems from 11 month-old endodormant chestnut plants grown under natural conditions in Madrid and subsequently transferred to controlled-environment chambers under conditions of 22°C and LD for 1 week. Samples were collected at 3-h intervals. (A) Northern blot analysis. The rRNA loading reference was detected by staining gels with ethidium bromide. (B) Quantitative RT-PCR analysis. Relative transcript abundances are shown in the graphs. Data are means from two biological replicates. Open and filled bars above each graph indicate lights on and lights off, respectively.
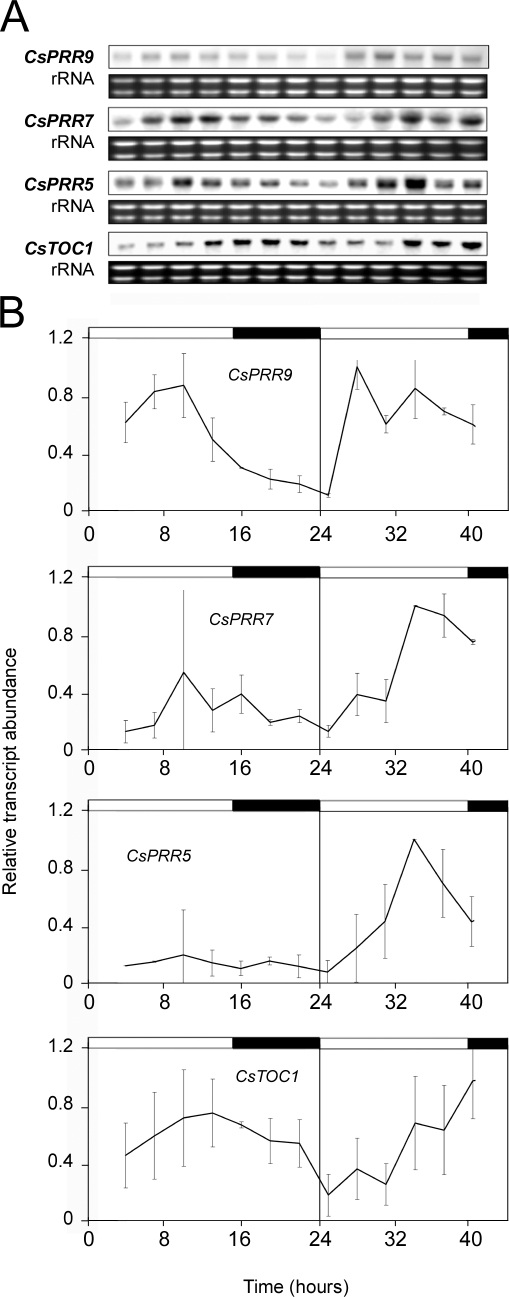
To establish whether exposure to low temperatures was sufficient to interrupt the circadian behavior of the CsPRR genes, experiments were performed on 16- to 24 week-old chestnut seedlings. CsPRR expression patterns in stems and leaves collected from seedlings grown under the conditions 22°C and LD were compared to expression patterns in seedlings subjected to 4°C and LD conditions for 1 week. At 22°C, the mRNAs of the 4 CsPRR genes indicated their cyclic expression with specific circadian oscillation both in stem and leaf samples. In contrast, in the plants exposed to low temperatures, this oscillatory behavior was interrupted (Figures 5 and S2). These effects of cold were characterized by the presence and not by the absence of the CsPRR mRNAs over time, as occurs in adult trees during winter.
Figure 5. CsPRR gene expression in the stems of chestnut seedlings grown under different temperature conditions.
The figure shows CsPRR blot analysis and mRNA abundances. (A and B) Stems from seedlings grown under conditions of LD and 22°C. (C and D) Stems from seedlings grown under standard conditions (LD, 22°C) and subsequently subjected to one week of LD at 4°C. Samples were collected at 3-h intervals. (A and C) CsPRR northern blot analysis. The rRNA loading reference was detected by staining gels with ethidium bromide. (B and D) Quantitative RT-PCR analysis. Relative transcript abundances are shown in the graphs. Data are means from two biological replicates. Open and filled bars above each graph indicate lights on and lights off, respectively.
These results indicate that low temperatures greatly modify the expression of at least 5 of the genes potentially involved in the oscillator mechanism of the chestnut circadian clock. This cold response in the chestnut may translate to all woody species. Indeed, expression levels of clock oscillator genes in Populus alba have also been observed to not show circadian cycling in winter (our unpublished results). In contrast, the Arabidopsis clock seems to exhibit different behavior in response to cold. Cycling of the mRNA levels of genes under circadian control (CAB and CCR2, cold-circadian rhythm-RNA binding 2) has been observed in Arabidopsis seedlings grown for 5 days at 4°C under a light/dark photoperiod [24] and, recently, Bieniawska et al. [25] have shown that under continuous light conditions, cold disrupts the circadian expression of Arabidopsis oscillator genes, while in normal diurnal light-dark conditions cold only reduces the amplitude of cycles of clock components. The different responses to low temperatures shown by the circadian clocks of Arabidopsis and the chestnut could be attributable to differences in the clock mechanism or in the regulation of a shared mechanism. It is difficult to predict which mechanisms of control could be affected by cold, and besides the possibility of an influence on transcriptional level control other alternatives should be considered. Studies on clock gene regulation mechanisms in plants at the posttranscriptional and posttranslational levels are currently underway [26]–[30]. In effect, of the four PRRs examined in this work, three (TOC1, PRR5 and PRR7) are now known to be regulated at the protein level in Arabidopsis [14], [26], [29]. Moreover, the circadian expression of TOC1 has been recently correlated with clock-controlled histone acetylation rhythms of its promotor [31].
Cold disruption of the chestnut circadian clock in normal diurnal light-dark conditions indicates that the cold acclimation process in woody plants may have special features compared to similar processes in herbaceous plants. Differences between species are hardly surprising when one considers that trees have had to adapt to two types of thermal stress: the fluctuating temperatures endured during the growing season and the continuous temperature drop that occurs in winter. Stopping of the clock is likely to have an impact on the general physiology of the plant, since this molecular oscillator is known to regulate major metabolic pathways, and over 10% of genes in Arabidopsis are under circadian control [32]–[34]. Moreover, the circadian clock participates in cold signaling pathways in Arabidopsis by gating the low-temperature induction of CBFs (CRT/DRE, C-repeat/drought-responsive, binding factor) and modulating low-temperature Ca2+ signals [35], [36]. Welling and Palva [37] have shown that in addition to their role in cold acclimation during the growing season, CBFs are involved in the regulation of cold tolerance during overwintering in birch.
It has long been established that the photoperiod controls the induction of winter dormancy processes in most trees growing in temperate climates [38], [39]. Recently Böhlenius et al. have shown that the CO/FT (constans/flowering time locus T) regulatory module, which controls flowering time in response to variations in daylength in annual plants, also regulates short-day-induced growth cessation [40]. However, in order to attain an advanced state of dormancy, low temperatures are also required and it is known that changes such as leaf senescence and abscission are not provoked by a short day photoperiod alone, but also require exposure to cold [41], [42]. In fact, in the apple tree and other species of the family Rosaceae, cold induces dormancy regardless of photoperiodic conditions [43]. Although winter clock disruption does not seem to maintain dormancy, since in endodormant plants transferred to conditions of 22°C and a LD, standard PRR gene expression cycling resumes, the initial changes induced by the first low autumn temperatures could trigger the onset of endodormancy. In addition, stopping of the circadian clock linked to low winter temperatures could in part explain the extensive remodeling of meristem transcriptome observed in the vascular cambium of poplar during the transition from growth to dormancy [44]–[47].
A similar circadian clock response to the cold has been observed in the ruin lizard (Podarcis sicula), a hibernating ectothermal vertebrate. At low temperatures (6°C), the cycling expression of two clock genes (Per2 and Clock) in this animal is diminished in peripheral clocks with a characteristic increase in basal expression levels [48], [49]. The basic mechanisms of clock function in plants and animals are similar, although their oscillator genes are unrelated. This parallelism between two such evolutionary distinct organisms suggests that the stopping of the circadian clock in response to cold could be part of a general adaptive strategy that enables living organisms that undergo dormancy or hibernation to survive the winter. Recently, the interruption of the molecular circadian clock in the European hamster during hibernation has also been described; the clock genes Per1, Per2, and Bmal1 being constantly expressed in the suprachiasmatic nucleus during deep torpor [50]. Interestingly, clock disruption in the three organisms results in increased expression of oscillator genes, suggesting a positive role of these components during low temperature periods.
In summary, our findings indicate that in the chestnut, low temperatures disrupt the canonical cyclic expression of the circadian oscillator genes CsPRR5, CsPRR7, and CsPRR9, as previously observed by us for CsTOC1/PRR1 and CsLHY. Concurrent changes in the expression of these five genes forming part of several oscillator feedback loops, point to control mechanisms not yet elucidated by current Arabidopsis circadian clock models.
Materials and Methods
Plant Material and Growth Conditions
In field experiments, stem samples (2-year-old branch internodes) and leaves were harvested from adult European chestnut trees (Castanea sativa Mill.) in Zarzalejo, Madrid (4°11′W, 40°35′N). Samples were collected in the months of June (22.8°C average temperature; 15 h, 5 min average day length) and December (4.9°C average temperature; 9 h, 16 min average day length). Controlled-environment experiments were performed using 16–24 week-old chestnut seedlings in growth chambers under the conditions described in Ramos et al. [18]. For the long day (LD) trials, seedlings were grown at 22°C and subjected to a 16 h light/8 h dark (16∶8) photoperiod. Exposure to cold (4°C) was undertaken for a week under the same light regime. Continuous light (LL) experiments were performed on plants that had been grown under conditions of LD and 22°C and thereafter subjected to LL from dawn. Endodormancy experiments were performed as in [18]. Chestnut plants grown in natural conditions enter a state of endodormancy at the end of November. Before satisfying the chilling requirement, plants were transferred to a growth chamber at 22°C under a 16 h light/8 h dark photoperiod and kept for 1 week in these conditions before sample collection. After subjecting the plants to the different treatments, specimens were collected at 3-h intervals. Each experiment was performed at least twice.
Isolation of cDNA Clones
Chestnut CsPRR5, CsPRR7 and CsPRR9 cDNA clones were isolated from a λUni-ZAP XR cDNA library following standard procedures [51]. The library was constructed using chestnut stem poly (A)+ RNA isolated from plants in winter [18]. To detect CsPRR clones other than TOC1, a full length CsTOC1/PRR1 clone was used as probe on two replicate membranes, one under high and the other under low stringency hybridization and washing conditions. After discarding the common spots as TOC1 clones, three different CsPRR clones were detected corresponding to fragments of the genes CsPRR9, CsPRR7 and CsPRR5. To obtain the CsPRR9 full-length cDNA clone, the corresponding fragment was used as probe under high-stringency hybridization and washing conditions. Probes were labeled with [α-32P] dATP using a random-primed DNA labeling kit (Roche Applied Science, Indianapolis). Full-length cDNA clones for CsPRR7 and CsPRR5 were obtained using the “BD SMART RACE cDNA Amplification” kit (Clontech, Mountain View, CA) according to the manufacturer's instructions.
Northern Blot Expression Analysis
Total RNA was obtained from chestnut stems and leaves as described in [52], separated on 1.2% agarose gels with 2.2 M formaldehyde [53], and subsequently transferred to Hybond-XL nylon membranes (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). 32P-labeled DNA probes used to detect each specific mRNA were designed to only span the non conserved region of each pseudo response regulator gene. The probes were PCR amplified using appropriate sets of forward and reverse primers:
CsPRR5 forward 5′-GGCAAATCGTTTCCAAGTGA-3′ and
CsPRR5 reverse 5′-TAGAAGAGTTGACAAGGACATA-3′,
CsPRR7 forward 5′-GAAGACATCGGGATGTGCAA-3′ and
CsPRR7 reverse 5′- CCTGAACACAGCTAGTGCC-3′,
CsPRR9 forward 5′-GCTTCCTCGCATTGCTACAG-3′ and
CsPRR9 reverse 5′-AACAACAAAGCCAGGCATCG-3′.
A gene fragment spanning CsTOC1 nucleotides 1380–1591 that specifically recognizes the TOC1/PRR1 member of the pseudoresponse regulator (PRR) gene family was used as in Ramos et al. [18]. Northern blot hybridizations were conducted according to the recommendations of the membrane manufacturer. Membranes were washed at high stringency, exposed on storage phosphor screens and visualized in a TYPHOON 9400 phosphorimage scanner (GE Healthcare, Bio-Sciences Corp.). The rRNA loading reference was estimated by staining gels with ethidium bromide.
Real time RT-PCR Expression Analysis
Total RNA was obtained from chestnut stems and leaves as described in [52] with a modification introduced after LiCl precipitation in which the RNeasy Plus Mini Kit columns from Qiagen were used. This kit includes one step to eliminate any contaminating genomic DNA in the total RNA sample. To check the lack of degradation, RNA was separated by electrophoresis on a formamide-formaldehyde denaturing agarose gel. RNA purity and quantity were checked with a Nanodrop Spectrophotometer. For each sample, single stranded cDNA was synthesized from one microgram of total RNA using the Superscript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen). This mix includes oligo (dT)20 and random hexamers. First-strand cDNA was synthesized in a 2720 thermocycler (Applied Biosystems). Gene-specific primers were designed using Primer Express 2.0 (Applied Biosystems) for the non conserved region of each pseudo response regulator gene as follows:
CsPRR5 forward 5′-GCAAAACAAAGAAGAATACTTG-3′ and
CsPRR5 reverse 5′-CTTCACTCCCATGCGTAAG-3′,
CsPRR7 forward 5′-ATTTGTTAAGTGCGTCCCTTG-3′ and
CsPRR7 reverse 5′- TTTCCATATTTGTTCCTGAAGC-3′,
CsPRR9 forward 5′-GAGGTTGTGCCCTTCGGAG-3′ and
CsPRR9 reverse 5′-ACAAGCATTTTCCTTCAATCTC-3′.
CsTOC1 forward 5′-ACTTGATGCTTCTGGCTTACCT-3′ and
CsTOC1 reverse 5′-ATTGTGCTGCTGATGGC-3′
Cs18S forward 5′-TCAACTTTCGATGGTAGGATAGTG-3′ and
Cs18S reverse 5′-CCGTGTCAGGATTGGGTAATTT-3′
Real time polymerase chain reactions were performed in an optical 384-well plate with an ABI PRISM 7900HT Sequence Detector System (Applied Biosystems), using SYBR Green to monitor dscDNA synthesis [54]. The reaction mixtures contained 1× Power SYBR Green Master Mix reagent (Applied Biosystems), 250 nM gene-specific primers (except Cs18S, for which 25 nM gene specific-primers were used) and 0.4 µl of the previously synthesized cDNA (except Cs18S for which 0.4 µl of a 25-fold fresh dilution of cDNA was used) in a final volume of 10 µl. The following standard conditions were used in all PCRs: 40 cycles of 95°C for 30 s and 60°C for 1 min. A dissociation step was performed after amplification to confirm the presence of a single amplicon. To estimate variation in the technique, three technical replicates were carried out for each biological replicate. Data were analyzed using SDS 2.2.2 software (Applied Biosystems). To generate a baseline-subtracted plot of the logarithmic increase in fluorescence signal (ΔR n) versus cycle number, baseline data and the R n threshold were detected automatically to obtain Ct (threshold cycle) values. Amplification efficiency for each gene was calculated based on four dilutions of template ranging from 500 ng to 0.5 ng and the equation E = 10−1/slope-1, with slopes in the range slope = −3.3±0.1 and E = 2. mRNA abundances for each candidate gene were calculated as: relative transcript abundance = 2 (−ΔΔCt). For the sample chosen as calibrator, ΔΔCt = 0 and therefore the fold-change = 1. Quantified data are shown in the graphs as relative amounts of mRNA. The sample with maximum expression (lower ΔCt) was used as calibrator and 18S ribosomal RNA was used as the reference gene to normalize data. The absence of genomic DNA contamination was checked using Non-Retrotranscriptase controls (RT-) and the absence of environmental contamination using Non-Template Controls (NTC).
Supporting Information
CsPRR gene expression in the leaves of adult chestnuts obtained in June. Leaves were collected in June at 3-h intervals. (A) CsPRR northern blot analysis. The rRNA loading reference was detected by staining gels with ethidium bromide. (B) Quantitative RT-PCR analysis. Relative transcript abundances are shown in the graphs. Data are means from two biological replicates. Open and filled bars above each graph represent natural day and night lengths, respectively, as provided by the National Institute of Meteorology, Madrid.
(0.33 MB TIF)
CsPRR gene expression in the leaves of chestnut seedlings grown under different temperature conditions. (A and B) leaves from seedlings grown under conditions of LD and 22°C. (C and D) leaves from seedlings grown under standard conditions (LD, 22°C) and subsequently subjected to one week of LD at 4°C. Samples were collected at 3-h intervals. (A and C) CsPRR northern blot analysis. The rRNA loading reference was detected by staining gels with ethidium bromide. (B and D) Quantitative RT-PCR analysis. Relative transcript abundances are shown in the graphs. Data are means from two biological replicates. Open and filled bars above each graph indicate lights on and lights off, respectively.
(0.62 MB TIF)
Acknowledgments
The authors thank J. Paz-Ares for helpful discussions and critical reading of the manuscript; and M. Ayllon and M. M. Salmean for allowing us to collect field samples in their estate. The help of P. González-Melendi in preparing the figures is also greatly appreciated.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the Spanish Ministerio de Educación y Ciencia, grant no. AGL2005-00080 and a predoctoral fellowship (A.C.). C.I. holds a fellowship awarded by the Chilean Government-BID.
References
- 1.Green RM, Tingay S, Wang Z, Tobin EM. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002;129:576–584. doi: 10.1104/pp.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dodd AN, Salathia N, Hall A, Kével E, Tóth R, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 3.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 4.Harmer SL, Panda S, Kay SA. Molecular bases of circadian rhythms. Annu Rev Cell Dev Biol. 2001;17:215–253. doi: 10.1146/annurev.cellbio.17.1.215. [DOI] [PubMed] [Google Scholar]
- 5.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 7.Mizuno T, Nakamichi N. Pseudo-Response Regulators (PRRs) or true oscillator components (TOCs). Plant Cell Physiol. 2005;46:677–685. doi: 10.1093/pcp/pci087. [DOI] [PubMed] [Google Scholar]
- 8.Gardner MJ, Hubbard KE, Hotta CT, Dodd AN, Webb AAR. How plants tell the time. Biochem J. 2006;397:15–24. doi: 10.1042/BJ20060484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClung CR. Plant circadian rhythms. Plant Cell. 2006;18:792–803. doi: 10.1105/tpc.106.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eriksson ME, Hanano S, Southern MM, Hall A, Millar AJ. Response regulator homologues have complementary, light-dependent functions in the Arabidopsis circadian clock. Planta. 2003;218:159–162. doi: 10.1007/s00425-003-1106-4. [DOI] [PubMed] [Google Scholar]
- 11.Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol. 2005;15:47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 12.Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005;46:686–698. doi: 10.1093/pcp/pci086. [DOI] [PubMed] [Google Scholar]
- 13.Salome PA, McClung CR. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell. 2005;17:791–803. doi: 10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farré EM, Kay SA. PRR7 protein levels are regulated by light and the circadian clock in Arabidopsis. Plant J. 2007;52:548–560. doi: 10.1111/j.1365-313X.2007.03258.x. [DOI] [PubMed] [Google Scholar]
- 15.Matsushika A, Makino S, Kojima M, Mizuno T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 2000;41:1002–1012. doi: 10.1093/pcp/pcd043. [DOI] [PubMed] [Google Scholar]
- 16.Locke JCW, Kozma-Bognár L, Gould PD, Fehér E, Nagy F, et al. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Sys Biol. 2006;2:59. doi: 10.1038/msb4100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeilinger MN, Farré EM, Taylor SR, Kay SA, Doyle FJ. A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Sys Biol. 2006;2:58. doi: 10.1038/msb4100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos A, Pérez-Solís E, Ibáñez C, Casado R, Collada C, et al. Winter disruption of the circadian clock in chestnut. Proc Natl Acad Sci USA. 2005;102:7037–7042. doi: 10.1073/pnas.0408549102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murakami M, Ashikari M, Miura K, Yamashino T, Mizuno T. The evolutionarily conserved OsPRR quintet: Rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 2003;44:1229–1236. doi: 10.1093/pcp/pcg135. [DOI] [PubMed] [Google Scholar]
- 20.Boxall SE, Foster JM, Bohnert HJ, Cushman JC, Nimmo HG, et al. Conservation and divergence of circadian clock operation in a stress-inducible crassulacean acid metabolism species reveals clock compensation against stress. Plant Physiol. 2005;137:969–982. doi: 10.1104/pp.104.054577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miwa K, Serikawa M, Suzuki S, Kondo T, Oyama T. Conserved expression profiles of circadian clock-related genes in two Lemna species showing long-day and short-day photoperiodic flowering responses. Plant Cell Physiol. 2006;47:601–612. doi: 10.1093/pcp/pcj027. [DOI] [PubMed] [Google Scholar]
- 22.Nakamichi N, Kita M, Niimura K, Ito S, Yamashino T, et al. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 2007;48:822–832. doi: 10.1093/pcp/pcm056. [DOI] [PubMed] [Google Scholar]
- 23.Murakami M, Tago Y, Yamashino T, Mizuno T. Characterization of the rice circadian clock-associated pseudo-response regulators in Arabidopsis thaliana. Plant Cell Physiol. 2007;71:1107–1110. doi: 10.1271/bbb.70048. [DOI] [PubMed] [Google Scholar]
- 24.Kreps JA, Simon AE. Environmental and genetic effects on circadian clock-regulated gene expression in Arabidopsis. Plant Cell. 1997;9:297–304. doi: 10.1105/tpc.9.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bieniawska Z, Espinoza C, Schlereth A, Sulpice R, Hincha DK, et al. Disruption of the Arabidopsis clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 2008;147:263–279. doi: 10.1104/pp.108.118059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Más P, Kim WY, Somers DE, Kay SA. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- 27.Daniel X, Sugano S, Tobin EM. CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci USA. 2004;101:3292–3297. doi: 10.1073/pnas.0400163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lidder P, Gutiérrez RA, Salomé PA, McClung CR, Green PJ. Circadian control of messenger RNA stability. Association with a sequence-specific messenger decay pathway. Plant Physiol. 2005;138:2374–2385. doi: 10.1104/pp.105.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiba T, Henriques R, Sakakibara H, Chua N-H. Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by a SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell. 2007;19:2516–2530. doi: 10.1105/tpc.107.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Portolés S, Más P. Altered oscillator function affects clock resonance and is responsible for the reduced day-length sensitivity of CKB4 overexpressing plants. Plant J. 2007;51:966–977. doi: 10.1111/j.1365-313X.2007.03186.x. [DOI] [PubMed] [Google Scholar]
- 31.Perales M, Más P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell. 2007;19:2111–2123. doi: 10.1105/tpc.107.050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harmer SL, Hogenesch LB, Straume M, Chang HS, Han B, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 33.Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, et al. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michael TP, McClung CR. Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol. 2003;132:629–639. doi: 10.1104/pp.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fowler SG, Cook D, Thomashow ME. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005;137:961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dodd AN, Jakobsen MK, Baker AJ, Telzerow A, Hou SW, et al. Time of day modulates low-temperature Ca signals in Arabidopsis. Plant J. 2006;48:962–973. doi: 10.1111/j.1365-313X.2006.02933.x. [DOI] [PubMed] [Google Scholar]
- 37.Welling A, Palva T. Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiol. 2008;147:1199–1211. doi: 10.1104/pp.108.117812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nitsch JP. Photoperiodism in woody plants. Proc Am Soc Hort Sci. 1957;70:526–544. [Google Scholar]
- 39.Wareing PF. Photoperiodism in woody plants. Annu Rev Plant Physiol. 1956;7:191–214. [Google Scholar]
- 40.Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- 41.Weiser CJ. Cold resistance and injury in woody plants. Science. 1970;169:1269–1278. doi: 10.1126/science.169.3952.1269. [DOI] [PubMed] [Google Scholar]
- 42.Arora R, Rowland LJ, Tanino K. Induction and release of bud dormancy in woody perennials: a science come of age. HortScience. 2003;38:911–921. [Google Scholar]
- 43.Heide OM, Prestrud AK. Low temperature, but not photoperiod, controls growth cessation and dormancy induction and release in apple and pear. Tree Physiol. 2005;25:109–114. doi: 10.1093/treephys/25.1.109. [DOI] [PubMed] [Google Scholar]
- 44.Schrader J, Moyle R, Bhalerao R, Herttzberg M, Lundeberg J, et al. Cambial meristem dormancy in trees involves extensive remodelling of the transcriptome. Plant J. 2004;40:173–187. doi: 10.1111/j.1365-313X.2004.02199.x. [DOI] [PubMed] [Google Scholar]
- 45.Druart N, Johansson A, Baba K, Schrader J, Sjödin A, et al. Environmental and hormonal regulation of the activity-dormancy cycle in the cambial meristem involves stage-specific modulation of transcriptional and metabolic networks. Plant J. 2007;50:557–573. doi: 10.1111/j.1365-313X.2007.03077.x. [DOI] [PubMed] [Google Scholar]
- 46.Rohde A, Bhalerao RP. Plant dormancy in the perennial context. Trends Plant Sci. 2007;12:217–223. doi: 10.1016/j.tplants.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 47.Ruttink T, Arend M, Morreel K, Storme V, Rombauts S, et al. A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell. 2007;19:2370–2390. doi: 10.1105/tpc.107.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiara-Magnone M, Jacobmeier B, Bertolucci C, Foà A, Albrecht U. Circadian expression of the clock gene Per2 is altered in the ruin lizard (Podarcis sicula) when temperature changes. Brain Res Mol Brain Res. 2005;133:281–285. doi: 10.1016/j.molbrainres.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 49.Vallone D, Frigato E, Vernesi C, Foà A, Foulkes NS, et al. Hypothermia modulates circadian clock gene expression in lizard peripheral tissues. Am J Physiol Regul Integr Comp Physiol. 2007;292:160–166. doi: 10.1152/ajpregu.00370.2006. [DOI] [PubMed] [Google Scholar]
- 50.Revel FG, Herwig A, Garidou ML, Dardente H, Menet JS, et al. The circadian clock stops ticking during deep hibernation in the European hamster. Proc Natl Acad Sci USA. 2007;104:13816–13820. doi: 10.1073/pnas.0704699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strauss WM. Screening of recombinant DNA libraries. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1987. pp. 6.3.1–6.3.6. [Google Scholar]
- 52.Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- 53.Allona I, Quinn M, Shoop E, Swope K, St. Cyr S, et al. Analysis of xylem formation in pine by cDNA sequencing. Proc Natl Acad Sci USA. 1998;95:9693–9698. doi: 10.1073/pnas.95.16.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bookout AL, Cummins CL, Kramer MF, Pesola JM, Mangelsdorf DJ. High-Throughput Real-Time Quantitative Reverse Transcription PCR. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley & Sons, Inc; 2006. pp. 15.8.1–15.8.28. [DOI] [PubMed] [Google Scholar]
- 55.Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CsPRR gene expression in the leaves of adult chestnuts obtained in June. Leaves were collected in June at 3-h intervals. (A) CsPRR northern blot analysis. The rRNA loading reference was detected by staining gels with ethidium bromide. (B) Quantitative RT-PCR analysis. Relative transcript abundances are shown in the graphs. Data are means from two biological replicates. Open and filled bars above each graph represent natural day and night lengths, respectively, as provided by the National Institute of Meteorology, Madrid.
(0.33 MB TIF)
CsPRR gene expression in the leaves of chestnut seedlings grown under different temperature conditions. (A and B) leaves from seedlings grown under conditions of LD and 22°C. (C and D) leaves from seedlings grown under standard conditions (LD, 22°C) and subsequently subjected to one week of LD at 4°C. Samples were collected at 3-h intervals. (A and C) CsPRR northern blot analysis. The rRNA loading reference was detected by staining gels with ethidium bromide. (B and D) Quantitative RT-PCR analysis. Relative transcript abundances are shown in the graphs. Data are means from two biological replicates. Open and filled bars above each graph indicate lights on and lights off, respectively.
(0.62 MB TIF)