Abstract
Distinct lipid compositions of intracellular organelles could provide a physical basis for targeting of membrane proteins, particularly where transmembrane domains have been shown to play a role. We tested the possibility that cholesterol is required for targeting of membrane proteins to the Golgi complex. We used insect cells for our studies because they are cholesterol auxotrophs and can be depleted of cholesterol by growth in delipidated serum. We found that two well-characterized mammalian Golgi proteins were targeted to the Golgi region of Aedes albopictus cells, both in the presence and absence of cellular cholesterol. Our results imply that a cholesterol gradient through the secretory pathway is not required for membrane protein targeting to the Golgi complex, at least in insect cells.
INTRODUCTION
The Golgi complex is instrumental in processing and sorting newly made proteins that move through the secretory pathway in all eukaryotic cells. Resident proteins that perform Golgi functions have been shown to contain important targeting information in their transmembrane domains (Machamer, 1993; Colley, 1997), but the mechanism of localization is still not understood. One hypothesis invokes specific protein–protein interactions within the lipid bilayer (Nilsson et al., 1993). Another is based on differences in lipid composition, particularly cholesterol and sphingolipids, across the Golgi stacks (Bretscher and Munro, 1993).
Sterols are ubiquitous components of cellular membranes in eukaryotes. In mammalian cells, the major site of cholesterol synthesis is the endoplasmic reticulum, but the highest concentration is at the plasma membrane (Liscum and Underwood, 1995). There is indirect evidence that an increasing gradient of cholesterol exists through the secretory pathway (Orci et al., 1981; Coxey et al., 1993). In addition to reducing membrane permeability, an increase in cholesterol content increases the thickness of model membranes (Levine and Wilkins, 1971; Nezil and Bloom, 1992; Bretscher and Munro, 1993). The plasma membrane is therefore expected to be thicker than other cellular membranes. Bretscher and Munro (1993) related this observation to protein sorting by analyzing the potential lengths of Golgi protein transmembrane domains. They found that, on average, Golgi proteins had shorter potential transmembrane domains than plasma membrane proteins with the same topology, and suggested that this might be an important factor in their sorting. If transport vesicles bud from cholesterol-rich regions of Golgi membranes, the short transmembrane domains of Golgi resident proteins may prevent them from entering these vesicles and leaving the Golgi complex. We tested the hypothesis that cholesterol provides a physical basis for the localization of Golgi proteins by examining their distribution in the absence of cholesterol.
Although mammalian cells have been used for most characterizations of the Golgi complex, they require cholesterol for growth. When deprived of exogenous cholesterol (normally obtained from lipoproteins in the form of cholesterol esters), mammalian cells up-regulate the biosynthesis of cholesterol in the endoplasmic reticulum, and, when biosynthesis is blocked, they increase endocytosis of lipoproteins to compensate (Brown and Goldstein, 1986). Thus, it is very difficult to manipulate the levels of cholesterol in mammalian cell membranes, and cellular levels can be depleted to only 30–40% of control levels. Insect cells are cholesterol auxotrophs, however, and can be depleted of detectable levels of cholesterol by growth in delipidated serum without compensatory synthesis of other sterols or major alterations in the phospholipid profile or fatty acid composition (Silberkang et al., 1983). We therefore used insect cells to determine whether cholesterol was required for targeting of Golgi membrane proteins. The cell line C6/36 derived from Aedes albopictus mosquito embryos was used because its growth in the absence of cholesterol has been well characterized (Silberkang et al., 1983; Phalen and Kielian, 1991; Marquardt et al., 1993; Marquardt and Kielian, 1996).
MATERIALS AND METHODS
Cell Culture
C6/36 cells were grown at 28°C in DMEM containing 10% heat-inactivated fetal bovine serum or 10% delipidated heat-inactivated fetal bovine serum prepared by adsorption on Cab-O-Sil as described (Phalen and Kielian, 1991; Marquardt et al., 1993; Marquardt and Kielian, 1996).
RNA Synthesis and Transfection
cDNA encoding murine α-mannosidase II (GlcNAc transferase I-dependent α1, 3[α1, 6] mannosidase, Moremen and Robbins, 1991), originally from K. Moremen was obtained from T. Hobman (University of Alberta, Edmonton, Alberta, Canada) already subcloned into the pSFV-1 (Life Technologies/BRL, Gaithersburg, MD) expression vector. cDNA encoding the “short” form of bovine β1,4-galactosyltransferase (UDP-galactose:b-d-N-acetylglucosaminide β1, 4 galactosyltransferase, Russo et al., 1990) was obtained from J. Shaper (Johns Hopkins Medical School, Baltimore, MD) and subcloned into the BamHI site of pSFV-1. cDNAs encoding Iip31 and 1–47GT/Iip31 (Nilsson et al., 1991) were obtained from T. Nilsson (EMBL, Heidelberg, Germany) and subcloned into pSFV-1 at the BamHI site. RNA was transcribed from plasmids linearized with SpeI using SP6 polymerase as recommended by Life Technologies/BRL with several modifications as described (Rolls et al., 1994). Cells were plated on glass coverslips 1–2 d before transfection and transfected with Semliki Forest virus (SFV) RNA using Lipofectin (Life Technologies/BRL) as described (Marquardt et al., 1993).
Fluorescence Labeling and Microscopy
Cells were labeled with fluorescent probes or conjugated antibodies and analyzed and photographed using a Nikon (Garden City, NY) Optiphot microscope as described (Machamer et al., 1993). For the experiment in Figure 4, a Bio-Rad (Richmond, CA) MRC600 laser scanning confocal microscope was used to view 1-μm optical sections, and images were analyzed and merged using Comos version 6.02 software.
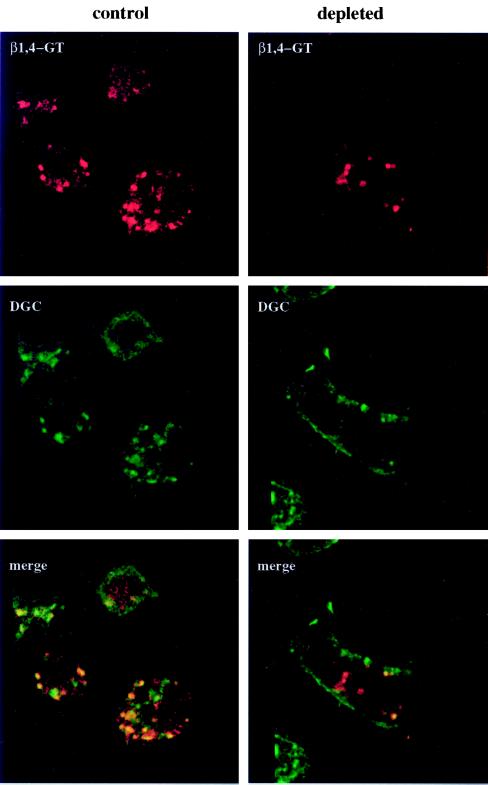
Figure 4.
β1,4GT colocalizes with a Golgi marker in normal and cholesterol-depleted C6/36 cells. Transfected normal or cholesterol-depleted cells expressing β1,4GT were double-labeled with rabbit anti-β1,4GT and Texas red-conjugated secondary antibody (red) and a monoclonal antibody that recognizes Drosophila Golgi membranes (anti-DGC) and fluorescein-conjugated secondary antibody (green). Analysis of 1-μm sections by confocal laser scanning microscopy demonstrated that there was substantial overlap in the two signals (yellow and orange), both in the presence and absence of cellular cholesterol. The decreased overlap (orange) in the Golgi region of depleted cells reflects an increased staining of the plasma membrane with anti-DCG, probably due to increased stability of proteins containing the carbohydrate epitope recognized by this antibody at the cell surface. Bar, 10 μm.
Fluorescent Probes
Cells were labeled with 50 μg/ml filipin complex (Sigma, St. Louis, MO) in PBS for 45 min at room temperature after fixation in 3% paraformaldehyde (Cadigan et al., 1990), and photographed using an UV filter. Live cells were stained by incubation in 5 μM N-[5-(5, 7-dimethyl BODIPY)-1-pentanoyl]-D-erythro-sphingosine (abbreviated C5-DMB-cer, Molecular Probes, Eugene, OR) in serum-free medium essentially as described (Pagano et al., 1991). Labeling was for 30 min at 2°C, followed by incubation in medium with normal or delipidated serum for 30 min at 28°C. Cells were fixed, mounted, and photographed using a rhodamine filter.
Indirect Immunofluorescence
Cells were fixed 16 h posttransfection in 3% paraformaldehyde and processed for indirect immunofluorescence after permeabilization with Triton X-100 as described (Machamer et al., 1993). Primary antibodies were rabbit anti-rat mannosidase II (Moremen and Touster, 1985; Velasco et al., 1993), affinity-purified rabbit anti-bovine β1,4-galactosyltransferase (Shaper et al., 1985), monoclonal anti-DGC, which recognizes a carbohydrate epitope in Drosophila Golgi complex membranes (obtained from V. Malhotra, University of California, San Diego, CA), and monoclonal antiIip31 [Clonab NL2 (Biotest Diagnostics, Denville, NJ), received from T. Nilsson (EMBL)]. Secondary antibodies were Texas red-conjugated goat anti-rabbit immunoglobulin G (IgG) and and goat anti-mouse IgG, and fluorescein-conjugated goat anti-mouse IgG (Jackson Immunoresearch, West Grove, PA).
Electron Microscopy
C6/36 cells grown in normal or delipidated serum for nine passages were fixed in 3% glutaraldehyde for 60 min, stained with 1% OsO4 in 0.1 M Na cacodylate, pH 7.2, and embedded in Epon. Thin sections were stained with 2% aqueous uranyl acetate for contrast and photographed at 60 kV using a Zeiss (Thornwood, NY) transmission electron microscope.
RESULTS
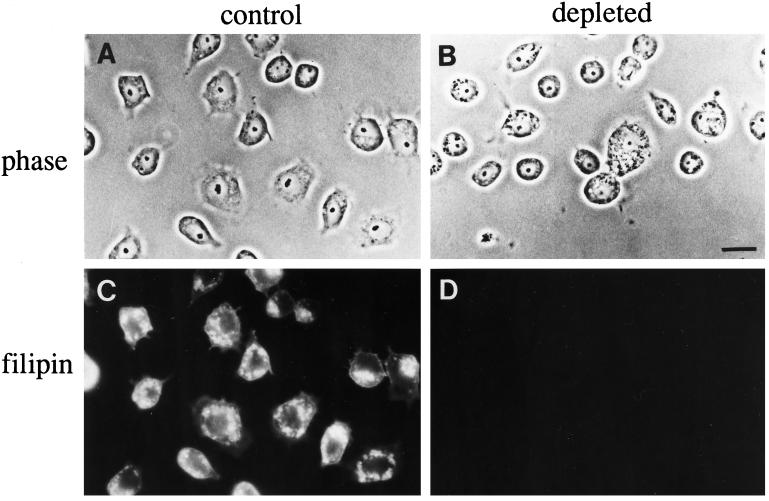
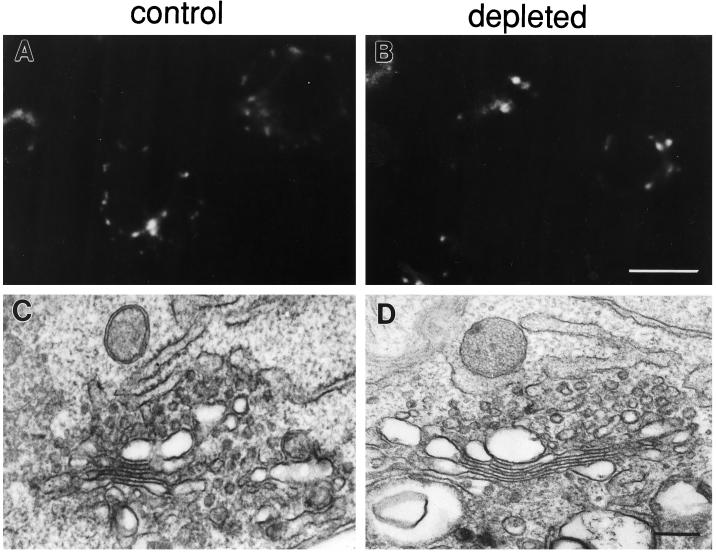
After four or more passages in lipoprotein-deficient serum, C6/36 cells had <2% the level of free and esterified cholesterol as cells grown in normal serum (Phalen and Kielian, 1991). Depleted cells failed to stain with the fluorescent compound filipin, which forms complexes with cholesterol in membranes (Figure 1). Even though the cells grown in delipidated serum lacked cholesterol, they had morphologically normal Golgi complexes. Staining cells with the fluorescent ceramide analog C5-DMB-cer, which labels the Golgi complex in mammalian cells (Pagano et al., 1991), revealed a number of punctate structures spread throughout the cytoplasm in both control and cholesterol-depleted cells (Figure 2, A and B). This dispersed, punctate labeling pattern is similar to that reported for a Golgi marker in a Drosophila cell line (Ripoche et al., 1994). By electron microscopy, the appearance of Golgi stacks in depleted cells was indistinguishable from that in control cells (Figure 2, C and D). At lower magnification (not shown), individual Golgi stacks were seen throughout the cytoplasm, consistent with the dispersed C5-DMB-cer staining pattern. Tight clustering of the stacks in the pericentriolar region was not observed, which suggests that Golgi complexes in insect cells may not associate with the microtubule-organizing center, as they do in mammalian cells. The observation that the morphology of Golgi stacks appears to be unaltered by cholesterol depletion is consistent with the finding that Golgi function is not impaired in these cells. There were no differences in rates of posttranslational processing or in transport to the plasma membrane for a surface glycoprotein in cholesterol-depleted C6/36 cells compared with normal cells (Marquardt et al., 1993).
Figure 1.
C6/36 cells from Aedes albopictus can be depleted of detectable levels of cholesterol. C6/36 cells were grown in normal serum (A and C) or lipoprotein-deficient serum for 12 passages (B and D), fixed, and stained with the fluorescent, cholesterol-binding dye filipin. Phase contrast and fluorescent images of the same fields are shown. The cells grown in delipidated serum fail to stain with filipin (D), unlike the control cells (C), where the plasma membrane and internal membranes are brightly labeled. Bar, 10 μm.
Figure 2.
Cholesterol-depleted C6/36 cells have morphologically normal Golgi complexes. Control (A) or depleted cells (B) were labeled with C5-DMB-cer (Pagano et al., 1991). Both contain similar punctate structures throughout the cytoplasm. When control (C) and depleted (D) cells were analyzed by electron microscopy, morphologically identical stacks of cisternal Golgi membranes were observed. Bar, 10 μm (A and B); 200 nm (C and D).
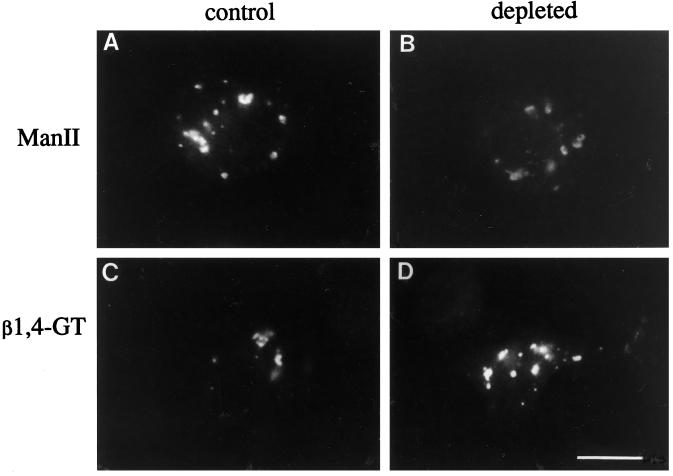
Mammalian Golgi proteins might have a more stringent cholesterol requirement than insect Golgi proteins for correct targeting, since mammalian cells require cholesterol for growth. We expressed two well-characterized mammalian Golgi proteins in C6/36 cells by transient transfection using the SFV expression system (Liljestrom and Garoff, 1991). SFV replicates in mosquito cells as part of its normal life cycle, and foreign genes cloned in place of the viral structural proteins can be expressed when RNA (transcribed in vitro) is transfected into C6/36 cells. Using this system, we expressed murine α-mannosidase II (ManII) and bovine β1,4-galactosyltransferase (β1, 4GT) in C6/36 cells. Punctate structures throughout the cytoplasm very similar to those seen in C5-DMB-cer–labeled cells were observed for both marker proteins in control and cholesterol-depleted cells (Figure 3). The punctate staining for ManII and β1,4GT did not colocalize with an endosomal tracer (fluorescent dextran) or a lysosomal marker (LysoTracker, Molecular Probes) (our unpublished observations). No plasma membrane staining was observed for either marker in the cholesterol-depleted cells, and the staining intensity of the punctate structures was similar in control and depleted cells. These results suggested that the two mammalian Golgi proteins were targeted to the Golgi complex in C6/36 cells, and that depletion of cholesterol had no effect on the targeting.
Figure 3.
Two mammalian Golgi proteins are targeted to the Golgi region of mosquito cells in the presence and absence of cholesterol. Control (A and C) or cholesterol-depleted C6/36 cells (B and D) were transfected with SFV RNA encoding either ManII (A and B) or β1,4GT (C and D), and fixed 16 h later. After permeabilization, cells were labeled with rabbit polyclonal sera specific for ManII or β1,4GT and Texas red-conjugated goat-anti-rabbit IgG. Both ManII and β1,4GT are targeted to punctate structures similar to those labeled by the Golgi marker, C5-DMB-cer (Figure 2). Qualitatively, there is no difference in targeting of either transfected marker in the presence or absence of cellular cholesterol. Bar, 10 μm.
To confirm that these mammalian proteins were targeted to the insect Golgi complex, we double-labeled transfected cells with a monoclonal antibody (anti-DGC) raised to purified Drosophila Golgi complex membranes. This antibody recognizes a carbohydrate epitope found on proteins in the Golgi and at lower levels at the plasma membrane (V. Malhotra, personal communication). Anti-DGC cross-reacts with mosquito cells and was the only feasible Golgi marker for double labeling because C5-DMB-cer staining is lost with permeabilization. Normal and depleted C6/36 cells expressing β1,4GT are shown in Figure 4. The structures labeled by anti-DGC (green) were similar to antiβ1,4GT-labeled structures (red), and there was substantial overlap (yellow). In cholesterol-depleted cells, an increased level of plasma membrane staining with anti-DGC was observed. However, the intracellular punctate structures labeled with anti-DGC in cholesterol-depleted cells still overlapped with expressed β1,4GT (orange and yellow). Therefore, it is likely that the punctate structures represent Golgi membranes in both control and cholesterol-depleted cells. Since the anti-DRG antibody recognizes a carbohydrate epitope on itinerant proteins rather than a protein epitope on a Golgi membrane protein, the decreased overlap in staining in the depleted cells does not imply a loss of retention of endogenous Golgi proteins in these cells. The increased surface staining with anti-DGC in cholesterol-depleted cells could reflect increased stability of the epitope at the plasma membrane, decreased synthesis in the Golgi, or both. It will be interesting to explore the distribution of the DGC epitope in normal and cholesterol-depleted cells when more is known regarding proteins that possess this carbohydrate epitope. In C6/36 cells expressing ManII, the overlap between anti-DGC- and anti-ManII–labeled structures was less pronounced than that with β1,4GT in both control and cholesterol-depleted cells (Machamer, unpublished results). This result suggested that the epitope recognized by anti-DGC is subcompartmentalized within Golgi membranes, consistent with the partially overlapping but distinct distributions of ManII and β1,4GT in mammalian cells (Rabouille et al., 1995). Unfortunately, the low efficiency of transfection precluded immunoelectron microscopy to localize the transfected markers within the Golgi complex. Thus, we could not rule out subtle differences in the targeting of the two marker proteins in the absence of cholesterol. Nevertheless, we could conclude that cholesterol was not required for targeting of ManII and β1,4GT to the Golgi region of C6/36 cells.
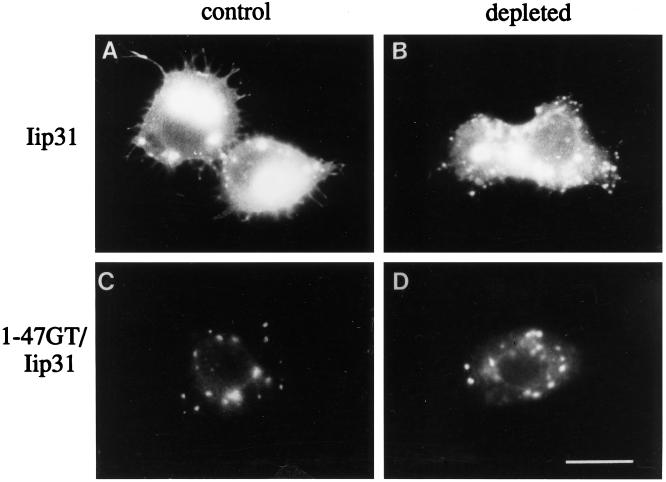
It was possible that the mechanism of protein targeting to Golgi membranes in insect cells is different than that in mammalian cells. To determine whether transmembrane domains of Golgi residents were likely to be involved in targeting, we expressed a chimeric reporter protein. In mammalian cells, Nilsson et al. (1991) showed that the membrane anchor of β1,4GT was sufficient to target the invariant chain (Iip31) to the Golgi complex. We found this to be the case in C6/36 cells as well (Figure 5). The amino terminus (cytoplasmic tail and transmembrane domain) of β1,4GT was sufficient to target Iip31 (1–47GT/Iip31) to the Golgi region instead of to endosomes and the plasma membrane, whether or not cholesterol was present (Figure 5, C and D). Thus, it is likely that Golgi protein transmembrane domains play an important role in proper targeting in insect cells, as they do in mammalian cells.
Figure 5.
The membrane anchor of β1,4GT can target a reporter protein to the Golgi region in C6/36 cells. Control (A and C) or cholesterol-depleted C6/36 cells (B and D) were transfected with SFV RNA encoding Iip31 (A and B) or a chimeric protein (C and D) with the membrane anchor of β1,4GT and the lumenal domain of Iip31 (1–47GT/Iip31), and fixed 16 h posttransfection. After permeabilization, cells were labeled with anti-Iip31 and Texas red-conjugated secondary antibody. The membrane anchor of β1,4GT is sufficient to redirect Iip31 from an endosomal/plasma membrane distribution (A and B) to a Golgi region distribution both in the presence (C) and absence (D) of cholesterol. Bar, 10 μm.
DISCUSSION
Our results imply that a cholesterol gradient through the Golgi cannot be required for targeting of membrane proteins in insect cells. Could another factor be responsible for progressively thickening membranes through the secretory pathway in these cells? One possibility might be an increase in the length or unsaturation of fatty acyl chains of phospholipids. Although we have not investigated this possibility in C6/36 cells, Silberkang et al. (1983) found that glycerolipid acyl chain length and degree of saturation remained unchanged in Drosophila Kc cells after depletion of cholesterol. Another possibility is that a gradient of sphingolipids (which favor thicker bilayers; Bretscher and Munro, 1993) might be sufficient for membrane thickening in cholesterol-depleted insect cells. Sphingolipids in cholesterol-depleted Drosophila or mosquito cells have not yet been characterized.
One prediction of the Bretscher and Munro hypothesis (Bretscher and Munro, 1993) is that altering the length of transmembrane domains could convert plasma membrane proteins into Golgi residents and vice versa. Some, but not all, Golgi membrane proteins are retained less efficiently when their potential transmembrane domains are lengthened (Munro, 1995; Colley, 1997). The plasma membrane protein CD8 was retained in the Golgi complex when its transmembrane domain was replaced with 17, but not 23, leucine residues (Munro, 1995). However, for another well-characterized plasma membrane protein (the vesicular stomatitis virus G protein), decreasing the potential transmembrane length from 20 to 14 amino acids did not prevent its transport to the cell surface (Adams and Rose, 1985). It is clear that transmembrane domain length may play an important role for targeting of some Golgi proteins, but cell type and protein-specific differences are likely to exist.
Is membrane thickness involved in protein sorting in the Golgi complex? Another prediction of the Bretscher and Munro hypothesis (Bretscher and Munro, 1993) is that Golgi membrane proteins with short transmembrane domains would be sequestered away from cholesterol-rich, thicker transport vesicles en route to the plasma membrane. However, a recent report that COPI- and COPII-coated transport vesicles budding from Golgi membranes have a thinner interleaflet clear space, compared with the surrounding membrane, is incompatible with this idea (Orci et al., 1996). Finally, there is no direct evidence for a cholesterol or cholesterol/sphingomyelin gradient through the secretory pathway. As pointed out by Allan and Kallen (1994), the cholesterol in the trans-Golgi network could be “exogenous” cholesterol released by hydrolysis of low-density lipoprotein in lysosomes. There is no evidence that cholesterol synthesized in the endoplasmic reticulum moves through the Golgi en route to the plasma membrane, and, in fact, there is evidence that it can bypass the Golgi (Urbani and Simoni, 1990). Although the idea that the lipid composition of Golgi membranes can influence protein localization within the organelle is an appealing one, more information on the actual lipid composition of Golgi subcompartment membranes will be required for its evaluation.
ACKNOWLEDGMENTS
We thank J. Shaper, T. Nilsson, T. Hobman, M. Farquhar, K. Moremen, and V. Malhotra for their generous gifts of antibodies and cDNAs, M. Delannoy for expert assistance with confocal microscopy, and M.G. Grim for help with the electron microscopy. This work was supported by the National Institutes of Health (grant GM-42522 to C.E.M.), the American Cancer Society (grant RPG93–013-05VM to M.K.), and the Pew Biomedical Scholars Program (to C.E.M. and M.K.).
REFERENCES
- Adams GA, Rose JK. Structural requirements of a membrane-spanning domain for protein anchoring and cell surface transport. Cell. 1985;41:1007–1015. doi: 10.1016/s0092-8674(85)80081-7. [DOI] [PubMed] [Google Scholar]
- Allan D, Kallen K-J. Is plasma membrane lipid composition defined in the exocytic or the endocytic pathway? Trends Cell Biol. 1994;4:350–353. doi: 10.1016/0962-8924(94)90076-0. [DOI] [PubMed] [Google Scholar]
- Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Spillane DM, Chang T-Y. Isolation and characterization of Chinese hamster ovary cell mutants defective in intracellular low density lipoprotein-cholesterol trafficking. J Cell Biol. 1990;110:295–308. doi: 10.1083/jcb.110.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley KJ. Golgi localization of glycosyltransferases: more questions than answers. Glycobiol. 1997;7:1–13. doi: 10.1093/glycob/7.1.1-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coxey RA, Pentchev PG, Campbell G, Blanchette-Mackie EJ. Differential accumulation of cholesterol in Golgi compartments of normal and Niemann-Pick type C fibroblasts incubated with LDL: a cytochemical freeze-fracture study. J Lipid Res. 1993;34:1165–1176. [PubMed] [Google Scholar]
- Levine YK, Wilkins MHF. Structure of oriented lipid bilayers. Nature New Biol. 1971;230:69–72. doi: 10.1038/newbio230069a0. [DOI] [PubMed] [Google Scholar]
- Liljestrom P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- Liscum L, Underwood KW. Intracellular cholesterol transport and compartmentation. J Biol Chem. 1995;270:15443–15446. doi: 10.1074/jbc.270.26.15443. [DOI] [PubMed] [Google Scholar]
- Machamer CE. Targeting and retention of Golgi membrane proteins. Curr Opin Cell Biol. 1993;5:606–612. doi: 10.1016/0955-0674(93)90129-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer CE, Grim MG, Esquela A, Chung SW, Rolls MM, Ryan K, Swift AM. Retention of a cis Golgi protein requires residues on one face of a predicted alpha-helix in the transmembrane domain. Mol Biol Cell. 1993;4:695–704. doi: 10.1091/mbc.4.7.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt MT, Kielian M. Cholesterol-depleted cells that are relatively permissive for Semliki Forest virus infection. Virology. 1996;224:198–205. doi: 10.1006/viro.1996.0521. [DOI] [PubMed] [Google Scholar]
- Marquardt MT, Phalen T, Kielian M. Cholesterol is required in the exit pathway of Semliki Forest virus. J Cell Biol. 1993;123:57–65. doi: 10.1083/jcb.123.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen KW, Robbins PW. Isolation, characterization and expression of cDNAs encoding murine alpha-mannosidase II, a Golgi enzyme that controls conversion of high mannose to complex N-glycans. J Cell Biol. 1991;115:1521–1534. doi: 10.1083/jcb.115.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen KW, Touster O. Biosynthesis and modification of Golgi mannosidase II in HeLa and 3T3 cells. J Biol Chem. 1985;260:6654–6662. [PubMed] [Google Scholar]
- Munro S. Sequences within and adjacent to the transmembrane segment of α2,6-sialyltransferase specify Golgi retention. EMBO J. 1995;14:4695–4704. doi: 10.1002/j.1460-2075.1991.tb04924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezil FA, Bloom M. Combined influence of cholesterol and synthetic amphiphilic peptides upon bilayer thickness in model membranes. Biophys J. 1992;61:1176–1183. doi: 10.1016/S0006-3495(92)81926-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Lucocq JM, Mackay D, Warren G. The membrane spanning domain of β-1,4-galactosyltransferase specifies trans Golgi localization. EMBO J. 1991;10:3567–3575. doi: 10.1002/j.1460-2075.1991.tb04923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Slusarewicz P, Hoe MH, Warren G. Kin recognition: a model for the retention of Golgi enzymes. FEBS Lett. 1993;330:1–4. doi: 10.1016/0014-5793(93)80906-b. [DOI] [PubMed] [Google Scholar]
- Orci L, Montesano R, Meda P, Mailaisse-Lagae F, Brown D, Perrelet A, Vassalli P. Heterogeneous distribution of filipin-cholesterol complexes across the cisternae of the Golgi. Proc Natl Acad Sci USA. 1981;78:293–297. doi: 10.1073/pnas.78.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Schekman R, Perrelet A. Interleaflet clear space is reduced in the membrane of COPI and COPII-coated buds/vesicles. Proc Natl Acad Sci USA. 1996;93:8968–8970. doi: 10.1073/pnas.93.17.8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano RE, Martin OC, Kang HC, Haugland RP. Molecular trapping of a fluorescent ceramide analogue at the Golgi apparatus of fixed cells: interaction with endogenous lipids provides a trans-Golgi marker for both light and electron microscopy. J Cell Biol. 1991;109:2067–2079. doi: 10.1083/jcb.109.5.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalen T, Kielian M. Cholesterol is required for infection by Semliki Forest virus. J Cell Biol. 1991;112:615–623. doi: 10.1083/jcb.112.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Hui N, Hunte F, Kieckbusch R, Berger EG, Warren G, Nilsson T. Mapping the distribution of Golgi enzymes involved in the construction of complex oligosaccharides. J Cell Sci. 1995;108:1617–1627. doi: 10.1242/jcs.108.4.1617. [DOI] [PubMed] [Google Scholar]
- Ripoche J, Link B, Yucell, Tokuyasu JK, K, Malhotra V. Location of Golgi membranes with reference to dividing nuclei in syncytial Drosophila embryos. Proc Natl Acad Sci USA. 1994;91:1878–1882. doi: 10.1073/pnas.91.5.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls MM, Webster P, Balba NH, Rose JK. Novel infectious particles generated by expression of the vesicular stomatitis virus glycoprotein from a self-replicating RNA. Cell. 1994;79:497–506. doi: 10.1016/0092-8674(94)90258-5. [DOI] [PubMed] [Google Scholar]
- Russo RN, Shaper NL, Shaper JH. Bovine β1,4-galactosyltransferase: two sets of mRNA transcripts encode two forms of the protein with different amino-terminal domains. J Biol Chem. 1990;265:3324–3331. [PubMed] [Google Scholar]
- Shaper NL, Mann PL, Shaper JH. Cell surface galactosyltransferase: immunochemical localization. J Cell Biochem. 1985;28:229–239. doi: 10.1002/jcb.240280305. [DOI] [PubMed] [Google Scholar]
- Silberkang, Havel M, CM, Friend DS, McCarthy BJ, Watson JA. Isoprene synthesis in isolated embryonic Drosophila cells. I. Sterol-deficient eukaryotic cells. J Biol Chem. 1983;258:8503–8311. [PubMed] [Google Scholar]
- Urbani L, Simoni RD. Cholesterol and vesicular stomatitis virus G protein take separate routes from the endoplasmic reticulum to the plasma membrane. J Biol Chem. 1990;265:1919–1923. [PubMed] [Google Scholar]
- Velasco A, Hendricks L, Moremen KW, Tulsiani DRP, Touster O, Farquhar MG. Cell type-dependent variations in the subcellular distribution of α-mannosidase I and II. J Cell Biol. 1993;122:39–51. doi: 10.1083/jcb.122.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]