Abstract
Over the past decade, apoptosis has emerged as an important field of study central to ongoing research in many diverse fields, from developmental biology to cancer research. Apoptosis proceeds by a highly coordinated series of events that includes enzyme activation, DNA fragmentation, and alterations in plasma membrane permeability. The detection of each of these phenotypic changes is accessible to advanced undergraduate cell and molecular biology students. We describe a 4-week laboratory sequence that integrates cell culture, fluorescence microscopy, DNA isolation and analysis, and western blotting (immunoblotting) to follow apoptosis in cultured human cells. Students working in teams chemically induce apoptosis, and harvest, process, and analyze cells, using their data to determine the order of events during apoptosis. We, as instructors, expose the students to an environment closely simulating what they would encounter in an active cell or molecular biology research laboratory by having students coordinate and perform multiple tasks simultaneously and by having them experience experimental design using current literature, data interpretation, and analysis to answer a single question. Students are assessed by examination of laboratory notebooks for completeness of experimental protocols and analysis of results and for completion of an assignment that includes questions pertaining to data interpretation and apoptosis.
Keywords: apoptosis, HL-60 cells, DNA laddering, western blot, fluorescence microscopy
INTRODUCTION
Training in molecular and cellular biology requires an abundance of theoretical knowledge and practical experience. Students may find it more enjoyable to reach their academic goals by learning theory as part of their empirical investigation (Odom and Grossel, 2002). After a combined 33 years of undergraduate teaching experience, we have come to believe strongly that it is more beneficial for the laboratory student to learn methodology in the context of how it is used in the “real world,” gaining valuable knowledge in the application of various techniques, and not just a “cookbook” survey of methods. This apoptosis project is part of a laboratory-based course similar to those described in educational reports such as Beyond Bio 101 from the Howard Hughes Medical Institute (1996) suggesting that cookbook laboratories be replaced with longer-term projects that more closely reflect research investigations performed by actual scientists. Such inquiry-based approaches to science education are well established and widely accepted by many science educators (National Research Council, 2003). To this end, we describe a series of experiments investigating mammalian cellular apoptosis induced via the p53 pathway with chemical inducers (though the protocol can be easily modified for serum withdrawal or radiation damage, for example.) We chose apoptosis because (1) it is a topic that incorporates and, therefore, reinforces much of the cellular and molecular information to which the students have been exposed in previous course work; (2) apoptosis is extremely topical owing to its relevance in developmental biology, cancer biology, virology, immunology, and degenerative conditions such as Huntington's and Parkinson's diseases, and it is expected that the resulting large body of current literature will be used by the students to assist them in the formulation of rational hypotheses and experimental designs; (3) many of the techniques used in the study of apoptosis are employed to investigate other cellular and molecular processes; and (4) since many of these methods take days to complete, the students will be occupied by several different activities in a single lab period as opposed to the less realistic “technique-of-the-day” approach common to many undergraduate laboratories. In addition, students will gain an opportunity to work in small groups, thereby honing their interactive, communicative, and organizational skills that are considered to be useful for success in postgraduate careers (Wright and Boggs, 2002).
Apoptosis is the process by which a cell purposefully self-destructs in response to endogenously derived signals. Cells undergoing apoptosis do so in a well-described pattern that is different from that of cells that are dying from necrosis due to physical injury, for example. Apoptosis is induced through either of two pathways: one that begins with extracellular death ligands binding receptors and another initiated by internal signals often mediated by the tumor suppressor p53. Both pathways involve the transduction of signals ultimately causing the activation of specialized proteolytic enzymes called caspases. Caspases are activated by being cleaved by other caspases (for example, caspase 9 cleaves and activates caspase 3) or by autoproteolysis, which is how caspases 8 and 9 self-activate. The self-activating caspases are generally known as initiators, and the caspases like caspase 3 are the effectors, also known as the executioners. Effector caspases, once activated, will cause the degradation or collapse of DNA, the nuclear lamina, the cytoskeleton, and other critical components of cellular integrity. Activation of effector caspases (and, therefore, the apoptotic program) may be detected by examining the proteolysis of their substrates. One commonly assayed substrate is poly(ADP-ribose)polymerase-1 (PARP-1), a DNA repair enzyme degraded by caspases 3 and 7. For excellent reviews of apoptosis and the functions of caspases, see Chang and Yang (2000) and Hengartner (2000).
Some of the objectives of this laboratory were for the students to be able to identify apoptotic cells, learning fluorescence microscopy in the process; they performed protein gel electrophoresis and western blots to identify the deactivation of an effector caspase substrate (PARP-1) in the execution of apoptosis. Additionally, they gained further experience in the isolation of genomic DNA and agarose gel electrophoresis while seeing firsthand the effects of apoptotic-induced nuclease activity and the pattern of fragmentation caused by deposition of the nucleosomes. After having made a hypothesis based on their review of the literature, the students' main goal was to determine, using the methods described above, the sequence of the following events in the apoptotic program: (1) activation of effector caspases, (2) chromatin condensation, (3) DNA fragmentation, and (4) increase in membrane permeability to vital stains. (For a review of these and other apoptotic markers, see Smyth et al., 2002). As educators, however, our main objective was not the actual answer to the sequence of events of apoptosis but, rather, to guide students through the scientific process of experimental design, data collection, analysis, and interpretation toward the resolution of a single biological question.
EXPERIMENTAL OUTLINE (SCHEDULE) OF STUDENT ACTIVITY
Though the schedule could be modified somewhat, this was a 4-week laboratory project requiring two meetings per week—1 day (induction) required student involvement throughout the day. Students worked in groups of three or four students, primarily to share the workload.
Two Weeks in Advance of Project Start
We made available to the students a selection of reference materials about apoptosis on library reserve and/or accessible in the laboratory or classroom (Appendix A). It was imperative that students research the subject on their own as is done in “real-life” situations. Our students had available several methods books such as Sambrook et al. (1989), Ausubel et al. (2002), Freshney (2000), and Martin (1994) that provide protocols for molecular biology and cell culture techniques. Studzinski (1999) is a particularly useful source of techniques and background information specific to apoptosis.
Additionally, we distributed an introduction to the methods and goals of the apoptosis project (Appendix B) and procedural information about cell culture maintenance (Appendix C) and enumeration (Appendix D). The initial apoptosis handout informed the students of the reference material, posed basic questions (e.g., What might explain why apoptosis does not induce inflammation, while necrosis does?), summarized apoptotic events very briefly, listed objectives and the main experimental goal (see above), and gave a brief summary of the techniques to be used in the project (i.e., western with anti-PARP-1 antibody, DNA staining with fluorescent stains such as Hoescht, and DNA laddering).
Also, the students were given an assignment to use the references to design a flowchart of the apoptosis project and include a set of protocols of their choosing that could be used to assay the four events of apoptosis described above. The students were to prepare an estimate of the number of cells needed for each assay, examples of apoptosis inducers including concentrations, the time points after induction at which they will harvest cells, and the hypothesis of how these events (DNA fragmentation and condensation, membrane permeability and PARP-1 cleavage) are ordered in the apoptosis program. This assignment was due the first day of the second week of the project (see below).
Week 1
Days 1 and 2
Days 1 and 2 involved an interactive lecture on cell culture maintenance, cell enumeration, and apoptosis (based on the handouts given earlier). The students were given extra details such as the minimum number of cells in total that each student group required by Week 2 and further information about the assays and their assignment. In the laboratory component of Week 1, the student groups were given a flask of actively growing HL-60 cells and medium that they had prepared themselves. They were shown how to perform vital staining, enumeration, and subculture of the suspension cells. The use of the inverted microscope, laminar flow hoods, and autoclave was demonstrated. Between Week 1 and Week 2, the students were required to maintain their cultures to ensure that a sufficient number of cells would be available by Week 2.
Week 2
Day 1
Students submitted their assignments and, in return, received the protocols and flowchart they actually would be using. (Out of sheer practicality, we had the students use our procedures instead of their own because using even their best protocols would have made advanced planning and the timely purchase of reagents impossible.) We reviewed the protocols that we had just distributed and answered the students' questions. Students were told to spend time coordinating the work planned for the second day of Week 2, which involved induction of apoptosis and harvesting of cells in minimally four time points over a period of at least 8 h (e.g., 0, 2, 4, and 8 h). Organization was key here, since students had to work around their class and job schedules to be sure that at least two of them could be present to harvest the cells, perform cell counts with vital and fluorescent staining, and begin protein and DNA isolations.
Day 2
This was the most challenging part of the experiment, as the students had to count their cells and distribute the needed number into two flasks (one for control [uninduced] cells, the other for induced cells). At different time points throughout the next 8 h or more, the students harvested a fraction of the cells from each flask, enumerated some for percentage viability and DNA condensation using light and fluorescence microscopy, incubated some with protein isolation buffer for subsequent western blot analysis, and treated a final aliquot with DNA extraction buffer, RNase A, and proteinase K to begin DNA isolation.
Week 3
Day 1
The students poured the separating portion of two denaturing polyacrylamide (SDS-PAGE) minigels for the protein electrophoresis.
Day 1 and/or Day 2
The protein analysis was continued, from pouring the stacking gels through electrotransfer and blocking the nitrocellulose blot of one gel and placing the second gel to stain in Coomassie blue. (Destaining occurred throughout the week.) While the students waited for electrophoresis, and then again for blotting, the DNA samples were processed through the ethanol precipitation step.
Week 4
Day 1
Western blots were treated with primary antibody and incubated overnight at 4°C.
Day 2
Western blots were completed with detection by secondary antibody, followed by chemiluminescence. Destained Coomassie gels were dried. DNA was size-separated through mini-agarose gels to detect DNA laddering. If cells stained with Hoescht had been fixed, they were assayed for condensed DNA.
MATERIALS AND METHODS
Cell Culture
HL-60 and Jurkat cells were purchased from the American Type Culture Collection (ATCC; Rockville, MD; catalog numbers CCL-240 and TIB-152, respectively). All cell culture media and reagents were purchased from Gibco-Invitrogen (Rockville, MD; see catalog numbers following each item below). The cells were grown in Iscove's minimal essential medium (12200) supplemented with 10–20% fetal bovine serum (26140) and HEPES (15630), as recommended by the ATCC, and penicillin–streptomycin (15140). The cells were maintained in a humidified CO2 incubator at 37°C, at a density of between 1 × 105 and 1 × 106 cells/ml. For all experiments, cells were used at a concentration of 1 × 106 cells/ml. Cell concentrations and viability were determined by hemacytometer counts of cells diluted in erythrosin B. Cells that stained with erythrosin B were considered nonviable.
Induction of Apoptosis and Harvesting and Processing of Cells
Etoposide (VP-16; Sigma E-1383; St. Louis, MO), a topoisomerase inhibitor, was dissolved in DMSO (Sigma D-2650) at a concentration of 100 mM, aliquoted, and stored at −20°C. Induction of apoptosis was accomplished by adding VP-16 to a final concentration of 500 μM or DMSO alone as a control. The cells were then incubated at 37°C. Immediately after the addition of the inducer or DMSO, and at appropriate time intervals, 2.5 × 106 cells (2.5 ml) were harvested and divided into three 1.5-ml microfuge tubes: two tubes with 1 × 106 cells each, one to be used for DNA and the other for protein extraction, and one tube with 0.5 × 106 cells for vital staining and fluorescence staining.
Vital Staining and Detection of Chromatin Condensation
For vital staining, 40 μl of cells were mixed with 160 μl of erythrosin B in a well of a microtiter dish. The cells were then counted using a hemacytometer. The remaining cells to be used for staining were pelleted briefly (8–10 s) at high speed (approximately 16,000g) in a microfuge. After removing the supernatant, the pellet was resuspended in 50 μl of a combination of freshly made Hoescht-33342 and propidium iodide at concentrations of 100 and 2 μg/ml in PBS, respectively, and incubated on ice for 5 min. The Hoescht-33342 was prepared from a 10 mg/ml solution (H-3570; Molecular Probes, Inc., Eugene, OR) and used to stain chromatin in all cells, whereas the propidium iodide (Sigma P-4170), prepared from a 2 mg/ml stock solution in ddH2O, was added to detect alterations in plasma membrane permeability, characteristic of very late apoptotic and necrotic cells. At this point, the cells were examined and counted immediately by fluorescence microscopy (emission at 492 nm with excitation at 356 nm for Hoescht and emission at 615 nm with excitation at 488 nm for propidium iodide) or fixed and stored for later examination of Hoescht stain only, since all fixed cells will stain with propidium iodide. To fix the cells, an equal volume of 10% formalin was added; the cells were stored for up to 1 week at 4°C without significant loss of staining. Cells with chromatin condensation (blue–green blebbing) and/or membrane permeability (bright red blebbing or homogeneous staining) were counted as a percentage of the total number of cells staining with Hoescht 33342.
Detection of DNA Fragmentation
The cells used for DNA isolation were pelleted briefly in a microfuge as described above and washed twice with 1 ml of cold phosphate-buffered saline, pH 7.2 (PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4). DNA extraction was performed as described by Yoshida et al. (1999). The washed cell pellet was then resuspended in 500 μl of DNA extraction buffer (50 mM Tris, pH 8.0, 20 mM Na2EDTA, 10 mM NaCl, 1% [w/v] SDS, and 20 μg/ml RNase A [Sigma R-5503]) and incubated at 37°C for 1 h. This was followed by the addition of proteinase K (Sigma P-2308) to a final concentration of 100 μg/ml and incubation at 65°C for 1 h. At this point, for convenience, the lysates were stored at 4°C. The DNA was later (week 3) extracted once with 500 μl of Tris-buffered phenol and once with chloroform/isoamyl alcohol (24:1) (Sambrook et al., 1989). The DNA was then precipitated by the addition of 5 M NaCl to a final concentration of 200 mM and 2 vol of ice cold 100% ethanol. After thorough mixing by inversion of the tubes, precipitated DNA was evident. The DNA was further precipitated at −80°C for 20 min or −20°C overnight. Following incubation, the DNA was pelleted by centrifugation in a microfuge at high speed for 20 min, washed once with ice cold 70% ethanol, and centrifuged again. The resulting DNA pellet was allowed to air-dry, and resuspended in 40 μl of TE, pH 7.5. More TE was added if the suspension was very viscous.
Ten microliters of each DNA sample were mixed with 2 μl of 5× loading dye (5× Tris–acetate–EDTA [TAE], 50% glycerol, 0.2% bromophenol blue), and the fragments were resolved by electrophoresis through a 1.2% agarose minigel (Sambrook et al., 1989) in 1× TAE buffer. Once the bromophenol blue had migrated three quarters of the gel length (approximately 70 min at 4 V/cm), the DNA was visualized by staining in ethidium bromide (1 μg/ml), followed by examination and photography on a UV transilluminator.
Protein Isolation and Detection of PARP by Western Blot Analysis
The cells were pelleted and washed twice with 1 ml of cold PBS as described above. Total cellular proteins were isolated essentially as described by Martins et al. (1997). Briefly, the washed cell pellet was resuspended in 33.3 μl of PMSF/glucose buffer (50 mM glucose, 25 mM Tris, pH, 8.0, 10 mM Na2EDTA, and 1 mM PMSF [Sigma P-7626]) and 16.7 μl of urea/SDS buffer (50 mM Tris, pH 6.8, 6 M urea, 6% β-mercaptoethanol, 3% SDS, and 0.003% bromophenol blue) and kept on ice. While still on ice, the cells were disrupted and DNA was sheared by passage of the suspension eight times through a 10-mm 26-gauge needle attached to a tuberculin syringe. The lysate was then immediately heated to 65°C for 15 min, and frozen at −20°C.
The extracted proteins were resolved by discontinuous 8% SDS–polyacrylamide gel electrophoresis in a Mini-PROTEAN II (Bio-Rad, Hercules, CA) apparatus, blotted, and blocked essentially as described by Sambrook et al. (1989) and Ausubel et al. (2002). In brief, each lane of the gels contained total protein from 3–5 × 105 cells. When desired, duplicate gels were made: one for Coomassie staining for total cell protein and the other for blotting. After electrophoresis, the proteins were electroblotted to a nitrocellulose membrane (Mighty Small Transphor apparatus, Hoefer Scientific Instruments) and blocked for 1 h at room temperature in 0.2% casein (Sigma C-5890) and 0.1% Tween-20 in Tris-buffered saline (TBS; 150 mM NaCl, 10 mM Tris, pH 7.5). After rinsing with deionized water, blots were either used immediately or wrapped in plastic wrap and stored at −20°C.
To detect PARP-1, blots were incubated overnight at 4°C with gentle agitation with an anti-PARP-1 monoclonal antibody (BD Pharmingen 556362; San Diego, CA), diluted 1:1,000 in blocking buffer. Blots were washed with 0.1% Tween-20 in TBS, followed by incubation for 1 h at room temperature with shaking in a goat anti-mouse IgG-HRP conjugate (Sigma A-4416) diluted 1:10,000 in blocking buffer. Blots were then thoroughly washed with 0.1% Tween-20 in TBS, followed by a 5-min incubation in a luminol and peroxide solution (West Pico chemiluminescent kit; Pierce 34079; Rockford, IL). Proteins were detected by exposure of the blot to X-ray film (Kodak BioMax MR) for about 10 to 30 min.
RESULTS AND DISCUSSION
This laboratory project exposed students to some of the assays used to survey the progression of apoptosis in cultured cells. Additionally, it provided a valuable opportunity for the students (mostly seniors) to enhance their skills in research preparation, data analysis and interpretation, and group collaboration while retaining laboratory independence. Although we (and our students) put forth considerable effort to optimize these exercises, there are still multiple opportunities in the form of independent research projects, for example, to embellish and/or enhance the apoptosis study presented here (see below).
In developing this series of apoptosis assays, we encountered many experimental variables. The variables we found to be most important included (1) the choice of cells, (2) the choice of chemical or other inducers of apoptosis, (3) the choice of apoptosis proteins detected, (4) the choice of conditions for isolating proteins for western blot analysis, and (5) the choice of antibodies (polyclonal vs. monoclonal) for immunodetection.
While designing this series of laboratory exercises, we tried several different methods and/or modifications for each of the assays described (fluorescent staining, DNA isolation for detection of DNA laddering, and protein isolation for western blot analysis) as well as two different cell lines (Jurkat and HL-60), to ascertain which cells and methods provided the most consistent results and were within the technical capabilities and time constraints of students in an undergraduate laboratory setting.
DNA Fragmentation
In the development of these laboratory exercises, we initially conducted a series of experiments using Jurkat and HL-60 cells exposed to either camptothecin or etoposide (VP-16). In these experiments, DNA laddering was used to detect apoptosis. Jurkat cells, while widely cited in the literature, consistently failed, in our hands, to exhibit clear DNA laddering compared to HL-60 cells with two different DNA isolation methods (data not shown). While both inducers caused DNA fragmentation in HL-60 cells, the laddering effect was more pronounced within 4 h using VP-16 at concentrations ranging from 100 μM to 1 mM. For these reasons, all of the results reported were obtained with HL-60 cells exposed to VP-16. Students obtained DNA laddering with distinct fragments ranging from about 200 bp to 1 kb (in approximately 200 bp increments) as early as 4 h after exposure to VP-16, while there was no evidence of significant degradation in the uninduced control cells (Figure 1).
Figure 1.
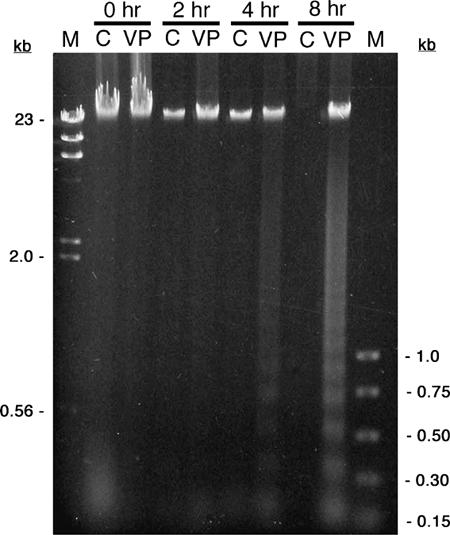
DNA laddering in HL-60 cells exposed to VP-16. HL-60 cells were exposed to either 500 μM VP-16 (VP) or DMSO (C) for the indicated times. Cells were lysed, and the DNA was isolated by phenol–chloroform extraction followed by ethanol precipitation. The DNA was resolved by electrophoresis through a 1.2% TAE–agarose gel and visualized with ethidium bromide. The outermost lanes contain size standards (M); relevant sizes are shown as kilobases (kb). DNA laddering is clearly evident within 4 h after exposure to VP-16. Note that the control DNA at 8 h was likely lost at some point during the isolation procedure, which occasionally occurs when students perform multiple DNA extractions.
Fluorescent Staining and Viability
Chromatin condensation was detected using the fluorescent DNA-binding stains Hoescht 33342 and propidium iodide. Since propidium iodide is normally excluded from healthy cells, while Hoescht 33342 penetrates all cells, the combination of these two stains allows for the distinction among healthy cells that stain only with Hoescht (compare Figure 2, A and D), early apoptotic cells that display condensed chromatin with Hoescht (Figure 2, A and C) but do not stain with propidium iodide (Figure 2, D and F), late apoptotic cells that display DNA condensation with Hoescht (Figure 2, B and C) and various degrees of propidium iodide staining (Figure 2, E and F), and necrotic cells that are distinguished by their homogeneous staining with both Hoescht and propidium iodide (Figure 2, A and D, respectively).
Figure 2.
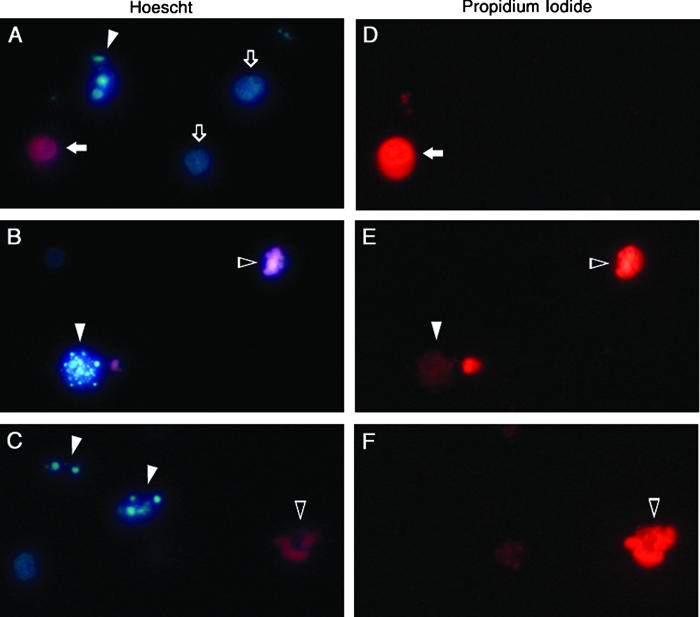
HL-60 cells displaying representative patterns of fluorescent staining with Hoescht and propidium iodide following induction of apoptosis with VP-16. Cells were stained with both Hoescht 33342 and propidium iodide and visualized for Hoescht fluorescence (A–C) and propidium iodide fluorescence (D–F) with pairs of panels (A and D, B and E, and C and F) representing three fields of view (original magnification, 400×). Chromatin of healthy cells stains homogeneously with Hoescht (open arrows; A) but not at all with propidium iodide (D). Cells with condensed chromatin but intact plasma membranes stain strongly with Hoescht (filled arrowheads; A and C) but not at all with propidium iodide (D and F), illustrating the very early stages of apoptosis. A cell further along the apoptotic program displays condensed chromatin with Hoescht (filled arrowhead; B) and slight uptake of propidium iodide (filled arrowhead; E), suggesting decreased membrane selectivity. As apoptosis progresses to completion, cells with condensed chromatin (open arrowheads; B and C) also stain intensely with propidium iodide (open arrowheads; E and F), indicating profound disruption of plasma membrane integrity. Less frequent necrotic cells are easily distinguished from healthy or apoptotic cells by homogeneous chromatin staining with both Hoescht (filled arrow; A) and propidium iodide (filled arrow; D).
In a typical experiment, we observed a small number of HL-60 cells (<10%) exhibiting condensed chromatin at the beginning of the experiment in both treated and untreated cultures (Table 1). These cells displaying condensed chromatin represent the background level of apoptosis in a normal cell culture. We found that a proportion of these cells (100% in this example) typically also stains with propidium iodide, indicating that the cells are in the later stages of apoptosis. The percentage of HL-60 cells displaying condensed chromatin characteristically increases dramatically beyond the second hour after induction with VP-16 and continues to increase for the remainder of the experiment, eventually exceeding 30% of the total cells counted. The percentage of cells with condensed chromatin that stains with both Hoescht and propidium iodide likewise increases after the second hour following induction; however, the percentage of these cells is always lower than that of the total number of cells with condensed chromatin, indicating that chromatin condensation occurs before alterations in plasma membrane permeability.
Table 1.
Fluorescent microscopy detects the progression of apoptosis in HL-60 cells following induction with VP-16a
| Control (%) | Induced (%) | |||||
| Time | ||||||
| (h) | Hoeschtb | PIc | PI/Hoeschtd | Hoeschtb | PIc | PI/Hoeschtd |
| 0 | 3.5 | 3.5 | 100 | 5.6 | 5.6 | 100 |
| 2 | 2.8 | 2.8 | 100 | 2.5 | 2.5 | 100 |
| 4 | 5.0 | 5.0 | 100 | 6.9 | 4.6 | 67 |
| 6 | 3.3 | 2.0 | 60 | 22.5 | 5.1 | 22 |
| 8 | 2.9 | 2.2 | 75 | 31.5 | 15.0 | 48 |
| 10 | 9.4 | 8.1 | 86 | 33.0 | 21.1 | 65 |
aAliquots of HL-60 cells harvested at the indicated time points were stained with a combination of the nuclear stain Hoescht 33342 and the vital stain propidium iodide (PI). Total number of cells (n > 100) was determined by fluorescence microscopy using Hoescht 33342.
bCells displaying condensed chromatin that stain with Hoescht as a percentage of the total number of cells counted.
cCells displaying condensed chromatin that stain with both Hoescht and PI as a percentage of the total number of cells counted.
dCells displaying condensed chromatin that stain with PI as a percentage of the cells displaying condensed chromatin that stain with Hoescht 33342.
Depending on time constraints or other considerations, it may be necessary to delay fluorescence microscopy until the end of the exercise or until a later day. In this case, the addition of an equal volume of 10% formalin to the cells allows for storage of the cells for at least 1 week at 4°C while maintaining good cell morphology and chromatin condensation by Hoescht 33342 fluorescence. In our experience, because of either fading or leakage and dilution, we were unable to detect propidium iodide staining after fixation and storage of cells. Since propidium iodide is a vital stain, it would be expected that fixation would result in uptake of propidium iodide by all cells. Indeed, when we attempted to restain the cells with propidium iodide after fixation and storage, all of the cells were stained.
Alternatively, changes in plasma membrane permeability can be detected with nonfluorescent vital stains such as erythrosin B and trypan blue. Though trypan blue is more commonly used as a vital stain, we found it to be much less sensitive than erythrosin B in detecting nonviable cells. We found that uptake of erythrosin B more closely corresponded with results obtained with propidium iodide. While enumerating cells stained with erythrosin B, blebbing of the cell membranes was obvious in a substantial fraction of the VP-16-treated cells within 4 h. The fraction of cells exhibiting blebbing increased for the duration of the experiment. Visualization of cell blebbing is even more striking under higher magnifications using phase-contrast microscopy (Figure 3).
Figure 3.
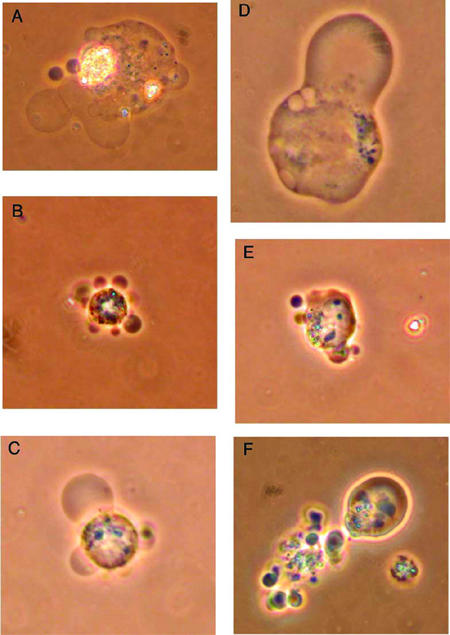
Phase-contrast microscopy of HL-60 cells showing membrane blebbing after exposure to etoposide. Six hours after students exposed HL-60 cells to VP-16, cells with membrane blebbing were visualized using a phase-contrast microscope and photographed with a digital camera. Original magnifications: B, C, E, and F, 400×; A and D, 1000× (oil immersion).
Immunodetection of PARP
Several proteins that participate in various apoptotic pathways and would be suitable for detection of apoptosis have been described in the literature. Caspases are a well-described family of proteins that are present in cells as zymogens and are cleaved during apoptosis to produce active enzymes that act on downstream protein targets, resulting in the execution of programmed cell death. For example, activated caspases 3 and 7 cleave and inactivate PARP-1. PARP-1 is a 116-kD enzyme that participates in DNA replication and repair in normal cells. During apoptosis, PARP-1 is cleaved into 85- and 24-kD inactive fragments.
Several antibodies, both polyclonal and monoclonal, specific for the different caspases are available from a wide variety of suppliers and may be reactive against the zymogen and/or the enzymatically active fragments. We initially chose caspase 3 for this laboratory exercise but were unable to detect the expected disappearance of the 32-kD zymogen and subsequent appearance of the 17- to 19-kD active caspase 3 fragment. Two different polyclonal antibodies specific for both inactive and active caspase 3 were tried, under a variety of experimental conditions, but the results were disappointing. The 32-kD procaspase 3 did not appear to decrease as apoptosis progressed, and the 17- to 19-kD active fragment would have migrated too rapidly to be retained using standard gel conditions. It is likely that a gradient gel is required to adequately resolve both active and inactive forms of caspase 3.
Our second choice for immunodetection was PARP-1. Since PARP-1 is cleaved by activated caspase 3, it can be used to determine indirectly whether cells are undergoing apoptosis via a caspase 3–dependent pathway. Both the 116- and the 85-kD forms of PARP-1 can be resolved using a 10% denaturing polyacrylamide gel, but we found that 8% polyacrylamide yielded better separation of the active and inactive forms of PARP-1. Analysis of cell lysates by western blot reveals obvious degradation of PARP-1 by the third hour of exposure of HL-60 cells to etoposide (VP-16), while the PARP-1 from the control cells remains intact (116-kD peptide) during the entire 6 h of this experiment (Figure 4). Note that while both the 116- and the 85-kD forms of PARP-1 were detectable in this blot, the band representing the inactive 85-kD fragment was far less intense than that of the active 116-kD protein. Student immunoblots tended to exhibit a less intense signal overall, and therefore the 85-kD fragment was not consistently detectable. When a student was successful in detecting this fragment, the signal was very weak (Figure 5), and assumption of PARP-1 cleavage was based on the disappearance of the band representing the 116-kD active protein.
Figure 4.
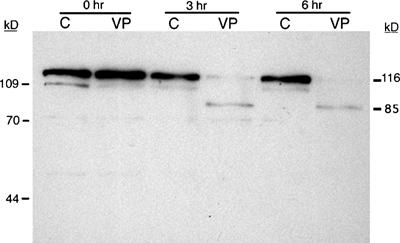
Immunodetection of PARP-1 cleavage during apoptosis in HL-60 cells exposed to VP-16. HL-60 cells were exposed to 500 μM VP-16 (VP) or DMSO (C) and incubated at 37°C for the indicated times. Proteins were isolated, resolved by 8% SDS-PAGE, and electroblotted to a nitrocellulose membrane. Each lane contained the total cellular proteins from approximately 4 × 105 cells. The membrane was probed with a monoclonal antibody against PARP-1, followed by a goat anti-mouse IgG–HRP conjugate. Bands were visualized by chemiluminescence. The apparent molecular masses of the active and inactive forms of PARP-1 are 116 and 85 kD, respectively.
Figure 5.
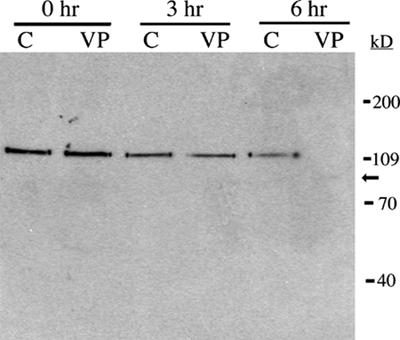
Student-generated immunoblot showing PARP-1 cleavage in Jurkat cells following exposure to VP-16. Jurkat cells were exposed to 500 μM VP-16 (VP) or DMSO (C) and incubated at 37°C for the indicated times. Proteins were isolated, resolved by 8% SDS-PAGE, and electroblotted to a nitrocellulose membrane. The membrane was probed with a monoclonal antibody against PARP-1, followed by a goat anti-mouse IgG-HRP conjugate. Bands were visualized by chemiluminescence. The active PARP-1 (116 kD) is present in cell lysates for up to 3 h but, in contrast to uninduced cell lysates, is no longer detected at 6 h following exposure to VP-16. Note that at 6 h after induction, a very faint band at approximately 85 kD (arrow), likely representing the cleaved form of PARP-1, is detected.
While both monoclonal and rabbit polyclonal antibodies allowed for detection of both active and inactive forms of PARP-1, the monoclonal antibody was much more specific, producing virtually no nonspecific binding or background. The polyclonal antibody, however, produced numerous bands in addition to those corresponding to PARP-1, making interpretation of the blots more difficult. For this reason, we decided that the greater expense of the monoclonal antibody was warranted.
We found that the method for isolating proteins for immunodetection of PARP-1 is critical for inhibition of protease activity that could result in cleavage of PARP-1 independent of apoptosis. Strong denaturing conditions and buffers containing concentrated urea produced the most consistent results. It is also important that the cells be kept on ice during aspiration through a 26-gauge needle, then immediately heated at 65°C, and frozen until electrophoresis is performed.
Evaluation and Assessment of Student Work
Since the course in which this apoptosis project is offered is very advanced and requires the use of laminar flow hoods and other equipment, the maximum size of our class is rather small, limited to 12 students. Students who performed this laboratory exercise were juniors and seniors who already had a course in molecular biology that extensively covered cloning and DNA analysis techniques in the laboratory and eukaryotic gene regulation in lecture, with little specific information on apoptosis. Consequently, we expected our students to be very challenged by this apoptosis project, yet capable of success with some effort and preparation on both our part and theirs. As in real-world investigations, the performance of this project required that students be organized and prepared but capable of troubleshooting and modifying protocols if necessary; collaborative with team members, yet independent enough to work alone on occasion; and focused on the task at hand, without losing sight of the main scientific question. Additionally, students needed to be disciplined and/or excited enough to persevere even when results would not be seen for another week or two.
Student Responses to Notebook Questions
Since this was a complex project, we gave the students some guidance for documentation and analysis with questions, to be answered in their notebooks, such as, “What information can you derive from the Coomassie-stained gel that cannot be determined by immunoblots alone?” “What differences are there between induced and uninduced protein lysates following western analysis?” “At what time point is DNA fragmentation first observed?” and “How do the percentages of cells displaying condensed chromatin staining with Hoescht compare with those staining with propidium iodide over time?” These questions and others (see Appendix E) covered all aspects of the project and required the students to perform detailed analyses of individual experimental results, as well as integrate results from different experimental procedures. With these questions, we hoped to encourage the students to make their own final summary and conclusions about the overall sequence of these events in the apoptotic program.
The students' laboratory notebooks were a major element to our evaluation of their work in the course in which this apoptosis project took place. The criteria we used for grading notebook entries were the following.
Entry had to include purposes for overall lab as well as individual exercises.
Entry had to include answers to questions posed throughout the handouts.
Entry had to include notes on how the exercises were performed (i.e., enough methodology so work could be repeated even by another person).
Entry had to include annotated documentation of results (i.e., well-labeled photos of gels and blots and tabulated data).
Entry had to include appropriate data analyses (i.e., standard curves, calculations, etc.)
Entry had to include interpretation of results and analysis and conclusions.
Notebook had to be organized with numbered and dated pages and written in ink.
As would be expected from a group of 12–15 students, the quality of their analysis ranged from very good to rather poor (see below) and was probably at least in part a reflection of the amount of time students spent reading the background materials provided to them and formulating hypotheses, as well as their willingness to spend time thinking about the results they obtained.
To guide the students through the thought process when they analyzed their results, questions (Appendix E) were posed by the instructors to be answered in the students' notebooks. The following are examples of students' responses to some of these questions. These responses, reflecting a range of quality, were transcribed directly from students' notebooks and were not edited for grammar, spelling, or content. To answer the questions “What is the value of staining cells with both Hoescht and propidium iodide (PI)?” and “Since both PI and erythrosin B are vital stains, what is the value of staining the cells with both?” one student wrote,
Hoescht is not a vital stain and therefore you can determine if the chromatin are condensed in both viable and non-viable cells. Propidium iodide allows us to determine which cells are not viable. Specifically we noticed Hoescht stained cells were easier to visualize than PI stained, but having both stains allows us to determine how many cells have condensed chromatin and are viable compared to how many have condensed chromatin and are not viable. We used both erythrosin B and PI because erythrosin appears to stain more cells. That is, it may pass through the membrane quicker than PI and possibly easier.
Another student provided a less complete answer that implied a misunderstanding of the activity of the PI stain:
Hoescht is a stain that can enter a cell and doesn't require the membrane to be compromised. Hoescht binds DNA to show condensation. Propidium iodide stains cells that have membranes that are leaky. It is a vital stain, if the cell is alive then it will allow the stain to enter. If both stains are present, the cell is not viable.
Another set of questions we asked was “Describe the appearance of DNA fragmentation observed in the induced and uninduced cells. What do these results tell you about how long after induction DNA fragmentation occurs? In analyzing the DNA gel, what is the range of sizes of the smaller fragments seen in some of the lanes?” In some cases, students' results did not allow for adequate interpretation. For this reason, in addition to their own results, students were provided a photograph of an agarose gel generated by another student group showing DNA laddering. In the following response, one student refers first to his own group's data: “As seen on our gel, we have no DNA laddering. This is a difficult conclusion considering the faintness of the smear we have. This problem arose because there were probably not enough cells to attain a reasonable amount of DNA.” Then, in referring to the gel provided, he writes, “DNA fragmentation begins to occur at around 4 hours (gel not shown). VP-16 cell's DNA has small fragments in a ladder formation while control DNA has only a smear (as seen on all of my lanes). The range of sizes of the smaller fragments is 200 bp, 400 bp, 600 bp, 800 bp, 1000 bp).” Another student, whose group generated the interpretable data that the latter student was just describing, appears to confuse DNA fragmentation with chromatin condensation, and writes, “DNA fragmentation in cells looked like the nucleus broke up into little fluorescent dots. Cells displaying unfragmented DNA had a solid fluorescent nucleus when stained with Hoescht. The induced cells displayed significantly more DNA fragmentation by the 4 hour interval.”
One question that we did not ask the students directly was, “What is the sequence of events of apoptosis in HL-60 cells following induction with VP-16?” While as instructors we felt that this was an obvious goal that was implied by the experimental design and the content of the introductory handout (Appendix B), disappointingly, only 3 of 13 of our most recent students wrote a conclusion in their notebooks that summarized the sequence of events in apoptosis according to their data. Of these three, the best (unedited) summary was,
These data collect indicate the order of events, for HL-60 cells, in apoptosis. PARP-1 cleavage occurs first, with active PARP-1 no longer visible in the western blot analysis 3 hr after apoptotic induction using VP16. Chromatin condensation occurs next, with approximately 50% of cells showing condensed chromatin at 4 hrs after apoptotic induction. DNA laddering is one of the last steps—we see it only 8 hrs after induction of apoptosis. This seems logical as DNA would first condense and then be digested to yield nucleosome sized fragments. Membrane selectivity (followed shortly by blebbing) appears to be the last step in apoptosis. 31% of HL-60 cells induced to undergo apoptosis were suseptable to Erythrocin B staining at 8 hrs.
While several other students did note the time courses of most of the apoptotic events, they neglected to integrate these individual findings into a concise conclusion such as that provided above.
Student Responses to Assignment Questions
As an addendum to their notebook entries, we asked another series of questions (Appendix E) leading the students to use their laboratory experience toward further exploration and possible future directions in experimentation of apoptosis. Examples of some of the highest- and poorest-quality student responses to selected assignment questions are provided here. To answer the questions “When observing cells by fluorescent microscopy, did the percentage of cells with condensed chromatin increase throughout the course of the experiment?” and “If you carried the assays out for a longer period of time, such as 24 hours, would you expect the percentage to continue to increase until all cells displayed condensed chromatin?” one student wrote,
The percentage of condensed chromatin did increase throughout the experiment as expected. If the experiment were carried out longer, a few things would happen. The percent chromatin condensation would increase for a while. After a certain time, cells with condensed chromatin would completely bleb out, and this number would decrease. There would also probably be cells that are not induced to undergo apoptosis by VP-16 via some molecular mechanism, therefore, not all of the cells would end up with condensed chromatin.
Another student's answer was not as astute, however: “During this experiment, the percentage of cells with condensed chromatin increased with time. I would expect a continued increase in the percent of condensed chromatin because it is a step in apoptosis. The chromatin condenses so that the cell can die in an orderly fashion.”
We also requested our students to comment on the observation that “in our experiment to induce apoptosis, not every cell responded to VP-16 by undergoing programmed cell death.” We asked that they “briefly propose a molecular mechanism for how response to apoptosis induction could be different between cells” and “considering their origin, speculate on whether or not HL-60 cells may have a different sensitivity to apoptosis induction than their normal counterparts.” A thoughtful student's answer to these questions was,
There are many possible molecular abnormalities that could lead to desensitization of cells to VP-16. Perhaps the membrane receptors are absent or there may be a defect in signal transduction or enzyme cascade functions. I would speculate that these cells (HL-60) may have differing sensitivities to apoptotic induction than their normal counterparts because these are cancer cells. These cells are human leukocytes from a leukemia patient, and the process of transformation may have altered their response to apoptotic stimuli.
The student who wrote the following, however, did not grasp the point of the question:
Depending on the overall function of the cell type would definitely make a difference in the response of the cell. Some cells, such as B-cells, could make antibodies to neutralize toxins. The HL-60's are not normal human cells. They are immortalized cells that can replicate an infinite amount of times. They would respond similarly to VP-16 but normal cells would probably be more susceptible to the dose given to these cells and they would die more easily.
It should be noted that, though some students did not reach the desired level of achievement in some of their answers, they all clearly learned enough vocabulary and basic theoretical and empirical knowledge to make a reasonable attempt at composing an answer.
In their responses to some of the questions, students expressed their opinions regarding certain aspects of the apoptosis project. For instance, one student commented on her impressions during the day of apoptosis induction and cell harvesting by writing that in the future she would “definitely limit the number of students in the cell culture room because the overcrowded room made working together rather annoying” and that “someone was always standing where others needed to be.” This student's suggested solution to this problem was to have groups perform the induction on different days. A couple of comments appeared to attribute stress to the group's size being too large when one student thought, for example, “that having three members doing the project is too many hands.” While it may be a good idea to keep the size of the group to a minimum, since at least two students are required at every time point on the day of apoptotic induction, groups of less than three members are impractical. Despite how hectic the day of induction was for some, several students thought that it would have been a good idea to include more time points in the experiment. Suggestions ranged from “observ[ing] the time between hours 4 and 8 to determine if membrane permeability is lost before DNA laddering occurs” to “assay[ing] apoptosis over a longer period of time” since “many sources induce apoptosis over a 1–2 day period, and in these assays they use a lower concentration of inducers.” Even with these concerns, comments such as “If I were to repeat the experiment a different time, I probably wouldn't change a thing!” and “Overall I think the experiment went well” suggest that the general impression of the students was positive. In fact, the project inspired some students to suggest ideas for further exploration, in addition to the different time points as discussed above, such as “another western blot analysis…to detect the activation of caspase-3 and try to compare it to the cleavage of PARP-1 to see when it does this relative to induction time.” Another student wrote, “I actually liked the experiment the way it is currently. I used it slightly as a model for my independent project and everything seemed to ‘work' or at least worked smoothly.”
Independent Research Projects
Each student, as part of the requirements for successful completion of this course, had to design and carry out an independent project of his/her choosing during the last 3 weeks of the semester. Additionally, a manuscript suitable for submission to Molecular Biology of the Cell had to be completed using the journal's “Instructions to Authors.” Underscoring the popularity of the apoptosis protocols, 6 of 15 of our most recent students chose to design and perform apoptosis-related experiments for their independent projects. These projects included (1) induction of apoptosis with apogenin, a bioactive flavenoid derived from plants, (2) comparison of the apoptotic response between lymphoid (Jurkat) and promyelocytic (HL-60) cell lines (Figure 5), (3) the effects of serum starvation of cells on etoposide-induced apoptosis, (4) induction of apoptosis in an attached cell line (Figure 6), and (5) comparison of apoptosis induced by topoisomerase inhibitors versus a protein synthesis inhibitor (Figure 7).
Figure 6.
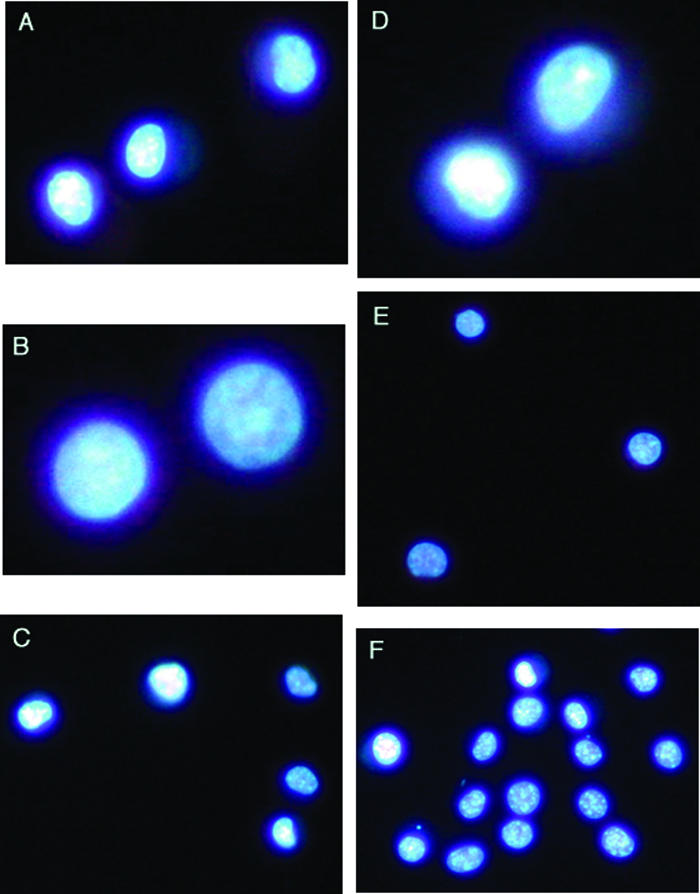
Student photographs of Hoescht-stained cells to study the effects of anchorage dependence on apoptosis. Anchorage-dependent mouse L929 cells were cultured in 24-well dishes with or without paraffin coating to prevent attachment (D and A, respectively). To determine whether paraffin directly induces apoptosis, anchorage-independent HL-60 cells were cultured in the same way (E and B, respectively). Additionally, L929 cells were exposed to VP-16, a known inducer of apoptosis (F), or medium alone (C) without paraffin. In all cases, chromatin condensation was used to indicate apoptosis. Interestingly, no chromatin condensation was evident in L929 cells under any condition even following exposure to VP-16. Original magnification, 400×.
Figure 7.
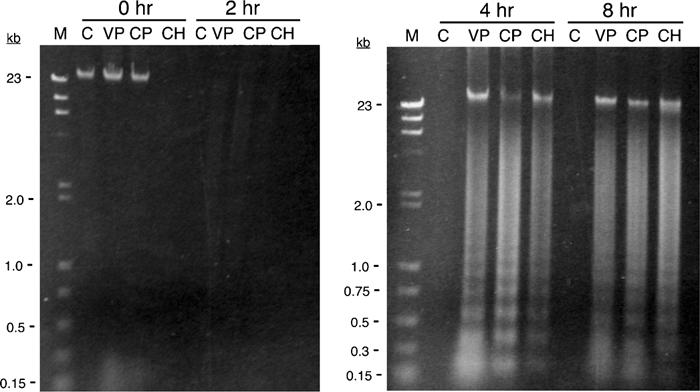
Student-generated DNA laddering in HL-60 cells exposed to three apoptosis-inducing agents. HL-60 cells were exposed to either DMSO alone (C), 500 μM VP-16 (VP), 11 μM camptothecin (CP), or 4 μM cycloheximide (CH) for the indicated times. Cells were lysed, and the DNA was isolated by phenol–chloroform extraction followed by ethanol precipitation. The DNA was resolved by electrophoresis through a 1.2% TAE–agarose gel and visualized with ethidium bromide. The relevant sizes of the standard markers (M) are shown as kilobases (kb). DNA laddering is clearly evident within 4 h after exposure to all three inducers. Note, however, that several DNA samples, especially at the 2-h time point, were likely lost during the isolation procedure.
Very popular with the students (and we, the instructors grading notebooks) were the cell counting data and protocol recording tables. We have included in Appendix F tables that are blank as well as a set that contains sample data to illustrate how the tables should be used. We highly recommend such tables as a way of helping the students keep track of their progress and to document their data throughout the experimental protocol on the day of apoptosis induction when the majority of sample processing and cell counting occurs. Modifications of the cell staining table could include additional columns for percentage of total cells with condensed chromatin and/or a column for the calculated ratio of cells staining with propidium iodide (PI) as a percentage of the cells displaying condensed chromatin.
In cell counting assays, objective documentation such as photography may be impractical. For this reason, the most variability (and therefore potential inaccuracy) may occur when assessing chromatin condensation and membrane permeability changes by fluorescent microscopy. Such variability may result from (1) students' inexperience in recognizing apoptotic cell morphology, (2) introduction of bias during cell counting, and (3) different students' counting cells at different time points (a necessity when students with other classes or job commitments are involved in a day-long experiment such as the one we describe). With regard to student bias while cell counting, it is recommended that the student actually counting the cells have no prior knowledge of whether the cells being counted are induced or uninduced; hence, requiring that another student in the group be the one to prepare the slides.
Instructor Preparation and Involvement
Surprisingly, for a project that extended over a 4-week period, the preparation for the instructors was minimal even while avoiding kits whenever possible due to their expense and because they discourage self-reliance and theoretical understanding of the techniques. In addition to the initial culturing and distribution of HL-60 cells, we prepared some reagents and buffers ourselves based primarily on their toxicity and/or complexity, including the DNA and protein extraction buffers, the saturated phenol, and stock solutions of etoposide, Hoescht, propidium iodide, and ethidium bromide stains, and acrylamide:bis monomer. Students were responsible for making the cell media and all of the protein gel electrophoresis and blotting buffers from stock solutions that they prepared either in this course or in their previous molecular biology course.
As instructors, we also had the responsibility of guiding and monitoring the progress of the students as they worked through the different assays. At least one instructor needed to be available during the day of the induction of apoptosis to help with questions and to prevent any major mistakes. It was also important to be sure that the students kept to a schedule, reminding them, if necessary, to perform certain tasks while they waited for incubations, for example.
Future Directions
The overall theme of apoptosis and the techniques performed in the exercises we have described suggest directions for future projects for class activities or independent research. For example, students could optimize the conditions of immunodetection for using the polyclonal anti-PARP-1 antibody discussed above including blocking and antibody dilutions. Other projects could include testing alternative inducers, both chemical and otherwise (e.g., serum deprivation) and/or inhibitors of apoptosis, or attempting to isolate cells that are resistant to apoptosis inducers. More advanced class activities could be added to this laboratory if time in the semester allows. One exciting and very current avenue is to monitor changes in gene expression by producing labeled cDNAs from induced and uninduced cells and hybridizing them to microarrays containing apoptotic genes.
ACCESSING MATERIALS
No additional materials available online.
Acknowledgments
ACKNOWLEDGMENTS
We are indebted to the students of Molecular and Cellular Techniques, Biology 466 (Spring 2002 and 2003), for their patience and hard work in performing these exercises. We also wish to thank Kurt Bachman and James G. Herman for technical advice. Some of the equipment used in this investigation was provided in part by funding from NSF ILI grants. Materials were purchased through the generosity of the Faculty Grants Committee and Department of Biology at Millersville University of Pennsylvania.
Appendix A: RECOMMENDED STUDENT READING
Chiarugi A., Moskowitz M. A. PARP-1—A perpetrator of apoptotic cell death? Science. 2002;297:200–201. doi: 10.1126/science.1074592.
Hengartner M. O. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710.
Leist M., Jaattela M. Four deaths and a funeral: From caspases to alternative mechanisms. Nature Rev. 2001;2:589–598. doi: 10.1038/35085008.
Mariani A. R., Columbaro M., Zauli G., Zamai L., Luchetti F., Gobbi P., Ghibellini D., Falcieri E., Vitale M. Lineage-related susceptibility of human hemopoietic cell lines to apoptosis. Anat Rec. 1999;254:1–6. doi: 10.1002/(SICI)1097-0185(19990101)254:1<1::AID-AR1>3.0.CO;2-6.
Martins L. M., Kottke T., Mesner P. W., Basi G. S., Sinha S., Frigon N., Jr., Tatar E., Tung J. S., Bryant K., Takahashi A., Svingen P. A., Madden B. J., McCormick D. J., Earnshaw W. C., Kaufmann S. H. Activation of multiple interleukin-1β converting enzyme homologues in cytosol and nuclei of HL-60 cells during etoposide-induced apoptosis. J Biol Chem. 1997;272(11):7421–7430. doi: 10.1074/jbc.272.11.7421.
Meng X. W., Fraser M. J., Feller J. M., Ziegler J. B. Caspase-3-dependent and caspase-3-independent pathways leading to chromatin DNA fragmentation in HL-60 cells. Apoptosis. 2000;5:61–67. doi: 10.1023/a:1009689710184.
Smyth P. G., Berman S. A., Bursztajn S. Markers of apoptosis: Methods for elucidating the mechanism of apoptotic cell death from the nervous system. BioTechniques. 2002;322:648–665. doi: 10.2144/02323dd02.
Studzinski G. P. Overview of Apoptosis. In: Studzinski G. P., editor. Apoptosis: A Practical Approach. Oxford University Press; New York: 1999. pp. 1–17.
Appendix B: INTRODUCTION TO APOPTOSIS
Introduction
When considering the growth and development of complex multi-cellular organisms, it is natural to think of robustly growing cells dividing and differentiating to complete the pre-planned architecture of a body. Over 100 years ago, however, developmental biologists noted that during development in many different organisms, some cells appear destined to live only a short while, and then die at precise times, and in a coordinated manner. The webbing between the fingers of a human fetus, and the tail of a tadpole are good examples of this phenomenon. In addition to the precise timing and coordination of events, this massive cell death does not induce an inflammatory response, which is a part of the innate immunity of most higher organisms. This form of programmed cell death, now referred to as apoptosis, is highly conserved between organisms separated by millions of years of evolutionary history. Indeed, much insight into the molecular genetics of this process has been elucidated using the nematode worm Caenorhabditis elegans as a model.
More recently, it has been found that programmed cell death also functions in mature stages of many organisms, including humans. In the thymus, for example, immature T-lymphocytes are selected based on their ability to recognize self-antigens displayed by dendritic cells. Immature lymphocytes that lack the antigen receptors allowing them to respond to self-antigens escape the thymus, while self-reactive lymphocytes receive a signal to undergo apoptosis. Several million precursor T-lymphocytes enter the thymus daily, with only 1 to 2% leaving to become functional T-lymphocytes.
Microscopically, apoptotic cells appear shrunken with membrane blebbing. Apoptosis presents a very different picture from necrosis, the other means by which cells die. Necrosis occurs in response to physical injury, such as that caused by physical trauma. Cells and cellular organelles undergoing necrosis typically appear swollen, due to a loss of membrane selectivity, which results in an influx of water into the cells. These cells eventually degrade, releasing their contents into the tissue. Many of the substances released, such as prostaglandins and tissue thromboplastin, are inducers of inflammation. Of what benefit is inflammation? What might explain why apoptosis does not induce inflammation, while necrosis does?
The significance given to apoptosis is underscored by the enormous body of literature that has arisen over the past few years (by one estimate 50,000 publications and growing). Many of the most recent publications describe how alterations in the induction, suppression, or regulation of apoptosis underlie many diseases, including some forms of autoimmunity and cancer. Several articles about apoptosis (both primary research articles, and reviews) are on reserve in the library. Every student is responsible for reading articles outside of class, in order to become familiar with apoptosis before coming to class. Be prepared to participate in a discussion on apoptosis. In general, scientific literature can be separated into two types of publications, review articles and primary research papers. Reviews often explore an area of research by summarizing the most recent results, and integrating new findings into the existing body of knowledge. For example, the review “Proteases for Cell Suicide: Functions and Regulation of Caspases” (Chang and Yang, 2000) provides a state of the art overview of what is known about the activity and functions of caspases, enzymes that are central players in programmed cell death.
Primary research papers, on the other hand, are more helpful when searching for appropriate protocols and applications. It is not necessary to read every article and review available. Rather, select papers to read based on their applicability to what you intend to do. For example, do not concern yourself too much with the use of caspase inhibitors in this particular study of apoptosis, since we will not be using inhibitors in our experiments. This type of judgement is exactly what you will be expected to use as a graduate student or a scientist.
Apoptosis can be induced experimentally by exposing cells to various stimuli, including chemicals or radiation. Topoisomerase inhibitors such as etoposide (also known, for some reason, as VP-16) are potent inducers of apoptosis, and are widely used in the study of programmed cell death. Within several hours following exposure of susceptible cells to an apoptosis inducer, a series of phenotypic changes occurs in the cells, including, but not necessarily in order of occurrence:
Alterations in the permeability of the plasma membrane
DNA fragmentation into nucleosome-sized fragments (laddering)
Condensation of chromatin
Cell membrane blebbing
Caspase activation
While virtually all types of cells can be induced to undergo apoptosis in culture, the apoptotic phenotype may differ depending on the lineage of cells being investigated. (Hint: there is an article on reserve that explores this topic). In this series of laboratory exercises, you will chemically induce apoptosis in HL-60 cells. HL-60 cells are a continuous cell line derived from a human promyelocytic leukemia. HL-60 cells, when induced to undergo apoptosis, typically display all of the apoptotic changes listed above, making these cells a good model for the study of this important cellular process.
The objectives for this series of experiments are:
To be able to describe the mechanisms of programmed cell death.
To perform techniques involved in the assay of apoptosis, including maintenance of cells in culture.
To determine experimentally the sequence of events of apoptosis in HL-60 cells.
This experiment will be performed in groups of 3 or 4. The general design will be as follows:
HL-60 cells will be exposed to an apoptosis inducer.
- At appropriate time intervals, cells will be harvested and processed for the following assays:
- Cleavage of poly ADP-ribosyl polymerase (PARP-1) by western blot analysis
- Alterations in plasma membrane permeability using vital and fluorescent stains
- DNA fragmentation by gel electrophoresis
- Chromatin condensation by fluorescence staining and microscopy
- Plasma membrane blebbing by phase-contrast microscopy
Detection of Apoptosis
1. Cleavage of PARP-1 as an indicator of caspase activation
A central player in apoptosis is a family of aspartyl proteases known as caspases. Caspases exist as zymogens in cells, and become activated following apoptosis induction. For several of the caspases, activation involves the cleavage of the zymogen into an enzymatically active fragment. Once activated, caspases go on to activate other caspases in a typical enzymatic cascade, as well as activate or degrade effector proteins whose activity, or loss thereof, leads to the destruction of the cell. One such protein is PARP-1, an enzyme that appears to be involved in DNA repair. The active enzyme is expressed in cells as a 116 kD protein. Activated caspases, most notably caspase-3 and caspase-7, cleave PARP-1 into an 85 kD inactive form. This cleavage is easily detected by western blot analysis using a monoclonal antibody specific for PARP-1.
In this experiment, western blot analysis will be performed using lysates obtained from both untreated HL-60 cells, and HL-60 cells exposed to an inducer of apoptosis. The cells will be lysed, the proteins separated by SDS polyacrylamide gel electrophoresis, blotted onto a nitrocellulose membrane, and probed with a monoclonal antibody specific for PARP-1. What is the difference between a polyclonal antibody and a monoclonal antibody? What are some potential advantages and disadvantages of each?
Much information on western blot analysis of PARP-1 and other proteins involved in apoptosis can be found in catalogues of the companies that produce antibodies and other reagents for the study of apoptosis. Santa Cruz Biotechnology and Pharmingen are two companies with a wide range of apoptosis products, and very informative catalogues.
What would you expect to see on a western blot of untreated versus treated cells if using an antibody specific for (1) both the active and inactive forms of PARP-1, and (2) the active enzyme only? Can you think of any advantage one antibody might have over the other antibody? In other words, which antibody might be the best one to use considering the objectives of this exercise?
2. Alterations in plasma membrane permeability
At some point during the progression of the apoptotic program, changes in the plasma membrane of the cell occur. These changes are both in the organization of membrane phospholipids, as well as functional changes that result in a loss of selective permeability. This change in the permeability of the plasma membrane can be detected by using vital stains. In general, vital stains are excluded from cells with intact plasma membranes, but are readily taken up by cells with altered plasma membranes. In this experiment, cells will be stained using (1) erythrosin B, and examined by light microscopy, and (2) propidium iodide, and examined by fluorescence microscopy. These results can then be compared.
3. DNA fragmentation
During apoptosis, the cell's DNA is degraded in a very specific manner, resulting in the formation of nucleosome-sized DNA fragments and multiples thereof that form a ladder of bands when separated by electrophoresis. While DNA laddering is not a universal phenomenon, it does occur in many types of cells undergoing apoptosis, including HL-60 cells. In this exercise, you will isolate genomic DNA from HL-60 cells and attempt to detect DNA laddering by agarose gel electrophoresis.
What is a nucleosome? On an ethidium bromide-stained gel, how does the appearance of DNA laddering compare to DNA that is non-specifically degraded?
4. Chromatin condensation by fluorescence microscopy
Cells undergoing apoptosis display a profound destruction of the nucleus that results in the formation of nuclear blebs containing DNA. Staining of apoptotic cells with a fluorescent DNA-binding stain allows for easy detection of this phenomenon, because the DNA-filled blebs stain brightly, and are easily distinguished from intact nuclei. Whether or not the stain can cross the plasma membrane of the cell is an important consideration when selecting from the wide variety of DNA-binding stains available from sources such as Molecular Probes Inc. Fluorescent stains such as DAPI and Hoescht 33342 easily cross the intact plasma membrane and nuclear envelope, and can be used to stain the DNA of live cells. Propidium iodide, on the other hand, is considered a vital stain, and only crosses the plasma membranes of non-viable cells. What is a fluorescent stain? How do fluorescent stains such as Hoescht 33342 work? Why is Hoescht 33342 green and propidium iodide red?
5. Plasma membrane blebbing
Membrane blebbing is observed as a ballooning out of the plasma membrane of apoptotic cells, endowing the cells with a bizarre appearance. An observant student will be able to detect membrane blebbing when performing cell counts using erythrosin B. This phenomenon, however, is more easily seen using phase-contrast microscopy that allows for higher magnification and greater resolution. Along with inverted and fluorescence microscopes, a phase-contrast microscope will be available for your use.
To prepare for this series of experiments, you must have the following information prior to coming to the next class:
A flow chart illustrating the experimental protocol your group plans to follow. This protocol will be designed by your group using the literature provided on reserve and any other sources that your group deems necessary. Searching for the most appropriate protocols can be time-consuming. You must consider many things when developing a protocol, including the type of cells to be used, how long the experiment might take to complete, whether a protocol might involve the use of very dangerous, or very expensive reagents, etc.
An estimation of the minimum number of cells your group will require to perform the entire experiment that you have designed, as well as the number of cells needed for each of the assays listed above.
Any reagents that you will require for the experiment (e.g., what apoptosis inducer you plan to use, what stains you plan to use, etc.), including concentrations, amounts, sources of reagents, etc.
A timeline for the experiment. At what point(s) after induction will you harvest cells, how many cells will be harvested at each time point, how will these harvested cells be processed or treated, etc.?
What is the expected order of events in apoptosis? For example, do you expect DNA fragmentation to occur before or after changes in membrane permeability?
Though members of a group may work together to design the protocol, each student will submit the above information in his/her own words upon arriving for class on March 10 for section 1, or February 25 for section 2. This information will be evaluated as part of the “Apoptosis” notebook entry.
Culture and Maintenance of HL-60 Cells
HL-60 cells are a continuous cell line isolated from a human promyelocytic leukemia. The cells grow unattached, in suspension. The most widely used medium for the maintenance of HL-60 cells is Iscove's Modified Dulbecco's Minimal Essential Medium (IMEM), supplemented with 20% fetal bovine serum. Occasionally references can be found using other modifications of MEM, as well.
HL-60 cells are sensitive to cell densities. It is crucial that cell densities are maintained between 0.2–1.5 × 106 cells/ml. The cells will rapidly lose viability if allowed to attain higher cell densities. Because of this sensitivity, it is important that you check your cells and care for them no less than every other day. FAILURE TO GIVE YOUR HL-60 CELLS THE PROPER LEVEL OF CARE WILL RESULT IN YOUR CULTURES CRASHING AND HAVING AN INSUFFICIENT NUMBER OF VIABLE CELLS TO PERFORM YOUR EXPERIMENTS.
It is the responsibility of each group to know how many cells will be needed for the experiment, and to have a sufficient number of HL-60 cells in order to perform the apoptosis experiments. It is also the responsibility of the class to have sufficient numbers of sterilized pipettes, tips, etc.
Appendix C: CELL CULTURE: INITIATION AND MAINTENANCE OF MAMMALIAN CELLS
Introduction
Prior to the discovery that individual animal cells could be grown in culture (in vitro), scientists were relegated to conducting studies on intact animals (in vivo). In vivo studies often suffered from lack of reproducibility due to genetic differences between animals, immune status, age, and a number of other variables, many of which were undefined and uncontrollable. The development of highly inbred strains of many animals, most importantly mice, eliminated many of the differences observed in outbred animals, but the use of animals in research still entails many difficulties. The development of tissue culture technology has allowed for the growth of virtually any specific type of cell under very precise and reproducible environmental conditions. This ability has had an enormous impact on many fields of study including health and medicine, molecular and cellular biology, and agriculture.
Types of Cell Cultures
(1) Primary cell cultures
A culture of cells may be established from most tissues that contain actively growing cells. In animals, a small plug of tissue is usually removed, and the cells are dissociated into a suspension of single cells by treatment with proteolytic enzymes. When placed in an appropriate nutrient medium, some of these cells eventually grow (divide) to establish a primary culture. Primary cell cultures are the closest to the cells found in the intact organism, and often contain more than one type of cell. Distinguishing characteristics of the source tissue often are retained, at least transiently, in primary animal cell cultures. Since different tissues have different capacities to replicate in culture, after time, usually a single cell type comes to dominate the culture. In primary cell cultures, however, cells lose the ability to divide, usually after 30–50 cell divisions, a characteristic known as the Hayflick limit. At this point, the cells senesce and die.
(2) Diploid cell strains
Diploid cell strains consist of a single type of cell, such as a kidney fibroblast. Diploid cells are normal cells, and often retain some characteristics found in the intact organism regarding metabolic activity and state of differentiation. Diploid cell strains are very useful for studying cellular processes specific to a particular cell type. Like primary cultures, however, diploid cell strains also have Hayflick limits.
(3) Continuous cell lines
Continuous cell lines consist of transformed cells. Continuous cell lines may be obtained from neoplasms, may be spontaneously generated by outgrowth of primary cell cultures, or may be induced by treatment of normal cells with oncogenic chemicals, ultraviolet light, or infection by oncogenic viruses. Continuous cell lines often retain only a slight resemblance to the parent cell or tissue type. These cells are often de-differentiated, and may not possess the same functional capacities as the parent cells from which they were derived. In addition, gene expression is altered, and continuous cell lines may express genes that were previously silent, or may silence genes that were expressed in their differentiated parent cells. These cell lines often display profoundly abnormal karyotypes, with polyploidy and chromosomal rearrangements and deletions being very common. Cells transformed by viruses express viral genes that are usually required for maintenance of the transformed phenotype. In addition, virus-transformed cells may or may not produce infectious virus.
Attached versus Suspension Cultures
Animal cells may be grown as suspension cultures or as attached cultures. Cells in suspension culture are dispersed throughout the medium. Primary lymphocyte and granulocyte cultures, as well as lymphocyte and granulocyte cell lines, grow in suspension. Suspension cultures can be harvested simply by centrifugation.
Many types of cells and cell lines are anchorage dependent, and require glass or plastic surfaces for attachment. These attached cells, covered with a thin layer of liquid medium, are able to grow until they form a confluent monolayer. At this point, normal cells (primary cultures and diploid cell strains) stop dividing (contact inhibition). Transformed cells, on the other hand, often pile up into three-dimensional foci due to loss of contact inhibition. Periodic detachment of the cells from the growing surface, and reduction in cell numbers are essential for continued growth of the cultures.
Harvesting of attached cells is possible only after the cells have been released from their growth surface. Scraping of cells from flask surfaces with a rubber “policeman” has been employed, but often leads to cell damage resulting in lower viability. Proteolytic enzymes such as trypsin are widely used as an alternative method of dislodging cells. Once the cells are in suspension, they can be manipulated easily.
Long-Term Storage of Cells
The ability to cryopreserve cells is an essential part of mammalian cell culture. Cryopreservation entails freezing and storage of cells at temperatures at or below −150°C. Cells grown in culture for prolonged periods of time eventually change as the selective pressures of in vitro growth select for those cells best able to replicate in culture. With primary cultures and diploid cell strains, these phenotypic changes occur well before the cells reach their Hayflick limit. Cryopreservation helps to offset these problems.
To cryopreserve mammalian cells, the cells are harvested and resuspended in culture medium containing 10% glycerol or dimethyl sulfoxide (DMSO), and placed in a sealed tube or ampule. The temperature of the cell suspension is then slowly lowered to at least −150°C. Viability of cells can be maintained indefinitely at this temperature.
Tissue Culture Media
There are many different media formulations available today for growing and maintaining a wide variety of eukaryotic cells. Cell culture systems have been described for various mammalian cell types, as well as insect cells, amphibian cells, and fish cells. Plant and algal cell culture encompasses another completely separate, and very extensive variety of media and techniques.
While all cells share some basic nutritional requirements, there are significant differences in the requirements of different types of cells that must be taken into account if one is to be successful in culturing cells. For example, primary lymphocyte cultures require interleukin-2, and will rapidly undergo apoptosis in the absence of this cytokine.
Tissue culture media can be purchased pre-made and sterile, or as powders that must be reconstituted and sterilized prior to use. Pre-made media are more convenient, but are also significantly more expensive. Many different media formulations are available. The formulations of Iscove's modified Dulbecco's and RPMI-1640 media can be found in the manufacturer's catalogue. Both of these media are suitable for the culture of HL-60 cells that will be used to study apoptosis.
Media components
All tissue culture media contain the following components:
Water—The water used for tissue culture media must be very high quality and free from impurities. The purity of water can be determined indirectly by measuring the electrical resistance of the water, measured in megaohms. All tissue culture laboratories have special water purification systems with resistance monitoring devices. The higher the purity of the water, the greater its electrical resistance. In general, water for tissue culture should have an electrical resistance of at least 16 megaohms.
Inorganic salts—CaCl2, KCl, MgSO4, NaCl, and NaH2PO4.
Amino acids—All media are supplemented with amino acids. Essential amino acids are those amino acids that cells cannot synthesize, so must be supplied in the media. Non-essential amino acids are those amino acids that cells can synthesize on their own, but when added to the media allow for more vigorous cell growth.
Other components—Glucose, succinic acid, and vitamins for metabolism. Additionally, sodium pyruvate is often added.
- Buffers—It is essential to buffer media to keep the pH within a narrow range of approximately pH 7–7.4. For this purpose, two buffers are often used:
-
Sodium bicarbonate—Since cells produce CO2 during respiration, a bicarbonate buffer system is useful for maintaining pH. This is one of the major buffer systems of plasma as well, and works in the following way:
- CO2 + H2O ↔ H2CO3 ↔ H+ + HCO3−
As cells produce CO2, the equilibrium of this buffer system shifts to the right, producing acid, which lowers the pH of the medium. Some of this CO2 will be lost by evaporation, helping to stabilize pH. Cells are often maintained in CO2 incubators an atmospheric CO2 tension of 2–5% that keeps the pH in the correct range for cell growth. - HEPES—HEPES is a second commonly used buffer that maintains the proper pH of media.
-
-
Phenol red—Phenol red is used as a pH indicator, and is useful in determining whether the pH of the medium is in the proper range. Phenol red is yellow in an acidic medium, and purple-red in a basic medium. If the medium is a salmon-red color, the pH will be suitable for most cells.
The cell culture medium is supplemented with the following components, usually prior to sterilization:- Non-essential amino acids and sodium pyruvate—Supplied as 100× sterile solutions.
-
Fetal bovine serum—All cells require serum of some sort to supply essential growth factors. Many of these growth factors are either unidentified, or are most conveniently supplied by the addition of serum. The highest quality serum for tissue culture is fetal bovine serum (FBS), and is used at a concentration of 5–20%, depending on the needs of the particular cells being grown. Recently, serum-free media have become available, and are very useful for special applications.Depending on the type of cells you are working with, and the growth rate of the cells, you will need to either feed the cells, or subculture them. In general, the serum concentration plays the largest role in the growth rate of the cells. Cells will have fast doubling times when the serum concentration is 10% or greater. Most continuous cell lines will grow well at serum concentrations of 10% or less.
- Penicillin/streptomycin—Supplied as a 100× sterile solution, and used to inhibit the growth of bacteria. Cell culture media are extremely rich, and will support the growth of many types of microorganisms. Good aseptic technique must be used at all times. It is important to remember that the addition of antibiotics reduces, but does not eliminate, the possibility of contamination. In addition, these antibiotics have no activity against yeasts and fungi.
Medium Preparation
To prepare 1 liter of medium:
Place approximately 900 ml of ultrapure water into an Erlenmeyer flask. Stir using a magnetic stirrer. Add one envelope of powdered medium. Add the remaining medium components (HEPES and NaHCO3, sodium pyruvate, amino acids, and penicillin/streptomycin.
Fetal bovine serum: Some investigators add the serum to the medium prior to sterilization. If serum is added at this point, it will be necessary to pre-filter the medium using a 0.45 μm filter prior to sterilization with a 0.22 μm filter. Alternatively, sterile serum can be added after the medium is 0.22 μm filter-sterilized and dispensed into sterile bottles.
Using a 1 liter graduated cylinder, q.s. the medium to 1 liter, and return the medium to the Erlenmeyer flask.
Under the laminar flow hood, filter-sterilize the medium using a bottle-top vacuum filter into sterile bottles. If the medium contains serum, pre-filter it using a 0.45 μm filter, followed by sterilization using a 0.22 μm filter. If the medium does not contain serum, only the 0.22 μm filtration is needed.
Culturing HL-60 Cells
Initiation of HL-60 cells from frozen stocks
When initiating cells from cryopreservation, the rapid removal or dilution of the DMSO or glycerol is important for the viability of thawed cells. For maximum recovery, it is important to thaw the cells quickly at 37°C.
Remove frozen cells from the freezer. While holding the cryovial by the lid, immerse the tube into the 37°C water bath. Shake gently until the cells are completely thawed. While thawing, do not immerse the tube below the level of the lid bottom, as this may lead to contamination of the cells.
Transfer the thawed cells to a sterile 15 ml conical centrifuge tube containing 10 mls of pre-warmed Iscove's medium. Centrifuge for 6–7 minutes at 1000 rpm (approximately 400 x g) using the table top centrifuge to pellet the cells.
Carefully decant the supernatant and resuspend the cell pellet in Iscove's medium plus 10% FBS.
Transfer this cell suspension to a T25 flask and incubate at 37°C in a 5% CO2 atmosphere for 24 hours.
Inspect the cell culture after 12–24 hours. Determine the viability of the cells using a vital stain, as described in the handout Enumeration of Mammalian Cells.
Feeding and subculturing HL-60 cells
Using a sterile pipet, mix the cell suspension. Pipet all of the cell suspension from the flask into a sterile 15 ml centrifuge tube.
Centrifuge the cells as described above.
Decant the medium into a waste container.
Add an appropriate amount of fresh, pre-warmed medium. Resuspend the cell pellet with a sterile pipet.
If you are seeding additional flasks, add the appropriate volume of the cell suspension to each flask.
Add additional medium. Loosen the flask caps and return the flasks to the incubator.
NOTE: Suspension cells are usually very sensitive to cell density (concentration). Many suspension cell lines cannot tolerate cell densities greater than 1.5 × 106 cells/ml. The concentration of HL-60 cells should not exceed 1 × 106 cells/ml. Prior to subculturing, count the cells so that you know how much to dilute them when seeding flasks. Final cell densities will depend on when you plan to use the cells, and how many cells you will need.
Feeding and Subculturing Attached Cells
Feeding attached cells
Some slow-growing cells may need to be fed (i.e., have the medium replaced) every 2–3 days for optimal growth. By looking at the color of the medium, you can determine the need for feeding. If the medium becomes yellow, it should be replaced with fresh medium.
Sanitize the hood prior to beginning work. Prewarm the medium to be used.
Decant the medium from the flask into a waste vessel containing a small amount of bleach.
Using a sterile pipet, add the appropriate amount of fresh medium to the flask, and return the flask to the incubator.
Sub-culturing attached cells
Attached cells must be split to reduce crowding if they reach confluency. Otherwise, growth will be retarded, and the cells will begin to die. Only work with one cell type at a time, to prevent contamination.
Subculturing cells requires the following:
Serum-free medium
Complete medium (with serum)
- Solution of trypsin or other proteolytic enzyme
- All solutions except trypsin should be pre-warmed prior to use.
- Decant the medium from the flask.
- Wash the flask twice with serum-free medium. For a 25 cm2 flask, about 4–5 mls of serum-free medium is sufficient for each wash. For 75 cm2 flasks, 8–10 mls of serum-free medium is sufficient. To wash the flask, add the serum-free medium, and cap the flask. Be sure not to pipet the medium directly onto the cells. Gently roll the flask so that the medium washes all of the internal surfaces. Do not allow medium to enter the neck of the flask. Decant the medium into a waste vessel, and repeat the process.
- Add trypsin (approximately 1 ml for a 25 cm2 flask, 1–2 mls for a 75 cm2 flask). Cap the flask, and incubate for 5–10 minutes at either room temp or at 37°C. Occasionally check the progress of the trypsinization. Gently tapping on the flask will help dissociate the cells from the flask surface.
- After the cells have detached, stop the action of the trypsin by adding medium with serum (approximately 3–4 mls). Pipet the cells up and down several times, washing the growing surface, and breaking up the cell clumps into a single-cell suspension.
- If the cells are being split into additional flasks, pipet an appropriate amount of cells to each new flask, and re-seed the original flask. If the cells are to be used for a protocol, transfer them into a sterile centrifuge tube. Count the cells using a vital stain (see the handout Enumeration of Mammalian Cells).
- Add fresh complete medium to each flask. The total volume of cells and medium after completion of subculturing should be approximately 5–6 mls for a 25 cm2 flask, and 10–12 mls for a 75 cm2 flask.
- Recap the flasks, and place them in the incubator. If you are using a CO2 incubator, leave the caps slightly loose to allow for gas exchange. If you are not using a CO2 incubator, tighten the caps to prevent gas exchange, and loss of CO2 from the media.
Appendix D: ENUMERATION OF CULTURED EUKARYOTIC CELLS
Most in vitro experiments require an accurate determination of cell numbers. When preparing cell lysates, for example, knowing how much protein to expect from a given number of cells allows for the production of consistent blots and results. In other assays where cells are grown in microtiter dishes, it is important to know how many cells to add to each well to achieve a confluent monolayer in a given number of days. Cells in suspension can be counted either manually using a hemacytometer, or electronically using a Coulter Counter or a cell sorter. In this exercise, we will count cells manually with a hemacytometer.
Hemacytometer Counting of Cells
Enumeration of cells in a well-dispersed cell suspension may be conveniently accomplished using a hemacytometer. These thick glass slides are divided into sections of calibrated area and depth (and therefore volume). Determining the number of cells in a particular area of the chamber allows for the calculation of cells per ml in the original suspension. Several sources of error are inherent with this procedure, including:
Inadequate mixing of the cell suspension.
Overfilling or underfilling the counting chamber.
Counting dust or debris as cells.
Micro Method with Vital Staining (Dye Exclusion Test)
When counting cells for use in experimental protocols, it is critical to determine not only the cell concentration, but cell viability as well. Cell viability is most often determined by mixing an aliquot of suspended cells with a vital stain such as trypan blue or erythrosin B. The dye exclusion test for cell viability depends upon the fact that viable cells do not take up these dyes whereas nonviable cells do. It has been shown that cells that take up the dye do not respire, undergo glycolysis, or extend cellular processes when cultured.
Using a pipettor, pipet 160 μl of trypan blue or erythrosin B into a well of a 96-well culture dish.
If the cells to be counted are attached cells, trypsinize and resuspend the cells in a sterile snap-cap culture tube in 3–5 mls of media with serum. Leave the tube in a rack at room temperature. If using suspension cells, thoroughly mix the cells with a pipet. Transfer a small amount of cells to a well adjacent to the well containing the vital stain.
Using a pipettor, transfer 40 μl of the well-mixed cell suspension to the well containing the vital stain, making a —— dilution of the cells. Mix the cells by gently pipetting up and down several times.
Using the pipettor, load each side of the hemacytometer chamber. Allow the cells to settle for 1–2 minutes, and count the cells in the 5 large squares on both sides of the hemacytometer.
-
Calculate the average cell count obtained in the 5 large squares on each side of the hemacytometer. The cell counts from the two sides should correspond to within 10–15%. If not, clean and reload the hemacytometer, and recount the cells. Using the average cell count of 5 large squares, calculate the cell concentration as follows:
(Cell count) × (dilution factor) × (1/5) × 104 = # of cells/ml
Appendix E: APOPTOSIS INVESTIGATION NOTEBOOK AND ASSIGNMENT
Answer in your notebook in a logical order as part of your analysis and conclusions:
What is the difference between detection of proteins using Coomassie-stained gels versus immunoblots? What important information can be derived from the Coomassie-stained gels (both blotted and unblotted gels) that cannot be determined by immunoblots alone?
Analyze your western blot membrane. Construct a standard curve from the standard marker proteins, and determine the sizes of all HL-60 proteins detected. Describe any differences in the sizes of the bands/proteins detected in induced and uninduced cells. Were these results expected given what you believe to be the identities of the proteins corresponding to these bands? Explain in terms of the apoptotic program. What do these results tell you about how long after induction that caspase activation occurs?
What is the value of staining cells with both Hoescht and Propidium iodide (PI)? (In other words, what unique information is obtained by each stain?) Since both PI and erythrosin B are vital stains, what is the value of staining the cells with both?
For each time point for both induced and uninduced cells, calculate the percent of cell viability obtained using both erythrosin B and PI. (Note: Percent viability using PI = 100 − [(# PI stained cells ÷ # Hoescht stained cells) x 100]. In general, are the percent viabilities you calculated using the two vital stains comparable over all? How do the percent viabilities obtained with the two vital stains compare over the course of the experiment? Were these results expected? Explain.
For each time point for both induced and uninduced cells, calculate the percentages of cells displaying condensed chromatin using each stain by dividing the number of condensed cells by the total number of cells staining with Hoescht and then multiplying by 100. How do the percentages of cells displaying condensed chromatin staining with Hoescht compare with those staining with PI over time? How do you explain these results in terms of the sequence of events of the apoptotic program?
Describe the appearance of DNA fragmentation observed in the induced and uninduced cells. What do these results tell you about how long after induction DNA fragmentation occurs? In analyzing the DNA gel, what is the range of sizes of the smaller fragments seen in some of the lanes?
Apoptosis Assignment (15 pts.)
For your assignment, answer 5 of the 6 questions posed below.
When observing cells by fluorescence microscopy, did the percentage of cells with condensed chromatin increase throughout the course of the experiment? If you carried the assays out for a longer period of time, such as 24 hours, would you expect the percentage to continue to increase until all cells displayed condensed chromatin? Explain.
While apoptosis is easily induced in vitro using chemicals such as VP-16, this process is also induced in vivo to prevent or limit the spread of disease. Describe an example in which apoptosis might be induced in vivo for this purpose. How is apoptosis induced in the situation you described? What is the significance of apoptosis in the example you chose? Cite references.
Chemical inhibitors of apoptosis are widely used in studying the activity of caspases. Name and describe one such apoptosis inhibitor used in a primary research article (cite the reference). What is the specific target of the inhibitor you chose? What question did the use of this inhibitor help answer in the article you cite? Would an apoptosis inhibitor have been useful for helping to determine the sequence of events of apoptosis? Explain.
In our experiment to induce apoptosis, not every cell responded to VP-16 by undergoing programmed cell death. Briefly propose a molecular mechanism for how response to apoptosis induction could be different between the cells. Considering their origin, speculate on whether or not HL-60 cells may have a different sensitivity to apoptosis induction than their normal counterparts. Hint: What might be the significance of these cells displaying altered sensitivity to apoptotic stimuli?
If you were to repeat the experiment a second time, are there any changes you would make to the protocol (different time frame, different types of assays, different type of cells, etc.)? Explain.
What other ways are known to induce apoptosis in vitro other than exposure to chemicals? How do these forms of induction relate to the induction of apoptosis in vivo?
Appendix F: DATA TABLES
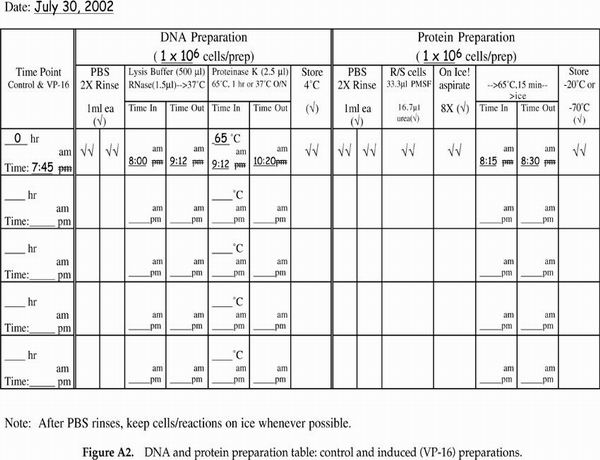
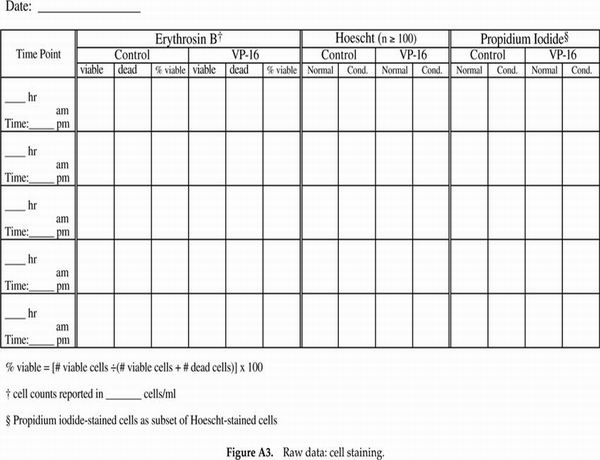
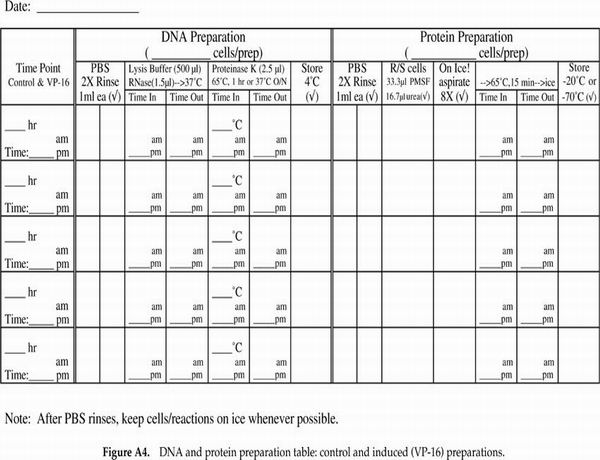
Footnotes
Alternatively, Hank's balanced salt solution or phosphate buffered saline may be used.
The volume of medium over 1 large square of the hemacytometer is 1 × 10−4 ml.
REFERENCES
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. Current Protocols in Molecular Biology. John Wiley and Sons; New York: 2002. [Google Scholar]
- Chang H. Y., Yang X. Proteases for cell suicide: Functions and regulation of caspases. Micro Mol Biol Rev. 2000;64(4):821–846. doi: 10.1128/mmbr.64.4.821-846.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshney R. I. Culture of Animal Cells: A Manual of Basic Technique. 4th ed. John Wiley and Sons; New York: 2000. [Google Scholar]
- Hengartner M. O. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- Howard Hughes Medical Institute . Beyond Bio 101: The Transformation of Undergraduate Biology Education. HHMI; Chevy Chase, MD: 1996. Available at http://www.hhmi.org/BeyondBio101. [Google Scholar]
- Martin B. M. Tissue Culture Techniques. Birkhauser; Boston, MA: 1994. [Google Scholar]
- Martins L. M., Kottke T., Mesner P. W., Basi G. S., Sinha S., Frigon N., Jr., Tatar E., Tung J. S., Bryant K., Takahashi A., Svingen P. A., Madden B. J., McCormick D. J., Earnshaw W. C., Kaufmann S. H. Activation of multiple interleukin-1β converting enzyme homologues in cytosol and nuclei of HL-60 cells during etoposide-induced apoptosis. J Biol Chem. 1997;272(11):7421–7430. doi: 10.1074/jbc.272.11.7421. [DOI] [PubMed] [Google Scholar]
- National Research Council of the National Academies . Bio 2010: Transforming Undergraduate Education for Future Research Biologists. National Academies Press; Washington, DC: 2003. [Google Scholar]
- Odom D. P., Grossel M. J. Using the two-hybrid screen in the classroom laboratory. Cell Biol Educ. 2002;1:43–62. doi: 10.1187/cbe.02-02-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Smyth P. G., Berman S. A., Bursztajn S. Markers of apoptosis: Methods for elucidating the mechanism of apoptotic cell death from the nervous system. Biotechniques. 2002;32(3):648–665. doi: 10.2144/02323dd02. [DOI] [PubMed] [Google Scholar]
- Studzinski G. P. Apoptosis: A Practical Approach. Oxford University Press; New York: 1999. [Google Scholar]
- Wright R., Boggs J. Learning cell biology as a team: A project-based approach to upper-division cell biology. Cell Biol Education. 2002;1:145–153. doi: 10.1187/cbe.02-03-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Shao R., Pommier Y. Assessment of DNA damage in apoptosis. In: Studzinski G. P., editor. Apoptosis: A Practical Approach. Oxford University Press; New York: 1999. pp. 41–55. [Google Scholar]


