Abstract
Background & Aims:
Hints, Histidine triad nucleotide-binding proteins, are adenosine monophosphate–lysine hydrolases of uncertain biological function. Here we report the characterization of human Hint2.
Methods:
Tissue distribution was determined by real-time quantitative polymerase chain reaction and immunoblotting, cellular localization by immunocytochemistry, and transfection with green fluorescent protein constructs. Enzymatic activities for protein kinase C and adenosine phosphoramidase in the presence of Hint2 were measured. HepG2 cell lines with Hint2 over expressed or knocked down were established. Apoptosis was assessed by immunoblotting for caspases and by flowcytometry. Tumor growth was measured in SCID mice. Expression in human tumors was investigated by microarrays.
Results:
Hint2 was predominantly expressed in liver and pancreas. Hint2 was localized in mitochondria. Hint2 hydrolyzed adenosine monophosphate linked to an amino group (AMP-pNA; kcat:0.0223 s-1;Km:128 μmol/L). Exposed to apoptotic stress, fewer HepG2 cells overexpressing Hint2 remained viable (32.2 ± 0 6% vs 57.7 ± 4.6%), and more cells displayed changes of the mitochondrial membrane potential (87.8 ± 2.35 vs 49.7 ± 1.6%) with more cleaved caspases than control cells. The opposite was observed in HepG2 cells with knock-down expression of Hint2. Subcutaneous injection of HepG2 cells over expressing Hint2 in SCID mice resulted in smaller tumors (0.32 ± 0.13 g vs 0.85 ± 0.35 g). Microarray analyses revealed that HINT2 messenger RNA is down regulated in hepatocellular carcinomas (−0.42 ± 0.58 log2 vs −0.11 ± 0.28 log2). Low abundance of HINT2 messenger RNA was associated with poor survival.
Conclusion:
Hint2 defines a novel class of mitochondrial apoptotic sensitizers down-regulated in hepatocellular carcinoma.
Histidine triad nucleotide-binding proteins (Hints) are highly conserved dimeric proteins that bind 2 purine nucleoside monophosphates per dimmer and hydrolyze model compounds such as adenosine monophospho-ramidate (AMPNH2) and adenosine monophosphate (AMP)-lysine, with the activity dependent on the middle Histidine in the conserved HIT motif (His-X-His-X-His).1-4 Activity on model compounds containing adenosine monophosphoramide linkages has been interpreted as evidence that Hint hydrolases reverse protein nucleotidylylation.5,6 Indeed, robust AMP-lysine hydrolase activities have been shown for Hint orthologs from yeast, rabbit,4,7 chicken,8 Escherichia coli, and humans.9 Lower specific activity AMP-lysine hydrolase activity is exhibited by Aprataxin, a human Hint-related enzyme inactivated in ataxia oculomotor apraxia 1.10-12 Genetic evidence indicates that in yeast, Hint1 functions as an active site-dependent positive regulator of the general transcription factor TFIIH.4,13 Human Hint1 has been proposed to be a negative regulator of the microthalmia transcription factor14 and of the β-catenin pathway.15 The human HINT1 gene is down-regulated by methyl-ation in the non–small cell lung cancer cell line NCI-H522, and reintroduction of Hint1 expression was shown to reduce tumorigenicity in vivo.16 Murine hint1 is not an essential gene in the mouse; however, hint1–1– mice were more prone to carcinogen-induced squamous tumors of the stomach than were wild-type mice.18 In addition, hint1–1–mice recover slowly from morphine-induced analgesia, which was attributed to a physical interaction between Hint1 and the μ-opioid G-protein coupled receptor.19 Analysis of the human genome has shown that, in addition to Hint1 and Aprataxin, there are 2 additional loci (HINT2 and HINT3) that encode Hint genes in humans.5 Here we show that Hint2 is a mammalian-specific, nuclear-encoded mitochondrial Hint hydrolase whose overexpression sensitizes cells to apoptosis and that it is down-regulated in hepatocellular carcinoma.
Materials and Methods
Cloning of HINT2 Complementary DNA and HINT2-H149A Complementary DNA
Human HINT2 complementary DNA (cDNA) was cloned into pControl vector obtained by digestion of the pEGFP-C1 plasmid with SalI to remove green fluorescent protein (GFP) cDNA. The H149A mutation was performed by site-directed mutagenesis using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The oligonucleotide primers TCTGTGTATCATCTGGCCAT-TCATGTACTTGGG and CCCAAGTACATGAATGGCCA-GATGATACACAGA, each complementary to opposite strands of the vector, were extended during temperature cycling by PfuTurbo DNA polymerase.
Antibody Production
Rabbits were immunized by repeated intradermal injections of purified Hint2 emulsified with Freund's adjuvant. Antibodies were affinity purified by absorption to antigen immobilized on Reacti-Gel (HW-65F) support (Pierce, Lausanne, Switzerland). Specific antibody was eluted with 100 mmol/L glycine, pH 2.5, and dialyzed against phosphate-buffered saline (PBS).
Immunoblot Analysis
Tissue distribution of Hint2 was analyzed on a commercially available membrane (Biochain, Lugano, Switzerland). Briefly, 50 μg tissue lysate from different human organs was loaded on a 4%–20% denaturing polyacrylamide gel. After electrophoresis, the gel was transferred and the blot was tested with rabbit anti-Hint2 antibody and, after stripping, with mouse anti-actin monoclonal antibody (Chemicon, Rueschlikon, Switzerland). Expression of proteins involved in apoptosis was determined after incubation with anti-Fas antibody at 100 ng/mL and actinomycin D at 50 ng/mL for 16 hours and after incubation with ethanol at 400 mmol/L for 24 hours. Whole cell lysates were obtained by lysis with PBS, pH 7.2, containing 1% Nonidet P-40, 0.4% sodium deoxycholate, and 1 mmol/L EDTA. Lysates were sonicated and centrifuged. Twenty-five micrograms of proteins was resolved by sodium dodecyl sulfate/polyacrylamide gel electrophoresis and subsequently transferred to protean nitrocellulose membranes. Membranes were blocked for 1 hour at 37°C with 5% nonfat milk. Membranes were then incubated with the appropriate primary antibody (Hint2, caspase-3, caspase-7, caspase-9, cleaved caspase-3, cleaved caspase-7, cleaved caspase-9, poly-(ADP-ribose) (PARP), cleaved PARP, Bid, cytochrome c, phospho-Bcl-2 [Ser 70], Bcl-xL, Bak) (Cell Signaling Technology, Allschwill, Switzerland) overnight at 4°C. The membranes were washed and incubated for 1 hour with peroxidase-conjugated secondary antibody donkey anti-rabbit immuno-globulin G 1:65,000(Pierce). Immunoblots were revealed using an enhanced chemiluminescence detection system (SuperSignal West Pico Chemiluminescent Substrate; Pierce) and exposure performed with a Fujifilm LAS-1000 CCD camera (Dielsdorf, Switzerland) coupled to a computer. Analysis of the signal was performed using the software AIDA 2.1 (Raytest, Urdorf, Switzerland). The expression of proteins was quantified using the expression of actin as calibrator and normalized against not stimulated condition value (value equal 1).
Immunolocalization in Huh7 Cells
Cells, seeded at a density of 104 cells/cm2, were grown on glass coverslips, rinsed with cold PBS (without Ca2+ and Mg2+ ,pH 7.4), and fixed on ice with 2% formaldehyde in PBS for 1 minute and subsequently permeabilized with methanol at 4°C for 10 minutes. For mitochondrial staining, living cells were incubated for 45 minutes in prewarmed medium containing 500 nmol/L Mito-Tracker-Red (Molecular Probes, Lucern, Switzerland), rinsed with cold PBS, and fixed. Fixed cells were incubated with single primary antibodies (Hint2 1:15) for 45 minutes at 37°C, washed 3 times in PBS, and incubated (20 minutes at 37°C) with the appropriate Alexa-conjugated secondary antibodies (Molecular Probes). After 3 washes in PBS, coverslips were mounted in PBS-glycerol mounting medium and examined with a Zeiss Axioskop fluorescence microscope (Feldbach, Switzerland). Confocal microscopy was performed using a Zeiss 510 confocal microscope and a 63× Plan-Apochromatic objective (NA = 1.4) oil immersion. Optical sections were collected at 0.2- to 0.3- μm intervals, and the images were processed using Photoshop 5.5software (Adobe, Paris, France).
HINT2-GFP Constructs and Localization in HEK-293 Cells
HINT2 cDNA was produced by polymerase chain reaction (PCR) and cloned in frame into the 2 GFP vectors, p-EGFP-C1 and pEGFP-N1 (Clontech, Basel, Switzerland), giving GFP-HINT2 or HINT2-GFP, respectively. The 2 constructs were transfected into HEK-293 cells grown on coverslips with TransFast (Promega) using 2 μg DNA per well. After 2 days, the cells on the coverslips were fixed for 10 minutes with 4% formaldehyde, washed 3 times with PBS, and mounted on glass slides with SlowFade Antifade Kit (Molecular Probes).
Real-Time Quantitative PCR
Quantification of the messenger RNA (mRNA) expression within different tissues was performed on a cDNA panel (Clontech) using CAGGTTGAACCTGCCAACTGA and GGGTCCATTTTTCCCTTTCC primers and TCCAAGCATCCAGAGTCTGGTGGTCCTT FAM-labeled probe for HINT2. Amplification and analysis of the result were performed using an ABI 7700 (Perkin-Elmer, Schwerzenbach, Switzerland). Normalization was performed against GAPDH with specific probe and primers (Assay on demand; Perkin-Elmer), and the tissue expressing the lowest level of mRNA for HINT2 was used as calibrator. mRNA quantification within tumors was performed using StrataScript Reverse Transcriptase (Stratagene) in combination with octamer priming on 1 μg total RNA. Diluted reverse-transcripted DNA was then amplified by real-time PCR using an Mx3005p (Stratagene) in combination with FullVelocity SYBR Green technology (Stratagene). Primer concentrations were optimized to achieve 100% ± 10% efficiency on standard curves without primer dimers. Primers sequences are available at http://medgen.ugent.be/rtprimerdb/index.php.
Production and Purification of HINT2 in Bacteria
Hint2 was expressed in Escherichia coli. Frozen and thawed cell pellets were lysed in 12.5 mL of buffer A (20 mmol/L Tris, pH 7.5, 150 mmol/L NaCl) containing EDTA-free protease inhibitor mix. Lysates were cleared by centrifugation and precipitation with protamine sulfate. Ammonium sulfate was added to 40% saturation and the pellet dialyzed against buffer A. For the enzymatic assay, Hint2 was purified by AMP-agarose affinity chromatography using a 4-mL column, washing with 24 column volumes of buffer A, and eluting with 3 column volumes of buffer A with 200 μmol/L adenosine.4 For the experiment with protein kinase C(PKC), HINT2 was expressed with a glutathione S-transferase tag and purified with glutathione beads. The glutathione S-transferase was then cleaved by factor X.
PKC Assays
The following reagents were mixed in an Eppendorf tube: 20 μL sonicated phospholipids (phosphatidylserine and 1,2-dioleolylglycerol in a 7:1 ratio in 20 mmol/L Tris, pH 7.5), 10 μL PKC rat brain preparation with >3 U/mL activity in 50% glycerol,20 either 330 ng histone or 40 ng myelin basic protein as substrate, 200 ng to 15 μg Hint2 as possible inhibitor, 10 μL of 10 mmol/L CaCl2, and 20 mmol/L Tris HCl to make up a final volume of 80 μL. To start the reaction, 20 μL of 100 mol/L adenosine triphosphate, 100 mmol/L MgCl2, and 13 μCi/mL [γ-32P] were added. The tubes were incubated for 5 minutes at room temperature, and 30 μL of 200 mmol/L EDTA and 200 mmol/L adenosine triphosphate were added. A sample (100 μL) was spotted on Whatman paper P81 (Bottmingen, Switzerland). After washing the filters twice for 1 minute in 10 mL H2O and once in ethanol, the radioactivity was counted in a beta counter.
Phosphorylation Assay
To investigate whether Hint2 is phosphorylated by PKC, the assay described previously was performed in the presence of increasing concentrations of purified Hint2 with or without 330 ng histone as an internal control. The reaction was incubated for 20 minutes. After addition of 30 μL 5× sample buffer, the samples were boiled for 5 minutes and subjected to sodium dodecyl sulfate/polyacrylamide gel elec-trophoresis on a 15% gel and then transferred to a membrane that was used for autoradiography and immunoblotting for Hint2.
Adenosine Phosphoramidase Assay
AMP-pNA assays were performed as described.7
Stable Transfection of HepG2 Cells
Human hepatoblastoma-derived HepG2 cells were grown in minimal essential medium with 10% fetal calf serum, 2 mmol/L glutamine, and penicillin/streptomycin. HepG2 cells were transfected with TransFast (Promega) using 1 μg of HINT2-pControl, HINT2 H149A mutated-pControl, and pControl constructs and a 2:1 ratio of liposomes. Stable selection was established in the presence of geneticin (2 mg/mL medium). Stable transfected cells were seeded on 96-well plates to establish single clones. The geneticin complete medium was changed everyday for 20 days.
Small Interference RNA
Small interference RNAs were designed with the 384–403 sense strand sequence of HINT2 coding sequence. After annealing of sense and antisense small interference RNA primers, the ligation of small interference RNA in linearized pSuppressor vector (Gene Suppressor System; Biocarta, SanDiego, CA) was performed with T4 DNA ligase overnight at 16°C. After transformation of DH5α bacteria and selection of positive clones by PCR, the plasmids were prepared with Wizard Plus SV Minipreps Kit (Promega). Twenty-four hours after the lipid-based transfection, geneticin was added to select the small interference RNA stable transfected cells.
Colorimetric 2,5-Diphenyltetrazolium Bromide Assay
HepG2-derived cell lines were seeded in triplicate 96-well plates at 1×105 cells/well. After 24 hours, cell wells were incubated with anti-Fas antibody at 100 ng/mL and actinomycin D at 50 ng/mL. After 16 hours, cells were incubated with 12.5 μL of 5 mg/mL 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide for 4 hours at 37°C. The medium was aspirated, and 100 μL of acidified isopropanol was added. After incubation for 15 minutes, the OD was measured at 570 and 655 nm as test and reference wave-lengths, respectively. The percentage of relative viability was determined by the following formula: % Viability =(OD Test [Stimulated Cells]/OD Control [Unstimulated Cells]) × 100.
Flow Cytometry for Detection of Apoptotic Cells
HepG2-derived cell lines were grown on 6-well plates and incubated with anti-Fas antibody at 100 ng/mL and actinomycin D at 50 ng/mL for different periods (0, 3, 6, 10, 16, and 24 hours). Cells were harvested with trypsin, centrifuged at 4°C at 500 rpm for 5 minutes, and resuspended at 2 × 107 cells/mL in PBS. After centrifugation, the cells were labeled with the fluorescent dye JC-1 (Molecular Probes) for 20 minutes. Cells were washed twice with PBS and resuspended in 200 μL of PBS. A flow cytometer (FACScan; BD Biosciences, Allschwil, Switzerland) was used to quantify JC-1–positive apoptotic cells using the software GeneQuant II (Amersham, Otelfingen, Switzerland).
Tumors in SCID Mice
A total of 3 × 106 HepG2-overexpressing Hint2 cells or 3 × 106 pControl transfected HepG2 cells were injected in the flank of 10 SCID mice (Charles River, WIGA, Sulzfeld, Germany) (n = 5 in each group). After 10 weeks, the tumors were removed and measured. Expression of Hint2, cleaved caspase-9, and cleaved PARP was assessed by immunoblot analysis, and apoptotic cells were identified by immunohistochemistry with an antibody recognizing a specific caspase cleavage site within cytokeratin 18 (M30 CytoDeath; Roche, Basel, Switzerland). The number of apoptotic cells was counted in 10 fields per tumor.
Human Tumor Samples
The acquisition of the hepatocellular carcinoma samples has been reported.21 Tissue banking was approved by the local institutional review board.
Microarray Analysis of Hepatocellular Carcinomas and Adjacent Tissue Samples
The materials and methods of this analysis have been reported.21 Survival was examined by Kaplan–Meier plot and log-rank test. For the survival curve, of the 90 patients who underwent liver resection for hepatocellular carcinoma (one patient twice), 13 patients were excluded: one because he died of sepsis immediately after surgery and 12 because they subsequently underwent transplantation.
Results
HINT2 (GenBank AF356515) at human 9p13.2 encodes a polypeptide of 163 amino acids with a 35–amino acid N-terminal extension when aligned to HINT1, which corresponds toa predicted mitochondria import signal (Figure 1A). Over 123 alignable residues, Hint2 is 61% identical to Hint1. Based on the conserved N-terminal extension and the overall sequence, HINT2 homologs are only found in mammals and are absent in other vertebrates such as fish and birds (Figure 1B). The tissue expression of HINT2 was determined by real-time quantitative PCR and immunoblot analysis. HINT2 mRNA and protein were predominantly detected in the liver and the pancreas but were also detected to a much lesser extent in the other human tissues analyzed (Figure 2A and B). Immunocyto-chemistry (Figure 2C), immunoblot analysis of isolated mitochondria (data not shown), and transfection with HINT2-GFP constructs (Figure 2D) established that Hint2 is a mitochondrial protein.
Figure 1.
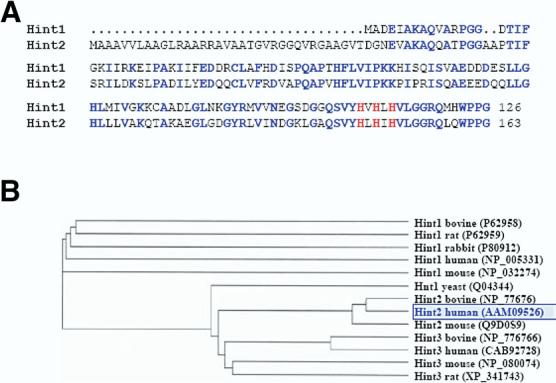
(A) Alignment of amino acid sequences of the human Hint1 and Hint2. Amino acids conserved between both sequences are indicated in blue, and the 3 histidines of the HIT motif are indicated in red. (B) Phylogenic tree established by comparison of interspecies sequences of Hint1, Hint2, and Hint3 proteins using ClustalW software (http://www.ebi.ac.uk/clustalw/).
Figure 2.
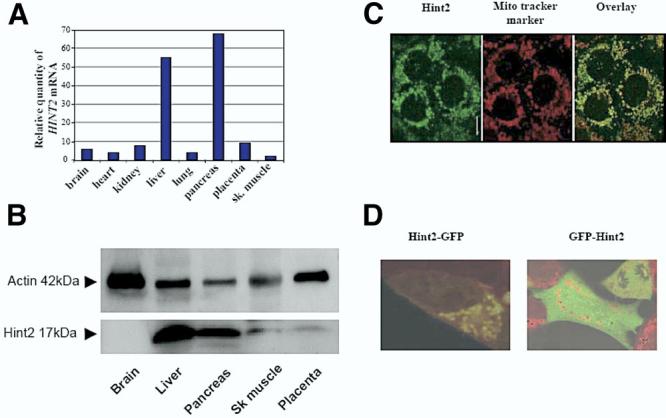
(A) Tissue distribution of HINT2 mRNA assessed by real-time quantitative PCR using a human cDNA panel (Clontech). Results were normalized to GAPDH expression and presented as fold variations comparatively with skeletal muscle. (B) Expression of Hint2 protein in different human tissues assessed by immunoblot analysis with anantibody against Hint2. (C) Immunocytochemistry for Hint2 in Huh-7 cells. Hint2 (in green, left panel, bar = 10μm) colocalized with the mitochondrial marker Mito Tracker (in red, middle panel) as shown in the overlay (in yellow, right panel). (D) Overlay confocal microscopy pictures of HEK-293 cells incubated with Mito Tracker to mark the mitochondria in red and expressing a chimeric protein made of Hint2 and GFP. When the GFP was at the C-terminus of Hint2, Hint2 localized to the mitochondria (left panel). When the GFP protein was at the N-terminus of Hint2, Hint2 remained in the cytoplasm (right panel; original magnification 40×).
Early reports described Hint1 as a PKC inhibitor22 or interactor (PKCI).1 We have monitored the phosphorylation of 2 PKC substrates in the presence of increasing amounts of purified Hint2. Hint2 did not inhibit the phosphorylation of histones (Figure 3A) or of myelin basic protein (data not shown). We further tested whether Hint2 could be a PKC substrate because its sequence contains a potential PKC phosphorylation site (122TAK), albeit within an α-helix. In contrast to histones, Hint2 was not phosphorylated by a rat brain fraction enriched in PKCs. Hint2 added at increasing concentration to the assay was not phosphorylated, whereas, as expected, histones were phosphorylated (Figure 3B). Neither overlay nor coimmunoprecipitation could reveal an interaction between Hint2 and PKC isoforms (data not shown). In contrast, Hint2 displayed robust, active site-dependent adenosine mono-phosphoramidase activity. Using the model compound AMP- pNA, in which a paranitroaniline reporter was linked by a phosphoramidate linkage to AMP,7 Hint2 had a kcat of 0.0223 ± 0.0031 s−1 and a Km of 128 ± 35 μmol/L, whereas rabbit Hint1 had a kcat of 0.00187 ± 0.00006 s−1 and a Km of 134 ± 11 μmol/L, endowing Hint2 with a kcat/Km 10 times larger than Hint1. When the middle Histidine of the HIT motif of Hint2 was mutated to an alanine (Hint2-H149A), the adenosine phosphoramidase activity was lost.
Figure 3.
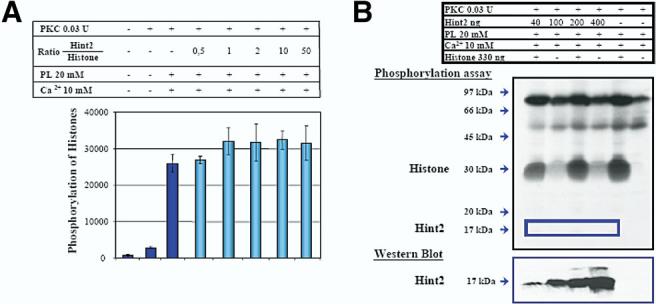
(A) The phosphorylation of histones (330 ng) by PKC was not affected by increasing amounts of Hint2 protein or by increasing amounts of bovine serum albumin (control, not shown). (B) Activated PKC phosphorylated histones but not Hint2 (upper panel). The presence of Hint2 in the reaction was confirmed by immunoblot analysis (lower panel).
Apoptotic signals, triggered either extrinsically by the activation of death receptors or intrinsically by the disruption of intracellular homeostasis, cascade through the mitochondria, which function as central apoptotic regulators.23 Because other HIT proteins, in particular Fhit, influence apoptosis 24,25 and because Hint2 is located in the mitochondria, the effect of Hint2 on apoptosis was investigated. Different HepG2 cell lines were generated: (1) HepG2 cells expressing a 20-fold excess of Hint2, (2) HepG2 cells overexpressing Hint2-H149A with no hydrolase activity (Figure 4A), and (3) HepG2 cells expressing less than 10% of native Hint2 due to small interference RNA knockdown (Figure 4B). When cells overexpressing Hint2 were exposed to anti-Fas antibody and actinomycin D, cell viability was reduced significantly compared with control cells (32.2% ± 0.6% vs 57.7% ± 4.6%; P < .02) (Figure 5A). In contrast, HepG2 cells transfected with the hydrolase-negative Hint2-H149A and exposed to the same stimulus did not display less viability. Permeabilization of the mitochondrial membranes dissipates the potential of the inner membrane and allows the leakage of caspase-activating proteins into the cytoplasm, which in turn mediates apoptosis. Incubation of HepG2 cells overexpressing Hint2 with an antibody against Fas and actinomycin D induced greater changes in the mitochondrial membrane potential measured by means of JC-1 fluorescence than in control HepG2 cells (87.8% ± 2.3% vs 49.7% ± 1.6% at 16 hours and 92.3% ± 2.6% vs 68.3% ± 1.8% at 24 hours; P<.05) (Figure 5B). The level of expression of Blc-2,Bcl- xL, Bak, Bid, and cytochrome c was not significantly different among the cell lines (Figure 5C). However, in response to incubation with anti-Fas antibody and actinomycin D, the abundance of cleaved PARP and cleaved caspase-3, -7, and -9 was higher in the cells over expressing Hint2 (Figure 5C). PARP cleavage could be prevented by apoptosis inhibitors in the cells over expressing Hint2 (Figure 5D). To confirm that these results were not specific for induction of apoptosis via anti-Fas antibody and actinomycin, cells were exposed to another apoptotic stress, ethanol.26 The abundance of cleaved PARP and cleaved caspase-3, -7, and-9 was higher in the cells overexpressing Hint2 in response to incubation with ethanol than in control cells (Figure 6). In the cell lines with less Hint2, exposure to anti-Fas antibody and actinomycin D resulted in less changes in the mitochondrial membrane potential determined by flow cytometry with JC-1 (42.4% ± 4.2% vs 60.9% ± 3.9% at 16 hours and 62.5% ± 8.6% vs. 85.8% ± 3.7% at 24 hours; P< .05) (Figure 7A) and less cleaved caspase-3 (Figure 7B).
Figure 4.
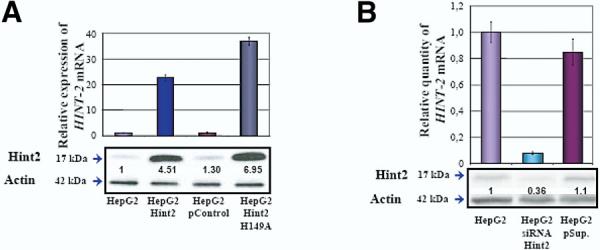
(A) HepG2 cells were stably transfected with HINT2cDNA, vector (pControl), and HINT2-H149A cDNA. mRNA abundance was assessed by real-time quantitative PCR (upper panel) and protein expression by immunoblot analysis (lower panel). (B) HepG2 cells were stably transfected to silence HINT2 mRNA ex-pression as assessed by real-time quantitative PCR (upper panel) and by immunoblot analysis (lower panel).
Figure 5.
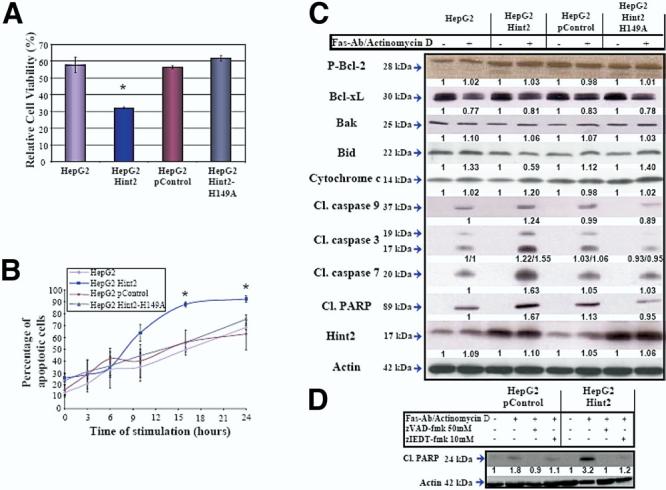
(A) Effect of Hint2 overexpression on cell viability. Cells were incubated for 16 hours with anti-Fas antibody (100 ng/mL) and actinomycin D (50ng/mL), and viability was assessed by the means of 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. Viability was significantly lower in cells overexpressing Hint2 (32.2% ± 0.6% vs 57.7% ± 4.6%; *P < .02). (B) Cells were labeled withJC-1, a mitochondrial membrane potential marker, and fluorescent properties were analyzed by flowcytometry after 0-, 3-, 6-, 10-, 16-, and 24-hour exposure to anti-Fas antibody (100 ng/mL) and actinomycin D (50ng/mL). The percentage of cells with a dissipated mitochondrial potential was significantly higher in the case of Hint2 overexpression (87.8% ± 2.3% vs 49.7% ± 1.6% at 16 hours and 92.3% ± 2.6% vs 68.3% ± 1.8% at 24 hours; *P < .05). (C) The expression of phosphorylated Blc-2, Bcl-xL, Bak, Bid, and cytochrome c was not affected by overexpression of Hint2. More cleaved caspase-9, caspase-3, and caspase-7 and PARP were found in HepG2 cells overexpressing Hint2 in response to incubation with anti-Fas antibody (100 ng/mL) and actinomycin D (50 ng/mL) for 16hours. The figure shows immunoblots representative of 3 independent experiments. (D) Incubation with the caspase inhibitors zVAD-fmk (50 mol/L) or zIETD-fmk (10 mol/L) prevented the anti-Fas antibody–and actinomycin D–induced cleavage of PARP as assessed by immunoblot analysis.
Figure 6.
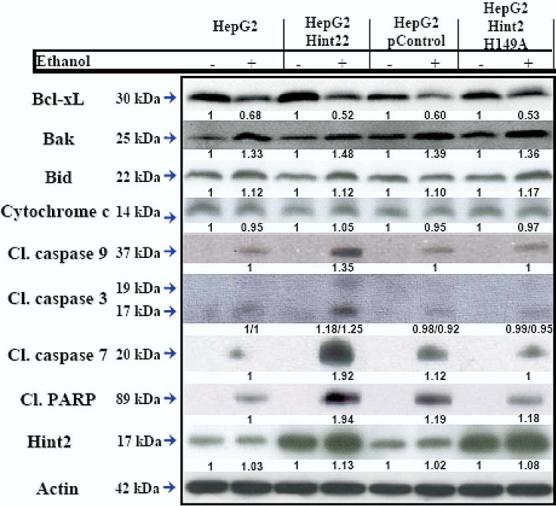
Effect of incubation with ethanol. The expression of phosphorylated Bcl-xL, Bak, Bid, and cytochrome c was not affected by overexpression of Hint2. More cleaved caspase-9, caspase-3, and caspase-7 and PARP were found in HepG2 cells overexpressing Hint2 in response to incubation with ethanol (400 mmol/L) for 24hours. The figure shows immunoblots representative of 3 independent experiments.
Figure 7.
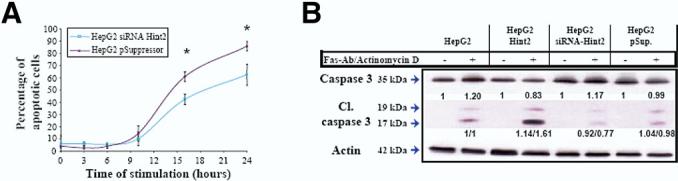
(A) Cells were labeled with JC-1, a mitochondrial membrane potential marker, and fluorescent properties were analyzed by flow cytometry after 0, 3, 6, 10, 16, and 24 hours of exposure to anti-Fas antibody (100 ng/mL) and actinomycin D (50 ng/mL). The percentage of apoptotic cells was significantly lower in the case of Hint2-silenced expression (42.4% ± 4.2% vs 60.9% ± 3.9% at 16 hours and 62.5% ± 8.6% vs 85.8% ± 3.7% at 24 hours; *P < .05). (B) Less cleaved caspase-3 was observed in HepG2 cells with silenced expression of Hint2 in response to anti-Fas antibody (100 ng/mL) and actinomycin D (50 ng/mL) for 16 hours compared with the other cell lines.
We performed in vivo experiments to assess the influence of Hint2 on tumor growth. HepG2 cells injected in the flank of SCID mice formed significantly smaller tumors when overexpressing Hint2 (0.32 ± 0.13 g vs 0.85 ± 0.35 g; P < .05) (Figure 8A). This was associated with more cleaved caspase-9 and less abundant uncleaved PARP, whereas the persistent overexpression of Hint2 in these tumors was confirmed by immunoblot (Figure 8B). The number of apoptotic cells was significantly higher in the tumors overexpressing Hint2 (11 ± 4 vs 4 ± 5 apoptotic figures/field; P < .05). This led us to analyze the expression of the HINT2 gene in human tumors. We characterized the expression of the HINT genes in 91 human primary hepatocellular carcinomas and 60 matched nontumor surrounding tissues. Microarray analysis revealed that HINT2 expression was significantly reduced in the hepatocellular carcinoma compared with surrounding liver tissue (−0.424 ± 0.579 vs −0.109 ± 0.277 log2; P < .0002) (Figure 9A). In contrast, the expression of HINT1 and HINT3 mRNAs was not reduced in human hepatocellular carcinoma. With this collection of samples, it was previously reported that using the Cox proportional hazards survival analysis, it was possible to identify a cell proliferation and antiapoptosis gene expression profile separating the patients in 2 subclasses with significantly different survival.14 Examination of HINT2 expression in these 2 subclasses of patients revealed that HINT2 mRNA was less abundant in the sub-class of hepatocellular carcinoma with a poor prognosis (n40) when compared with the subclass with better prognosis (n = 51) (−0.728 ± 0.522 vs −0.187 ± 0.506 log2; P < 1.0 × 10−5) (Figure 9A). Moreover, 77% of the hepatocellular carcinomas had a lower level of expression of HINT2 mRNA than in normal liver, and when the survival of the patients was analyzed according to this relative level of expression of HINT2, the patients with a lower level of expression of HINT2 had a significantly worse prognosis than those with a higher level (P = .01; Figure 9B).
Figure 8.
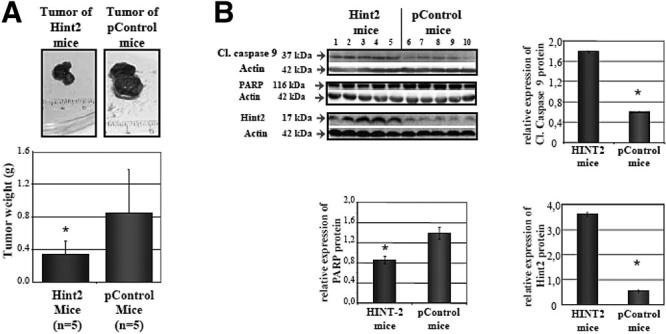
Hint2 protein expression and tumor burden. (A) Effect of Hint2 overexpression on tumor size in SCID mice after subcutaneous injection of 3 × 106HepG2 cells stably transfected with pControl vector or HINT2cDNA (0.32 ± 0.13 g vs 0.85 ± 0.35g; *P < .05; n = 5 in each group). (B) Expression of cleaved caspase-9, PARP, and Hint2 was assessed by immunoblot analysis in the tumors. The level of expression of these proteins was significantly different between the Hint2 tumors and the pControl tumors (1.79 ± 0.07 vs 0.65 ± 0.06, 0.84 ± 0.09 vs 1.38 ± 1.1, and 3.62 ± 0.08 vs 0.54 ± 0.5, respectively; *P < .05).
Figure 9.
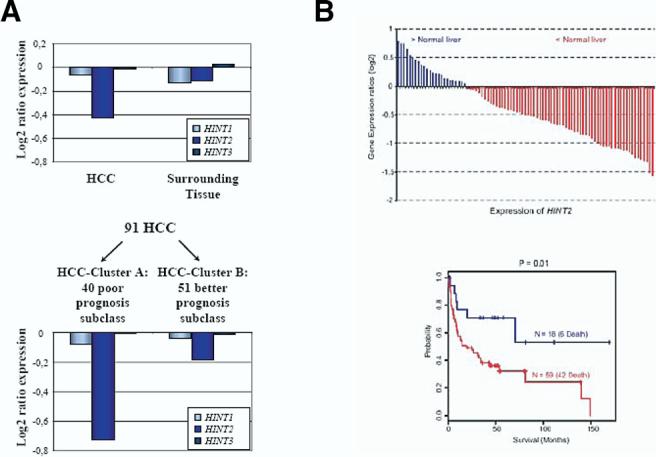
(A) Expression of HINT1, HINT2, and HINT3 in 91 hepatocellular carcinomas and their surrounding tissues analyzed by microchip array (upper panel) and in the 91 hepatocellular carcinoma samples divided according to prognosis (lower panel). HINT2 mRNA was less abundant in hepatocellular carcinoma than in adjacent tissue (−0.424 ± 0.579 vs −0.109 ± 0.277 log2; P < .0002) and less abundant in tumors with a poor prognosis than in those with a better prognosis (−0.728 ± 0.522 vs −0.187 ± 0.506 log2; P < 1.0 × 10−5). Such changes were not observed for HINT1 or HINT3. (B) Survival difference of patients with hepatocellular carcinoma based on the expression patterns of HINT2. Patients were ranked on the basis of HINT2 mRNA levels in tumoral tissue using normal liver as reference and separated into 2 groups as indicated (upper panel). Kaplan–Meier plot of overall survival of patients with hepatocellular carcinoma grouped on the basis of HINT2 mRNA expression level (lower panel). The difference between groups was significant (P = .01, log-rank test).
Discussion
Our results show that Hint2 is predominantly expressed in the liver and in the pancreas and establish mammalian Hint2 as the first Hint protein to be localized to the mitochondria. Furthermore, we show that Hint2 sensitizes to apoptosis and that its expression is down-regulated in human hepatocellular carcinoma.
Hint proteins are AMP-lysine hydrolases and may remove AMP adducts from proteins.5 Protein adenylylation, the covalent link of the AMP moiety of adenosine triphosphate to an amino acid residue of a protein, is a potentially reversible covalent modification of proteins. This posttranslational modification occurs in bacteria, for example to inactivate glutamine synthetase.27 Mammalian cells can adenylylate proteins; in particular, casein kinase II can adenylylate herpes simplex virus I pro-teins.6,28 Several proteins in the liver have been reported to be adenylylated.29,30 The activity of Hint2 is higher than that of Hint1, a fact of relevance in the mitochon-drial environment where the concentrations of adenosine nucleotides are high. It is not yet known whether mito-chondrial AMP adducts are the endogenous substrates of Hint2. Another possible substrate is adenosine monophosphoramidate (AMPNH2), a compound that is found in eukaryotic cells31 and is synthesized from AMPSO4 and ammonia in a wide variety of organisms.32 However, the biological significance of adenosine monophosphoramidate and, in particular, in respect to apoptosis is unknown.
According to our results, overexpression of the enzymatic activity of Hint2 sensitizes cells to apoptosis. Regulators of apoptosis play a pivotal role in tumor biology. Decreased sensibility to apoptosis characterizes most cancer cells and correlates with tumor aggressiveness. Indeed, we found that the expression of HINT2 was down-regulated in hepatocellular carcinomas.The reason for this down-regulation could be methylation of the HINT2 gene promoter. Other HIT proteins such as Hint1 and Fhit have been reported to be down-regulated in tumors by methylation of their promoter.16,33,34 Reduced expression of tumor suppressors in cancer can also be accounted by loss of heterozygosity. In hepatocellular carcinoma, loss of heterozygosity on chromosome 9p is not rare35 and is associated with a poor prognosis.36 Whether such mechanisms are involved in the case of HINT2 gene remains to be elucidated.
In conclusion, this work identifies mitochondrial Hint2 enzymatic activity as a novel apoptotic sensitizer that is down-regulated in hepatocellular carcinoma.
References
- 1.Lima CD, Klein MG, Weinstein IB, Hendrickson WA. Three-dimen-sional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins. Proc Natl Acad Sci USA. 1996;93:5357–5362. doi: 10.1073/pnas.93.11.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner C, Garrison P, Gilmour J, Peisach D, Ringe D, Petsko GA, Lowenstein JM. Crystal structures of HINT demonstrate that his-tidine triad proteins are GalT-related nucleotide-binding proteins. Nat Struct Biol. 1997;4:231–238. doi: 10.1038/nsb0397-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lima CD, Klein MG, Hendrickson WA. Structure-based analysis of catalysis and substrate definition in the HIT protein family. Science. 1997;278:286–290. doi: 10.1126/science.278.5336.286. [DOI] [PubMed] [Google Scholar]
- 4.Bieganowski P, Garrison PN, Hodawadekar SC, Faye G, Barnes LD, Brenner C. Adenosine monophosphoramidase activity of hint and hint1 supports function of kin28, ccl1, and tfb3. J Biol Chem. 2002;277:10852–10860. doi: 10.1074/jbc.M111480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner C. Hint, Fhit, and GalT: function, structure, evolution, and mechanism of three branches of the Histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry. 2002;41:9003–9014. doi: 10.1021/bi025942q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell C, Blaho JA, McCormick AL, Roizman B. The nucleotidylylation of herpes simplex virus 1 regulatory protein alpha22 by human casein kinase II. J Biol Chem. 1997;272:25394–25400. doi: 10.1074/jbc.272.40.25394. [DOI] [PubMed] [Google Scholar]
- 7.Krakowiak A, Pace HC, Blackburn GM, Adams M, Mekhalfia A, Kaczmarek R, Baraniak J, Stec WJ, Brenner C. Biochemical, crystallographic, and mutagenic characterization of hint, the AMP-lysine hydrolase, with novel substrates and inhibitors. J Biol Chem. 2004;279:18711–18716. doi: 10.1074/jbc.M314271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parks KP, Seidle H, Wright N, Sperry JB, Bieganowski P, Howitz K, Wright DL, Brenner C. Altered specificity of Hint-W123Q supports a role for Hint inhibition by ASW in avian sex determination. Physiol Genomics. 2004;20:12–14. doi: 10.1152/physiolgenomics.00204.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou TF, Bieganowski P, Shilinski K, Cheng J, Brenner C, Wagner CR. 31P NMR and genetic analysis establish hint as the only Escherichia coli purine nucleoside phosphoramidase and as essential for growth under high salt conditions. J Biol Chem. 2005;280:15356–15361. doi: 10.1074/jbc.M500434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Date H, Onodera O, Tanaka H, Iwabuchi K, Uekawa K, Igarashi S, Koike R, Hiroi T, Yuasa T, Awaya Y, Sakai T, Takahashi T, Nagatomo H, Sekijima Y, Kawachi I, Takiyama Y, Nishizawa M, Fukuhara N, Saito K, Sugano S, Tsuji S. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat Genet. 2001;29:184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 11.Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, Watanabe M, Ikeda Y, Aoki M, Nagata T, Coutinho P, Sequeiros J, Koenig M. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 12.Seidle HF, Bieganowski P, Brenner C. Disease-associated mutations inactivate AMP-lysine hydrolase activity of aprataxin. J Biol Chem. 2005;280:20927–20931. doi: 10.1074/jbc.M502889200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korsisaari N, Makela TP. Interactions of Cdk7 and Kin28 with Hint/PKCI-1 and Hnt1 histidine triad proteins. J Biol Chem. 2000;275:34837–34840. doi: 10.1074/jbc.C000505200. [DOI] [PubMed] [Google Scholar]
- 14.Lee YN, Nechushtan H, Figov N, Razin E. The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcepsilonRI-activated mast cells. Immunity. 2004;20:145–151. doi: 10.1016/s1074-7613(04)00020-2. [DOI] [PubMed] [Google Scholar]
- 15.Weiske J, Huber O. The Histidine triad protein Hint1 interacts with Pontin and Reptin and inhibits TCF-beta-catenin-mediated transcription. J Cell Sci. 2005;118:3117–3129. doi: 10.1242/jcs.02437. [DOI] [PubMed] [Google Scholar]
- 16.Yuan BZ, Jefferson AM, Popescu NC, Reynolds SH. Aberrant gene expression in human non small cell lung carcinoma cells exposed to demethylating agent 5-aza-2′-deoxycytidine. Neoplasia. 2004;6:412–419. doi: 10.1593/neo.03490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korsisaari N, Rossi DJ, Luukko K, Huebner K, Henkemeyer M, Makela TP. The Histidine triad protein hint is not required for murine development or Cdk7 function. Mol Cell Biol. 2003;23:3929–3935. doi: 10.1128/MCB.23.11.3929-3935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su T, Suzui M, Wang L, Lin CS, Xing WQ, Weinstein IB. Deletion of histidine triad nucleotide-binding protein 1/PKC-interacting protein in mice enhances cell growth and carcinogenesis. Proc Natl Acad Sci USA. 2003;100:7824–7829. doi: 10.1073/pnas.1332160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guang W, Wang H, Su T, Weinstein IB, Wang JB. Role of mPKCI, a novel mu-opioid receptor interactive protein, in receptor desensitization, phosphorylation, and morphine-induced analgesia. Mol Pharmacol. 2004;66:1285–1292. doi: 10.1124/mol.66.5.. [DOI] [PubMed] [Google Scholar]
- 20.Schechtman D, Mochly-Rosen D. Isozyme-specific inhibitors and activators of protein kinase C. Methods Enzymol. 2002;345:470–489. doi: 10.1016/s0076-6879(02)45039-2. [DOI] [PubMed] [Google Scholar]
- 21.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, Durnez A, Demetris AJ, Thorgeirsson SS. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 22.McDonald JR, Walsh MP. Ca2-binding proteins from bovine brain including a potent inhibitor of protein kinase C. Biochem J. 1985;232:559–567. doi: 10.1042/bj2320559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez-Checa JC, Garcia-Ruiz C. Apoptosis and mitochondria. In: Dufour J-F, Clavien PA, editors. Signaling pathways in liver diseases. Springer: 2005. pp. 367–376. [Google Scholar]
- 24.Nan KJ, Ruan ZP, Jing Z, Qin HX, Wang HY, Guo H, Xu R. Expression of fragile Histidine triad in primary hepatocellular carcinoma and its relation with cell proliferation and apoptosis. World J Gastroenterol. 2005;11:228–231. doi: 10.3748/wjg.v11.i2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sevignani C, Calin GA, Cesari R, Sarti M, Ishii H, Yendamuri S, Vecchione A, Trapasso F, Croce CM. Restoration of fragile histidine triad (FHIT) expression induces apoptosis and suppresses tumorigenicity in breast cancer cell lines. Cancer Res. 2003;63:1183–1187. [PubMed] [Google Scholar]
- 26.Katz GG, Shear NH, Malkiewicz IM, Valentino K, Neuman MG. Signaling for ethanol-induced apoptosis and repair in vitro. Clin Biochem. 2001;34:219–227. doi: 10.1016/s0009-9120(01)00218-1. [DOI] [PubMed] [Google Scholar]
- 27.van Heeswijk WC, Rabenberg M, Westerhoff HV, Kahn D. The genes of the glutamine synthetase adenylylation cascade are not regulated by nitrogen in Escherichia coli. Mol Microbiol. 1993;9:443–457. doi: 10.1111/j.1365-2958.1993.tb01706.x. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell C, Blaho JA, Roizman B. Casein kinase II specifically nucleotidylylates in vitro the amino acid sequence of the protein encoded by the alpha 22 gene of herpes simplex virus 1. Proc Natl Acad Sci USA. 1994;91:11864–11868. doi: 10.1073/pnas.91.25.11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.San Jose E, Benguria A, Villalobo A. A novel adenylylation process in liver plasma membrane-bound proteins. J Biol Chem. 1990;265:20653–20661. [PubMed] [Google Scholar]
- 30.San Jose E, Villalobo E, Gabius HJ, Villalobo A. Inhibition of the adenylylation of liver plasma membrane-bound proteins by plant and mammalian lectins. Biol Chem Hoppe Seyler. 1993;374:133–141. doi: 10.1515/bchm3.1993.374.1-6.133. [DOI] [PubMed] [Google Scholar]
- 31.Fankhauser H, Berkowitz GA, Schiff JA. A nucleotide with the properties of adenosine5′ phosphoramidate from Chlorella cells. Biochem Biophys Res Commun. 1981;101:524–532. doi: 10.1016/0006-291x(81)91291-2. [DOI] [PubMed] [Google Scholar]
- 32.Fankhauser H, Schiff JA, Garber LJ. Purification and properties of adenylyl sulphate:ammonia adenylyltransferase from Chlorella catalysing the formation of adenosine5′-phosphoramidate from adenosine 5′-phosphosulphate and ammonia. Biochem J. 1981;195:545–560. doi: 10.1042/bj1950545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maruyama R, Sugio K, Yoshino I, Maehara Y, Gazdar AF. Hyper-methylation of FHIT as a prognostic marker in nonsmall cell lung carcinoma. Cancer. 2004;100:1472–1477. doi: 10.1002/cncr.20144. [DOI] [PubMed] [Google Scholar]
- 34.Foja S, Goldberg M, Schagdarsurengin U, Dammann R, Tannapfel A, Ballhausen WG. Promoter methylation and loss of coding exons of the fragile Histidine triad (FHIT) gene in intrahepatic cholangiocarcinomas. Liver Int. 2005;25:1202–1208. doi: 10.1111/j.1478-3231.2005.01174.x. [DOI] [PubMed] [Google Scholar]
- 35.Li SP, Wang HY, Li JQ, Zhang CQ, Feng QS, Huang P, Yu XJ, Huang LX, Liang QW, Zeng YX. Genome-wide analyses on loss of het-erozygosity in hepatocellular carcinoma in Southern China. J Hepatol. 2001;34:840–849. doi: 10.1016/s0168-8278(01)00047-2. [DOI] [PubMed] [Google Scholar]
- 36.Laurent-Puig P, Legoix P, Bluteau O, Belghiti J, Franco D, Binot F, Monges G, Thomas G, Bioulac-Sage P, Zucman-Rossi J. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology. 2001;120:1763–1773. doi: 10.1053/gast.2001.24798. [DOI] [PubMed] [Google Scholar]


