Abstract
The increasing prevalence of BK virus (BKV)-associated diseases in immunosuppressed patients has prompted an investigation of the immune response to BKV, especially the role of cytokines in regulating viral replication. We examined the effect of TGF-β, a cytokine that is stimulated by certain immunosuppressive therapies, on BKV gene expression during lytic infection of renal proximal tubule epithelial cells. Viral gene expression, and specifically the activity of the BKV early promoter, is regulated by TGF-β in a strain-dependent manner. Promoter activity is upregulated in the presence of TGF-β for the TU strain of BKV, and not for the Dik, Dunlop, or Proto-2 strains. Using site-directed mutagenesis, we have identified a small segment of the TU promoter that is required for stimulation in response to TGF-β. These results demonstrate that BKV strains can respond differently to cytokine treatment and suggest that TGF-β may play a role in the reactivation of BKV.
Keywords: polyomavirus, BK virus, TGF-β, Smad3
INTRODUCTION
BK virus (BKV) is a human polyomavirus that has a well-established role in complications following transplantation and immunosuppression (Comoli et al., 2006; Nickeleit and Mihatsch, 2006). In particular, BKV is the causative agent of polyomavirus nephropathy (PVN) in up to 10% of kidney transplant recipients, resulting in loss of graft function in 10–80% of those affected (Hirsch et al., 2006). BKV reactivation is also associated with hemorrhagic cystitis (HC) in bone marrow transplant recipients, with approximately 50% of patients with active BKV viruria progressing to HC (Bedi et al., 1995; Pavlakis et al., 2006). Despite extensive investigation, there has been limited success in identifying antiviral treatments for BKV reactivation and lytic infection (Josephson et al., 2006; Trofe et al., 2006). Typically, upon diagnosis of BKV reactivation and PVN, the immunosuppressive regimen of the patient is reduced to allow the immune system to fight the viral infection. This approach, however, increases the risk of graft rejection. Thus it is important to investigate the immune response to BKV to identify immune components that control viral replication.
BKV was first isolated from the urine of a kidney transplant patient with ureteric stenosis in 1971 (Gardner et al., 1971). Primary infection with BKV occurs early in childhood, with seroconversion occurring by the age of 10 in up to 80% of the human population (Knowles, 2006). Following the primary infection, BKV undergoes viremic dissemination and establishes a lifelong persistent infection primarily in the cells of the kidney and urinary tract (Chesters et al., 1983; Heritage et al., 1981). More specifically, tubular epithelial cells of the kidney and epithelial cells of the urinary tract are major sites of BKV persistence and reactivation (Doerries, 2006; Nickeleit and Mihatsch, 2006).
BKV has a non-enveloped, icosahedral virion composed of three proteins, VP1, VP2, and VP3, which encapsidate a circular double-stranded DNA genome of approximately 5.2 Kb. The genome can be divided into three distinct regions: the early region, which contains the coding sequences for large tumor antigen (TAg) and small tumor antigen (tAg); the late region, which contains the coding sequences for VP1, VP2, VP3, and agnoprotein; and the non-coding control region (NCCR), which contains the origin of replication and the viral early and late promoters. The NCCR is used to distinguish one strain of BKV from another because of the propensity of this region to acquire point mutations and structural rearrangements (Moens and Van Ghelue, 2005). BKV strains can be divided into two classes: archetypal (pre-rearranged) strains, presumed to be the infectious and transmissible virus, and rearranged strains, which are predominately isolated from tissue biopsy samples (Cubitt, 2006). The NCCRs of archetypal strains are structurally divided into blocks of transcription factor binding sites, arbitrarily designated O (142 bp, containing the origin of replication, TATA box, and TAg binding sites), P (68 bp), Q (39 bp), R (63 bp), and S (63 bp) (Markowitz and Dynan, 1988; Moens et al., 1995). Rearranged BKV strains result from the restructuring of the archetypal NCCR, such that certain blocks (primarily P, Q, and R) are, in whole or in part, duplicated or deleted (Moens and Van Ghelue, 2005).
It is commonly reported that the predominant NCCR configuration actively shed in urine is archetypal (Markowitz et al., 1991; Negrini et al., 1991; Sharma et al., 2007; Sundsfjord et al., 1999; Takasaka et al., 2004), and that NCCR rearrangements seem to be isolated more frequently from the tissues and sera of patients with high viral loads (Boldorini et al., 2001; Gosert et al., 2008; Stoner et al., 2002). In addition, changes in the NCCR structure arise spontaneously in tissue culture and these rearrangements enhance the ability of the virus to replicate and transform cells (Rubinstein et al., 1991; Watanabe and Yoshiike, 1985; Watanabe and Yoshiike, 1986). It is possible that changes in the viral promoter region can also result in altered pathogenesis, such as a heightened ability to reactivate or disseminate. Furthermore, rearrangements may affect the cell tropism of BKV, allowing infection of other cell types in addition to kidney and urinary epithelial cells. The confounding observation, however, is that as of yet there is no apparent correlation between BKV NCCR structure and clinical outcome (Sharma et al., 2007).
TGF-β is a secreted cytokine having three isoforms in mammals (TGF-β1, TGF-β2, TGF-β3), all with similar functions involved in the regulation of cell proliferation, differentiation, and immune suppression (Feng and Derynck, 2005; Li et al., 2006). TGF-β is produced by many different cell types, including renal epithelial cells, a major site of BKV reactivation. Interestingly, the expression of TGF-β is enhanced in the presence of immunosuppressive therapies commonly administered to renal transplant patients (Khanna et al., 1999a; Khanna et al., 1999b; McMorrow et al., 2005; Shihab et al., 1996). We hypothesized that TGF-β would have an upregulatory effect on BKV gene expression and replication, correlating with the evident reactivation of BKV in kidney transplant recipients.
TGF-β is initially expressed as an inactive complex of precursor polypeptides that undergoes activation by proteolytic cleavage. Upon maturation, TGF-β can bind to TGF-β receptor II dimers, resulting in the recruitment and phosphorylation of TGF-β receptor I dimers (Shi and Massague, 2003). Once activated, TGF-β receptor I recruits and phosphorylates Smad2 and Smad3 proteins, the primary components of the TGF-β signaling cascade (Massague et al., 2005). Phosphorylated Smad2 and Smad3 proteins can then form a complex with Smad4, resulting in the nuclear translocation of these proteins. This complex has some weak intrinsic DNA binding activity of its own, but is more effective in regulating TGF-β-dependent gene expression in conjunction with cellular transcriptional co-activators that have high DNA binding activity and specificity (Brown et al., 2007; Massague and Wotton, 2000; Shi et al., 1998).
In this report, we characterize the effect of TGF-β on the lytic infection of BKV in renal proximal tubule epithelial (RPTE) cells and on the activity of the viral early region promoter. We demonstrate that the response to TGF-β-mediated regulation is dependent on the strain of BKV and thus the NCCR structure. We show that upregulation by TGF-β maps to a short region of the promoter that most likely contains two distinct transcription factor binding sites. These findings demonstrate transcriptional regulation of BKV by a cytokine that is found at elevated levels in transplant patients.
MATERIALS AND METHODS
Cell culture and reagents
Primary human renal proximal tubule epithelial cells (RPTE cells, Cambrex) were maintained in renal epithelial cell basal medium (REBM, Cambrex) supplemented with human epidermal growth factor, fetal bovine serum (FBS), hydrocortisone, epinephrine, insulin, triiodothyronine, transferrin, and GA-1000 as indicated for renal epithelial cell growth medium (REGM, supplements obtained as REGM SingleQuots, Cambrex). HT-1080 cells (ATCC CCL-121) were maintained in Dulbecco’s Modified Eagle Medium (Gibco) containing 10% FBS (Cambrex), 100 units/ml penicillin, and 100 µg/ml streptomycin (Cambrex). Both RPTE and HT-1080 cells were grown at 37°C with 5% CO2 in a humidified incubator. Recombinant human TGF-β1, produced in A293 cells (Peprotech, Inc.), was reconstituted according to manufacturer’s recommendations and used at a concentration of 10 ng/ml.
Viruses
BKV stocks were prepared from genomic clones of TU (cloned into the EcoRI site of pGEM-7Zf(-)), and Dunlop and Proto-2 (cloned into the BamHI site of pBR322, gift of Peter Howley), as previously described (Abend et al., 2007). The resulting crude viral stocks were titrated by fluorescent focus assay as previously described (Abend et al., 2007) and the integrity of the NCCR was confirmed by sequencing of PCR products.
Infections
RPTE cells at 70% confluence were infected with the TU, Dunlop, or Proto-2 strains of BKV in REGM at an MOI of 0.5 IU/cell (infectious units per cell), incubating for one hour at 37°C. Viral lysates used for the infection were replaced with fresh REGM. TGF-β was added at three to four hours post-infection (hpi).
Western blotting
Total cell protein was harvested at three days post-infection (dpi) using E1A lysis buffer (Harlow et al., 1986) supplemented with 5 µg/ml PMSF, 5 µg/ml aprotinin, 5 µg/ml leupeptin, 0.05 M sodium fluoride, and 0.2 mM sodium orthovanadate. The Bio-Rad protein assay was used to determine the protein concentration of each lysate and 10 µg of protein were electrophoresed on an 4–20% Tris-glycine polyacrylamide gel (Lonza) and analyzed by Western blotting for the expression of viral early protein TAg and GAPDH as previously described (Abend et al., 2007).
RNA extraction and cDNA synthesis
Total cell RNA was harvested at 24, 36, 48, 72, and 96 hpi using TRIzol reagent (Invitrogen) according to manufacturer’s instructions. Samples were treated with DNase I (Promega) to reduce contaminating DNA, and RNA integrity was confirmed by electrophoresis on an agarose gel. To generate cDNA, reverse transcription reactions were performed on 1 µg of input RNA using the iScript cDNA Synthesis Kit (Bio-Rad), according to manufacturer’s instructions.
Real-Time PCR: TaqMan Assay
Primers and probes used to assay TAg and GAPDH transcript levels are reported previously (Abend et al., 2007). PCR reactions were performed in a total volume of 25 µl using TaqMan Universal PCR 2x master mix (Applied Biosystems), 2.5 µl cDNA template, 500 nM of each primer, and 200 nM probe. The iCycler iQ5 Real-Time Detection System (Bio-Rad) was used for amplification with the following PCR program: 2 min at 50°C; 10 min at 95°C; 40 cycles of denaturation at 95°C for 15 sec and annealing and extension at 56°C for 1 min. Results are presented as the fold change in TAg transcript levels, with the relative level observed at 24 hpi, untreated, arbitrarily set to one. Results are normalized to the levels of GAPDH transcripts present using the 2−ΔΔC(T) (Livak) method (Livak and Schmittgen, 2001).
Generation of luciferase constructs
The NCCRs of four strains of BKV were amplified from their genomic clones using the primers Agno1 (5’ AGTGCTAGCGCCTTTGTCCAGTTTAACT 3’) and LTAg2 (5’ AGTCTCGAGAAATAGTTTTGCTAGGCCTCA 3’), which contain the restriction endonuclease sites for NheI and XhoI, respectively (underlined). Polymerase chain reactions utilizing these primers produced 350–400 bp fragments spanning the NCCR from the start codon of agnoprotein to 35 bp before the TAg start codon. These fragments were first cloned into the pGEM-T Easy vector (Promega) and then subcloned into the luciferase vector pGL2-basic (Promega) by means of the NheI and XhoI sites. In these resulting luciferase constructs (pGL2-TU, pGL2-Dik, pGL2-Dunlop, pGL2-Proto-2), the BKV early promoter drives the expression of the firefly luciferase gene.
Site-directed mutagenesis
The following primers were synthesized and HPLC purified to introduce a point mutation (underlined) at nucleotide 362 (GenBank accession no. DQ305492) in the BKV TU NCCR: TU-SmtFOR (5’ TCGCAAAACATGTCTGTGTGGCTGCTTTCCGG 3’), TU-SmtREV (5’ CCGGAAAGCAGCCACACAGACATGTTTTGCGA 3’). The following primers were synthesized and HPLC purified to insert 6 bp normally present only in the TU NCCR into the BKV Dik NCCR (underlined): Dik+6TUFOR (5’ AAACATGTCTGTCTGGCTGCTTTCCGGTTTCACTCCTTTGG 3’), Dik+6TUREV (5’ CCAAAGGAGTGAAACCGGAAAGCAGCCAGACAGACATGTTT 3’). Mutagenesis was performed following the protocol for the QuikChange II Site-Directed Mutagenesis Kit (Stratagene) using the primer pairs at 1.25 nM each, 100 ng of pGL2-TU or pGL2-Dik as template, 1 mM dNTPs, and 1.25 U Native Pfu DNA Polymerase (Stratagene) in 25 µl total reaction volume. The following two-step PCR program was used: 3 min at 95°C; 18 cycles of denaturation at 95°C for 15 sec and annealing and extension at 68°C for 12 min.
Luciferase assays
RPTE or HT-1080 cells were grown in 12-well tissue culture-treated plates to 60% confluence. Firefly luciferase constructs (pGL2-BKV strain) and a promoterless control Renilla luciferase plasmid (pRL-Null, Promega) were cotransfected into RPTE cells at a ratio of 9:1, with a total of 0.6 µg of DNA per well, using the Effectene Transfection reagent (Qiagen) according to manufacturer’s recommendations. TGF-β was added at three to four hours post-transfection (hpt) and total cell lysates were harvested at 48 hpt in 1x Passive Lysis Buffer (Promega). Luciferase assays were performed on triplicate samples using the Dual-Luciferase Reporter Assay (Promega), according to manufacturer’s recommendations. Results are expressed as relative light units (RLU) of firefly luciferase activity normalized to RLU of Renilla luciferase activity. TGF-β did not significantly affect the levels of Renilla luciferase activity (data not shown). Statistical significance was determined using a two-tailed Student’s t test assuming unequal variance, and P values < 0.01 were considered significant.
RESULTS AND DISCUSSION
TGF-β-mediated regulation of BKV gene expression during infection
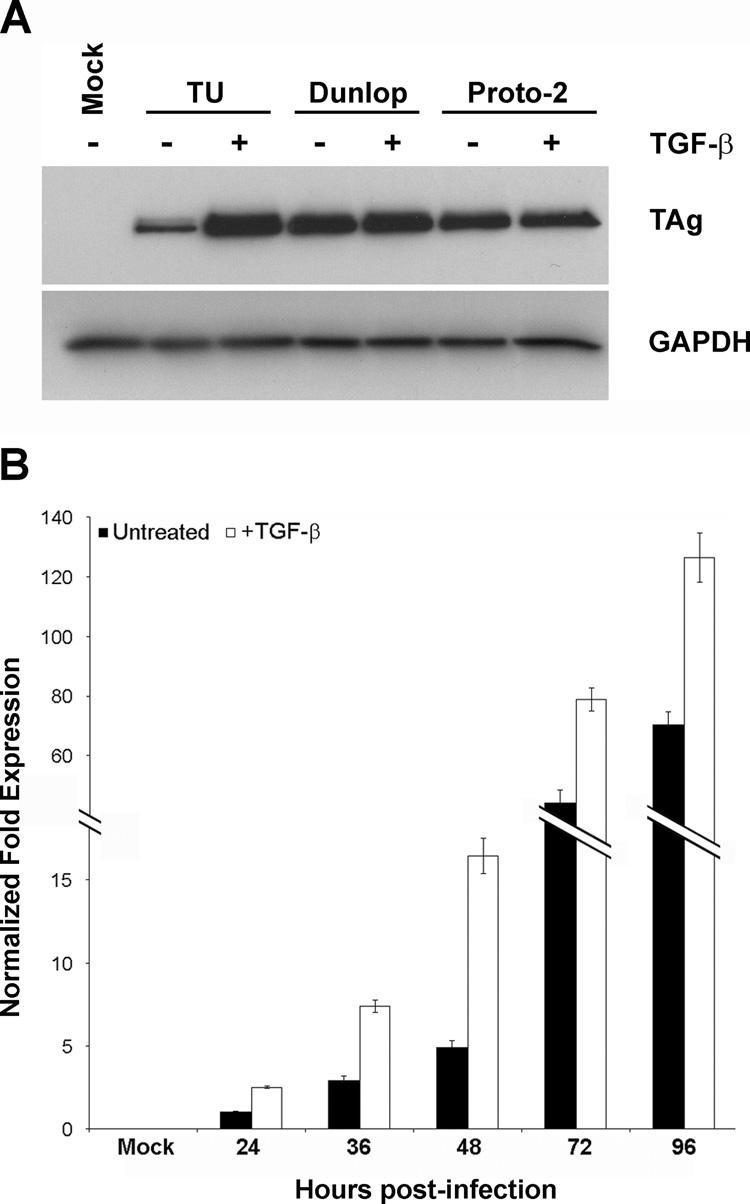
We first investigated the regulatory potential of TGF-β on BKV gene expression during lytic infection of RPTE cells. We examined the levels of TAg at three dpi in cells infected with three strains of BKV that differ significantly in their NCCR structures: TU (O-P-Q-R1–12-P16–68-Q1–35-R52–63-S), Dunlop (O-P-P1–7;26–68-P1–64-S), and Proto-2 (O-P-P1–7;26–68-P-Q1–28-S7–63). TGF-β only significantly affected the expression of viral proteins in cells infected with the TU strain of BKV, as shown by the upregulation of TAg levels (Figure 1A). TAg expression remained relatively unchanged in cells infected with the Dunlop and Proto-2 strains. To demonstrate that the effect was specific to viral gene expression and not a result of TGF-β-mediated inhibition of epithelial cell proliferation, samples were also analyzed for levels of GAPDH, a cellular housekeeping gene. These results indicate that the presence of TGF-β affects BKV gene expression in a strain-specific manner.
FIGURE 1. TGF-β upregulates BKV TU gene expression during infection.
A) RPTE cells were infected with the indicated strains of BKV at an MOI of 0.5 IU/cell and treated with 10 ng/ml TGF-β at three to four hpi. Total cell protein was harvested at three dpi and analyzed by Western blot, probing for TAg and GAPDH. Mock, mock-infected samples with no TGF-β treatment. B) RPTE cells were infected with the TU strain of BKV at an MOI of 0.5 IU/cell in the presence or absence of 10 ng/ml TGF-β, and total cell RNA was prepared at 24, 36, 48, 72, and 96 hpi. Relative TAg transcript levels were determined using real time RT-PCR, normalizing to levels of GAPDH transcripts in each sample. One representative experiment is shown; triplicate samples were analyzed in the same assay. Fold expression of TAg at 24 hpi, untreated was arbitrarily set to one. Mock, mock-infected samples with no TGF-β treatment.
To determine whether TGF-β-mediated regulation occurs during transcription or translation, we examined the levels of early gene transcripts during the course of BKV TU infection. Total cell RNA was harvested at 24, 36, 48, 72, and 96 hpi and analyzed by a real-time RT-PCR assay to detect TAg transcripts in the absence or presence of TGF-β (Figure 1B). Results were normalized to the levels of GAPDH mRNA present in the samples. TGF-β had a prominent effect on TAg transcription at early time points, with 2.5-fold upregulation in transcript levels at 24 and 36 hpi, and 3.3-fold upregulation over untreated cells at 48 hpi. At the later stages of infection the effect of TGF-β was less pronounced, with only 1.8-fold upregulation in TAg transcripts at both 72 and 96 hpi. These results suggest that TGF-β-mediated regulation occurs at the level of transcription. Furthermore, the timing and limited duration of the upregulation suggest that transcription factors involved in the TGF-β signaling cascade are responsible for the effect.
BKV early promoter activity in the presence of TGF-β
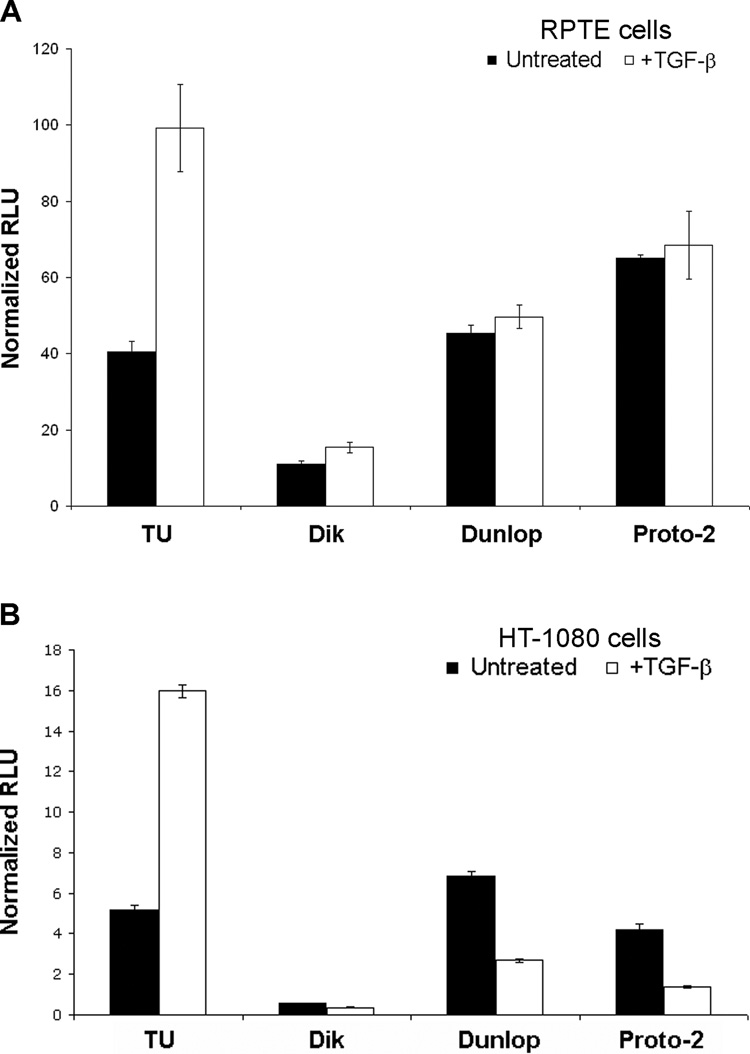
We next set out to examine the effect of TGF-β on the viral early promoter. We cloned the NCCRs of four BKV strains [TU, Dunlop, Proto-2, Dik (archetypal)] into a luciferase reporter plasmid, such that the viral early promoter drives the expression of the firefly luciferase gene. This assay permitted us to examine the effect of TGF-β on promoter activity in the absence of viral gene expression and other BKV genomic sequences. RPTE cells were cotransfected with the BKV promoter-driven luciferase plasmids and a promoterless control Renilla luciferase plasmid in the absence or presence of TGF-β, and lysates were assayed for luciferase activity at 48 hpt (Figure 2A). The results confirmed that only the TU early promoter was affected by TGF-β treatment, while the other three viral promoters showed no significant change in activity in the presence of TGF-β (1.4-, 1.1-, and 1.1-fold change for Dik, Dunlop, and Proto-2 promoters, respectively). The 2.5-fold upregulation (P = 0.009) in TU promoter activity was similar to that seen during BKV infection of RPTE cells (Figure 1). The results of this assay suggest that the effect of TGF-β is mediated solely at the promoter and is therefore driven by specific transcription factors.
FIGURE 2. BKV early promoter activity in the presence of TGF-β.
A) RPTE cells were cotransfected with BKV early promoter-firefly luciferase constructs and a promoterless control Renilla luciferase plasmid. TGF-β was added at three to four hpt and total cell lysates were harvested at 48 hpt. Luciferase assays were performed on triplicate samples and data are represented as relative light units (RLU) of firefly luciferase activity, normalized to RLU of Renilla luciferase activity. Data shown represent results obtained from three independent experiments. B) HT-1080 cells were cotransfected with BKV early promoter-firefly luciferase constructs and a promoterless control Renilla luciferase plasmid and assayed as described in (A). Data shown represent results obtained from three independent experiments.
The regulation of BKV early promoter activity by TGF-β signaling may also depend on the cell type examined. The luciferase assay described above was also performed in HT-1080 cells, a human fibrosarcoma cell line (Figure 2B). The TU and Dik promoters responded similarly to TGF-β treatment in both cell types: TU promoter activity was upregulated by 3.1-fold (P < 0.001) and Dik activity remained relatively unchanged (1.6-fold decrease, P < 0.001) in the HT-1080 cells. However, the Dunlop and Proto-2 promoters were repressed in the presence of TGF-β in HT-1080 cells by 2.6-and 3.1-fold respectively (P < 0.001 and P = 0.002, respectively). In addition to cells of the kidney and urinary tract, BKV sequences have been isolated from peripheral blood mononuclear cells (Chatterjee et al., 2000; Doerries et al., 1994), tonsils (Goudsmit et al., 1982), and brain (Elsner and Doerries, 1992). Thus, TGF-β-mediated signaling may differentially regulate BKV during infections of other cell types.
Having mapped the effect of TGF-β to the BKV promoter, we wanted to examine the differences between the TU NCCR and the other three NCCRs used in our studies. We performed an alignment of the four BKV NCCRs and observed that there was one region (7 bp, starred nucleotides) of the TU NCCR that did not align to any region of the Dik, Dunlop, or Proto-2 NCCRs (Figure 3). We used the MatInspector transcription factor binding site prediction program (Cartharius et al., 2005) to analyze the potential binding sites in the viral promoters. Within this unique sequence in the TU NCCR, the program predicted a binding site for the transcription factor ZEB-1/AREB6 with a high probability. There were no predicted ZEB-1 binding sites in the other three NCCRs. ZEB-1 has been reported to interact with Smad3 and mediate TGF-β-dependent gene regulation (Postigo, 2003; Postigo et al., 2003). Interestingly, in both the TU and Dik NCCRs the MatInspector program also predicted an adjacent binding site for the transcription factor Smad3 with a high probability. The proximity of the predicted ZEB-1 site to the predicted Smad3 site in the TU NCCR suggests that a Smad3-ZEB-1 complex could actively bind to the promoter and regulate early gene transcription.
FIGURE 3. Alignment of BKV NCCRs.
The segments of the NCCRs (TU, Dik, Dunlop, Proto-2) from the nucleotide before the start codon of agnoprotein through the P block directly adjacent to the O block are shown. The O block is not shown because it is highly homologous between strains. Bold type indicates nucleotides that are identical in at least three of the strains. Underlined regions indicate the predicted Smad3 and ZEB-1 binding sites. Dots indicate nucleotides not found within a sequence. The starred nucleotides indicate the region of the TU NCCR that does not align anywhere in the Dik, Dunlop, or Proto-2 NCCRs. The early promoter is read from left to right (top to bottom), in the direction of the early coding region. The late promoter is read from right to left (bottom to top), in the direction of the late coding region.
TU promoter sequences required for TGF-β-mediated regulation
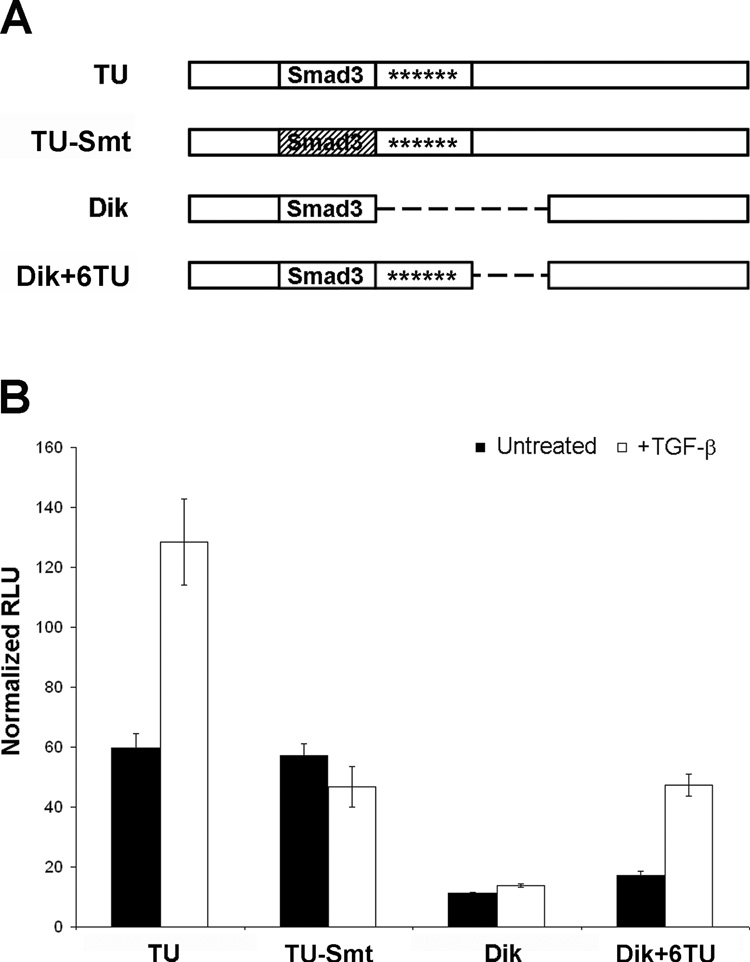
We used site-directed mutagenesis to modify the luciferase plasmids to define the exact sequences required for TGF-β-dependent activation of the early promoter (Figure 4A). Starting with the pGL2-TU plasmid as a template, we introduced a single base change (C to G) in the core of the predicted Smad3 binding site, resulting in the pGL2-TU-Smt plasmid. This mutation has previously been shown to effectively disrupt the ability of Smad proteins to bind to the DNA (Jonk et al., 1998). In a similar manner, we modified the pGL2-Dik plasmid, which contains a predicted Smad3 binding site identical to that found in the TU NCCR, lacks a predicted ZEB-1 binding site, and is not affected by TGF-β. Using site-directed mutagenesis, we introduced six nucleotides (GGTTTC) to place a predicted ZEB-1 binding site in the same relative position and orientation as in the TU NCCR (pGL2-Dik+6TU).
FIGURE 4. Regions of the TU promoter required for TGF-β-mediated regulation of BKV.
A) Schematic representation of promoter constructs. TU-Smt has a single base change in the core of the predicted Smad3 binding site (hatched). Dik+6TU has a 6 bp insertion that creates the predicted ZEB-1 binding site. The early promoter is read from left to right, with the start codon for the firefly luciferase gene following the right end of the promoter. B) RPTE cells were cotransfected with wildtype or mutant BKV early promoter-firefly luciferase constructs and a promoterless control Renilla luciferase plasmid. TGF-β was added at three to four hpt and total cell lysates were harvested at 48 hpt. Luciferase assays were performed on triplicate samples and data are represented as relative light units (RLU) of firefly luciferase activity, normalized to RLU of Renilla luciferase activity. Data shown represent results obtained from three independent experiments.
The wildtype and mutated luciferase reporter plasmids were transfected into RPTE cells in the absence or presence of TGF-β, and lysates were assayed for luciferase activity at 48 hpt (Figure 4B). Similar to previous results, the TU early promoter was upregulated by 2.2-fold (P = 0.009) in the presence of TGF-β, while the Dik promoter showed little change in activity (1.2-fold increase). The mutant TU promoter was also unresponsive to TGF-β treatment (1.2-fold decrease), suggesting that the putative Smad3 site is required for TGF-β-dependent promoter activation and that the putative ZEB-1 site alone is not sufficient. In addition, the mutant Dik promoter was upregulated by 2.7-fold (P = 0.002) in the presence of TGF-β, suggesting that both the predicted Smad3 and ZEB-1 sites are necessary for upregulation. It is important to note that the mutations did not significantly change the basal activities of the promoters (Figure 4B: compare untreated, TU to TU-Smt; untreated, Dik to Dik+6TU). These results suggest that regulation depends directly on the presence of binding sites for transcription factors involved in TGF-β signaling in the viral early promoter, and that the absence of such sites results in unresponsiveness to TGF-β.
The MatInspector binding site predictions and the results of the luciferase assays strongly suggested that Smad3 and ZEB-1 regulate the TU promoter. However, our attempts to show Smad3 and ZEB-1 binding to the viral promoter using three different assays, electrophoretic mobility shift assay (EMSA), anchored transcriptional promoter assay (Ravichandran and Major, 2006), and siRNA-mediated knockdown of Smad3 and ZEB-1, failed to demonstrate conclusively Smad3 and ZEB-1 binding to, or activation of, the TU promoter in the presence of TGF-β (data not shown). It remains possible that Smad3 is acting with another transcription factor, or that the assays simply were not well suited for demonstration of binding of these particular factors in our system. It will be of interest to define the nature of the factors mediating the TGF-β response in the future.
Previous studies have shown the relevance of Smad-dependent TGF-β signaling during viral infections. For example, JC virus (JCV)-infected oligodendrocytes have elevated levels of TGF-β, Smad3, and Smad4, and chloramphenicol acetyl transferase assays demonstrate the activating effect of Smad protein overexpression on the early and late viral promoters (Enam et al., 2004). Recently, it was shown that TGF-β stimulates JCV replication and that MAPK kinase (MEK) inhibitors can block this effect, indicating a role for activated downstream transcription factors in TGF-β-mediated upregulation (Ravichandran et al., 2007). Epstein-Barr virus (EBV), a ubiquitous human gammaherpesvirus that, like BKV, has both latent and lytic stages of infection, is also regulated by TGF-β-mediated signaling. Studies have shown that TGF-β treatment drives the reactivation of EBV lytic infection from latently-infected B cells (di Renzo et al., 1994) and epithelial cells (Fukuda et al., 2001). These and other reports have established the relevance of TGF-β and Smad3 in viral regulation (Li et al., 2006; Reed, 1999).
Our studies of cytokine-mediated regulation of BKV gene expression and replication are targeted at understanding the process of viral reactivation in immunosuppressed transplant patients. Previously, it has been shown that common immunosuppressive therapies used in renal transplantation patients, such as cyclosporine and tacrolimus, result in upregulation of TGF-β expression in various cell types, including kidney epithelial cells (Khanna et al., 1999a; Khanna et al., 1999b; McMorrow et al., 2005; Shihab et al., 1996). Therefore, we hypothesized that TGF-β-mediated upregulation of the TU strain of BKV would result in an enhanced ability to reactivate in renal transplant recipients and, consequently, an increase in virulence of this particular strain. However, there are few reports of the TU strain in clinical isolates (Sundsfjord et al., 1994; Sundsfjord et al., 1990), and we have been unable to identify other naturally occurring strains of BKV that contain both the sequences in the TU promoter that are required for TGF-β-mediated activation. Nevertheless, the predicted Smad3 site occurs in a number of other strains [MAN10B (GenBank accession no. DQ176633), WWT (M34048), URO1 (U33549), TC-3 (AF164514), AS (M23122), TW-2 (AB213487), SJH85B (DQ176634), T2R.1BKreg-3-4 (AF442893), SA090600 (AF356532)], and Smad3 is known to have many different functional binding partners, including AP-1, c- MYC, NF-κB, and SP1 (Brown et al., 2007), all of which also have binding sites in the NCCR. The results of the luciferase assays using mutated promoters (Figure 4) support the likelihood that there are two transcription factors involved in regulating the response to TGF-β. Thus, there is potential for TGF-β-mediated activation or repression of other BKV strains.
Our studies provide evidence for the transcriptional regulation of BKV early gene expression by as yet unidentified components of the TGF-β signaling cascade. However, the postulated contribution of TGF-β to BKV reactivation in transplant patients must be considered in the context of the overall immune status of those individuals. Previously, we also described the inhibition of BKV replication by IFN-γ, in a strain-independent manner (Abend et al., 2007). It is therefore reasonable to posit that reactivation of BKV in immunocompromised patients may require multiple signals, such as the enhancement of viral gene expression by TGF-β and the absence of IFN-γ-mediated inhibition of replication, as well as others. Understanding the contributions of each signal to the outcome of the infection will be critical for building a complete picture of the interaction between BKV and the host immune system.
ACKNOWLEDGMENTS
We thank members of the Imperiale lab for help with this work, Mark Day and members of the Day lab for assistance with luciferase assays, John Lednicky for pBR322-Dik, and Kathy Spindler for critical review of the manuscript. This work was supported by AI060584 awarded to M.J.I. from the NIH and in part by CA46592 awarded to the University of Michigan Cancer Center from the NIH. J.R.A. was supported by the NIH National Research Service Award T32-GM07544 and the F. G. Novy Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abend JR, Low JA, Imperiale MJ. Inhibitory effect of gamma interferon on BK virus gene expression and replication. J Virol. 2007;81:272–279. doi: 10.1128/JVI.01571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedi A, Miller CB, Hanson JL, Goodman S, Ambinder RF, Charache P, Arthur RR, Jones RJ. Association of BK virus with failure of prophylaxis against hemorrhagic following bone marrow transplantation. J Clin Oncol. 1995;13:1103–1109. doi: 10.1200/JCO.1995.13.5.1103. [DOI] [PubMed] [Google Scholar]
- Boldorini R, Omodeo-Zorini E, Suno A, Benigni E, Nebuloni M, Garino E, Fortunato M, Monga G, Mazzucco G. Molecular characterization and sequence analysis of polyomavirus strains isolated from needle biopsy specimens of kidney allograft recipients. Am J Clin Pathol. 2001;116:489–494. doi: 10.1309/GAUE-92W7-ACDV-X46M. [DOI] [PubMed] [Google Scholar]
- Brown KA, Pietenpol JA, Moses HL. A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling. J Cell Biochem. 2007;101:9–33. doi: 10.1002/jcb.21255. [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Weyandt TB, Frisque RJ. Identification of archetype and rearranged forms of BK virus in leukocytes from healthy individuals. J Med Virol. 2000;60:353–362. [PubMed] [Google Scholar]
- Chesters PM, Heritage J, McCance DJ. Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissues. J Infect Dis. 1983;147:676–684. doi: 10.1093/infdis/147.4.676. [DOI] [PubMed] [Google Scholar]
- Comoli P, Binggeli S, Ginevri F, Hirsch HH. Polyomavirus-associated nephropathy: update on BK virus-specific immunity. Transpl Infect Dis. 2006;8:86–94. doi: 10.1111/j.1399-3062.2006.00167.x. [DOI] [PubMed] [Google Scholar]
- Cubitt CL. Molecular genetics of the BK virus. Adv Exp Med Biol. 2006;577:85–95. doi: 10.1007/0-387-32957-9_6. [DOI] [PubMed] [Google Scholar]
- di Renzo L, Altiok A, Klein G, Klein E. Endogenous TGF-beta contributes to the induction of the EBV lytic cycle in two Burkitt lymphoma cell lines. Int J Cancer. 1994;57:914–919. doi: 10.1002/ijc.2910570623. [DOI] [PubMed] [Google Scholar]
- Doerries K. Human polyomavirus JC and BK persistent infection. Adv Exp Med Biol. 2006;577:102–116. doi: 10.1007/0-387-32957-9_8. [DOI] [PubMed] [Google Scholar]
- Doerries K, Vogel E, Gunther S, Czub S. Infection of human polyomaviruses JC and BK in peripheral blood leukocytes from immunocompetent individuals. Virology. 1994;198:59–70. doi: 10.1006/viro.1994.1008. [DOI] [PubMed] [Google Scholar]
- Elsner C, Doerries K. Evidence of human polyomavirus BK and JC infection in normal brain tissue. Virology. 1992;191:72–80. doi: 10.1016/0042-6822(92)90167-n. [DOI] [PubMed] [Google Scholar]
- Enam S, Sweet TM, Amini S, Khalili K, Del Valle L. Evidence for involvement of transforming growth factor beta1 signaling pathway in activation of JC virus in human immunodeficiency virus 1-associated progressive multifocal leukoencephalopathy. Arch Pathol Lab Med. 2004;128:282–291. doi: 10.5858/2004-128-282-EFIOTG. [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R. Specificity and versatility in TGF-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Ikuta K, Yanagihara K, Tajima M, Kuratsune H, Kurata T, Sairenji T. Effect of transforming growth factor-beta1 on the cell growth and Epstein-Barr virus reactivation in EBV-infected epithelial cell lines. Virology. 2001;288:109–118. doi: 10.1006/viro.2001.1071. [DOI] [PubMed] [Google Scholar]
- Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1:1253–1257. doi: 10.1016/s0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- Gosert R, Rinaldo CH, Funk GA, Egli A, Ramos E, Drachenberg CB, Hirsch HH. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J Exp Med. 2008;205:841–852. doi: 10.1084/jem.20072097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudsmit J, Wertheim-van Dillen P, van Strien A, van der Noordaa J. The role of BK virus in acute respiratory tract disease and the presence of BKV DNA in tonsils. J Med Virol. 1982;10:91–99. doi: 10.1002/jmv.1890100203. [DOI] [PubMed] [Google Scholar]
- Harlow E, Whyte P, Franza BR, Jr, Schley C. Association of adenovirus early-region 1A proteins with cellular polypeptides. Mol Cell Biol. 1986;6:1579–1589. doi: 10.1128/mcb.6.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heritage J, Chesters PM, McCance DJ. The persistence of papovavirus BK DNA sequences in normal human renal tissue. J Med Virol. 1981;8:143–150. doi: 10.1002/jmv.1890080208. [DOI] [PubMed] [Google Scholar]
- Hirsch HH, Drachenberg CB, Steiger J, Ramos E. Polyomavirus-associated nephropathy in renal transplantation: critical issues of screening and management. Adv Exp Med Biol. 2006;577:160–173. doi: 10.1007/0-387-32957-9_11. [DOI] [PubMed] [Google Scholar]
- Jonk LJ, Itoh S, Heldin CH, ten Dijke P, Kruijer W. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-beta, activin, and bone morphogenetic protein-inducible enhancer. J Biol Chem. 1998;273:21145–21152. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- Josephson MA, Williams JW, Chandraker A, Randhawa PS. Polyomavirus-associated nephropathy: update on antiviral strategies. Transpl Infect Dis. 2006;8:95–101. doi: 10.1111/j.1399-3062.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- Khanna A, Cairns V, Hosenpud JD. Tacrolimus induces increased expression of transforming growth factor-beta1 in mammalian lymphoid as well as nonlymphoid cells. Transplantation. 1999a;67:614–619. doi: 10.1097/00007890-199902270-00021. [DOI] [PubMed] [Google Scholar]
- Khanna AK, Cairns VR, Becker CG, Hosenpud JD. Transforming growth factor (TGF)-beta mimics and anti-TGF-beta antibody abrogates the in vivo effects of cyclosporine: demonstration of a direct role of TGF-beta in immunosuppression and nephrotoxicity of cyclosporine. Transplantation. 1999b;67:882–889. doi: 10.1097/00007890-199903270-00016. [DOI] [PubMed] [Google Scholar]
- Knowles WA. Discovery and epidemiology of the human polyomaviruses BK virus (BKV) and JC virus (JCV) Adv Exp Med Biol. 2006;577:19–45. doi: 10.1007/0-387-32957-9_2. [DOI] [PubMed] [Google Scholar]
- Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Markowitz RB, Dynan WS. Binding of cellular proteins to the regulatory region of BK virus DNA. J Virol. 1988;62:3388–3398. doi: 10.1128/jvi.62.9.3388-3398.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz RB, Eaton BA, Kubik MF, Latorra D, McGregor JA, Dynan WS. BK virus and JC virus shed during pregnancy have predominantly archetypal regulatory regions. J Virol. 1991;65:4515–4519. doi: 10.1128/jvi.65.8.4515-4519.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMorrow T, Gaffney MM, Slattery C, Campbell E, Ryan MP. Cyclosporine A induced epithelial-mesenchymal transition in human renal proximal tubular epithelial cells. Nephrol Dial Transplant. 2005;20:2215–2225. doi: 10.1093/ndt/gfh967. [DOI] [PubMed] [Google Scholar]
- Moens U, Johansen T, Johnsen JI, Seternes OM, Traavik T. Noncoding control region of naturally occurring BK virus variants: sequence comparison and functional analysis. Virus Genes. 1995;10:261–275. doi: 10.1007/BF01701816. [DOI] [PubMed] [Google Scholar]
- Moens U, Van Ghelue M. Polymorphism in the genome of non-passaged human polyomavirus BK: implications for cell tropism and the pathological role of the virus. Virology. 2005;331:209–231. doi: 10.1016/j.virol.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Negrini M, Sabbioni S, Arthur RR, Castagnoli A, Barbanti-Brodano G. Prevalence of the archetypal regulatory region and sequence polymorphisms in nonpassaged BK virus variants. J Virol. 1991;65:5092–5095. doi: 10.1128/jvi.65.9.5092-5095.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickeleit V, Mihatsch MJ. Polyomavirus nephropathy in native kidneys and renal allografts: an update on an escalating threat. Transpl Int. 2006;19:960–973. doi: 10.1111/j.1432-2277.2006.00360.x. [DOI] [PubMed] [Google Scholar]
- Pavlakis M, Haririan A, Klassen DK. BK virus infection after non-renal transplantation. Adv Exp Med Biol. 2006;577:185–189. doi: 10.1007/0-387-32957-9_13. [DOI] [PubMed] [Google Scholar]
- Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. EMBO J. 2003;22:2443–2452. doi: 10.1093/emboj/cdg225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 2003;22:2453–2462. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran V, Jensen PN, Major EO. MEK1/2 inhibitors block basal and transforming growth factor-beta1-stimulated JC virus multiplication. J Virol. 2007;81:6412–6418. doi: 10.1128/JVI.02658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran V, Major EO. Viral proteomics: a promising approach for understanding JC virus tropism. Proteomics. 2006;6:5628–5636. doi: 10.1002/pmic.200600261. [DOI] [PubMed] [Google Scholar]
- Reed SG. TGF-beta in infections and infectious diseases. Microbes Infect. 1999;1:1313–1325. doi: 10.1016/s1286-4579(99)00252-x. [DOI] [PubMed] [Google Scholar]
- Rubinstein R, Schoonakker BC, Harley EH. Recurring theme of changes in the transcriptional control region of BK virus during adaptation to cell culture. J Virol. 1991;65:1600–1604. doi: 10.1128/jvi.65.3.1600-1604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma PM, Gupta G, Vats A, Shapiro R, Randhawa PS. Polyomavirus BK non-coding control region rearrangements in health and disease. J Med Virol. 2007;79:1199–1207. doi: 10.1002/jmv.20909. [DOI] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang YF, Jayaraman L, Yang H, Massague J, Pavletich NP. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- Shihab FS, Andoh TF, Tanner AM, Noble NA, Border WA, Franceschini N, Bennett WM. Role of transforming growth factor- beta 1 in experimental chronic cyclosporine nephropathy. Kidney Int. 1996;49:1141–1151. doi: 10.1038/ki.1996.165. [DOI] [PubMed] [Google Scholar]
- Stoner GL, Alappan R, Jobes DV, Ryschkewitsch CF, Landry ML. BK virus regulatory region rearrangements in brain and cerebrospinal fluid from a leukemia patient with tubulointerstitial nephritis and meningoencephalitis. Am J Kidney Dis. 2002;39:1102–1112. doi: 10.1053/ajkd.2002.32795. [DOI] [PubMed] [Google Scholar]
- Sundsfjord A, Flaegstad T, Flo R, Spein AR, Pedersen M, Permin H, Julsrud J, Traavik T. BK and JC viruses in human immunodeficiency virus type 1-infected persons: prevalence, excretion, viremia, and viral regulatory regions. J Infect Dis. 1994;169:485–490. doi: 10.1093/infdis/169.3.485. [DOI] [PubMed] [Google Scholar]
- Sundsfjord A, Johansen T, Flaegstad T, Moens U, Villand P, Subramani S, Traavik T. At least two types of control regions can be found among naturally occurring BK virus strains. J Virol. 1990;64:3864–3871. doi: 10.1128/jvi.64.8.3864-3871.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundsfjord A, Osei A, Rosenqvist H, Van Ghelue M, Silsand Y, Haga HJ, Rekvig OP, Moens U. BK and JC viruses in patients with systemic lupus erythematosus: prevalent and persistent BK viruria, sequence stability of the viral regulatory regions, and nondetectable viremia. J Infect Dis. 1999;180:1–9. doi: 10.1086/314830. [DOI] [PubMed] [Google Scholar]
- Takasaka T, Goya N, Tokumoto T, Tanabe K, Toma H, Ogawa Y, Hokama S, Momose A, Funyu T, Fujioka T, Omori S, Akiyama H, Chen Q, Zheng HY, Ohta N, Kitamura T, Yogo Y. Subtypes of BK virus prevalent in Japan and variation in their transcriptional control region. J Gen Virol. 2004;85:2821–2827. doi: 10.1099/vir.0.80363-0. [DOI] [PubMed] [Google Scholar]
- Trofe J, Hirsch HH, Ramos E. Polyomavirus-associated nephropathy: update of clinical management in kidney transplant patients. Transpl Infect Dis. 2006;8:76–85. doi: 10.1111/j.1399-3062.2006.00166.x. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Yoshiike K. Decreasing the number of 68-base-pair tandem repeats in the BK virus transcriptional control region reduces plaque size and enhances transforming capacity. J Virol. 1985;55:823–825. doi: 10.1128/jvi.55.3.823-825.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Yoshiike K. Evolutionary changes of transcriptional control region in a minute-plaque viable deletion mutant of BK virus. J Virol. 1986;59:260–266. doi: 10.1128/jvi.59.2.260-266.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]