Abstract
Ontologies help to identify and formally define the entities and relationships in specific domains of interest. Bio-ontologies, in particular, play a central role in the annotation, integration, analysis, and interpretation of biological data. Missing from the number of bio-ontologies is one that includes phenotypic trait information found in livestock species. As a result, the Animal Trait Ontology (ATO) project being carried out under the auspices of the USDA-National Animal Genome Research Program is aimed at the development of a standardized trait ontology for farm animals and software tools to assist the research community in collaborative creation, editing, maintenance, and use of such an ontology. The ATO is currently inclusive of cattle, pig, and chicken species, and will include other livestock species in the future. The ATO will eventually be linked to other species (e.g., human, rat, mouse) so that comparative analysis can be efficiently performed between species.
Keywords: ontology, trait, phenotype, animal, cattle, chicken
INTRODUCTION
Technological advances in the past decade have dramatically increased the rate with which biological and genetic information can be gathered. This has resulted in a proliferation of large biological databases designed to facilitate access to these data. For example, the 2008 database issue of Nucleic Acids Research lists 1,078 such databases (or 110 more than the previous year; Galperin, 2008). Terminological differences (different words being used to mean the same thing), syntactic differences (different representations of the same term; e.g., due to differences in spelling), and semantic differences (the same words being used to mean different things in different sources) present significant hurdles in sharing data and knowledge between disparate researchers and research groups (Greally, 2007).
The terminological, syntactic, and semantic gaps between data sources need to be overcome for it to be possible for researchers to have seamless access to disparate, independently developed, yet interrelated data sources (e.g., genetic data, trait data, different types of experimental data) in exploring specific scientific questions (e.g., through data mining). Consequently, there is a growing awareness of the need for ontologies in the life sciences (Schulze-Kremer and Smith, 2005). There has also been a need to share a wealth of knowledge between disparate researchers and research groups. As a result, a growing number of biological ontologies are being constructed (Blake and Bult, 2006). These ontologies, which are composed of terms and the relationships between them, range from a few hundred to thousands of terms. Until now, an ontology for domesticated farm animal phenotypic traits was missing from the number of biological ontologies. Previously, the concept of an animal trait ontology was introduced in the building of the pig QTL and animal QTL databases (Hu et al., 2005, 2007) and implemented as a simple hierarchy structure. Obviously, the wide utility of this implementation is limited in terms of universal applications and meeting the challenges of community collaborative interactions.
With the large amount of biological and genomic information associated with farm animal traits, it is imperative that a standard nomenclature be created so that animal science researchers may communicate consistently and unambiguously. The need for an animal trait ontology has risen because of several farm animal databases and journals that cater to animal scientists. These databases and journals contain important biological, genomic, and phenotypic information, but they are located in disparate locations. The Animal Trait Ontology (ATO) is the first large-scale ontology effort that will deal with the standardization and centralizing of livestock animal traits.
The word ontology is derived from the Greek words ontos, meaning “to be” and logos, meaning “word.” An ontology is defined as “a formal specification of a shared conceptualization” (Borst, 1997). In other words, an ontology is a controlled vocabulary that describes objects and the relations between them in a formal way. Ontologies have become useful in recent years due to their ability to allow for the sharing of information among people and software agents. Experts in particular domains are now able to share descriptions of concepts. Also, software agents are able to manipulate this data through resources such as the World Wide Web Consortium (W3C), which is developing technologies that encode knowledge on web pages so that underlying software agents may understand it (World Wide Web Consortium, 2007). This ability allows the underlying agents to make inferences or discoveries about the data.
Bio-Ontologies
Bio-ontologies are emerging in several biological domains. These ontologies contain from a few hundred to thousands of terms. The most prevalent bio-ontology is the Gene Ontology (GO; Ashburner et al., 2000), which is a controlled vocabulary that describes gene and gene products in several model organisms. The Mammalian Phenotype Ontology describes mammalian phenotypes that are used as models of human biology and disease (Smith et al., 2005). Other bio-ontologies include the Plant Ontology (Ilic et al., 2007), the Zebrafish Anatomy and Development Ontology (Sprague et al., 2003), and the FlyBase Controlled Vocabulary (FBcv; Crosby et al., 2007). The ATO will be instrumental in standardizing traits descriptions within livestock and will contribute to the wealth of knowledge in bio-ontologies.
IMPACT OF AN ANIMAL TRAIT ONTOLOGY
The ATO currently contains data for 3 domesticated farm animal species: Bos taurus (cattle), Gallus gallus (chicken), and Sus scrofa (pig). The original goal of the ATO was to create a medium for the standardization, annotation, retrieval, integration, and analysis of animal trait information; in particular, traits with associated QTL. However, it has become evident, with the large amount of research being conducted by animal science researchers, that a trait ontology is instrumental in forming a standard so that researchers may communicate with each other more consistently and effectively.
Why Do We Need an Animal Trait Ontology?
The need for an ATO was evident for several reasons. First, there was no central repository for trait information. Trait information is currently spread among journal articles, books, local researcher archives, and other miscellaneous sources. These disparate sources of phenotypic information further contributed to the inconsistency of trait terms (e.g., daily gain and average daily gain). By analogy, in the early days of gene discovery, several labs would simultaneously identify a gene and put forth different names in the published literature (Hamerton, 1977). If a researcher was familiar with all of the relevant literature this did not present a huge issue. However, anyone new to the field would have a very hard time finding all of the relevant literature. To solve this problem, the HUGO gene nomenclature system, which is responsible for creating a gene name and symbol for every known human gene (McAlpine and Shows, 1990), was created. The HUGO system has allowed researchers to communicate about genes without inconsistency in the naming of genes and symbols. It has also facilitated the retrieval of electronic data from publications.
Second, on a global perspective, trait information is sometimes inconsistent between different regions of the world. For instance, in Europe, it is common to see the term “meat colour” used for describing a meat quality trait, but in the United States, “meat color” is used to describe the trait (Keokamnerd et al., 2007; Wimmers et al., 2007). Also, there is the issue of variations for the same traits. For instance, “ribeye area,” “rib eye area,” and “muscle area” all share the same semantics or meanings, but different spellings. Although this problem is easy for humans to overcome in that we learn over time that these terms are equivalent, computer agents on the other hand do not recognize that these terms are equivalent, unless they are instructed otherwise, and thus treat them as independent terms. With the development of the ATO, the relatedness of terms will be electronically curated for the first time. Thus, the ATO should help bridge the gap between nomenclatures in different parts of the world and between different variations of the same trait.
Third, by having an ATO, it will be possible to perform computational analysis of the traits using the semantic web (Berners-Lee et al., 2001) that is composed of machine-understandable data and knowledge for the automatic discovery, integration, and reuse of those data and knowledge across several applications. The trait information in the ATO, for instance, is expected to be linked to quantitative trait information in the future, which will allow inferences to be made by linking different QTL regions to traits. Such a feature will allow comparative phenotype/QTL studies to be performed between species, including humans, rats, and mice. Alternatively, it may be used to link information across disciplines (e.g., nutrition and genetics). The ATO could also be beneficial to animal scientists in that it can be interconnected with other ontologies, which will allow the transfer of knowledge across species and scientific disciplines. For example, the traits in the ATO could be linked to an anatomy ontology or an ontology that stores genomic information such as the GO. It may also be used to improve the searching of electronic publications through the inclusion of all relevant synonyms for a particular trait name.
Examples of ATO Annotations
The importance of the ATO can be seen with the following examples. In the first example, the problem of semantics of farm animal traits is illustrated. Semantics involves the meanings of words and, in this case, multiple trait names for the same trait. During the digital curation of several research journals for the trait name “ribeye area,” several alternative forms of the trait were found (e.g., rib eye area, rib muscle area). Each of these trait names shares the same semantics (meanings), but different spellings. Computers recognize these terms as being independent and not as the interrelated terms that they are unless the relationships between terms (e.g., that 2 terms are synonyms of each other, or one term is more general than another) are explicitly specified. The ATO will help alleviate this problem through standardization of terms used to refer to animal traits and explicit specification of relationships between terms that refer to the same or related traits. There will still be an issue with historical data, but incorporating synonyms in the ATO will aid in the inclusion of such data.
Another issue that the ATO will encounter is the variation in trait information. Variations occur when a phenotype is measured in several different ways. For example, the trait “backfat” is measured in 3 different measurement types: methods (e.g., ruler, ultrasound), time (e.g., 14 wk of age), and locations (e.g., shoulder, tenth rib; Figure 1). With the ATO, such measurement variations will be better contained and understood by researchers. The standardization of trait information variations will further aid in the eradication of ambiguities in trait names.
Figure 1.
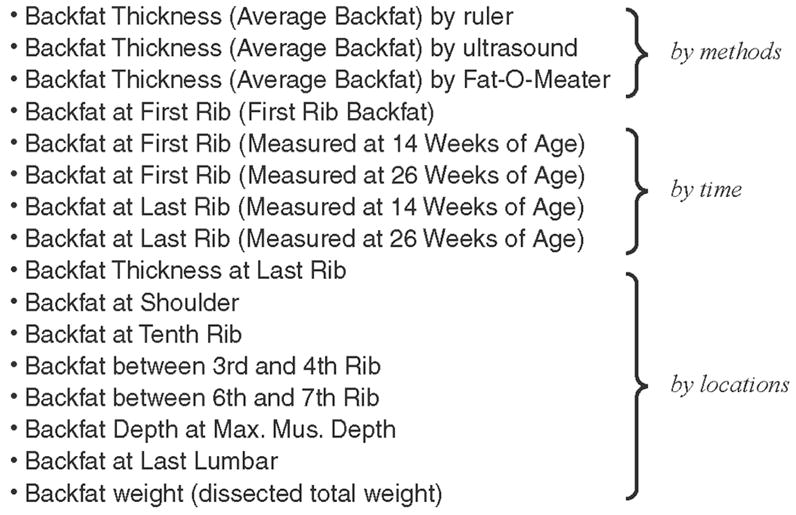
An example of the variation of trait measurements for some traits. In this example, backfat is used to show how the trait is measured according to different methods, times, and locations. Also, within each measurement type, there are different measurement specifications.
Structure of the ATO
The trait information in the ATO is obtained from published papers and reports, books, private researcher archives, and other miscellaneous sources. We define a trait as that which is specifically measured. For example, femur length would be a trait. In contrast, diabetes is a disease, but not a trait. Insulin level or blood glucose level would be a trait that is measured to quantify the level of diabetes observed. We further differentiate trait from phenotype in that a phenotype is a scalar trait. To illustrate this point, let us look at femur length, which we define as a trait; in contrast, increased femur length would be a phenotype as it now associates directionality to a trait. The trait information is organized into different trait classes by “categories” and “types.” A “trait category” is used to describe very general aspects of animal products or the processes by which the product is made (Figure 2). The top-level trait categories include:
Figure 2.

The trait categories associated with the Animal Trait Ontology (ATO), which include development, exterior, immune function, product quality, production, and reproduction. Each species (pig, cattle, and chicken) has the same 6 trait categories.
Development traits (growth) pertain to the physical growth of species;
Exterior traits (e.g., behavioral, anatomical) deal with traits that can be observed over time;
Immune function pertains to traits associated with the health of a species;
Product quality (e.g., marbling, milk traits) traits measure the quality of the animal products;
Production traits describe products (e.g., meat, milk, eggs) that are produced by the species; and
Reproduction traits are associated with the production of offspring.
A “trait type” describes physical or chemical properties of the animal products or features that can influence the process by which an animal product is made, or it describes types of measurements within each trait category; for example, fat deposition, flavor, and growth. “Trait names” are then defined with each trait type with more detailed information, known as “properties.” The current properties include: trait name, synonym, trait description, scale unit, measurement (how the trait was measured), custom name (lay or common name), and abbreviation (Figure 3).
Figure 3.

An example of the properties associated with the trait name “age at puberty.” The properties describe detailed information about a trait name.
Current ATO Statistics
The ATO is currently composed of 3 integrated ontologies, along with a global ontology that represents each of the 3 ontologies (pig, cattle, and chicken). The ontology currently comprises 809 terms. Most of the terms, a total of 463, are associated with the pig. At 223 terms, the cattle portion has the second largest number of terms. The chicken portion has the smallest number of associated terms with 124.
Methodology of the ATO
To date, we have used the following methodical process to determine which traits need to be stored in the ATO. First, it was critical to determine the top-level terms (described above) that were inclusive of all the prospective traits and livestock species that would be added to the ATO. The top-level terms were then broken down into sub categories to accommodate the various branching of the top-level terms. This process continued until the lower level terms were included.
At each level of the ATO, specific trait type information (e.g., synonym, trait description) was included for each term. The ATO allows 2 types of relationships between terms: “is_a” [e.g., backfat is a (kind of) fat] and “part_of” (e.g., the intestine is a part of the digestive system). Most of the information was obtained from published reports. At times, multiple published reports were used to annotate a single term. The methodology for entering data has been choosing a category such as reproduction in one species and upon completion, going to the same category in the next species. This strategy helps to create consistency between terms that are associated with multiple species. We draw on the expertise of the broader animal science community in specific areas (e.g., dairy science) to help validate the term and term information that is entered into the ATO.
Application of the Usefulness of the ATO
The usefulness of the ATO is demonstrated in the following hypothetical use case. A group of animal scientists is interested in the annotation of a new gene that they have been studying. This gene is associated with a reproductive trait in the pig that has been shown to have an effect on the “onset to puberty.” They are interested in the characterization of the phenotype associated with this gene as it corresponds to previous literature.
They begin their search with the GO Web site, but there is no information with regards to puberty in this ontology. The ATO is used to browse and search livestock species traits, so the group uses it to search for traits associated with the onset of puberty in the pig. From their previous experience of searching for this trait in the scientific literature, they had noticed several ambiguities in the naming (e.g., puberty, age at puberty, and onset of puberty) and definition of the term. They perform a search using the search interface and find the term “age at puberty” and they also notice that “onset to puberty” is listed as a synonym. They agree that the definition (the stage of adolescence in which an animal becomes physiologically capable of sexual reproduction) of the term corresponds to the phenotype of their trait. The group decides to use “age at puberty” as the phenotype of the new gene, which is the accepted standard in animal science for defining this trait.
During their search of the ATO, they also find that “age at puberty” is associated with pigs and cattle. Because the trait is stored in the ATO, it is likely that it has QTL associated with it in the other species because several of the traits have been mapped to particular QTL. This leads them to the literature, where they find QTL that have been mapped to this trait in the other species. This information gives them the insight to formulate hypotheses that could form the basis for a comparative study between the species. This scenario could be repeated with other traits that are different or similar to this trait, resulting in enhanced research.
DISCUSSION
Contributing to the ATO
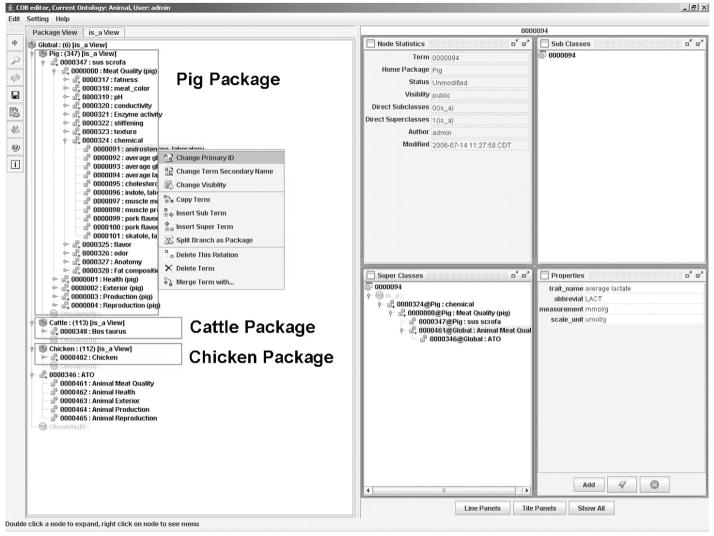
To develop ontologies, ontology editors must be used to facilitate the process. Ontology editors allow curators to browse, search, visualize, and edit ontologies. The OBO edit is the most commonly used bio-ontology editor (Day-Richter et al., 2007), but it was developed to support bio-ontologies that have a similar structure to the GO. For example, it supports fields such as trait name, definition, and synonyms, but it lacks the support for fields such as abbreviation, custom name, and scale unit. Consequently, the ATO is being developed by a different tool. The collaborative ontology building (COB) tool (Figure 4), which was developed in the Artificial Intelligence Laboratory of Iowa State University is an ontology editor with additional capabilities (Bao et al., 2006). The COB editor can support ontology building efforts among researchers in disparate locations by allowing curators to check out packages (or certain parts of the ontology). It also has a concurrent access and locking mechanism that prevents curators from editing a particular term at the same time. This COB editor can be downloaded to a local machine, which then gives a user access to a centrally located database. Depending on access rights, the user may be in edit or view-only mode.
Figure 4.
A snapshot of the interface of the collaborative ontology building (COB) editor.
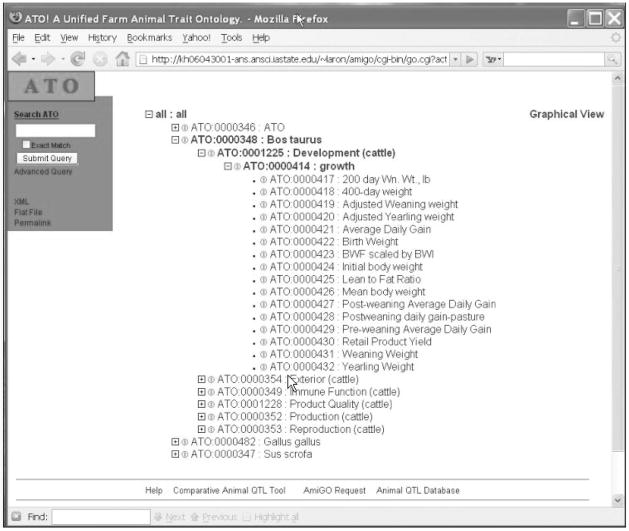
An ontology browser has been created to allow public access to the ATO (http://www.animalgenome.org/atoamigo; Figure 5). Ontology browsers allow users to browse and query ontology information. The browser is modeled after the AmiGO tool created by the GO consortium. The browser interface is linked to the ATO ontology, which is stored in a MySQL (version 12.22) relational database management system.
Figure 5.
Diagram of the Animal Trait Ontology (ATO) browser home page.
Future Growth
The ontology is expected to grow significantly in the upcoming years as more terms are created and more researchers become involved with the project. The home page for the ATO aims to serve as a central hub for the exchange of information, progress coordination, and as an end-product portal to the community. Additional features are expected to be implemented in the future such as hyperlinks to original sources of data and links to figures of traits that can be represented pictorially.
Eventually, the developers of the ATO would like to expand it and become a member of the Phenotype and Trait Ontology (PATO; The OBO Foundry, 2007) effort through the National Center for Biomedical Ontologies (NCBO). The goal of PATO is to deal with the difficulties and challenges of standardizing phenotype trait information across several species databases. Standardizing phenotypic trait information can help shed light into the relationships between genes, the environment, and phenotypes (C. Mungall, Lawrence Berkeley National Laboratory, Berkeley, CA, unpublished work).
The PATO is characterized by an entity and quality. An entity is described as a bearer of a quality (e.g., compound eye, blood, and wing growth). A quality is a property or attribute of an entity (e.g., cold, squamous, light sensitivity). The ATO and PATO will be mapped to each other by decomposing the trait names from the ATO into more elemental terms that are derived from PATO. For instance, the ATO term “ovulation rate” would be decomposed into the PATO term for “rate” (PATO:0000161) and the GO term for “ovulation” (GO:0001542). The advantage of this system is that the querying of phenotypes will become more comprehensive by forming a logical definition through the combination of the GO and PATO information. Unfortunately, decomposition of animal traits into PATO terms would render many terms unrecognizable by the livestock community. Thus, there is a need to link the ATO and PATO by establishing semantics-preserving mappings between PATO and ATO to maximize the utility of both systems.
A unified phenotype ontology would likely facilitate a smoother comparison of genes and phenotypes across organisms by integrating ontologies associated with species such as the mouse and rat. Researchers interested in contributing to the ATO are encouraged to visit the Web site (http://www.animalgenome.org/atoamigo) or contact James Reecy (jreecy@iastate.edu).
Conclusions
The ATO is the first ontology that deals with domesticated animal phenotypic traits. The ATO group will also collaborate with other ontology efforts such as PATO. The ATO will grow in the future by including more livestock species. The ATO is expected to have a profound impact on the descriptions of phenotypes associated with livestock species by forming a standard of nomenclature for animal scientist around the world. The ATO will enable scientist to browse and search current and future terms, allowing scientists to contribute individually to a large community effort.
Acknowledgments
This project was supported by funds from the USDA Cooperative State Research, Education, and Extension Service. This journal paper of the Iowa Agriculture and Home Economics Experiment Station, Ames, Iowa (Project No. NRSP-8) was supported by Hatch Act and State of Iowa funds.
LITERATURE CITED
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Hu ZL, Caragea D, Reecy J, Honavar V. A tool for collaborative construction of large biological ontologies. In: Database and Expert Systems Applications, 2006. DEXA ’06; Proc. 17th International Conference on Database and Expert Systems Applications.2006. pp. 191–195. [Google Scholar]
- Berners-Lee T, Hendler J, Lassila O. The semantic web. Sci Am. 2001;284:34–43. [Google Scholar]
- Blake JA, Bult CJ. Beyond the data deluge: Data integration and bio-ontologies. J Biomed Inform. 2006;39:314–320. doi: 10.1016/j.jbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Borst WN. Construction of engineering ontologies for knowledge sharing and reuse. PhD Diss. Dutch Graduate School for Information and Knowledge Systems, Enschede; Netherlands: 1997. [Google Scholar]
- Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM FlyBase Consortium. Flybase: Genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day-Richter J, Harris MA, Haendel M, Lewis S The Gene Ontology OBO Edit Working Group. Obo-edit—An ontology editor for biologists. Bioinformatics. 2007;23:2198–2200. doi: 10.1093/bioinformatics/btm112. [DOI] [PubMed] [Google Scholar]
- Galperin MY. Database issue. Nucleic Acids Res. 2008;36:D2–D4. doi: 10.1093/nar/gkm1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greally JM. Genomics: Encyclopaedia of humble DNA. Nature. 2007;447:782–783. doi: 10.1038/447782a. [DOI] [PubMed] [Google Scholar]
- Hamerton JL. IVth International Workshop on Human Gene Mapping. Hum Genet. 1977;36:I–I. [Google Scholar]
- Hu ZL, Dracheva S, Jang W, Maglott D, Bastiaansen J, Rothschild MF, Reecy JM. A qtl resource and comparison tool for pigs: PigQTLDB. Mamm Genome. 2005;16:792–800. doi: 10.1007/s00335-005-0060-9. [DOI] [PubMed] [Google Scholar]
- Hu ZL, Fritz ER, Reecy JM. AnimalQTLdb: A livestock QTL database tool set for positional QTL information mining and beyond. Nucleic Acids Res. 2007;35:D604–D609. doi: 10.1093/nar/gkl946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic K, Kellogg EA, Jaiswal P, Zapata F, Stevens PF, Vincent LP, Avraham S, Reiser L, Pujar A, Sachs MM, Whitman NT, McCouch SR, Schaeffer ML, Ware DH, Stein LD, Rhee SY. The plant structure ontology, a unified vocabulary of anatomy and morphology of a flowering plant. Plant Physiol. 2007;143:587–599. doi: 10.1104/pp.106.092825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keokamnerd T, Acton JC, Han IY, Dawson PL. Effect of ethanol rinse, Lactobacillus fermentum inoculation, and modified atmosphere on ground chicken meat quality. Poult Sci. 2007;86:1424–1430. doi: 10.1093/ps/86.7.1424. [DOI] [PubMed] [Google Scholar]
- McAlpine PJ, Shows TB. What’s in a name? Nature. 1990;346:616. doi: 10.1038/346616a0. [DOI] [PubMed] [Google Scholar]
- Schulze-Kremer S, Smith B. Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics. Vol. 4. John Wiley and Sons; New York, NY: 2005. Ontologies for the Life Sciences. [Google Scholar]
- Smith CL, Goldsmith CA, Eppig JT. The mammalian phenotype ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J, Clements D, Conlin T, Edwards P, Frazer K, Schaper K, Segerdell E, Song P, Sprunger B, Westerfield M. The Zebrafish Information Network (ZFIN): The zebrafish model organism database. Nucleic Acids Res. 2003;31:241–243. doi: 10.1093/nar/gkg027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The OBO Foundry. The OBO Foundry. [Accessed Sept. 19, 2007];2007 http://obofoundry.org/
- Wimmers K, Murani E, Te Pas MF, Chang KC, Davoli R, Merks JW, Henne H, Muraniova M, da Costa N, Harlizius B, Schellander K, Bll I, Braglia S, de Wit AA, Cagnazzo M, Fontanesi L, Prins D, Ponsuksili S. Associations of functional candidate genes derived from gene-expression profiles of prenatal porcine muscle tissue with meat quality and muscle deposition. Anim Genet. 2007;38:474–484. doi: 10.1111/j.1365-2052.2007.01639.x. [DOI] [PubMed] [Google Scholar]
- World Wide Web Consortium. The World Wide Web consortium (W3C) [Accessed Feb. 20, 2008];2007 http://www.W3.org/