Abstract
Human Cathepsin D (hCatD) is an aspartic peptidase with a low pH optimum. X-ray crystal structures have been solved for an active, low pH (pH 5.1) form (CatDlo) [Baldwin ET, Bhat, T.N., Gulnik, S., Hosur, M.V., Sowder, R.C., Cachau, R.E., Collins, J., Silva, A.M. and Erickson J.W. (1993) Proc. Natl. Acad. Sci. 90:6796–6800] and an inactive, high pH (pH 7.5) form (CatDhi) [Lee AY, Gulnik SV, Erickson JW. (1998) Nat Struct Biol 5:866–871]. It has been suggested that ionizable switches involving the carboxylate side chains of E5, E180, and D187 may mediate the reversible interconversion between CatDhi and CatDlo and that Y10 stabilizes CatDhi [Lee AY, Gulnik SV, Erickson JW. (1998) Nat Struct Biol 5:866–871]. To test these hypotheses, we generated single point mutants in “short” recombinant human pseudocathepsin D (srCatD), a model kinetically similar to hCatD [Beyer BM and Dunn BM. (1996) J Biol Chem 271:15590–15596]. E180Q, Y10F, and D187N exhibit significantly higher kcat/Km values (2, 3, and 6-fold, respectively) at pH 3.7 and pH 4.75 compared to srCatD indicating that these residues are important in stabilizing the CatDhi. E5Q exhibits a 2-fold lower kcat/Km compared to srCatD at both pH values indicating the importance of E5 in stabilizing the CatDlo. Accordingly, full time-course “pH-jump” (pH 5.5 to 4.75) studies of substrate hydrolysis indicate that E180Q, D187N, and Y10F have shorter kinetic lag phases that represent the change from CatDhi to CatDlo compared to srCatD and E5Q. Intrinsic tryptophan fluorescence reveals that the variants have a native-like structure over the pH range of our assays. The results indicate that E180 and D187 participate as an electrostatic switch that initiates the conformational change of CatDlo to CatDhi and Y10 stabilizes CatDhi by hydrogen bonding to the catalytic Asp 33. E5 appears to play a less significant role as an ionic switch that stabilizes CatDlo.
Cathepsin D (CatD) is a member of the pepsin family of aspartic peptidases. It has an acidic pH optimum (pH 2.8 to 4.0) (1, 2) and was originally detected in the endosomal and lysosomal compartments of higher eukaryotes (3). Traditionally, CatD was believed to play an important role in general proteolysis during bulk turnover at acidic pH (4). However, numerous studies showed that CatD plays other important physiological roles (5) some of which may involve restricted proteolysis. These include parathyroid hormone (6) processing, antigen processing (7–8), and the generation of angiostatin from plasminogen (9). CatD has also been implicated in pathological events including tumor growth and metastasis in breast and colorectal cancer (10–14). Consequently, CatD is a valuable target for the design of inhibitors.
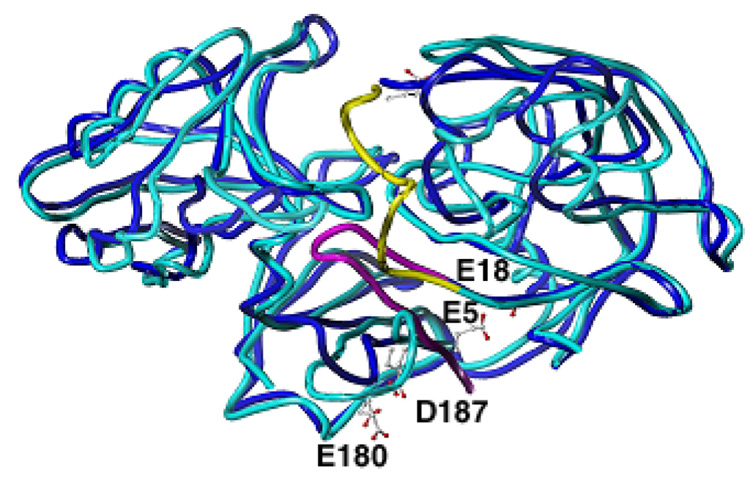
X-ray crystal structures obtained at pH 5.1 (CatDlo) of both native and pepstatin-inhibited forms of human CatD have been solved to design effective CatD inhibitors (15–16). More recently, the crystal structure of a catalytically inactive form of cathepsin D (CatDhi) was determined to 2.5 Å resolution obtained at pH 7.5. Structural comparison of CatDhi and CatDlo revealed significant conformational differences (Figure 1) (17).
Figure 1.
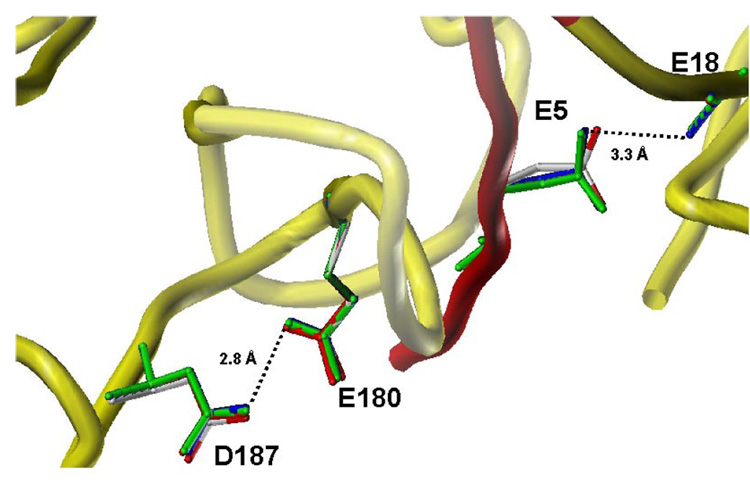
Superposition of the X-ray crystal structures of CatDhi and CatDlo. CatDhi is shown in cyan/yellow and CatDlo, blue/magenta. E5, E180, and D187 are shown in capped-stick.
One of the most pronounced structural differences between CatDlo and CatDhi is the relocation of the N-terminal segment (residues 3–7). This segment constitutes the first interdomain β-strand of a 6-stranded β-sheet that forms the base of the molecule in CatDlo. In CatDhi, however, it is rearranged to a flexible coil and inserted into the active site. It is suggested that this relocation of nearly 180° can be achieved by rotation about a hinge at Ala 13. An important consequence of N-terminal relocation into the active site is that access by substrate or inhibitor is prevented (17).
Since the structures of CatDhi and CatDlo were obtained at different pH values, the conformational change was proposed to be regulated by ionic switches. It was suggested that possible switch residues in CatDlo involve interactions between the two carboxylates of Glu 180 and Asp 187, and between the carboxylate of Glu 5 and the carbonyl oxygen of Glu 18. The carboxylate side chains of Glu 180 and Asp 187, as well as Glu 5 and the carbonyl of Glu 18, are within hydrogen bonding distance in CatDlo but are over 8 Å apart in CatDhi. Also, Glu 180, Asp 187, and Glu 5 are conserved among CatD sequences from many different species ranging from invertebrates to mammals.
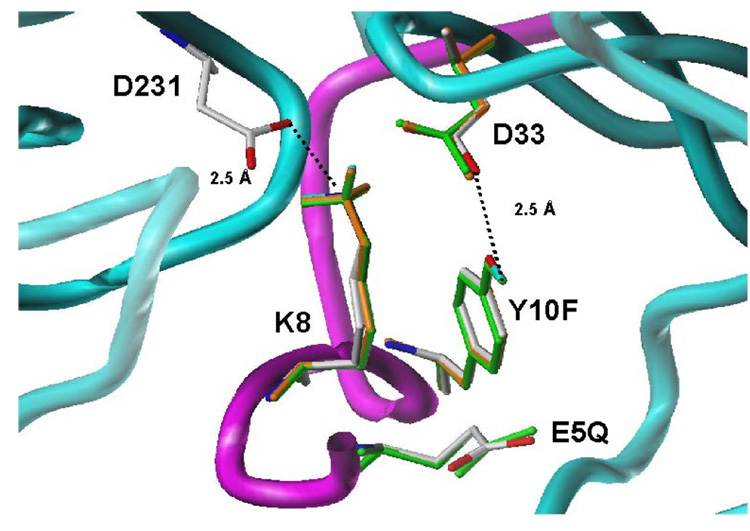
It was hypothesized that at low pH, hydrogen bonding between the switch residues would help stabilize CatDlo. However, an increase in pH would result in the loss of protons and charge repulsion between the carboxylates of Glu 180 and Asp 187, as well as between the carboxylate of Glu 5 and carbonyl of Glu 18. This would destabilize the active, low pH conformation. Consequently, the mechanism suggests that an increase in pH induces an electrostatic repulsion between the proposed switch carboxylates that results in a concerted set of conformational changes including the relocation of the N-terminus into the active site. The rearrangement of the N-terminal strand would eliminate the unfavorable electrostatic interactions between the charged carboxylates. In CatDhi, the N-terminal segment is inserted into the active site, where both catalytic aspartates are likely ionized. The positively charged Nε atom of Lys 8 and the hydroxyl group of Tyr 10 form a salt bridge and hydrogen bond interaction, respectively, with the catalytic aspartates to stabilize the inactive, CatDhi. Reactivation by acidification could occur due to the protonation of one of the two catalytic aspartates, thus weakening their electrostatic interactions with Lys 8. This would permit the release of the N-terminal peptide allowing it to relocate to its position as the sixth strand of the antiparallel β–sheet. The result is an active site that has full accessibility to substrate (17).
To test the role of the putative ion switch residues, E5, E180, and D187, in mediating the pH-dependent conformational change of CatD, we generated single point mutations in srCatD in which nonionizable residues were introduced at these positions. This recombinant model retains six N-terminal amino acids after zymogen processing compared to the native enzyme, and it is kinetically similar to the native enzyme (18). Replacing E5, E180, and D187 with nonionizable residues effectively removes charge repulsion due to ionization at the putative electrostatic switches. Additionally, the role of hydrogen bonding between Y10 and Asp 33 in stabilizing CatDhi was examined by replacing Y10 with phenylalanine. Here we present the results that indicate that E5, E180, and D187 function as ion switches that mediate the pH-dependent conformational change of srCatD. Finally, we show that Y10 forms an important hydrogen bond to Asp 33 that stabilizes the high pH, inactive CatDhi.
Material and Methods
Reagents
Native human liver Cathepsin D was purchased from Athens Research and Technology, Inc. (Athens). Competent cells were purchased from Stratagene (La Jolla, CA). Sequenase kit was purchased from United State Biochemical Corp (Cleveland). Quik-change™ Site Directed Mutagenesis Kits were from Stratagene. Restriction enzymes were from New England Biolabs (Beverly, M A). Synthetic oligonucleotides were obtained from LifeTechnologies, Inc. (Gaithersberg, MD). Pepstatin and oxidized glutathione were purchased from Sigma (St. Louis) and other biochemicals came from Fisher Scientific (Pittsburgh, PA). Chromogenic substrates were synthesized by the solid phase method using an Applied Biosystems Model 430A or 432A in the UF-ICBR Protein Chemistry Core Facility. The purity of the peptides (≥95%) was verified by HPLC and by using a Perceptive Biosystems Voyager™ RP mass spectrometer (MALDI-TOF). N-terminal sequence analysis was also performed by the UF-ICBR Protein Chemistry Core Facility with Applied Biosystems 470A and 473A Protein Sequencers. Q-Sepharose Hi-trap ion-exchange chromatography columns were purchased from Amersham-Pharmacia. Pepstatinyl-agarose was synthesized according to the method of Huang et al. (19). Stock peptide solutions were made in filtered distilled water and quantified by amino acid analysis. The purity of the peptides (≥95%) was verified by HPLC and MALDI-TOF analysis. Native human liver cathepsin D was dissolved in ice cold distilled water and aliquots were stored in 10% glycerol at −20°C for future use.
All routine DNA manipulation procedures were performed according the manufacturer’s protocol (Stratagene). All plasmid and PCR purification procedures employed kits purchased from Quiagen. These kits utilize a proprietary anion exchange column to efficiently purify plasmid DNA.
Mutagenesis of “Short” Recombinant Pseudocathepsin D
Mutagenesis of “short” recombinant pseudocathepsin D was carried out using the Quik-change™ Site Directed Mutagenesis Kit from Stratagene using PfuTurbo™ DNA polymerase II. The plasmid, pTCPSD2, that contains the entire coding region for srCatD was used as the template DNA (18). The following oligonucleotide primers and their reverse complements were used: E5Q 5’GGCCCATTCCCCAGGTGCTCAA3’, E180Q 5’TGGGGGTCAGCTGATGCT3’, D187N 5’TGGCACAAACTCCAAGTATTA3’, Y10F 5’CCCATTCCCGAGGTGCTCAAGAACTTCATGGAC3’. The mutagenesis reactions contained 5 to 50 ng of double stranded DNA template, 125 ng of the forward oligonucleotide primer, 125 ng of reverse oligonucleotide primer, dNTP mix, reaction buffer, and 1 µL PfuTurbo DNA polymerase in a final volume of 50 µL. The PCR reaction was carried out in a Gene Machine II Programmable Thermal Controller (USA Scientific Plastics). The following temperature cyles were used: 94°C, 30 seconds; 55°C, 60 seconds; 68°C,10 minutes. These steps were repeated for 17 cycles. To confirm the presence of the intended mutation and to ensure against presence of spurious mutations, the entire region encoding for “short” recombinant pseudocathepsin D was sequenced at the University of Florida ICBR DNA Sequencing Core Laboratory using ABI Prism Big Dye Terminator cycle sequencing protocols (part number 4303153) developed by Applied Biosystems (Perkin-Elmer Corp., Foster City, CA). Constructs were resequenced after all transformation steps.
Expression, refolding, and purification
For overexpression, mutant constructs were transformed into E. coli BL21(DE3)pLysS cells (Stratagene). Protein expression was induced by the addition of IPTG as previously reported (18). To obtain active, mature srCatD, inclusion bodies were isolated, solubilized, and refolded as previously described (18) with the following exceptions. The solubilized inclusion bodies were refolded without stirring and time of refolding was experimentally determined. Additionally, activation of the refolded material to convert the zymogen to the mature form was done for predetermined times. The refolded/activated protein solution was filtered by a slow vacuum to remove particulate material prior to loading it onto the column. The C 16/20 column (Amersham-Pharmacia) containing 1 mL of resuspended pepstatinyl-agarose was equilibrated with 0.1 M sodium formate pH 3.7, 0.4 M NaCl, 0.05% Brij-35. The refolding solution was then applied to the affinity matrix column at room temperature by gravity flow. The column was washed with at least 5 mL of 0.01 M sodium formate pH 3.7, 0.4 M NaCl, 0.05% Brij-35 then eluted with cold 20 mM Tris-HCl pH 8.0, 0.4 M NaCl and 0.5% Brij 35. Column fractions exhibiting catalytic activity were pooled after analysis and stored in 10% glycerol at −20°C. To desalt the active protein after affinity chromatography, the sample was dialyzed against 4 L of 10 mM Tris-HCl pH 8.0, for 4 hours, then overnight at 4°C in fresh buffer. An ion-exchange chromatography step was included to purify the mature form of the enzyme. A 1 mL HiTrap Q Sepharose HP (Amersham Pharmacia), an anion exchanger, was equilibrated with at least 5 column volumes of 10 mM Tris-HCl pH 8.0 (buffer A). The desalted material was then applied to the column at a flow rate of 1 mL/min. Next, the column was washed with 5 mL buffer A. The protein was eluted with a salt gradient of 0 to 80 mM. Column fractions exhibiting catalytic activity were pooled after analysis and stored in 10% glycerol at −20°C.
Intrinsic tryptophan fluorescence
All fluorescence experiments were conducted on a T-format QuantaMaster QM-1 Luminescence Spectrometer equipped with a 75 watt xenon lamp, computer controlled excitation/emission monochromators (0.2-meter Czerny-Turner type) tunable from 0 to 1150 nm, and a Hamamatsu R928 photomultiplier tube with low light level detection from 185 to 900 nm (kindly provided by Dr. Linda Bloom, University of Florida). The protein sample was centrifuged at 14,000 r.p.m. for 20 minutes, 4°C to pellet any precipitate. 200 nM enzyme was pre-incubated for 5 minutes, 37°C in 0.2 M buffer (sodium formate pH 2.5 to 4; sodium acetate pH 4.5 to 6.0). An excitation wavelength of 295 nm was used to selectively excite tryptophan residues thus excluding the excitation of tyrosine moieties. Emission spectra were collected between 300 to 400 nm using a 3 nm band-pass. All measurements were made using freshly purified protein that had not been subjected to freeze/thaw or additives. Blank spectra were recorded and subtracted from measurements. Additionally, the buffers were shown not to exhibit pH-dependent spectra. Next, the sample was placed into an 80 µL quartz cuvette (Hellma) housed in a jacket maintaining a constant temperature of 37°C and the spectrum recorded.
Spectrofluorometric analysis
Enzyme hydrolysis of the fluorogenic substrate Mca-Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys(Dnp)-D-Arg-NH2 at the Phe-Phe bond was spectrofluorometrically monitored on a PerSeptive Biosystems CytoFluor Series 4000/TC Multi-Well Plate Reader. The substrate contains a fluorophore, 7-methoxycoumarin-4-yl acetyl) (Mca) moiety and a quenching 2,4-dinitrophenyl (Dnp) group (20). For all kinetic assays, eight wells containing buffer, water, and enzyme were preincubated for 3 minutes at 37°C. At the end of the incubation, the mixtures were transferred to wells containing varied substrate concentrations. After a three second mixing time of enzyme, buffer and substrate, samples were excited at 360 nm (40 nm bandwith) and emission monitored at 460 nM (40 nm bandwidth). The Km and Vmax values were determined from the initial rates using Marquardt analysis (21) and the equation v=Vmax[S]/(Km+[S]). The observed rates in AU*s−1 were converted to M*s−1 by dividing by the total change in fluorescence for complete hydrolysis of a known concentration of substrate.
Active site titration and kcat
Inhibitor stock solutions used for active site titrations were made in ethanol:acetic acid (90:10) and quantitated by amino acid analysis. The amount of active enzyme in each assay was determined by competitive inhibition with the potent active-site inhibitor, pepstatin (Isovaleryl-Val-Val-Sta-Ala-Sta) (Sigma). The resulting curve was fitted with the Dixon equation on the Enzyme Kinetics module of SigmaPlot.
kcat values (sec−1) were determined by the equation kcat=Vmax(AU/sec)/(ΔF/[S]*[E]) were ΔF = total change in fluorescence (ΔF = total change in fluorescence with 10 µM fluorescent substrate ([S] = substrate concentration, [E] = total enzyme concentration (molar)). The standard deviations of the kcat and kcat/Km values were propagated using equations derived by standard procedures for independent errors (22).
pH-dependent stability
Enzyme was preincubated for 10 minutes at 37°C in the following 100 mM buffers: glycine-HCl pH 2.0 to 3.0; sodium formate pH 3.75; sodium acetate pH 4.5; MES pH 5.5 to 7.0; Tris-HCl pH 8.3 to 9.3; CAPS pH 10.7 to 11.7. After 10 minutes, the pH was recorded using a microelectrode pH probe, calibrated with two pH points with 37°C standard solutions (Microelectrodes, Inc.), and the enzyme was added to 0.2 M sodium formate pH 3.8 at 37°C. The solution was incubated at 37°C for an additional 3 minutes and then added to 30 µM substrate. Initial rates of substrate cleavage were monitored for at least six identical reactions. The final pH was recorded for all points. All pH points had a final pH between pH 3.8 and 4.0.
pH-equilibrium jump
Enzyme was preincubated in 0.5 M sodium formate buffer pH 5.5, 37°C, 3 minutes. It was determined that these conditions are sufficient to equilibrate the enzyme to the closed form. The reaction was initiated by addition of 100 nM enzyme to 0.2 M buffers (sodium formate pH 3.5 to 4.0; sodium acetate pH 4.2 to 5.0) containing 14 µM substrate, 37°C. Hydrolysis of substrate was monitored as previously mentioned. The final pH of the reaction was immediately recorded. For “no jump” reactions, enzyme was preincubated in 0.5 M sodium formate pH 3.7 and then added to 0.2 M sodium formate pH 3.7 and assayed, as previously described.
Product analysis
Reactions were terminated in the lag phase, linear phase, and at completion of substrate consumption by addition of Tris-HCl, pH 9.0 at a final concentration of 300 mM. 50 % acetonitrile was added and the reactions were incubated overnight at 25°C. Products of substrate digestion were separated by HPLC and peaks were analyzed by mass spectroscopy (UF-ICBR Protein Chemistry Core Facility).
Molecular modeling
Models were built with the high resolution three-dimensional structure of human liver cathepsin D (15) using SYBYL (version 6.7, supplied by Tripos). Point mutants were generated by using the “Modify Substructure” command. A 4 Å radius about the point mutation was energy minimized for 100 iterations to minimize steric clashes and unfavorable interactions. Distance measurements were made using the “Measure” command in the analyze menu of SYBYL. Atoms within a 4 Å radius of the point mutation were inspected for possible interactions.
Results
Purification of srCatD and variants
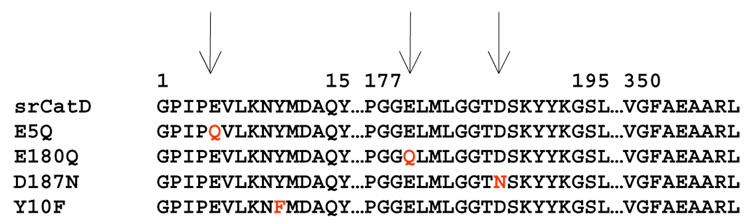
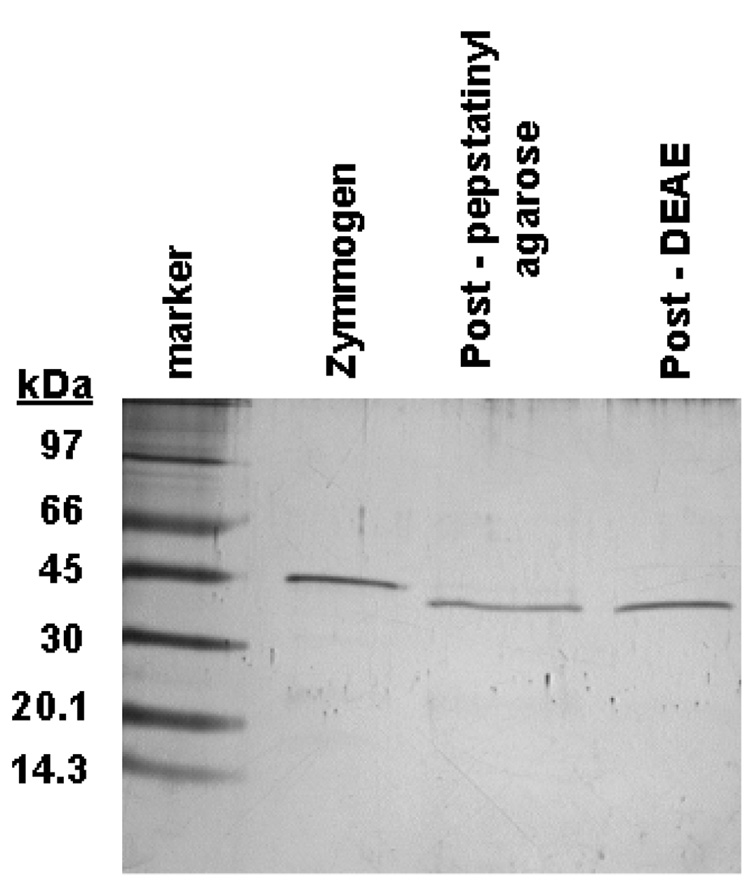
The proenzyme form of srCatD and variants (Figure 2) was over-expressed in E. coli and isolated as insoluble inclusion bodies. Refolding time-course experiments and activation time-course experiments were performed to optimize the purification yield of mature, active enzyme (data not shown). Due to poor refolding efficiency, the investigation was limited to the variants E5Q, E180Q, D187N, and Y10F. A very pure sample of mature enzyme was obtained following affinity and ion exchange chromatography, as indicated by a single band on a silver stained SDS-PAGE (Figure 3) and N-terminal sequencing. Analysis of N-terminal processing for srCatD and variant enzymes shows that srCatD, E5Q, E180Q, D187N, and Y10F all processed at the same cleavage site during the conversion of the zymogen form the mature form (Table 1).
Figure 2.
Amino acid sequence alignment of srCatD and variants.
Figure 3.
SDS-PAGE analysis of purification of refolded "short" rHuCatD from the refolding mixture. The far left lane corresponds to molecular weight markers. Lane #1 represents full length rHuCatD from the refolding mixture Lane #2 shows the pepstatinyl-agarose fraction. Lane #3 illustrates the ion-exchange purified “short” pseudo rHuCatD migrating to about 42 kDa. The single band indicates purity to near homogeneity as judged by silver staining.
Table 1.
N-terminal sequence alignment for srCatD and variants.
| Enzyme | N-terminal Sequence |
|---|---|
| “short” rHuCatD | …FRLVTEG |
| E5Q | …FRLVTEG |
| E180Q | …FRLVTEG |
| D187N | …FRLVTEG |
| Y10F | …FRLVTEG |
Kinetic Analysis
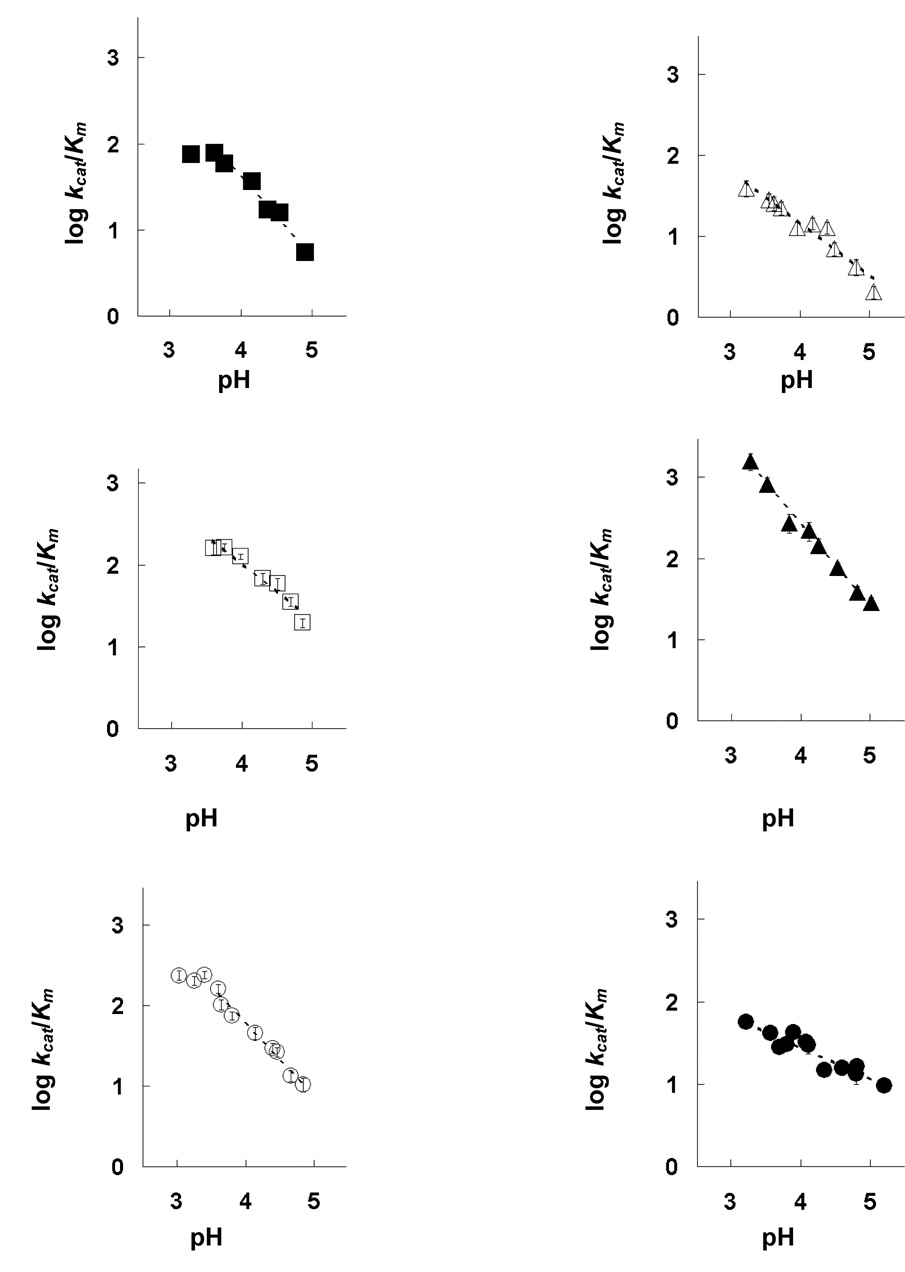
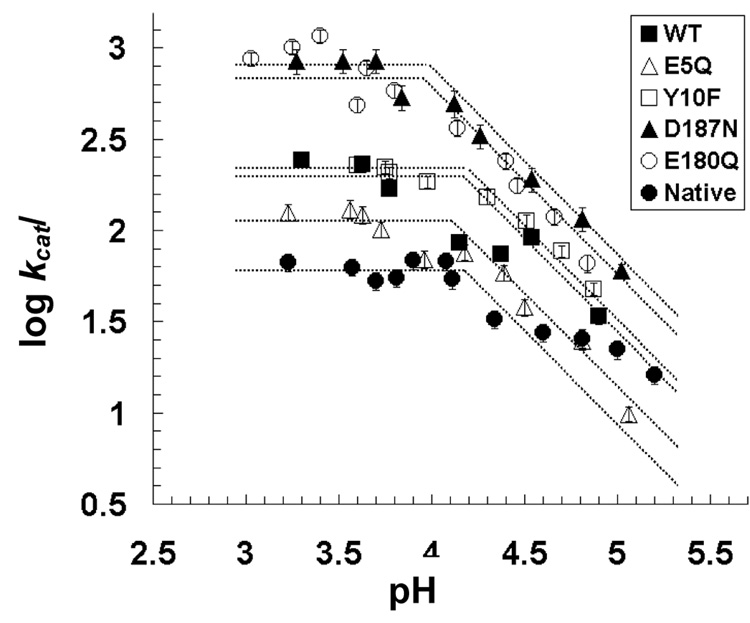
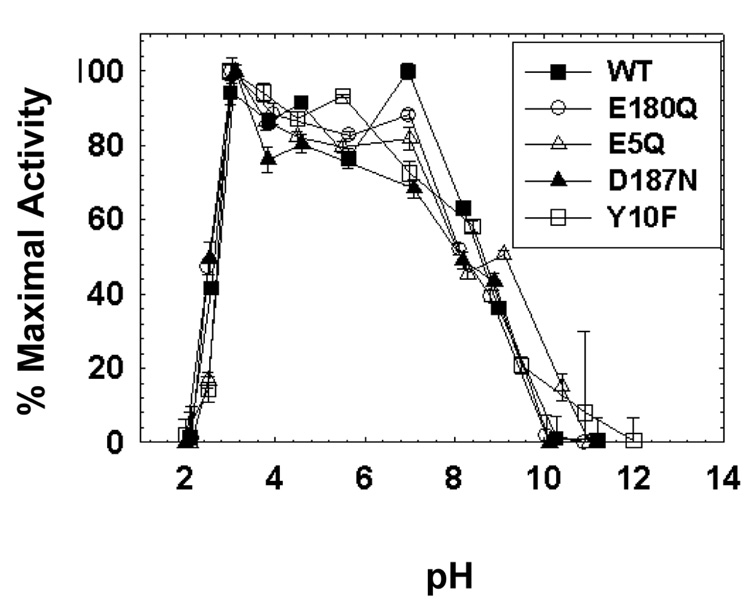
The pH-dependent Michaelis-Menten parameters were determined for srCatD and variants. These data demonstrate that srCatD and variants exhibit significant differences in their specificity constants, kcat/Km, as a function of pH (Figure 4). Kinetic characterization indicates that E180Q, Y10F, and D187N have a higher specificity constant, kcat/Km, (two-fold, three-fold, and six-fold, respectively) at pH 3.7 and pH 4.75 compared to srCatD. Conversely, E5Q exhibits a two-fold lower kcat/Km compared to srCatD at both pH values. Interestingly, native hCatD exhibits a two-fold lower kcat/Km at pH 3.7 but a two-fold higher kcat/Km at pH 4.75. Additionally, the logarithmic plot of kcat reveals that srCatD and mutants all share unique pH-profiles (Figure 5). Table 1 shows the Michaelis-Menten parameters at pH 3.7 and pH 4.75. At pH 3.7, the open conformation of the enzyme should be favored and at pH 4.75, the closed conformation should be favored. The values for kcat were determined by fitting the data with straight lines and extrapolating to the y-axis. From this plot pKa values are defined for a single ionization event with pKa values in the range of 4.0 to 4.2 (Table 3).
Figure 4.
The pH-dependency of kcat/Km for srCatD, nHuCatD, and variants. srCatD is plotted as closed squares; E5Q, open triangles; Y10F open squares; D187N, closed triangles; E180Q, open circles; native cathepsin D, closed circles.
Figure 5.
The pH-dependency of kcat for srCatD, nHuCatD, and variants. srCatD is plotted as closed squares; E5Q, open triangles; Y10F open squares; D187N, closed triangles; E180Q, open circles; native cathepsin D, closed circles.
Table 3.
pKa values for srCatD and variants.
pH-equilibrium jump kinetics
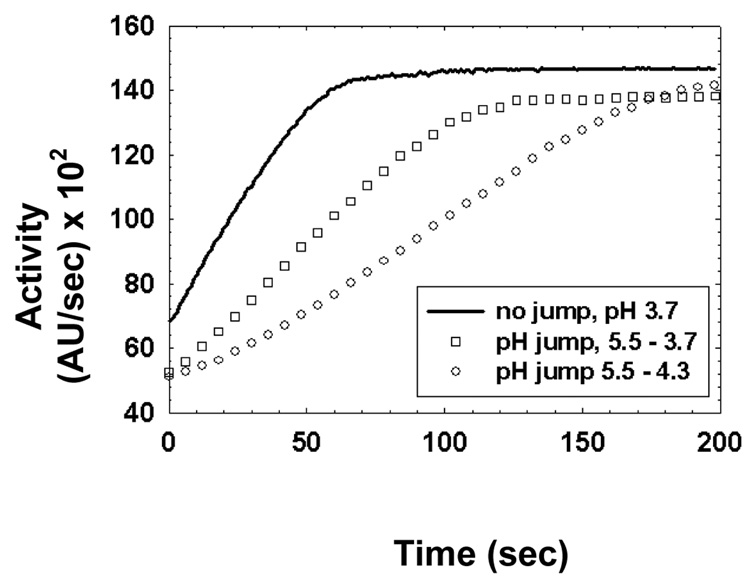
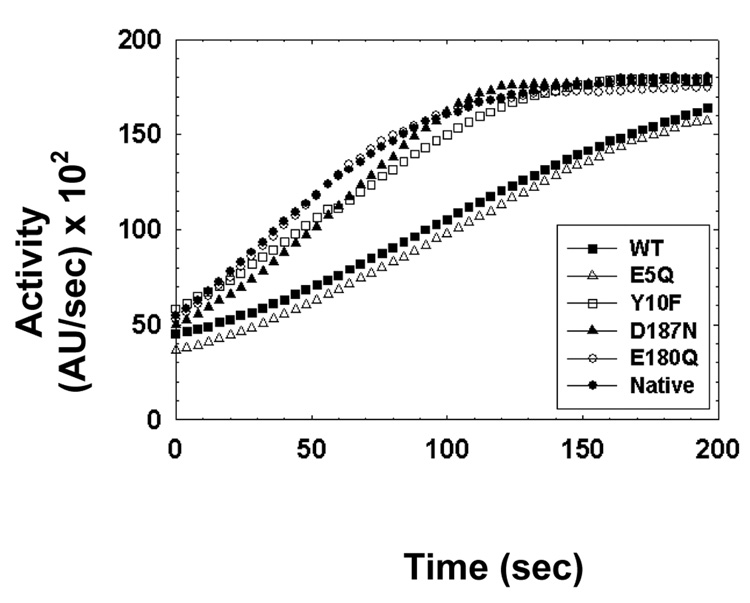
A kinetic lag phase was discovered by performing pH-equilibrium jump kinetic reactions. Figure 6 shows that srCatD exhibits a kinetic lag phase. Moreover, as the final pH of the reaction is increased, the kinetic lag lengthens indicating the length of the lag phase is pH-dependent. Most importantly, however, is that when the enzyme is pre-incubated at low pH (no “jump” reaction) such that the majority of the molecules are in the open conformation, the lag phase is abolished. Figure 7 shows the pH-equilibrium jump studies conducted with the variants. These data show that native CatD and the variants E180Q, D187N, Y10F all exhibit shorter lag phases compared to srCatD and the E5Q variant.
Figure 6.
Full progress pH-“jump” kinetics for srCatD. The pH-jump reactions were jumped from pH 5.5 to either pH 3.9 (open square) or pH 4.5 (closed square). The “no jump” reaction (solid line), enzyme was preincubated at pH 3.7 and assayed at pH 3.7. Data points at 5 second intervals were omitted for clarity.
Figure 7.
The pH-“jump” kinetics for srCatD, nHuCatD, and variants. The reactions were jumped from pH 5.5 to pH 4.5.
Product analysis
To rule out alternative cleavage of the fluorescent substrate during the lag phase of the kinetic reaction and to demonstrate correct substrate cleavage at the Phe-Phe bond, product analysis was performed. srCatD showed correct substrate cleavage at the Phe-Phe bond during the kinetic lag phase and during the linear phase of the reaction (data not shown).
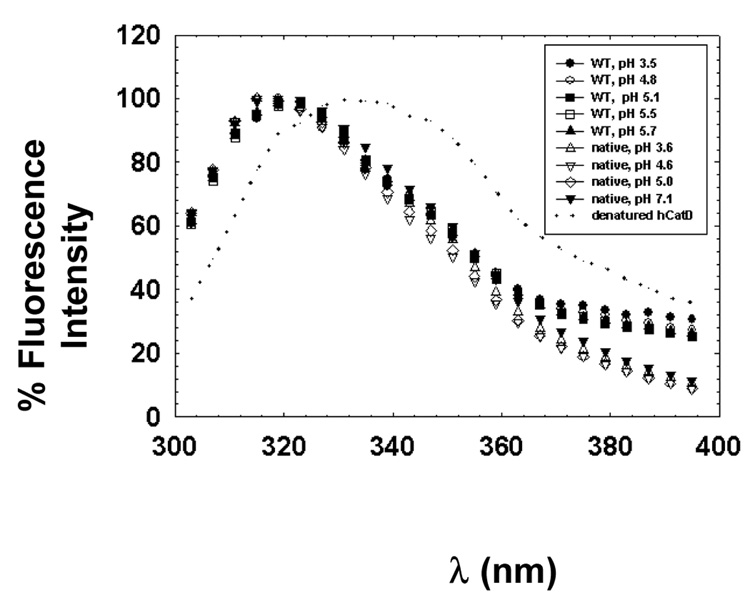
Intrinsic tryptophan fluorescence
Intrinsic tryptophan fluorescence pH-titration experiments were utilized to assay the pH-dependent stability of srCatD. Figure 8 shows the pH-dependent emission spectra for srCatD, native CatD, and denatured enzyme. These data indicate a clear similarity in the peak maxima of about 320 nm between the recombinant and native material. Additionally, the denatured material exhibits a red shift in the emission spectrum.
Figure 8.
Tryptophan fluorescence of srCatD and nHuCatD. srCatD is shown in heavy solid lines, native CatD is represented as thin lines, and denatured CatD is shown as a dotted line.
pH-dependent stability
The pH-dependent stability of srCatD and variant enzymes was explored to corroborate the stability of the enzymes over the pH range of titration experiments. Figure 9 shows that srCatD and variants share virtually identical pH-stability profiles. These data show that srCatD and variant enzymes are stable over the pH range of 3.0 to 7.5 with 50% recovery of activity at pH 2.5 and 8.5.
Figure 9.
The pH-dependent stability of srCatD and variants. srCatD is plotted as closed squares; E5Q, open triangles; Y10F open squares; D187N, closed triangles; E180Q, open circles.
Molecular modeling of variants
Molecular modeling aided in the interpretation of the effect of mutation on the structure of the enzyme. Both the low (Figure 10) and high pH form (Figure 11) of the variants were modeled and the resulting structure energy minimized using the program SYBYL. The models were superimposed on either the high pH or low pH crystal structure. Inspection of the superpositions of the low pH structure indicates that a hydrogen bonding distance of 2.8 Å between position 180 and 187 is maintained in E180Q, D187N, and E5Q. However, the hydrogen bonding distance between the carbonyl of E18 and the side chain of position 5 is increased from 3.1 Å to 3.3 Å in all of the variants. Inspection of the high pH model indicates that the Lys 8 to Asp 231 distance is maintained at 2.5 Å with theY10F variant. Examination of the models suggests that no new interactions are made as a result of mutation. Figure 10 and Figure 11 show the retention of native-like structure and interactions exhibited by the variants.
Figure 10.
Molecular models generated for E5Q, E180Q, and D187N. E5Q, shown in green, E180Q, red, and D187N, blue, are superimposed on the crystal structure for CatDlo (grey with red caps). All residues rendered are within a 4 Å radius of the mutated residue.
Figure 11.
Molecular models generated for E5Q andY10F. E5Q is shown in green and Y10F, orange, are superimposed on the crystal structure for CatDhi (grey with red caps). All residues rendered are within a 4 Å radius of the mutated residue.
Discussion
This study examined the roles that E5, E180, D187, and Y10 play in mediating the pH-dependent conformational change of srCatD. In the case of E5Q, E180Q, and D187N, a carboxylate was substituted with carboxamide functionality, thus effectively removing a negative charge at those positions. This was done to remove charge repulsion between the putative ion switch residues at higher pH values. This permitted addressing whether charge repulsion between the ionic switches is involved in the mechanism of the pH-dependent conformational change. The Y10F variant was generated to test the function of Y10 as a H-bond donor to Asp 33 in the high pH, inactive conformation. Substituting Y10 with phenylalanine removes the hydroxyl moiety of Tyr10 thereby removing H-bonding to Asp 33.
Kinetic characterization of the variants showed interesting differences in the specificity constant, kcat/Km. This revealed that the introduced mutations altered substrate binding and/or catalytic turnover. This finding is consistent with the hypothesis that the introduced mutations will favor one conformation over the other. Considering that the relocation of the N-terminus, as a consequence of conformational change, either completely occludes substrate from the active site (high pH conformation) or vacates the active site resulting in catalytic machinery that is free for substrate binding and turnover, it is reasonable to predict that mutants with a modified equilibrium will exhibit changes in kcat/Km relative to srCatD. For example, if the mutation results in an enzyme with a conformational equilibrium shifted to the open form, then it may yield a higher specificity constant at a particular pH compared to srCatD. A mutation that results in an enzyme with a conformational equilibrium shifted to the closed form may exhibit a lower specificity constant at a particular pH.
Comparison of kcat/Km values at pH 3.7 and pH 4.75 (Table 2) indicates that D187N, E180Q, and Y10F are more catalytically efficient enzymes compared to srCatD and native CatD. D187N shows about a six-fold increase in kcat/Km; E180Q shows approximately a two-fold increase, and Y10F exhibits about a three- fold increase over srCatD at both pH 3.7 and 4.75. Conversely, E5Q shows a two to three-fold decrease in kcat/Km compared to srCatD. Native hCatD exhibits a two-fold lower kcat/Km at pH 3.7 but a two-fold higher kcat/Km at pH 4.75. The differences are mainly reflected in the values for kcat which are dramatically different. The increased kcat/Km values for D187N, E180Q, and Y10F indicated that the conformational equilibrium for these variants is shifted to the open form. This results in more molecules that are open and available to react with substrate. Consequently, an increase in kcat is manifested with D187N, E180Q, and Y10F. The fact that D187N, E180Q, and Y10F exhibit higher kcat/Km values at pH 3.7 compared to srCatD suggests that even at the pH optimum for the enzyme, the conformational equilibrium for srCatD is not shifted completely to the open conformation. The opposite phenomenon is apparent with the E5Q. The lower kcat/Km value for this variant indicates it has a conformational equilibrium shifted to the closed form compared to srCatD.
Table 2.
pH-dependent Michaelis-Menten parameters for srCatD and variants.
| Enzyme | kcat pH 3.7 (s−1) | kcat/Km pH 3.7 (µM−1s−1) | kcat pH 4.75 (s−1) | kcat/Km pH 4.75 (µM−1s−1) |
|---|---|---|---|---|
| “short” rHuCatD | 200 ± 20 | 79 ± 10 | 46 ± 10 | 8 ± 1 |
| E5Q | 117 ± 20 | 24 ± 5 | 22 ± 5 | 4 ± 1 |
| E180Q | 708 ± 90 | 126 ± 20 | 89 ± 10 | 13 ± 5 |
| D187N | 813 ± 130 | 562 ± 70 | 123 ± 20 | 50 ± 10 |
| Y10F | 240 ± 20 | 257 ± 20 | 62 ± 10 | 26 ± 5 |
| Native CatD | 66 ± 10 | 40 ± 5 | 18 ± 5 | 15 ± 5 |
Noteworthy is the interesting pH-dependent kinetic behavior of srCatD compared to the native enzyme. srCatD exhibits a two-fold higher kcat/Km at pH 3.7 but a two-fold lower kcat/Km at pH 4.75 compared to native hCatD. A possible explanation for this is the presence of 6 additional N-terminal residues on srCatD that are absent in native hCatD. We speculate that these residues may interact with the enzyme thereby stabilizing both the high pH and low pH structures of srCatD. In other words, the additional stability at both high and low pH may shift the conformational equilibrium of srCatD to CatDlo, at low pH, and CatDhi at high pH, relative to native hCatD. This would result in srCatD exhibiting higher kcat/Km values at low pH and a lower kcat/Km at high pH.
Effects of pH on kcat also reflect acid-base group involvement in the catalytic steps of substrate to product conversion; that is, the ionization of the enzyme-substrate complex (23). By analyzing the plot of the value of the kinetic constant, kcat, on a logarithmic scale as a function of pH, additional information regarding ionization events may be determined. For a single titration event, the plot will appear as the superposition of two linear functions, one with a slope of zero and the other with a unit slope value. The pKa is defined by the point of intersection for the two straight lines drawn through the data point in the regions of minimal curvature of the plot. Additionally, the number of acid-base groups can be estimated. The slope of the line in the transition region of the plot reflects the number of ionizable groups that are titrated. A change in slope with a −1 value is representative of a single ionization event (24). The pKa value of approximately 4.0, determined from Figure 5, most likely reflects the titration of a single carboxylate. We believe that these values represent the ionization of the second catalytic aspartic acid, since the substrate does not contain ionizable groups in this pH range. Additionally, this value is consistent with the pKa values determined for the second active site carboxylate of closely related aspartic peptidases gastricsin (25), rhizopuspepsin, and pepsin (26). The native material, however, exhibits a unique pH profile. Points above pH 5.0 could define a second ionization event; however, data collection in this region is very difficult and we were unable to obtain sufficient data to unambiguously define a second pKa.
A kinetic lag phase was discovered when srCatD was pre-incubated at a high pH then assayed at a low pH. Moreover, when the enzyme is pre-incubated at a low pH, in order to favor the open conformation of the enzyme, no lag phase is apparent. This strongly suggests that the lag phase represents the time it take the enzyme to undergo the conformational change from the closed, inactive form to the open, active form. Additionally, product analysis indicated that the fluorescent substrate was not cleaved during the lag phase. This ruled out the possibility that the enzyme cleaved the substrate at a bond that would not yield a detectable change in fluorescence. The variants were also tested for a kinetic lag phase. These experiments lucidly show that the variants E180Q, D187N, and Y10F, as well as native hCatD all exhibit shorter kinetic lag phases compared to srCatD and E5Q.
The pH-jump experiments provide insightful qualitative data regarding the conformational equilibrium constant, Keq, for the isomerization of CatDhi and CatDlo. The results of the pH-jump experiment were consistent with the pH-dependent Michaelis-Menten data that clearly showed a significant difference in the conformational equilibrium between CatDhi and CatDlo. For example, E180Q, D187N, Y10F, and native CatD all exhibit much shorter kinetic lag phases compared to srCatD and E5Q. These data indicate that E180Q, D187N, and native CatD have a conformational equilibrium shifted to the open form. It is interesting that native CatD exhibited no lag phase compared to srCatD when jumped to pH 4.5. It should be noted that the native material exhibits a kinetic lag phase when the reaction pH is increased to pH 5.5 (data not shown). There are several possible explanations for the difference in the lag phase duration. For example, the native material is a two-chain molecule that is glycosylated whereas srCatD is a single chain, non-glycosylated enzyme. A more likely explanation is that native CatD lacks the additional 6 N-terminal residues (Phe-Arg-Leu-Val-Thre-Glu) that perhaps stabilize the open and closed form of “short” hCatD at both low and high pH values, respectively. These data are consistent with the pH-dependent Michaelis-Menten data that indicate that E180Q, D187N, Y10F, and native exhibit a more open conformation at pH 4.75 compared to srCatD. However, disparity exists between the two experiments for the E5Q variant. The two-fold lower kcat/Km value at pH 4.75 is not readily evident in the pH-jump experiment that shows E5Q with a full-progress kinetic trace similar to srCatD. Perhaps the high substrate concentration used in the experiment pulls the equilibrium towards the open conformation, thus masking the effect of the E5Q mutation.
Another unexpected aspect of this kinetic hysteresis is the lengthy half-times of the kinetic lag phase that is on the time scale of seconds. This might suggest that the nature of the conformational change is not simple. Importantly, half-times for lag phases on the second time scale that represent isomerization events have been documented. These include isomerization events for threonine deaminase (B. subtilis), DPNH oxidase (M. tuberculosis), UDPG hydrolase (E. coli), and glyceraldehyde 3-phosphate dehydrogenase (yeast) (27). Perhaps the lengthy duration of the lag phase implies that a complex set of molecular interactions are involved in the transition from one conformation to the other. This is consistent with the high degree of segmental movement, a rigid-body rotation of 7.2° and a translation of 1.5 Å that accompanies the relocation of the N-terminal segment (18).
The first indication that the variants adopt a similar structure to srCatD was that they all bound to pepstatinyl-agarose. Binding of the enzyme to the resin is generally believed to require a well formed active site cleft that has both N- and C- domain contributions. Additionally, N-terminal sequencing revealed that E5Q, E180Q, D187N, and Y10F all process at the correct cleavage junction during the conversion of the proenzyme to the mature form.
Since the enzyme unfolding then refolding as a consequence of the pH-jump could explain the kinetic lag phase, structural characterization of srCatD and variants over a pH-range was conducted. The pH-dependent tryptophan fluorescence emission maxima for srCatD were virtually identical to the native enzyme and both were characteristic of folded protein within the pH range 3.5 to 7. These experiments argue that the enzyme is not exhibiting any appreciable unfolding during the pH range of the pH-equilibrium jump experiment.
Molecular modeling was used to aid in interpretation of the kinetic data. Modeling of the variants in both the high pH form and low pH form was conducted. The models of the variants E5Q, E180Q, and D187N in the low pH conformation suggest that the introduction of the point mutations at position 5, 180, and 187 does not affect the hydrogen bonding distance between the E180 and D187 ion switch residues. However, an increase in the distance between the residues in the second proposed switch, E5/E18, from 3.1 to 3.3 Å is evident, and suggests that the interaction between E5 and E18 is weakened. Therefore, the observed differences in the kinetic parameters for E180Q and D187N cannot be unequivocally attributed to the lost of charge repulsion at this switch since collateral loss of the E5/E18 interaction may result. However, the E5Q variant also exhibits the same distance increase between Q5 and Q18 and results in an enzyme with slower kinetics compared to srCatD. Conversely, D187N and E180Q exhibit faster kinetics relative to srCatD. This argues that loss of the E5/E18 is an artifact of modeling or, its effect is small and is masked by the dominating effect of loss of charge repulsion at the E180/D187 position.
The molecular models for the high pH form of Y10F and E5Q indicated that the substitution at these positions do not modify the interactions between Lys 8, Tyr 10 and the catalytic Asp residues. Likewise, the position of E5 is not significantly altered in the Y10F model. Models for all of the variants in both the high pH and low pH structure suggest that no new interactions are created as a result of the introduced mutations.
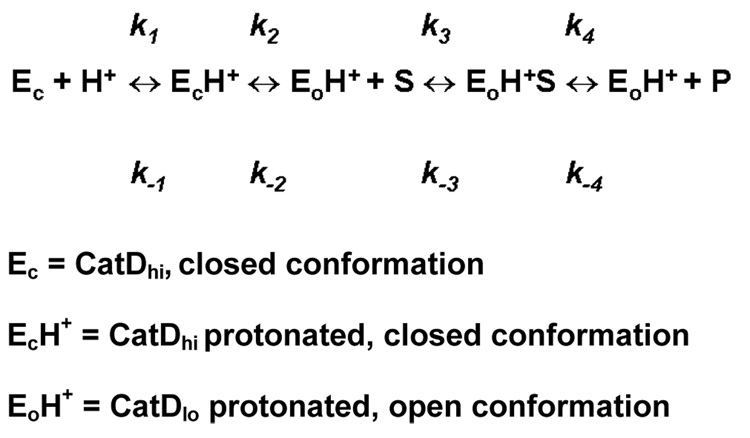
We present evidence that explains a novel molecular mechanism for the pH-dependent autoregulation of an aspartic peptidase that is described by the proposed scheme (Scheme 1). Specifically, the results indicate that E180 and D187 participate as an electrostatic switch that initiates the conformational change of CatDlo to CatDhi through charge repulsion. Additionally, Y10 stabilizes CatDhi by hydrogen bonding to the catalytic Asp 33. Finally, E5 appears to play a less significant role as an ionic switch that stabilizes CatDlo through its hydrogen bonding interaction with the carbonyl of E18. These results are substantiated by a study by Alexov (28) wherein the proton uptake/release and the binding energy for three inhibitor-enzyme complexes with available experimental data were numerically studied. The results for cathepsin D indicated that Glu 180 and Asp 33, but not Glu 5, are calculated to contribute to proton uptake and release, and that these groups may have specific roles considering their conservation (28). Indeed, our data show that Glu 180, Asp 33, and to a lesser extend, Glu 5, participate in ionization events that modulate the pH-dependent conformational change of cathepsin D.
Scheme 1.
Possible equilibria involved in the pH dependence of cathepsin D activity. At low pH, the enzyme exists in an open conformation as shown by crystal structures (15,16), while at high pH the enzyme exists in the closed conformation (19).
As for the physiological relevance of conformational switching, it likely plays an important role in the cellular location-dependent activity of cathepsin D. This may include modulation of substrate specificity (18) which is consistent with its function in both general and restricted proteolysis, as well as its function as both a protease and a ligand (29). Additionally, the kinetic hysteresis of cathepsin D implies it may be an important checkpoint for metabolic processes (27). Future studies will undoubtedly unravel the interestingly complex behavior of cathepsin D, thus providing an exciting new chapter in molecular regulation and recognition.
Acknowledgments
The authors would like to thank the UF-ICBR Protein Chemistry Core Facility for protein and peptide sequence and analysis. The authors would also like to thank Dr. Linda Bloom for her helpful discussions.
Footnote for abbrevs
- CatD
cathepsin D
- srCatD
“short” recombinant human pseudocathepsin D
- CatDlo
low pH, active form of cathepsin D
- CatDhi
high pH, inactive form of cathepsin D
- Open form
CatDlo
- Closed form
CatDhi
Footnotes
NIH DK18865
References
- 1.Barret AJ. Cathepsin D and other carboxyl proteinases. In: Barrett AJ, editor. Proteinases in Mammalian cells and Tissues. Amsterdam: North-Holland Biomed. Press; 1977. pp. 209–248. [Google Scholar]
- 2.Offermann MK, Chlebowski JF, Bond JS. Action of cathepsin D on fructose-1,6-bisphosphate aldolase. Biochem J. 1983;211:529–534. doi: 10.1042/bj2110529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett AJ. Human cathepsin D. Adv Exp Med Biol. 1977;95:291–300. doi: 10.1007/978-1-4757-0719-9_17. [DOI] [PubMed] [Google Scholar]
- 4.Dean RT. Direct evidence of importance of lysosomes in degradation of intracellular proteins. Nature. 1975;257:414–416. doi: 10.1038/257414a0. [DOI] [PubMed] [Google Scholar]
- 5.Saftig P, Hetman M, Schmahl W, Weber K, Heine L, Mossmann H, Koster A, Hess B, Evers M, von Figura K, et al. Mice deficient for the lysosomal proteinase cathepsin D exhibit progressive atrophy of the intestinal mucosa and profound destruction of lymphoid cells. Embo J. 1995;14:3599–3608. doi: 10.1002/j.1460-2075.1995.tb00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diment S, Martin KJ, Stahl PD. Cleavage of parathyroid hormone in macrophage endosomes illustrates a novel pathway for intracellular processing of proteins. J Biol Chem. 1989;264:13403–13406. [PubMed] [Google Scholar]
- 7.Maric MA, Taylor MD, Blum JS. Endosomal aspartic proteinases are required for invariant-chain processing. Proc. Natl. Acad. Sci. 1994;91:2171–2175. doi: 10.1073/pnas.91.6.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizuochi T, Yee ST, Kasai M, Kakiuchi T, Muno D, Kominami E. Both cathepsin B and cathepsin D are necessary for processing of ovalbumin as well as for degradation of class II MHC invariant chain. Immunol Lett. 1994;43:189–193. doi: 10.1016/0165-2478(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 9.Morikawa W, Yamamoto K, Ishikawa S, Takemoto S, Ono M, Fukushi J, Naito S, Nozaki C, Iwanaga S, Kuwano M. Angiostatin generation by cathepsin D secreted by human prostate carcinoma cells. J Biol Chem. 2000;275:38912–38920. doi: 10.1074/jbc.M005402200. [DOI] [PubMed] [Google Scholar]
- 10.Ikeguchi M, Fukuda K, Oka S, Yamaguchi K, Hisamitsu K, Tsujitani S, Sakatani T, Ueda T, Kaibara N. Clinicopathological significance of cathepsin D expression in gastric adenocarcinoma. Oncology. 2001;61:71–78. doi: 10.1159/000055356. [DOI] [PubMed] [Google Scholar]
- 11.Garcia M, Augereau P, Briozzo P, Capony F, Cavailles V, Freiss G, Maudelonde T, Montcourrier P, Vignon F, Rochefort H. Regulation, clinical and biological significance of cathepsin D in breast cancer. Rev Esp Fisiol. 1990;46:39–41. [PubMed] [Google Scholar]
- 12.Glondu M, Coopman P, Laurent-Matha V, Garcia M, Rochefort H, Liaudet-Coopman E. A mutated cathepsin-D devoid of its catalytic activity stimulates the growth of cancer cells. Oncogene. 2001;20:6920–6929. doi: 10.1038/sj.onc.1204843. [DOI] [PubMed] [Google Scholar]
- 13.Liaudet E, Derocq D, Rochefort H, Garcia M. Transfected cathepsin D stimulates high density cancer cell growth by inactivating secreted growth inhibitors. Cell Growth Differ. 1995;6:1045–1052. [PubMed] [Google Scholar]
- 14.Kokkonen N, Rivinoja A, Kauppila A, Suokas M, Kellokumpu I, Kellokumpu S. Defective acidification of intracellular organelles results in aberrent secretion of cathepsin D in cancer cells. J. Biol. Chem. 2004;279:39982–39988. doi: 10.1074/jbc.M406698200. [DOI] [PubMed] [Google Scholar]
- 15.Baldwin ET, Bhat TN, Gulnik S, Hosur MV, Sowder RC, Cachau RE, Collins J, Silva AM, Erickson JW. Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design. Proc. Natl. Acad. Sci. 1993;90:6796–6800. doi: 10.1073/pnas.90.14.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metcalf P, Fusek M. Two crystal structures for cathepsin D: the lysosomal targeting signal and active site. Embo J. 1993;12:1293–1302. doi: 10.1002/j.1460-2075.1993.tb05774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee AY, Gulnik SV, Erickson JW. Conformational switching in an aspartic proteinase. Nat Struct Biol. 1998;5:866–871. doi: 10.1038/2306. [DOI] [PubMed] [Google Scholar]
- 18.Beyer BM, Dunn BM. Self-activation of recombinant human lysosomal procathepsin D at a newly engineered cleavage junction, "short" pseudocathepsin D. J Biol Chem. 1996;271:15590–15596. doi: 10.1074/jbc.271.26.15590. [DOI] [PubMed] [Google Scholar]
- 19.Huang JS, Huang SS, Tang J. Cathepsin D isozymes from porcine spleens. Large scale purification and polypeptide chain arrangements. J Biol Chem. 1979;254:11405–11417. [PubMed] [Google Scholar]
- 20.Yasuda Y, Kageyama T, Akamine A, Shibata M, Kominami E, Uchiyama Y, Yamamoto K. Characterization of new fluorogenic substrates for the rapid and sensitive assay of cathepsin E and cathepsin D [published erratum appears in J Biochem (Tokyo) 1999 Jul;126(1):260] J Biochem (Tokyo) 1999;125:1137–1143. doi: 10.1093/oxfordjournals.jbchem.a022396. [DOI] [PubMed] [Google Scholar]
- 21.Marquardt D. An algorithm for least-square estimation of non-linear parameters. J. Soc. Indust. Appl. Mathe. 1963;11:411–441. [Google Scholar]
- 22.Meyer S. Data Analysis for Scientists and Engineers. New York: John Wiley and Sons; 1975. pp. 121–145. [Google Scholar]
- 23.Cornish-Bowden A. Fundamentals of Enzyme Kinetics. London: Butterworths; 1979. [Google Scholar]
- 24.Dixon M, Webb C. Enzymes. New York: Academic Press; 1979. p. 1116. [Google Scholar]
- 25.Auffret CA, Ryle AP. The catalytic activity of pig pepsin C towards small synthetic substrates. Biochem J. 1979;179:239–246. doi: 10.1042/bj1790239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Y, Fusek M, Lin X, Hartsuck JA, Kezdy FJ, Tang J. pH dependence of kinetic parameters of pepsin, rhizopuspepsin, and their active-site hydrogen bond mutants. J Biol Chem. 1992;267:18413–18418. [PubMed] [Google Scholar]
- 27.Frieden C. Kinetic aspects of regulation of metabolic processes. J Biol Chem. 1970;245:5788–5799. [PubMed] [Google Scholar]
- 28.Alexov E. Calculating proton uptake/release and binding free energy taking into account ionization and conformation changes induced by protein-inhibitor association application to plasmepsin, cathepsin D and endothiapepsin-pepstatin complexes. Proteins: Structure, Function, and Bioinformatics. 2004;56:572–584. doi: 10.1002/prot.20107. [DOI] [PubMed] [Google Scholar]
- 29.Berchem G, Glondu M, Gleizes M, Brouillet JP, Vignon F, Garcia M, Liaudet-Coopman E. Cathepsin-D affects multiple tumor progression steps in vivo: proliferation, angiogenesis and apoptosis. Oncogene. 2002;21:5951–5955. doi: 10.1038/sj.onc.1205745. [DOI] [PubMed] [Google Scholar]