Abstract
In the growth plate, the reserve and perichondral zones have been hypothesized to have similar functions, but their exact functions are poorly understood. Our hypothesis was that significant differential gene expression exists between perichondral and reserve chondrocytes that may differentiate the respective functions of these two zones. Normal Sprague-Dawley rat growth plate chondrocytes from the perichondral zone (PC) and reserve zone (RZ) were isolated by laser microdissection and then subjected to microarray analysis. In order to most comprehensively capture the unique features of the two zones, we analyzed both the most highly expressed genes and those that were most significantly different from the proliferative zone (PZ) as a single comparator. Confirmation of the differential expression of selected genes was done by quantitative real time RT-PCR. A total of 8 transcripts showing high expression unique to the PC included translationally-controlled tumor protein (Tpt1), connective tissue growth factor (Ctgf), mortality factor 4 (Morf4l1), growth arrest specific 6 (Gas6), type V procollagen (Col5a2), frizzled-related protein (Frzb), GDP dissociation inhibitor 2 (Gdi2) and Jun D proto-oncogene (Jund). In contrast, 8 transcripts showing unique high expression in the RZ included hyaluronan and proteoglycan link protein 1 (Hapln1), hemoglobin beta-2 subunit, type I procollagen (Col1a2), retinoblastoma binding protein 4 (LOC685491), Sparc related modular calcium binding 2 (Smoc2), and calpastatin (Cast). Other genes were highly expressed in cells from both PC and RZ zones, including collagen II, collagen IX, catenin (cadherin associated protein) beta 1, eukaryotic translation elongation factor, high mobility group, ribosomal protein, microtubule-associated protein, reticulocalbin, thrombospondin, retinoblastoma binding protein, carboxypeptidase E, carnitine palmitoyltransferase 1, cysteine rich glycoprotein, plexin B2 (Plxnb2), and gap junction membrane channel protein. Functional classification of the most highly expressed transcripts were analyzed, and the pathway analysis indicated that ossification, bone remodeling, and cartilage development were uniquely enriched in the PC whereas both the PC and RZ showed pathway enrichment for skeletal development, extracellular matrix structural constituent, proteinaceous extracellular matrix, collagen, extracellular matrix, and extracellular matrix part pathways. We conclude that differential gene expression exists between the RZ and PC chondrocytes and these differentially expressed genes have unique roles to play corresponding to the function of their respective zones.
Keywords: Growth plate, microarray, chondrocytes, rat, bone
Introduction
Longitudinal bone growth results from a differentiation cascade of growth plate chondrocytes through a series of morphologic changes along with provisional calcification, apoptosis, and metaphyseal bone deposition. Cells in the reserve zone give way to flattened stacks of proliferative zone cells which go through a transitional phase before ceasing cell division and going through a hypertrophic stage just before they yield to the terminal changes in the growth plate. Perichondral chondrocytes surround the periphery of the growth plate. Although perichondral and reserve chondrocytes are well characterized histomorphologically, an understanding of their contribution in regulating progression in the growth plate is not fully explained. Both endocrine effects and a local regulatory loop involving Indian hedgehog (Ihh) and parathyroid hormone-related protein (PTHrP) have been described [1, 2,3]. The local regulatory loop involves epiphyseal and perichondrial chondrocyte production of PTHrP, which maintains chondrocyte proliferation [4,5]. PTHrP promotes expression of B-cell leukemia-2 protein (Bcl-2), inhibiting apoptosis by blocking the pro-apoptotic effects of Bcl-2-associated X protein (BAX). Beginning in the transitional zone, Ihh promotes chondrocyte differentiation [3]. The regulatory role in the perichondral and reserve chondrocytes for insulin-like growth factor I (IGF-I), fibroblastic growth factor (FGF), transforming growth factor β (TGF-β) and the bone morphogenic proteins (BMP) is less well-defined [6,7,8].
Whether the pathways currently implicated in control of longitudinal growth are the primary ones responsible is unknown. Other pathways involving genes not previously described in the growth plate may also play a contributory role. Transcriptional analysis by DNA microarray can enable a more comprehensive investigation of these complex interactions. In addition, laser microdissection allows isolation of chondrocytes from individual growth plate zones for comparative analysis. In this study, we combined both approaches to identify in vivo differential gene expression between chondrocytes in the perichondral zone (PC) and reserve zone (RZ) in the tibial growth plates of the adolescent Sprague-Dawley rat. Our hypothesis was that significant differential gene expression exists between perichondral and reserve chondrocytes that would provide clues at the transcriptional level as to their functions.
Materials and Methods
Tissue Preparation
All animal procedures were reviewed and approved by the Committee for Humane Use of Animals (CHUA). Three male Sprague-Dawley rats were euthanized at 42, 46 and 51 days of age, respectively, all by carbon-dioxide asphyxiation. The tibias were dissected and halved sagittally immediately after euthanasia. Tissues were then molded in pre-chilled Tissue Tek O.C.T. compound (Sakura-finetek, Tokyo, Japan) and rapidly frozen in liquid nitrogen. Tissue samples were then stored at −70°C in sealed bags. Tissue blocks were equilibrated to −21°C for one hour in the cryostat cabinet before cryo-microtomy. Blocks were trimmed with a razor blade to remove excessive OCT embedding media and mounted for sectioning with a thin layer of OCT on the cutting head, and slowly warmed to −18°C. Subsequently, 6 um sections cut on a Leica CM3050 cryostat were collected and mounted onto RNase-free stainless steel framed PEN-foiled slides (Leica) for laser capture microdissection. All sectioned tissue was briefly brought to room temperature for 30 sec to enhance tissue adherence to the slides, and returned to the cryomicrotome. Slides with sectioned tissue were then stored in a desiccator at −70°C until further use or laser capture microdissection.
Laser Microdissection
The PC, RZ and PZ proximal tibial growth plate chondrocytes were laser microdissected separately by the Leica Application Solution Laser Microdissection instrument (Leica Microsystems, Bannockburn, IL) as previously described [9]. The reserve zone was defined as the region from the epiphysis (excluding all cells related to the secondary center of ossification) to the first flattened chondrocyte at the base of a cell column. The hypertrophic zone extended from the transitional zone to the chondro-osseous junction. The transitional zone was defined as extending from the first shape change away from flattened cell morphology to the first level of fully enlarged chondrocytes. The perichondral zone was referred to the ring of chondrocytes surrounding the growth plate. In the growth plate, we identified chondrocytes close to the epiphysis including regenerating columns and non-regenerating column chondrocytes as PZ and chondrocytes close to metaphyseal side as HZ chondrocytes.
RNA Extraction and Gene Chip Hybridization Procedures
The dissected piece of film with the captured tissue fell directly into 40 µl of RLT Lysis buffer and total RNA was extracted using RNeasy mini kit (Qiagen, Valencia, CA). The quality and concentration of the total RNA were analyzed on an Agilent Bioanalyzer as previously described [9]. Initial RNA samples used in the array study contained approximately 10 ng per sample. Based on the Bioanalyzer concentrations, we pooled approximately equal amounts of RNA from three rats samples of each cell type (PC, RZ or PZ) at three time points (42 days, 46 and 51 days of age) to create 9 pooled samples. Each pooled sample contained 30–50 ng total RNA. The RNA samples were prepared for hybridization using the Ovation™ Biotin RNA Amplification and Labeling System (NuGen). After the amplification and labeling, equal amounts (2.2 ug) of labeled single-stranded cDNA product was then added to a Rat RAE 230 2.0 GeneChip (Affymetrix) according to manufacturer instructions. After hybridization, washing, and scanning the GeneChips, the Affymetrix software (GeneChip Operating System, Santa Clara, CA) calculated the intensity of the signal from each perfect match probe relative to the signal for the mismatch probe and determined whether or not the gene was present in the sample, and also provided a measure of the expression level of the gene. The overall chip intensities for each sample were scaled by linear adjustment to the same target value (500), and subsequently normalized by the RMA algorithm The experimental series of data files has been deposited into the Gene Expression Omnibus (GEO) at NCBI (accession GSE9537).
Analyses of Gene Expression
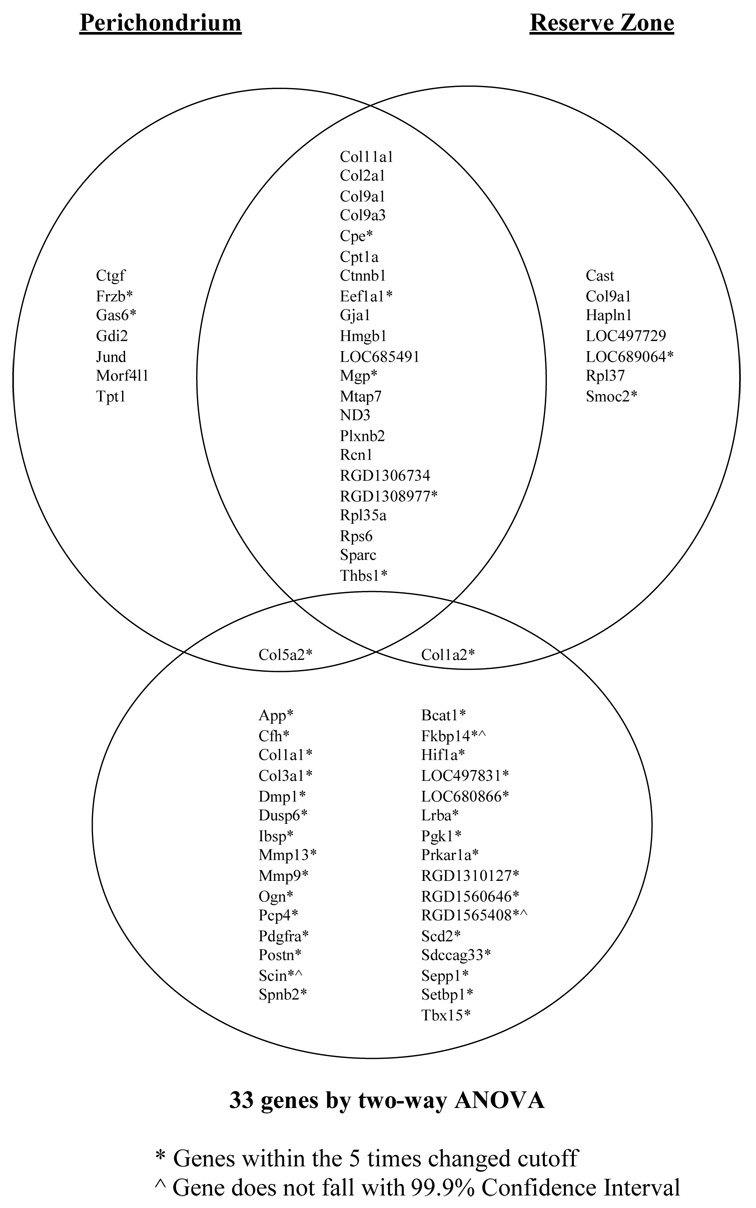
Affymetrix GCOS/MAS 5 was used to generate a list of genes designated as “present” in PC, RZ and PZ. GeneSpring GX (Agilent Technologies, Palo Alto, CA) was used to identify differential expression and create a raw dataset using the Robust Multiarray analysis (RMA) method. Using the raw data set, the 30 probe sets with the highest expression within both RZ and PC were identified, excluding unnamed transcribed locus probes and duplicate gene names (Figure 1). To identify the significant differential gene expression between perichondral and reserve chondrocytes, the proliferative zone was used as a comparator. Normalized data was filtered on expression using a cutoff of five-fold increased in PC, RZ and PZ. A total of 471 probe sets passed this filter and were cross-referenced with the top 30 genes identified by expression level alone. Of these 471 probes, eight overlapped with the top 30 in RZ and eight overlapped with the top 30 in PC. The differential expression of these latter genes ranked among the top 0.1% of all changes (i.e., exceeded the 99.9% confidence level) when using PZ as a comparator. Differential expression between zones was further analyzed using a two-way ANOVA (Zone × Age), to create a list of 49 probe sets with statistically significant differences in expression. Of these 49, 44 exceeded the 99.9% confidence level of all changes when using PZ as a comparator. After eliminating the unnamed transcribed locus probes and duplicate gene names, this list of 49 was reduced to only 33 genes (Table 1). When comparing this list of 33 significantly different genes with the list containing the top 30 by expression level in both PC and RZ, there was one gene in common for each of the zones between the lists (Figure 2).
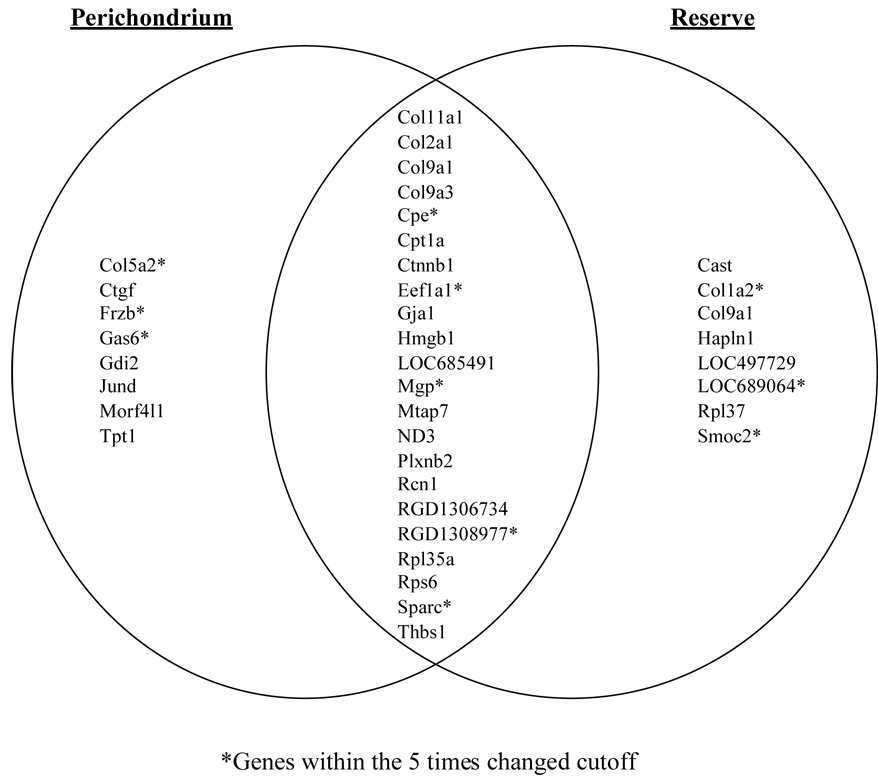
Figure 1. Top 30 genes showing the greatest expression in PC and RZ.
Venn diagram of differential expression according to growth plate zone. A list of genes was designated as “present” in PC and RZ using Affymetrix GCOS/MAS 5 software. Differential gene expressions within PC and RZ were identified by GeneSpring GX and a raw dataset was created using the Robust Multiarray analysis (RMA) method. The top 30 genes by highest expression within both RZ and PC (individually) were identified, excluding unnamed transcribed locus probes and duplicate gene names. Twenty-two genes from the top 30 genes expressed within the PC were also present among the top 30 genes expressed within the RZ. Only 8 genes from each list were unique to the respective zone.
Table 1.
Log2 Ratio of 33 genes showing differential expression in PC and RZ by two-way ANOVA and Expression level cutoff of 5 times changed. Red indicates presence in positive 99.9% confidence level while green indicates presence in negative 99.9% confidence level.
| 33 Genes by 2-Way ANOVA Showing Differential Expression in RZ and PC | ||||||||
|---|---|---|---|---|---|---|---|---|
| 99.9% Cl | 99.9% Cl | |||||||
| Mean Log2 | 0.01 | 0.02 | 0.03 | 0.00 | 0.01 | 0.01 | ||
| StDev | 0.71 | 0.68 | 0.57 | 0.56 | 0.52 | 0.57 | ||
| +99.9 CL | 2.42 | 2.30 | 1.97 | 1.89 | 1.77 | 1.93 | ||
| # Incr 99.9CL | 384 | 495 | 122 | 321 | 355 | 308 | ||
| −99.9 CL | −1.38 | −1.31 | −1.10 | −1.09 | −1.02 | −1.11 | ||
| # Decr 99.9CL | 618 | 303 | 1112 | 781 | 496 | 715 | ||
| Log2 RZ/PZ | Log2 PC/PZ | |||||||
| Probe ID | Gene Name | Gene Symbol | 7 days | 11days | 16 days | 7 days | 11days | 16 days |
| 1388204_at | --- | Mmp13 | 7.43 | 6.69 | 4.55 | 4.42 | 0.85 | −1.46 |
| 1368416_at | integrin binding sialoprotein | Ibsp | 5.88 | 7.37 | 4.62 | 3.95 | 6.43 | 1.59 |
| 1370864_at | procollagen, type 1, alpha 1 | Col1a1 | 4.26 | 6.87 | 6.02 | 3.91 | 4.83 | 6.41 |
| 1398275_at | matrix metallopeptidase 9 | Mmp9 | 6.32 | 4.89 | 1.97 | 0.87 | −0.25 | 0.62 |
| 1370959_at | procollagen, type III, alpha 1 | Col3a1 | 3.79 | 1.03 | 0.58 | 4.92 | 2.84 | 6.15 |
| 1393756_at | dentin matrix protein 1 | Dmp1 | 3.71 | 5.87 | 3.44 | −0.27 | 1.15 | 2.75 |
| 1370155_at | procollagen, type I, alpha 2 | Col1a2 | 5.65 | 5.85 | 2.56 | 4.42 | 2.63 | 5.81 |
| 1370895_at | procollagen, type V, alpha 2 | Col5a2 | 4.09 | 4.68 | 3.35 | 4.25 | 4.84 | 5.50 |
| 1387029_at | complement component factor H | Cfh | 5.13 | 5.44 | 3.22 | 3.90 | 2.60 | 4.91 |
| 1370941_at | platelet derived growth factor receptor, alpha polypeptide | Pdgfra | 3.02 | 2.99 | 2.07 | 3.02 | 2.81 | 4.48 |
| 1382778_at | Dual specificity phosphatase 6 | Dusp6 | 2.27 | 4.28 | 2.66 | 0.65 | 2.38 | −1.14 |
| 1373911_at | periostin, osteoblast specific factor | Postn | −0.22 | 0.63 | 0.33 | 3.09 | 1.56 | 3.96 |
| 1371571_at | amyloid beta (A4) precursor protein | App | 2.57 | 1.75 | −0.01 | 3.95 | 2.92 | 1.90 |
| 1371419_at | spectrin beta 2 | Spnb2 | 3.74 | 3.40 | 1.45 | 1.64 | 0.15 | 0.51 |
| 1368145_at | Purkinje cell protein 4 | Pcp4 | 3.41 | 0.40 | 0.59 | 3.66 | 2.33 | 2.64 |
| 1390450_a_at | osteoglycin (predicted) | Ogn | 2.72 | 2.45 | 0.03 | 2.95 | 3.22 | 3.40 |
| 1368806_at | selenoprotein P, plasma, 1 | Sepp1 | 2.84 | 3.28 | 1.49 | 1.98 | 0.87 | 2.33 |
| 1383916_at | T-box 15 | Tbx15 | 2.09 | 1.85 | 0.40 | 1.77 | 2.33 | 2.86 |
| 1389562_at | SET binding protein 1 | Setbp1 | 1.10 | 0.00 | 0.12 | 2.47 | 1.43 | 2.82 |
| 1391537_at | similar to SERTA domain containing 4 | RGD1565408 | 2.22 | −0.05 | 0.55 | 2.02 | 0.01 | 2.80 |
| 1373951_at | Protein kinase, cAMP dependent regulatory, type I, alpha | Prkar1a | 0.98 | 2.77 | 0.33 | 1.39 | 2.73 | 1.43 |
| 1378988_at | LPS-responsive beige-like anchor | Lrba | 1.94 | 0.20 | 0.06 | 2.63 | 0.40 | 0.63 |
| 1383573_at | Serologically defined colon cancer antigen 33 | Sdccag33 | −0.26 | 1.13 | 0.93 | 1.73 | 1.72 | 2.12 |
| 1384195_at | --- | LOC680866 | 0.75 | 0.91 | −1.73 | 1.45 | 1.87 | 0.70 |
| 1387076_at | hypoxia inducible factor 1, alpha subunit | Hif1a | 0.15 | 1.35 | −1.21 | 1.15 | 1.86 | 0.73 |
| 1367668_a_at | stearoyl-Coenzyme A desaturase 2 | Scd2 | 0.53 | 1.86 | −2.01 | 0.22 | 1.29 | −0.90 |
| 1389330_at | similar to cDNA sequence BC017158 | RGD1310127 | 1.75 | 0.75 | 0.74 | 0.08 | 0.11 | −0.47 |
| 1372525_at | FK506 binding protein 14 | Fkbp14 | 1.62 | 0.93 | −0.36 | 1.31 | 0.46 | 1.37 |
| 1388318_at | phosphoglycerate kinase 1 | Pgk1 | 1.17 | 1.58 | −1.56 | 0.74 | 0.64 | −0.52 |
| 1370869_at | branched chain aminotransferase 1, cytosolic | Bcat1 | 0.51 | 0.94 | −1.43 | 0.25 | 0.58 | 1.28 |
| 1391279_at | Scinderin | Scin | 0.90 | −0.17 | −0.94 | −0.27 | −0.20 | 0.80 |
| 1372897_at | hypothetical gene supported by NM_175869 | LOC497831 | −0.80 | 0.75 | −2.12 | −0.16 | 0.23 | −0.73 |
| 1376804_at | similar to Myosin VI | RGD1560646 | −0.60 | −0.23 | −1.36 | −0.54 | −0.89 | −2.03 |
| Falls within +99.9% Cl | ||||||||
| Falls within −99.9% Cl | ||||||||
Figure 2. Top 30 genes in PC and RZ by greatest expression compared to 33 genes by two-way ANOVA and 5 times changed.
To identify the significant differential gene expression between PC and RZ chondrocytes, the PZ was used as a comparator. Normalized data was filtered on expression using a cutoff of five times changed up or down in PC, RZ and PZ described as in the methods section. Differential expression between zones was analyzed using two-way ANOVA and created a gene list of 33 significant differentially expressed genes. When comparing the gene list of 33 significantly different than the PZ to the top 30 by expression in both PC and RZ, there was one gene in common for each of the zones.
Real-time RT-PCR
Real-time quantitative RT-PCR was performed using an ABI Prism 7000 Sequence Detection System (PE Applied Biosystems, CA). The 25 µl reaction consisted of SYBR Green PCR Master Mix (PE Applied Biosystems, CA), a set of rat specific primers and template cDNA generated by reversed-transcripted PCR (Table 2). The primers were designed to genes of matrix Gla protein (Mgp), Growth arrest specific 6 (Gas6), Thrombospondin 1 (Thbs1), Frizzled-related protein (Frzb), Procollagen type IX alpha 1 (Col9a1), Procollagen type X alpha 1 (Col10a1), secreted acidic cysteine rich glycoprotein (Sparc), Cyclin-dependent kinase inhibitor 1C (Cdkn1c), hemoglobin beta-2 subunit (LOC689064), Procollagen type I alpha 2 (Col1a2), Connective tissue growth factor (Ctgf), Stearoyl-coenzyme A desaturase 2 (Scd2), Procollagen type XXVII alpha 1 (Col27a1), Sparc-related modular calcium binding 2 (Smoc2), Procollagen type V alpha 2 (Col5a2). All samples were run in triplicate along with 18S rRNA as reference gene and no template controls. The threshold cycle (CT) was normalized to that of 18S rRNA to account for differences in cDNA loading. The 18S rRNA was chosen because there was no differential expression between samples from different days. The comparative threshold (CT) method was utilized for relative quantitative analysis of gene expression. In addition to the melting point analysis that is routinely provided by the ABI Prism unit, the size and identity of the amplicons from the PCR reaction were then directly verified by gel electrophoresis (using a 2% agarose gel).
Table 2.
Primers and targeted genes in real time PCR
| Systematic Name | Gene symbol | Gene Title | Amplicon Size (bp) | Sequences (5’ – 3’) | |
|---|---|---|---|---|---|
| 18 srRNA | 149 | Forward: | GGTCATAAGCTTGCGTTGAT | ||
| Reverse: | TCAAGTTCGACCGTCTTCTC | ||||
| 1367568_a_at | Mgp | matrix Gla protein | 127 | Forward: | GCCTACAACCGCTACTTCAG |
| Reverse: | CGCACACGAATCTGTGTATC | ||||
| 1383047_at | Gas6 | Growth arrest specific 6 | 129 | Forward: | GTACATAGTGGGACTGATAC |
| Reverse: | TTCTTCAGATAGTCCCGCTC | ||||
| 1374529_at | Thbs1 | Thrombospondin 1 | 122 | Forward: | CTTAAAACTACTGTCGTGTC |
| Reverse: | ACCAGAACTTTGAGGAGTTG | ||||
| 1375961_at | Frzb | Frizzled-related protein | 127 | Forward: | ATTTGGCTCGCTGTATTGAC |
| Reverse: | GTGCACATTTCACTGTCCTG | ||||
| 1392915_at | Col11a1 | Procollagen type IX alpha 1 | 156 | Forward: | GCCAATCCATTTTATGCCAC |
| Reverse: | ATGAGCCCTGTTGCCATCTC | ||||
| 1370944_at | Col10a1 | Procollagen type X alpha 1 | 141 | Forward: | GAAACAGGTGTCTGACTTAC |
| Reverse: | TACTTCCAGTGGAATAGAAG | ||||
| 1367562_at | Sparc | Secreted acidic cysteine rich glycoprotein | 147 | Forward: | TGGGCTACAGGAAAGTGAGA |
| Reverse: | TGGCTTGGAAGTTTCTCTTG | ||||
| 1372299_at | Cdkn1c | Cyclin-dependent kinase inhibitor 1C | 125 | Forward: | ACCAGCTTCAGATTACCCAC |
| Reverse: | TCTAATAATGTGGAGGACAC | ||||
| 1371245_a_at | LOC689064 | hemoglobin beta-2 subunit | 108 | Forward: | CCTTTCCTGCTTGTCTATGC |
| Reverse: | TTTATTAGATAGAAGCTAGC | ||||
| 1387854_at | Col1a2 | Procollagen type I alpha 2 | 136 | Forward: | GAAGACTCGATCCTCCATAG |
| Reverse: | ATGCTGAATCTAGAGAGGAG | ||||
| 1381925_x_at | LOC497729 | hypothetical gene supported by NM_172157 | 125 | Forward: | GAAGAAACACAACTTCAAAG |
| Reverse: | GAAAAGGAGGAATTTACATC | ||||
| 1367631_at | CTGF | Connective tissue growth factor | 154 | Forward: | GACGTTTGTGCCTATTGTTC |
| Reverse: | CAGACAGTCACTCAGGTTAC | ||||
| 1367668_a_at | Scd2 | Stearoyl-coenzyme A desaturase 2 | 131 | Forward: | GCACATTTTATAGTAACCG |
| Reverse: | TTCCATAGCAATGAGTATAC | ||||
| 1374870_at | Col27a1 | Procollagen type XXVII alpha 1 | 126 | Forward: | AACCCTTCCCTGTATACTGC |
| Reverse: | AAGGAGACAGCGCAAGACAC | ||||
| 1392965_a_at | Smoc2 | Sparc-related modular calcium binding 2 | 124 | Forward: | AAACTGTTCATGGTCCCCAG |
| Reverse: | AGCGTTGATGTGAGCTGGTC | ||||
| 1373463_at | Col5a2 | Procollagen type V alpha 2 | 109 | Forward: | CCATCAATGACCAACACAAG |
| Reverse: | GGTCAGGCACTTCAGATCAT | ||||
| 1371226_at | Col2a1 | Procollagen type II alpha 1 | 104 | Forward: | CCGGACTGTGAGGTTAGGAT |
| Reverse: | AACCCAAAGGACCCAAATAC | ||||
Pathway Analysis
For both the RZ and PC, overall functional pathway analysis was performed for the top 30 genes by expression level using Gene Ontology (GO) annotations provided by the NetAffx browser (Affymetrix). Enrichment of GO functional groups was determined to be meaningful when the number of probe sets in our list that mapped to a specific GO pathway was greater than 1, a hypergeometric p-value estimating the probability that this amount of overlap could have occurred due to chance alone based on the size of the list and the annotated content of the array was less than 0.05, and the fold enrichment of overlap with a specific pathway compared to what would be expected by chance alone exceeded 2. Because of the large number of pathways that met these criteria, however, we utilized two additional screens to narrow the field. First, we separated out those pathways which had a p-value less than 0.05, ≥5 probe sets (rather than >1 probe set) and showed a fold enrichment of ≥5.0. Second, we identified those from the narrowed list that relate to bone, cartilage, matrix, and/or skeletal development (BCMSD). The latter screen was determined based on a search utilizing AmiGO, a search engine for the GeneOntology database (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi).
Results
Highly Expressed Genes According to Growth Plate Zone
Of the 31,099 probe sets arrayed on the RAE 230.2.0 chips, the 30 genes showing the greatest absolute expression levels within the PC (Table 3) and RZ (Table 4) were selected for further analysis. Twenty-one genes from the top 30 genes expressed within the PC were also present among the top 30 genes expressed within the RZ (Figure 1), so only 8 genes from each list were unique to the respective zone. The 37 total genes in both zones at 42, 46 and 51 days of age for the Sprague-Dawley rat were comprised of 8 genes known to be involved in chondrocyte function and 33 (9 in PC, 9 in RZ, 15 shared) not previously identified within chondrocytes.
Table 3.
Top 30 Genes in Perichondrium by highest expression
| 7 days | 11 days | 16 days | |||||
|---|---|---|---|---|---|---|---|
| Gene Title | Gene Symbol | MC | PCR | MC | PCR | MC | PCR |
| procollagen, type II, alpha 1 | Col2a1 | 10414 | 4672.6 | 5941 | 677.93 | 10595 | 7643.4 |
| procollagen, type IX, alpha 3 | Col9a3 | 5082 | 3097 | 2785 | |||
| Similar to RIKEN cDNA 1110017I16 | RGD1308977 | 2951 | 1092 | 5071 | |||
| secreted acidic cysteine rich glycoprotein | Sparc | 2291 | 4.91 | 927 | −37.92 | 4972 | 3.25 |
| eukaryotic translation elongation factor 1 alpha 1 | Eef1a1 | 1659 | 622.3 | 4296 | |||
| NADH dehydrogenase subunit 3 | ND3 | 3765 | 3314 | 2957 | |||
| high mobility group box 1 | Hmgb1 | 3598 | 3739 | 2064 | |||
| matrix Gla protein | Mgp | 1068 | 53.08 | 324.2 | 17.15 | 3665 | 411.57 |
| tumor protein, translationally-controlled 1 | Tpt1 | 2110 | 1748 | 3647 | |||
| connective tissue growth factor | Ctgf | 1159 | 4.06 | 1714 | 6.52 | 3618 | 524.57 |
| ribosomal protein L35a | Rpl35a | 2593 | 3057 | 1513 | |||
| mortality factor 4 like 1 | Morf4l1 | 945.2 | 732.1 | 2911 | |||
| procollagen, type V, alpha 2 | Col5a2 | 1267 | 10.20 | 528.7 | 28.25 | 2897 | 92.73 |
| growth arrest specific 6 | Gas6 | 273 | 1.13 | 113.3 | 1.55 | 2816 | 151.17 |
| Plexin B2 | Plxnb2 | 2277 | 2112 | 2730 | |||
| Catenin (cadherin associated protein), beta 1 | Ctnnb1 | 2600 | 2185 | 2717 | |||
| Microtubule-associated protein 7 | Mtap7 | 2330 | 2640 | 1127 | |||
| reticulocalbin 1 | Rcn1 | 1996 | 1892 | 2504 | |||
| Thrombospondin 1 | Thbs1 | 763.2 | 22.16 | 328.4 | 36.25 | 2360 | 461.44 |
| Similar to hypothetical protein FLJ32743 | RGD1306734 | 1539 | 2359 | 917.8 | |||
| procollagen, type IX, alpha 1 | Col9a1 | 1824 | 710.9 | 2358 | |||
| similar to retinoblastoma binding protein 4 | LOC685491 | 2340 | 1873 | 1292 | |||
| ribosomal protein S6 | Rps6 | 1725 | 858.7 | 2321 | |||
| procollagen, type XI, alpha 1 | Col11a1 | 2183 | 59.71 | 1013 | 28.35 | 2112 | 269.66 |
| carboxypeptidase E | Cpe | 661 | 209.4 | 2132 | |||
| Carnitine palmitoyltransferase 1a, liver | Cpt1a | 1285 | 2130 | 1699 | |||
| frizzled-related protein | Frzb | 479.1 | 1.61 | 252.3 | 3.43 | 2093 | 54.00 |
| gap junction membrane channel protein alpha 1 | Gja1 | 1288 | 2047 | 822.6 | |||
| GDP dissociation inhibitor 2 | Gdi2 | 769.4 | 460 | 2019 | |||
| Jun D proto-oncogene | Jund | 1148 | 1969 | 394.5 | |||
| Correlation coefficient R | 0.9773 | 0.9523 | 0.9336 | ||||
Note: Among the top 30 genes in PC by highest expression in microarray (MC) analysis, nine selected representative genes were confirmed by real time PCR (PCR) with high correlation coefficient (R > 0.9). The comparative threshold (ΔΔCT) method was utilized for relative quantitative analysis of gene expression in real time PCR. The threshold cycle (CT) was normalized to the reference gene and the numbers represent relative quantitation for gene expression level.
Table 4.
Top 30 Genes in Reserve Zone (RZ) by highest expression
| 7 days | 11 days | 16 days | |||||
|---|---|---|---|---|---|---|---|
| Gene Title | Gene Symbol | MC | PCR | MC | PCR | MC | PCR |
| procollagen, type II, alpha 1 | Col2a1 | 11250 | 15076 | 4313 | 916.5 | 6432 | 594.3 |
| high mobility group box 1 | Hmgb1 | 2951 | 3613 | 4480 | |||
| procollagen, type IX, alpha 3 | Col9a3 | 4269 | 3031 | 4477 | |||
| NADH dehydrogenase subunit 3 | ND3 | 3689 | 2909 | 3270 | |||
| Similar to RIKEN cDNA 1110017I16 | RGD1308977 | 3629 | 757.1 | 884.7 | |||
| Microtubule-associated protein 7 | Mtap7 | 2038 | 2110 | 3236 | |||
| secreted acidic cysteine rich glycoprotein | Sparc | 3225 | 13.00 | 619.7 | 1.53 | 1071 | −22.09 |
| Similar to hypothetical protein FLJ32743 | RGD1306734 | 1326 | 1735 | 3007 | |||
| ribosomal protein L35a | Rpl35a | 2131 | 2127 | 2708 | |||
| Plexin B2 | Plxnb2 | 1972 | 2693 | 2219 | |||
| Catenin (cadherin associated protein), beta 1 | Ctnnb1 | 2399 | 2653 | 2418 | |||
| reticulocalbin 1 (predicted) | Rcn1 | 2498 | 1974 | 2566 | |||
| Hyaluronan and proteoglycan link protein 1 | Hapln1 | 1164 | 991.7 | 2516 | |||
| procollagen, type XI, alpha 1 | Col11a1 | 2506 | 93.70 | 1072 | 66.26 | 1686 | 48.84 |
| Carnitine palmitoyltransferase 1a, liver | Cpt1a | 2307 | 1502 | 1529 | |||
| gap junction membrane channel protein alpha 1 | Gja1 | 1178 | 2157 | 1114 | |||
| similar to Hemoglobin beta-2 subunit | LOC689064 | 935.8 | 28.05 | 2119 | 21.11 | 154 | 152.70 |
| procollagen, type I, alpha 2 | Col1a2 | 2019 | 272.48 | 1822 | 13 | 1207 | 109.50 |
| procollagen, type IX, alpha 1 | Col9a1 | 1969 | 867.1 | 1008 | |||
| similar to retinoblastoma binding protein 4 | LOC685491 | 1490 | 1950 | 1639 | |||
| eukaryotic translation elongation factor 1 alpha 1 | Eef1a1 | 1907 | 1140 | 738.5 | |||
| SPARC related modular calcium binding 2 | Smoc2 | 1901 | 67.18 | 107.1 | 15.30 | 38.31 | 13.78 |
| carboxypeptidase E | Cpe | 1879 | 691.9 | 534.8 | |||
| ribosomal protein S6 | Rps6 | 1858 | 772.9 | 567.6 | |||
| ribosomal protein L37 | Rpl37 | 1759 | 803.5 | 578.2 | |||
| hypothetical gene supported by NM_172157 | LOC497729 | 670.6 | 10.93 | 1759 | 4.96 | 962.8 | 21.48 |
| Calpastatin | Cast | 789 | 942.5 | 1756 | |||
| matrix Gla protein | Mgp | 1742 | 129.34 | 162.6 | 36.33 | 82.63 | 3.04 |
| similar to DnaJ homolog subfamily B member 6 | LOC686213 | 676 | 1699 | 1473 | |||
| ribosomal protein L13A | Rpl13a | 1612 | 656.3 | 428.9 | |||
| Correlationi coefficient R | 0.9720 | 0.8228 | 0.9152 | ||||
Note: Among the top 30 genes in RZ by highest expression in microarray (MC) analysis, eight selected representative genes were confirmed by real time PCR (PCR) with high correlation coefficient (R > 0.8). The comparative threshold (ΔΔCT) method was utilized for relative quantitative analysis of gene expression in real time PCR. The threshoyld cycle (CT) was normalized to the reference gene and the numbers represent relative quantitation for gene expression level..
Highly Expressed Genes in Both PC and RZ
Among the mRNAs highly expressed within both the PC and RZ were 21 genes, including Col2a1, Col9a3, Ctnnb1 (Catenin β1), Eef1a1 (eukaryotic translation elongation factor 1 α1), ND3 (NADH dehydrogenase subunit 3), Hmgb1 (high mobility group box 1), Rpl35a (ribosomal protein L35a), Plxnb2 (Plexin B2), Mtap7 (microtubule-associated protein 7), Rcn1 (reticulocalbin 1), Col9a1, Rps6 (ribosomal protein S6), Col11a1, Cpe (carboxypeptidase E), Cpt1a (carnitine palmitoyltransferase 1a), Sparc (secreted acidic cysteine rich glycoprotein), Gja1 (gap junction membrane channel protein 1), RGD 1308977 (similar to RIKEN cDNA), RGD 1306734 (similar to hypothetical protein FLJ32743), and LOC 685491 (similar to retinoblastoma binding protein 4), Mgp (matrix Gla protein). Eight of these play a known role in growth plate extracellular matrix (ECM) function: Col2a1, Ctnnb1, Eefla1, Col9a1, Col11a1, Cpt1a, Sparc, and Mgp. The most abundant ECM mRNA in both PC and RZ zones by microarray was Col2a1 at 42, 46 and 51 days, and its absolute expression level was greater in the proliferative zones at all three time points.
Differential Expression According to Growth Plate Zone
Of the top 30 most highly expressed gene transcripts within the PC, 8 transcripts (Tpt1, Ctgf, Morf4l1, Col5a2, Gas6, Frzb, Gdi2, Jund,) were uniquely highly expressed only within the PC chondrocytes (Figure 1). Five (Ctgf, Col5a2, Gas6, Frzb, Jund) of these transcripts had previously been identified in bone or cartilage.
Eight transcripts (Hapln1, LOC689064, Col1a2, Col9a1, Smoc2, Rpl37, LOC497729, Cast) were uniquely highly expressed within the RZ (Figure 1). Only 3 (Cola2, Col9a1 and Cast) of these transcripts had previously been identified in bone or cartilage.
Real time RT-PCR
To confirm the microarray result, real-time quantitative RT-PCR was performed with a set of rat specific primers and template cDNA generated by reverse-transcription PCR. The primers were designed to selected representative genes among the top 30 most highly expressed genes in each zone as determined by microarray filters described previously. These genes included matrix Gla protein (Mgp), growth arrest specific 6 (Gas6), thrombospondin 1 (Thbs1), frizzled-related protein (Frzb), procollagen type IX alpha 1 (Col9a1), procollagen type X alpha 1 (Col10a1), secreted acidic cysteine rich glycoprotein (Sparc), cyclin-dependent kinase inhibitor 1C (Cdkn1c), hemoglobin beta-2 subunit (LOC689064), procollagen type I alpha 2 (Col1a2), connective tissue growth factor (Ctgf), stearoyl-coenzyme A desaturase 2 (Scd2), procollagen type XXVII alpha 1 (Col27a1), Sparc-related modular calcium binding 2 (Smoc2), procollagen type V alpha 2 (Col5a2). All samples were run in triplicate along with 18S rRNA as reference gene and controls containing no template. The LOG2 ratios of microarray and real time PCR data for both the PC and RZ zones were analyzed, indicating that the real time PCR results highly correlated with the microarray and confirmed these gene expression levels in both the PC and RZ (Table 5).
Table 5.
LOG2 Ratio of microarray and real time PCR between PC/RZ, RZ/PZ, PC/PZ
| PC/RZ | RZ/PZ | PC/PZ | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7days | 11days | 16days | 7days | 11days | 16days | 7days | 11days | 16days | |||||||||||
| Gene Title | Gene Symbol | MC | PCR | MC | PCR | MC | PCR | MC | PCR | MC | PCR | MC | PCR | MC | PCR | MC | PCR | MC | PCR |
| procollagen, type II, alpha 1 | Col2a1 | −0 11 | −1.69 | 0.46 | −0.44 | 0.72 | 3.69 | 0.68 | 4.67 | −0.17 | −0.40 | −1.27 | 0.57 | 2.98 | 0.30 | 0.32 | 2.42 | ||
| secreted acidic cysteine rich glycoprotein | Sparc | −0.49 | −1.52 | 0.58 | −2.64 | 2.21 | 6.17 | 2.98 | 3.64 | 0.47 | 2.70 | −1.32 | −4.64 | 2.49 | 2.12 | 1.05 | 1.30 | 0.90 | 1.53 |
| matrix Gla protein | Mgp | −0.71 | −1.29 | 1.00 | −1.10 | 5.47 | 7.08 | 3.36 | 5.13 | 4.26 | 9.10 | −0.92 | −1.89 | 2.66 | 3.85 | 5.26 | 8.01 | 4.55 | 5.19 |
| connective tissue growth factor | Ctgf | −0.29 | −1.70 | 0.89 | 0.07 | 1.51 | 3.53 | 0.06 | 3.42 | 1.20 | 1.60 | 0.14 | 0.41 | −0.23 | 1.72 | 2.09 | 7.31 | 1.65 | 3.94 |
| procollagen, type V, alpha 2 | Col5a2 | 0.75 | −0.36 | −0.91 | −0.64 | 2.55 | 2.76 | 2.05 | 4.58 | 5.30 | 3.72 | 2.28 | 2.80 | 4.22 | 4.39 | 6.27 | 5.03 | ||
| growth arrest specific 6 | Gas6 | −0.41 | −1.48 | −0.13 | −2.77 | 5.00 | 7.14 | 1.58 | 4.21 | 2.93 | −0.16 | −1.37 | 1.17 | 2.73 | 2.80 | 4.84 | 5.78 | ||
| Thrombospondin 1 | Thbs1 | 1.25 | 1.58 | 0.81 | 0.81 | 2.91 | 4.95 | −0.37 | 0.45 | 1.49 | −1.67 | −2.40 | 0.87 | 2.03 | 2.30 | 1.24 | 2.55 | ||
| procollagen, type XI, alpha 1 | Col11a1 | −0.20 | −0.70 | −0.08 | −1.23 | 0.33 | 2.47 | −0.14 | −0.97 | −0.07 | −2.04 | −3.40 | −0.34 | 1.34 | −0.15 | −1.72 | −0.94 | ||
| frizzled-related protein | Frzb | −0.85 | −0.74 | 2.85 | 0.25 | 6.11 | 5.67 | 4.88 | 7.11 | 0.09 | −3.19 | −1.63 | 4.03 | 7.11 | 2.94 | 2.92 | 4.05 | ||
| procollagen, type I, alpha 2 | Col1a2 | −0.22 | −1.31 | −0.96 | −5.47 | 0.29 | 2.07 | 3.65 | 5.95 | 5.19 | 5.23 | 3.62 | 3.43 | 4.64 | 4.23 | 5.52 | 5.68 | ||
| Procollagen, type XXVII, alpha 1 | Col27a1 | −1.22 | −2.08 | −0.29 | −2.86 | 1.44 | 2.58 | 2.71 | 5.35 | −0.86 | −1.60 | −2.08 | 1.50 | 3.27 | −1.15 | −0.16 | 0.49 | ||
| hypothetical gene supported by NM_172157 | LOC497729 | 0.52 | 2.66 | −0.46 | 1.79 | −1.52 | 1.36 | −0.39 | −0.53 | 2.37 | 3.50 | 0.80 | −0.71 | 0.13 | 2.13 | 1.92 | 1.75 | −0.73 | 0.65 |
| SPARC related modular calcium binding 2 | Smoc2 | −0.80 | −1.47 | 1.77 | 1.02 | 4.93 | 3.30 | 5.85 | 5.15 | 2.59 | −0.98 | −1.00 | 5.06 | 3.68 | 4.36 | 3.95 | 2.30 | ||
| cyclin-dependent kinase inhibitor 1C (P57) | Cdkn1c | 0.69 | 0.02 | 0.46 | 0.50 | 0.92 | 0.22 | 0.52 | 3.39 | −0.24 | −3.31 | −3.00 | 1.21 | 3.41 | 0.22 | −2.39 | −2.78 | ||
| stearoyl-Coenzyme A desaturase 2 | Scd2 | −0.32 | −1.47 | −0.57 | −1.79 | 1.11 | 2.78 | 0.53 | 2.84 | 1.86 | −2.01 | −2.51 | 0.22 | 1.37 | 1.29 | −0.90 | 0.27 | ||
| procollagen, type X, alpha 1 | Col10a1 | 2.20 | 1.96 | 3.90 | 2.82 | 5.48 | 5.36 | −0.49 | 1.52 | −0.38 | −8.00 | −4.64 | 1.71 | 3.47 | 3.52 | −2.52 | −0.91 | ||
| similar to Hemoglobin beta-2 subunit | LOC689064 | −6.01 | −6.93 | −8.47 | −11.69 | −4.06 | −2.86 | 7.66 | 8.72 | 9.20 | −1.11 | −0.04 | 1.64 | 1.79 | 0.73 | −5.16 | −2.90 | ||
| Correlation coefficient R | 0.9136 | 0.8792 | 0.8486 | 0.8482 | 0.95 | 0.8549 | 0.7471 | 0.7572 | 0.9414 | ||||||||||
Note: LOG2 ratios of microarray (MC) and real time PCR (PCR) between zones (PC/RZ, RZ/PZ and PC/PZ) show high correlation coefficient ( R = 0.7471~0.9414) and confirmed the microarray data.
Pathway Analysis According to Zone
Functional analysis using the 30 most highly expressed genes from each zone revealed 37 pathways that showed enrichment with a minimum of 5 probe sets per pathway from our data set and a minimum enrichment score of 5. Eighteen (49%) of the 37 pathways overlapped the two zones, but there were 13 pathways (35%) unique to the PC and 6 pathways (16%) unique to the RZ. Nine (24%) of the 37 pathways involved bone, cartilage, matrix, and/or skeletal development (BCMSD). Of those, three pathways were unique to the PC: GO ID 1503 ossification, 46849 bone remodeling, and 51216 cartilage development; none were unique to the RZ, the rest overlapping the two zones: 1501 skeletal development, 5201 extracellular matrix structural constituent, 5578 proteinaceous extracellular matrix, 5581 collagen, 31012 extracellular matrix, 44420 extracellular matrix part. Other overlapping and unique pathways not related to BCMSD are shown in Table 6.
Table 6.
Enriched pathways related to other than bone, cartilage, matrix and/or skeletal development comprised of 5 or more probe sets from our data and their corresponding zones
| GO ID | GO Term | GP Zone(s) | |
|---|---|---|---|
| 902 | cell morphogenesis | PC | |
| 3735 | structural constituent of ribosome | RZ | |
| 5198 | structural molecule activity | PC | RZ |
| 5509 | calcium ion binding | PC | |
| 5576 | extracellular region | PC | RZ |
| 5615 | extracellular space | PC | RZ |
| 5829 | cytosol | RZ | |
| 5840 | ribosome | RZ | |
| 6412 | translation | RZ | |
| 6811 | ion transport | PC | RZ |
| 6817 | phosphate transport | PC | RZ |
| 6820 | anion transport | PC | RZ |
| 7155 | cell adhesion | PC | RZ |
| 8283 | cell proliferation | PC | |
| 9059 | macromolecule biosynthetic process | RZ | |
| 9887 | organ morphogenesis | PC | |
| 9888 | tissue development | PC | RZ |
| 15698 | inorganic anion transport | PC | RZ |
| 16337 | cell-cell adhesion | PC | |
| 22610 | biological adhesion | PC | RZ |
| 30529 | ribonucleoprotein complex | RZ | |
| 31214 | biomineral formation | PC | |
| 32989 | cellular structure morphogenesis | PC | |
| 44421 | extracellular region part | PC | RZ |
| 48513 | organ development | PC | RZ |
| 48731 | system development | PC | |
| 48771 | tissue remodeling | PC | |
| 65008 | regulation of biological quality | PC | |
Pathway analysis of the list of 33 genes which were derived from RZ and PC by filtering for an expression level of five times changed and significance by ANOVA showed 30 enriched pathways (minimum 2 probe sets, fold enrichment score 5 or greater, p≤0.05). Twelve pathways involved bone, cartilage, matrix, and/or skeletal development, 8 of which overlapped with those pathways identified by similar pathway analysis of the 30 most highly expressed genes (1503 ossification, 46849 bone remodeling, 1501 skeletal development, 5201 extracellular matrix structural constituent, 5578 proteinaceous extracellular matrix, 5581 collagen, 31012 extracellular matrix, 44420 extracellular matrix part) described earlier. Only four additional but closely related pathways (5583 fibrillar collagen, 5584 collagen type I, 8147 structural constituent of bone, 5588 collagen type V) were uncovered by this analysis (Table 7).
Table 7.
Perichondral zone (PC) and Reserve Zone (RZ) enriched pathways of 33 genes by two-way ANOVA showing hypergeometric p values ≤ 0.05, FER ≥ 5 and ≥ 2 probe sets. Gray indicates pathways association with bone, cartilage, matrix and/or skeletal development.
| GO Category | GO ID | GO Term | % of Array | % of list | Fold Enrichment | P value |
|---|---|---|---|---|---|---|
| Biological | 48513 | organ development | 5.52% | 34.69% | 6.29 | 0.000000 |
| Biological | 1501 | skeletal development | 1.09% | 24.49% | 22.53 | 0.000000 |
| Cellular | 5578 | proteinaceous extracellular matrix | 0.90% | 24.49% | 27.11 | 0.000000 |
| Cellular | 31012 | extracellular matrix | 0.92% | 24.49% | 26.64 | 0.000000 |
| Biological | 9888 | tissue development | 1.52% | 20.41% | 13.40 | 0.000000 |
| Biological | 1503 | ossification | 0.59% | 16.33% | 27.66 | 0.000000 |
| Biological | 31214 | biomineral formation | 0.59% | 16.33% | 27.66 | 0.000000 |
| Biological | 46849 | bone remodeling | 0.68% | 16.33% | 24.10 | 0.000000 |
| Biological | 48771 | tissue remodeling | 0.75% | 16.33% | 21.82 | 0.000000 |
| Biological | 6817 | phosphate transport | 0.35% | 14.29% | 41.01 | 0.000000 |
| Biological | 15698 | inorganic anion transport | 0.66% | 14.29% | 21.60 | 0.000000 |
| Biological | 6820 | anion transport | 0.80% | 14.29% | 17.93 | 0.000000 |
| Cellular | 5583 | fibrillar collagen | 0.06% | 14.29% | 246.03 | 0.000000 |
| Cellular | 5581 | collagen | 0.18% | 14.29% | 79.08 | 0.000000 |
| Cellular | 44420 | extracellular matrix part | 0.43% | 14.29% | 33.05 | 0.000000 |
| Molecular | 5201 | extracellular matrix structural constituent | 0.33% | 14.29% | 43.85 | 0.000000 |
| Molecular | 5198 | structural molecule activity | 2.59% | 14.29% | 5.51 | 0.000221 |
| Biological | 30198 | extracellular matrix organization and biogenesis | 0.25% | 10.20% | 41.62 | 0.000000 |
| Biological | 43062 | extracellular structure organization and biogenesis | 0.53% | 10.20% | 19.17 | 0.000006 |
| Biological | 6800 | oxygen and reactive oxygen species metabolic process | 0.60% | 10.20% | 16.92 | 0.000011 |
| Cellular | 5584 | collagen type I | 0.01% | 8.16% | 632.65 | 0.000000 |
| Molecular | 8147 | structural constituent of bone | 0.03% | 8.16% | 316.33 | 0.000000 |
| Molecular | 5506 | iron ion binding | 1.03% | 6.12% | 5.97 | 0.012311 |
| Cellular | 5588 | collagen type V | 0.02% | 4.08% | 210.88 | 0.000036 |
| Molecular | 8133 | collagenase activity | 0.02% | 4.08% | 253.06 | 0.000024 |
| Molecular | 4222 | metalloendopeptidase activity | 0.32% | 4.08% | 12.65 | 0.010439 |
| Molecular | 16705 | oxidoreductase activity, acting on paired donors | 0.45% | 4.08% | 8.97 | 0.019554 |
| Molecular | 4866 | endopeptidase inhibitor activity | 0.55% | 4.08% | 7.44 | 0.027229 |
| Molecular | 30414 | protease inhibitor activity | 0.55% | 4.08% | 7.40 | 0.027509 |
| Molecular | 8237 | metallopeptidase activity | 0.65% | 4.08% | 6.30 | 0.036338 |
Discussion
Our findings support the hypothesis that significant differential gene expression exists between perichondral and reserve chondrocytes. A number of genes never reported previously to play a role in the growth plate were identified as showing differential expression in either the PC or RZ, suggesting unique functions for these two zones.
Highly Expressed Genes and Pathways in Both PC and RZ
By the presence of 8 genes known to be involved in growth plate extracellular matrix (ECM) function among the 21 most highly expressed genes in both the PC and RZ, it would appear that support of the extracellular matrix is an important function of both zones. The most highly expressed single gene in both zones was Col2a1, which is the single gene coding for collagen type II protein, the major protein of extracellular cartilage matrix. Genes for the two minor components of growth plate collagen, collagen types IX and XI, were also highly expressed in both the RZ and PC. Col9a3 may contribute to the three-dimensional integrated structure of type II collagen molecules [10]. Col9a1 is required for long term tissue stability by mediating interactions between fibrillar and extrafibrillar macromolecules [11]. Type XI collagen (Col11a1) is a component of the collagen fibrillar network found in cartilage and consists of three genetically distinct polypeptide chains: α1, α2 and α3 [12]. Matrix GLA protein (Mgp) is a mineral binding extracellular matrix protein synthesized by growth plate cartilage chondrocytes [13]. In mammalian growth plate, Mgp has been reported to be expressed by proliferative and late hypertrophic chondrocytes. Coordinately regulated levels of Mgp during chondrocyte differentiation are crucial for chondrocyte survival and mineralization [14]. Secreted Protein Acidic and Rich in Cystein/osteonectin (Sparc) is a nonstructural matricellular protein involved in cell-matrix interaction during tissue remodeling and embryonic development. Sparc modulates ECM synthesis and turnover through its effect on collagen and extracellular proteases [15]. Thrombospondin 1 (Thbs1) is an extracellular modular glycoprotein that—like Sparc—modulates cell-matrix interactions. It is pleiotropic in function and affects processes as disparate as bone growth and hemostasis. Pericellular levels of the matrix metalloproteinase, Mmp2, are controlled to a large extent by Thbs1 and Thbs2 [16]. High expression of both Sparc and Thbs1 in both zones was confirmed by RT-PCR.
Pathway analysis further supported the results from the individual gene expression. Of the six enhanced pathways shared by zones and involving bone, cartilage, matrix, or skeletal development (BCMSD), five of the six were directly related to some aspect of the extracellular matrix. These included extracellular matrix structural constituent, proteinaceous extracellular matrix, collagen, extracellular matrix, and extracellular matrix part pathways. The only other shared enhanced pathway involved skeletal development. Also highly expressed in both the RZ and PC was catenin beta-1 (Ctnnb1), which has a dual role in stabilizing cell-cell adhesion and transducing canonical Wnt signaling [17]. Its role of repressing chondrocytic differentiation in the Wnt/β-catenin pathway also interacts with the Ihh pathway in distinct ways to control chondrocyte proliferation, hypertrophy, and survival during enchondral bone growth [18]. Catenins have been reported previously to reside in the murine growth plate, mostly in the zone of hypertrophy, but to our knowledge this is the first report of catenins in the RZ or PC [19].
Three additional highly expressed genes in both the RZ and the PC are predominately involved in protein synthesis. Eukaryotic elongation factor 1A (Eef1A1) is a core component of the protein synthesis machinery involved at the onset of cell transformation. Ribosomal protein L35a (Rpl35a) is phosphorylated and activated by Rpsk 1, a major protein kinase involved in translation initiation, resulting in selectively increased translation of mRNA encoding for elongation factors and ribosomal proteins [20]. Carboxypeptidase E (Cpe) is a major enzyme in the biosynthesis of numerous neuroendocrine peptides that are involved in a wide variety of physiological processes [21].
The remainder of the highly expressed genes common to the two zones are more ubiquitous and not previously described specifically in growth plate chondrocytes to our knowledge. NADH dehydrogenase subunit 3 (ND3) belongs to the intricate membrane-bound enzyme family of the mitochondrial respiratory chain. High mobility group box 1 (Hmgb 1) is a nuclear DNA-binding protein that is widely distributed [22]. Although described previously in primary osteoblasts and osteoclasts, it has not yet been reported in the growth plate [23]. Reticulocalbin (Rcn1), which is distributed predominantly in endocrine and exocrine organs, is one member of the Ca2+-binding proteins in the secretory pathway [24]. Carnitinine palmitoyltransferase 1A (Cpt1a) is the key regulatory enzyme of hepatic long-chain fatty acid β-oxidation [25]. Cpt1A is also known to regulate apoptotic processes [26]. Gja1, also known as connexin 43, is a member of the larger family of connexins, the subunits of gap junctions, and it’s mutation has been shown to cause skeletal malformations [27].
Highly Expressed Genes and Pathways Unique to PC
Very few of the highly expressed genes unique to the PC have been previously described to have a major role in the growth plate. Of those that have are Ctgf and Frzb. Two additional genes (JunD and Gas6) are related to the Mapk pathway. All of those except JunD were examined and confirmed by RT-PCR in the current manuscript to be uniquely highly expressed in the PC.
Connective tissue growth factor (Ctgf, Ccn2) is a secreted protein that mediates interactions with growth factors, integrins and extracellular matrix components. Ctgf is suggested to mediate collagen deposition during wound healing and is important for cell proliferation and matrix remodeling during chondrogenesis. In the growth plate, Ctgf is a key regulator coupling extracellular matrix remodeling to angiogenesis. Ctgf deficiency leads to skeletal dysmorphisms as a result of impaired chondrocyte proliferation and extracellular matrix composition within the hypertrophic zone, but it has not been previously isolated to the PC [28, 29]. Frzb encodes for secreted frizzled-related protein 3 (sFRP3), a glycoprotein that antagonizes the signaling of wingless (wnt) ligands through the frizzled membrane-bound receptors that are required for maintaining cartilage integrity [30]. Through its influence on Wnt signaling, Frzb is a powerful and direct modulator of chondrocyte maturation. In the growth plate, Frzb-1 expression was reported previously to be strong in prehypertrophic chondrocytes. [31].
Two of the genes highly expressed by the PC and not by the RZ that also are regulated in part by the Mapk pathway are JunD, and Gas6. JunD is a member of the Jun family proteins which play critical roles in cell growth and cell apoptosis, bone remodeling, and apoptotic phenomena [32]. Much is known about JunD signaling in osteoblasts, but little is known regarding its function in chondrocytes [33]. Growth arrest-specific gene 6 (Gas6) was originally identified in fibroblasts as a gene whose expression is upregulated in growth arrest. Its role in the growth plate is not established. Gas6 acts directly on mature osteoclasts through activation of Tyro 3 and p42/p44 Mapk, possibly contributing to the bone loss associated with estrogen deficiency [34]. Gas6 has been shown to negatively regulate chondrogenic differentiation through the Mapk pathway. The gene coding for the α2 chain of human type V collagen (Col5a2) accounts for a relatively minor component of tissues rich in collagen I, such as bones, blood vessels, and tendons, where collagen V copolymerizes with collagens I and III to form heterotopic I/III/V fibrils [35]. In this manuscript, Col5a2 high expression in the PC was confirmed by RT-PCR. Tpt1 encodes the Translationally Controlled Tumor Protein (TCTP), which interacts with translation factor Eef-1a, a gene that is reported in this study to be highly expressed in both the RZ and PC [36]. Mortality factor on chromosome 4 (Morf4) is a member of the MRG (Morf-related genes) protein family, which plays a vital role in embryonic development, cell proliferation and cellular senescence [37]. Rho GDP-dissociation inhibitor 2 (Gdi2) binds to various Rho proteins and regulates their function in cell adhesion and migration, as well as multiple cellular activities including proliferation, apoptosis, and transcription [38]. The three pathways identified to be unique to the perichondrium by pathway analysis included those for ossification, bone remodeling, and cartilage development.
Highly Expressed Genes Unique to RZ
Similar to the PC, the RZ has only a few of the most highly expressed genes that have been previously described in the growth plate. These include Hapln1 and Col9a1. In addition, three genes highly expressed in this zone (Col1a2, SMOC-2, Cast) have previously been described in bone. The others are reported here as previously undescribed growth plate genes which, based upon their high level of expression in the RZ, may play an important role in the growth plate. There were no pathways identified as being uniquely enriched in the RZ by our analysis filters.
Hyaluronan and proteoglycan link protein (Hapln1) is an unsulfated glycosaminoglycan consisting of a single repeating disaccharide unit that comprises up to 10% of cartilage glycosaminoglycans [39]. Col9a1 is one of the main proteoglycan components of hyaline cartilage. The major proteinaceous component of the bone matrix, collagen I, is encoded for in part by Col1a2, which has been previously identified in the HZ of the growth plate in our own work [Error! Bookmark not defined.]. Secreted modular calcium-binding protein (SMOC-2) is a Sparc related protein that is expressed in nearly all tissues and has been demonstrated to show an up-regulation in response to injury [40]. Calpastatin (Cast) is an endogenous inhibitor protein acting specifically on calpain that has a known role in bone in the regulation of osteoclastogenesis, pre-osteoblastic proliferation and differentiation [41].
Of the three genes shown by microarray to be most highly expressed uniquely in the RZ, one (LOC497729) was confirmed by real-time PCR in the current work. Gene 6A3-5 (Transcription factor 1, 6A3-5) (LOC497729) is a member of the ARID transcription factor family involved in control of gene expression during cell growth, cell cycle, and organism development [42]. Ribosomal protein L37 (Rpl37) is the gene for ribosomal protein, the expression of which is likely determined mainly by cellular growth and proliferation [43].
In summary, the current work identified a number of highly expressed genes in both the RZ and PC which related to extracellular matrix and skeletal development. In the perichondrium, Ctgf is uniquely highly expressed along with components of the Mapk signaling cascade. Wnt/b-catenin signaling is a common theme among genes highly expressed within both zones and some (Frzb) uniquely expressed in the PC. While no single pathway was identified among the highly expressed genes unique to the RZ, additional matrix components and genes related to Sparc were identified. These pathways should be the targets of additional evaluation to determine their roles not only in the normal growth plate but in response to injury.
Acknowledgements
The authors wish to acknowledge the contributions of Joseph Spadaro, Judy Strauss, Bryan Margulies and Karen Gentile for their assistance with this project.
Funding Sources: National Institutes of Health (National Cancer Institute) grant R01-CA83892 and David G. Murray Endowed Professorship
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ballock RT, O’Keefe RJ. Physiology and pathophysiology of the growth plate. Birth Defects Res Part C Embryo Today. 2003;69:123–143. doi: 10.1002/bdrc.10014. [DOI] [PubMed] [Google Scholar]
- 2.der Eerden BC, Karperien M, Wit JM. Systemic and local regulation of the growth plate. Endocr Rev. 2003;24:782–801. doi: 10.1210/er.2002-0033. [DOI] [PubMed] [Google Scholar]
- 3.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 4.Rabie AB, Tang GH, Xiong H, Hagg U. PTHrP regulates chondrocyte maturation in condylar cartilage. J Dent Res. 2003;82:627–631. doi: 10.1177/154405910308200811. [DOI] [PubMed] [Google Scholar]
- 5.Schipani E, Provot S. PTHrP, PTH, and the PTH/PTHrP receptor in endochondral bone development. Birth Defects Res Part C Embryo Today. 2003;69:352–362. doi: 10.1002/bdrc.10028. [DOI] [PubMed] [Google Scholar]
- 6.Yakar S, Rosen CJ, Beamer WG, Ackert-Bicknell CL, Wu Y, Liu JL, et al. Circulating levels of IGF-1 directly regulates bone growth and density. J Clin Invest. 2002;110:771–781. doi: 10.1172/JCI15463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 8.Serra R, Karaplis A, Sohn P. Parathyroid hormone-related peptide (PTHrP)-dependent and -independent effects of transforming growth factor beta (TGF-beta) on endochondral bone formation. J Cell Biol. 1999;145:783–794. doi: 10.1083/jcb.145.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Middleton F, Horton JA, Reichel L, Farnum CE, Damron TA. Microarray analysis of proliferative and hypertrophic growth plate zones identifies differentiation markers and signal pathway. Bone. 2004;35:1273–1293. doi: 10.1016/j.bone.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Asamura K, Abe S, Imamura Y, Aszodi A, Suzuki N, Hashimoto S, Takumi Y, Hayashi T, Fassler R, Nakamura Y, Usami S. Type IX collagen is crucial for normal hearing. Neuroscience. 2005;132:493–500. doi: 10.1016/j.neuroscience.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Hagg R, Hedbom E, llers UM, Aszo' di A, Faässler R, Bruckner P. Absence of the alpha 1(IX) chain leads to a functional knock-out of the entire collagen IX protein in mice. J Biol Chem. 1997;272:20650–20654. doi: 10.1074/jbc.272.33.20650. [DOI] [PubMed] [Google Scholar]
- 12.Matsuo N, Wang Y, Sumiyoshi H, Sakata-Takatanit K, Nagato H, Sakai K, Sakurai M, Yoshioka H. Transcription factor CCAAT-binding Factor CBF/NF-Y regulates the proximal promoter activity in the human alpha 1 (XI) collagen gene (COL11A1) J Biol Chem. 2003;278:32763–32770. doi: 10.1074/jbc.M305599200. [DOI] [PubMed] [Google Scholar]
- 13.Hale JE, Fraser JD, Price PA. The identification of matrix Gla protein in cartilage. J Biol Chem. 1988;263:5820–5824. [PubMed] [Google Scholar]
- 14.Newman B, Gigout LI, Sudre L, Grant ME, Wallis GA. Coordinated expression of matrix Gla protein is required during endochondral ossification for chondrocyte survival. J Cell Biol. 2001;154:659–666. doi: 10.1083/jcb.200106040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alford AI, Hankenson KD. Matricellular proteins: Extracellular modulators of bone development, remodeling, and regeneration. Bone. 2006;38:749–757. doi: 10.1016/j.bone.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 16.Bornstein P, Agah A, Kyriakides TR. The role of thrombospondins 1 and 2 in the regulation of cell-matrix interactions, collagen fibril formation, and the response to injury. Int J Biochem Cell Biol. 2004;36:1115–1125. doi: 10.1016/j.biocel.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryu JH, Kim SJ, Kim SH, Oh CD, Hwang SG, Chun CH, Oh SH, Seong JK, Huh TL, Chun JS. Regulation of the chondrocyte phenotype by beta-catenin. Development. 2002;129:5541–5550. doi: 10.1242/dev.129.23.5541. [DOI] [PubMed] [Google Scholar]
- 19.Sampson HW, Dearman AC, Akintola AD, Zimmer WE, Parrish AR. Immunohistochemical localization of cadherin and catenin adhesion molecules in the murine growth plate. J Histochem Cytochem. 2007;55:845–852. doi: 10.1369/jhc.7A7184.2007. [DOI] [PubMed] [Google Scholar]
- 20.Suryawan A, Davis TA. Developmental regulation of protein kinase B activation is isoform specific in skeletal muscle of neonatal pigs. Pediatric Research. 2006;59:175–179. doi: 10.1203/01.PDR.0000180536.51032.AB. [DOI] [PubMed] [Google Scholar]
- 21.Leiter EH. Carboxypeptidase E and obesity in the mouse. Journal of Endocrinology. 1997;155:211–214. doi: 10.1677/joe.0.1550211. [DOI] [PubMed] [Google Scholar]
- 22.Yang H, Tracey KJ. High mobility group box 1 (HMGB1) Crit Care Med. 2005;33:S472–S474. doi: 10.1097/01.ccm.0000187005.81616.a9. [DOI] [PubMed] [Google Scholar]
- 23.Charoonpatrapong K, Shah R, Robling AG, Alvarez M, Clapp DW, Chen S, Kopp RP, Pavalko FM, Yu J, Bidwell JP. HMGB1 expression and release by bone cells. J. Cell. Physiol. 2006;207:480–490. doi: 10.1002/jcp.20577. [DOI] [PubMed] [Google Scholar]
- 24.Fukuda T, Oyamada H, Isshiki T, Maeda M, Kusakabe T, Hozumi A, Yamaguchi T, Igarashi T, Hasegawa H, Seidoh T, Suzuki T. Distribution and variable expression of secretory pathway protein reticulocalbin in normal human organs and non-neoplastic pathological conditions. J Histochemistry & Cytochemistry. 2007;55:335–345. doi: 10.1369/jhc.6A6943.2006. [DOI] [PubMed] [Google Scholar]
- 25.Gobin S, Thuillier L, Jogl G, Faye A, Tong L, Chi M, Bonnefont J, Girard J, Prip-Buus C. Functional and structural basis of carnitine palmitoyltransferase 1A deficiency. J Biol Chem. 2003;278:50428–50434. doi: 10.1074/jbc.M310130200. [DOI] [PubMed] [Google Scholar]
- 26.Karlic H, Louda N, Pfeilstocker M, Keil F, Lohninger A, Pittermann E, Paukovits J. Effect of the hemoregulatory peptide (Peedck)2 (PYROgLU-Glu-Asp-Cys-Lys)2 and MIP-1alpha is reduced in bone marrow cultures from patients with chronic myeloid leukemia (CML) Stem Cells. 2001;19:321–328. doi: 10.1634/stemcells.19-4-321. [DOI] [PubMed] [Google Scholar]
- 27.Iovine MK, Higgins EP, Hindes A, Coblitz B, Johnson SL. Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Developmental Biology. 2005;278:208–219. doi: 10.1016/j.ydbio.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Ivkovic S, Yoon BS, Popoff SN, Safadi FF, Libuda DE, Stephenson RC, Daluiski A, Lyons KM. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development. 2003;130:2779–2791. doi: 10.1242/dev.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukunaga T, Yamashiro T, Oya S, Takeshita N, Takigawa M, Takano-Yamamoto T. Connective tissue growth factor mRNA expression pattern in cartilages is associated with their type I collagen expression. Bone. 2003;33:911–918. doi: 10.1016/j.bone.2003.07.010. [DOI] [PubMed] [Google Scholar]
- 30.Loughlin J, Dowling B, Chapman K, Marcelline L, Justafa Z, Southam L, Ferreira A, Ciesielski C, Carson DA, Corr M. Functional variants within the secreted frizzled-related protein 3 gene are associated with hip osteoarthritis in females. Proc Natl Acad Sci USA. 2004;101:9757–9762. doi: 10.1073/pnas.0403456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamamura Y, Otani T, Kanatani N, Koyama E, Kitagaki J, Komori T, Yamada Y, Costantini F, Wakisaka S, Pacifici M, Iwamoto M, Enomoto-Iwamoto M. Developmental regulation of Wnt/beta-catenin signals is required for growth plate assembly, cartilage integrity, and endochondral ossification. J Biol Chem. 2005;280:19185–19195. doi: 10.1074/jbc.M414275200. [DOI] [PubMed] [Google Scholar]
- 32.Papachristou D, Pirttiniemi P, Kantomaa T, Agnantis N, Basdra EK. Fos- and Jun-related transcription factors are involved in the signal transduction pathway of mechanical loading in condylar chondrocytes. Eur J Orthod. 2006;28:20–26. doi: 10.1093/ejo/cji101. [DOI] [PubMed] [Google Scholar]
- 33.Lai CF, Cheng SL. Signal transductions induced by bone morphogenetic protein-2 and transforming growth factor-beta in normal human osteoblastic cells. J Biol Chem. 2002;277:15514–15522. doi: 10.1074/jbc.M200794200. [DOI] [PubMed] [Google Scholar]
- 34.Katagiri M, Hakeda Y, Chikazu D, Ogasawara T, Takato T, Kumegawa M, Nakamura K, Kawaguchi H. Mechanism of stimulation of osteoclastic bone resorption through Gas6/Tyro 3, a receptor tyrosine kinase signaling k in mouse osteoclasts. J Biol Chem. 2001;276:7376–7382. doi: 10.1074/jbc.M007393200. [DOI] [PubMed] [Google Scholar]
- 35.Chanut-Delalande H, Bonod-Bidaud C, Cogne S, Malbouyres M, Ramirez F, Fichard A, Ruggiero F. Development of a functional skin matrix requires deposition of collagen V heterotrimers. Mol Cell Biol. 2004;24:6049–6057. doi: 10.1128/MCB.24.13.6049-6057.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andree H, Thiele H, Fahling M, Schmidt I, Thiele BJ. Expression of the human TPT1 gene coding for translationally controlled tumor protein (TCTP) is regulated by CREB transcription factors. Gene. 2006;380:95–103. doi: 10.1016/j.gene.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 37.Zhang P, Du J, Sun B, Dong X, Xu G, Zhou J, Huang Q, Liu Q, Hao Q, Ding J. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Research. 2006;34:6621–6628. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishizaki H, Togawa A, Tanaka-Okamoto M, Hori K, Nishimura M, Hamaguchi A, Imai T, Takai Y, Miyoshi J. Defective chemokine-directed lymphocyte migration and development in the absence of Rho guanosine diphosphate-dissociation inhibitors alpha and beta. J Immunol. 2006;177:8512–8521. doi: 10.4049/jimmunol.177.12.8512. [DOI] [PubMed] [Google Scholar]
- 39.Laurent TC, Laurent UB, Fraser JR. The structure and function of hyaluronan; an overview. Immunol Cell Biol. 1996;74:A1–A7. doi: 10.1038/icb.1996.32. [DOI] [PubMed] [Google Scholar]
- 40.Rocnik EF, Liu P, Sato K, Walsh K, Vaziri C. The novel SPARC family member SMOC-2 potentiates anglogenic growth factor activity. J Biol Chem. 2006;281:22855–22864. doi: 10.1074/jbc.M513463200. [DOI] [PubMed] [Google Scholar]
- 41.Goll DE, Thompson VF, Li H, Wei W, Cong J. The Calpain system. Physiol Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 42.Ito T, Yamauchi M, Nishina M, Yamamichi N, Mizutani T, Ui M, Murakami M, Iba H. Identification of SWI. SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J Biol Chem. 2001;276:2852–2857. doi: 10.1074/jbc.M009633200. [DOI] [PubMed] [Google Scholar]
- 43.Bévort M, Leffers H. Down regulation of ribosomal protein mRNAs during neuronal differentiation of human NTERA2 cells. Differentiation. 2000;66:81–92. doi: 10.1046/j.1432-0436.2000.660203.x. [DOI] [PubMed] [Google Scholar]