Abstract
Transcriptional dysregulation has emerged as a central pathogenic mechanism in Huntington's disease (HD), which is associated with neuropathological changes predominantly in the striatum. Here we demonstrate that expression of Bcl11b (a.k.a. CTIP2), a transcription factor exhibiting highly-enriched localization in adult striatum, is significantly decreased in HD cells, mouse models and human subjects and that overexpression of Bcl11b attenuates toxic effects of mutant huntingtin in cultured striatal neurons. We show that Bcl11b directly activates the proximal promoter regions of striatal-enriched genes and can increase mRNA levels of striatal-expressing genes. We further demonstrate an interaction between Bcl11b and huntingtin protein in cultured cells and brain homogenates from HD R6/1 and YAC72 transgenic mice. We propose that sequestration and/or decreased expression of Bcl11b in HD is responsible, at least in part, for the dysregulation of striatal gene expression observed in HD and may contribute to the specificity of pathology observed in this disease.
Keywords: Huntington's disease, striatum, transcription factor, gene expression, promoter, interaction, specificity
Introduction
Huntington's disease (HD) is an inherited, neurodegenerative disorder caused by expansion of a trinucleotide (CAG) repeat region in the HD gene, which results in an expanded polyglutamine stretch in the encoded protein, huntingtin (htt) (Group 1993). A major enigma exists regarding the pathology in HD: although the mutated htt protein is expressed ubiquitously throughout the brain and body, the most striking neurodegenerative changes are observed in the striatum, a brain region that regulates movement and directed motor behavior, as well as cognitive function (Graybiel 2005). Since the distribution of htt is itself inadequate to explain the regional patterns of pathology that ensues in HD, a major question exists regarding the mechanism(s) for this specificity.
Diverse models for HD cellular pathogenesis have been proposed (Ross 2004). However, increasing evidence implicates transcriptional dysregulation as a central pathogenic mechanism in HD (Cha 2000; Okazawa 2003; Sugars and Rubinsztein 2003). The exact roles of htt in the regulation of transcription are not clear; however interactions between htt and several transcription factors have been demonstrated. Studies have shown that mutant htt interacts with ubiquitous transcription factors such as the cAMP response element-binding protein- (CREB)-binding protein (CBP), Specificity protein 1 (Sp1), TATA binding box protein (TBP) and widely expressed components of the basal polymerase II transcriptional machinery such as the transcription initiation factor complexes TFIID and TFIIF, and the TATA box binding protein (TBP)-associated factor (TAF4; a.k.a. TAFII130) (McCampbell et al., 2000; Shimohata et al., 2000; Dunah et al., 2002; Li et al., 2002; van Roon-Mom et al., 2002; Zhai et al., 2005). However, other mechanisms of transcription factor disruption due to mutant htt have been proposed (Chen-Plotkin et al., 2006; Sadri-Vakili et al., 2007). Overall, there is much evidence that mutant htt disrupts transcriptional regulation; consequential of this concept are gene profiling studies that have revealed dramatic expression alterations of genes, especially those exhibiting enriched expression in the striatum, in the brains of HD mouse models and human subjects with HD (Luthi-Carter et al., 2000; Zucker et al., 2005; Desplats et al., 2006; Hodges et al., 2006; Thomas 2006). While this model is widely accepted, it does not establish how only specific neuronal populations are affected in HD.
In our previous microarray studies focusing on the expression of striatal-enriched genes in HD, we identified a transcription factor, Bcl11b (a.k.a. CTIP2), that showed enriched expression in the striatum and was decreased in the brains of R6/1 transgenic mice (Desplats et al. 2006). Bcl11b is a C2H2 zinc finger DNA binding protein that has been shown to alter transcription in vitro by interacting with chicken ovalbumin upstream promoter transcription factor (COUP-TF) family members (Avram et al., 2000), or via a sequence-specific DNA binding element that is related to the canonical GC box (Avram et al., 2002). Bcl11b is a critical developmental protein. Studies on Bcl11b null mice, which die shortly after birth of unknown causes, have demonstrated important roles for Bcl11b in the immune system, where it controls T cell subtype specification and survival in the developing thymus (Wakabayashi et al., 2003). Studies in the CNS have demonstrated essential roles in the development of corticospinal motor neurons (Arlotta et al., 2005), and the differentiation of medium spiny neurons and establishment of cellular architecture of the striatum (Arlotta et al., 2008). Despite important developmental roles, its functional role in adult CNS remains unclear. Due to its highly enriched expression in adult striatum, we hypothesize that Bcl11b may regulate striatal gene transcription and hence, may represent a candidate transcription factor associated with dysregulation of striatal gene expression in HD.
In this study, we have investigated the potential role for Bcl11b in the control of striatal gene expression and further characterized Bcl11b in HD cells and transgenic mouse models expressing mutant htt, as well as in brains from human HD subjects. We present evidence indicating that Bcl11b is responsible for transcriptional control of important genes in the striatum and suggest that its dysregulation and/or aggregation caused by expanded htt protein may contribute to the striatal transcriptional deficits observed in HD.
Material and Methods
Animals and human subjects
HD R6/1 transgenic mice, expressing exon 1 of the human HD gene carrying 115 CAG repeats (Mangiarini et al., 1996) and YAC72 mice, which express a full-length human HD gene carrying 72 CAG repeats (Hodgson et al., 1999) were obtained from Jackson Laboratories. At the age of 4 weeks, mice were genotyped according to the Jackson Laboratories protocol to determine hemizygousity of the HD transgene. Mice were housed in groups of four on a standard 12/12hr light-dark cycle with ad libitum access to standard laboratory chow and tap water. All procedures were in strict accordance with the NIH Guidelines for the Care and Use of Laboratory Animals. Human postmortem caudate samples were obtained from Harvard Brain Tissue Resource Center (McLean Hospital, Belmont, MA, USA). Caudate samples consisted of three HD samples, grade 3, a 53 years old female, postmortem interval (PMI)= 16.90 h and two males, 46 and 54 years old, PMIs= 22.25 h and 15.92 h respectively, and three controls, a 58 years old female, PMI= 24.25 h and two males, 49 and 55 years old, PMIs= 27.13 h and 14.50 h respectively. The pH values of the samples ranged from 6.1 to 6.5. All samples were stored at -70°C until use.
Real-Time PCR Analysis
Real-time PCR experiments were performed using the ABI PRISMs 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA) as described previously (Desplats et al. 2006). Amplification was performed on a cDNA amount equivalent to 25 ng total RNA with 1 x SYBR® Green universal PCR Master mix (Applied Biosystems) containing deoxyribonucleotide triphosphates, MgCl2, AmpliTaq Gold DNA polymerase, and forward and reverse primers. PCR reactions were performed on two independent sets of template (n= 6 mice or human samples per condition and STHdh cells). Specific primers for each studied sequence and for mouse and human endogenous controls were designed using Primer Express 1.5 software and their specificity for binding to the desired sequences was searched against NCBI database (Supp. Table 1). Standard curves were generated for each gene of interest using serial dilutions of mouse or human cDNAs. Experimental samples and no-template controls were all run in triplicate. The PCR cycling parameters were: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 94°C for 15 s, 60°C for 1 min. Finally, a dissociation protocol was also performed at the end of each run to verify the presence of a single product with the appropriate melting point temperature for each amplicon. The amount of studied cDNA in each sample was calculated using SDS2.1 software by the comparative threshold cycle (Ct) method and expressed as 2exp(Ct) using hypoxanthine guanine phosphoribosyl transferase (Hprt) as an internal control for mice sequences, whereas β2-microglobulin (B2M) was used for the human sequences. For calculations applying the Ct method, age-matched wild-type littermate control mice, control, non-affected human individuals or mock-transfected cells were used as calibrator samples.
Statistics
Student's t test was used to determined differences in expression of Bcl11b in HD R6/1 mice and human HD caudate. One-way ANOVAs with Bonferroni post-tests for multiple comparisons followed by student's t test for determination of exact p-values were used to determined significant effects of Bcl11b on promoter activity, cell viability and gene expression in HD cells. Fisher exact probability test was used to assess the statistical significance of the overrepresentation of Bcl11b binding sites in proximal promoter regions of striatal-expressed vs. non-striatal-expressed genes, using a 2x2 contingency table. All statistical tests were performed using GraphPad software (GraphPad Prism, San Diego, CA).
Immunohistochemical/In situ hybridization analyses
Immunohistochemical experiments were performed using an anti-Bcl11b (1:2,000 dilution; Bethyl Laboratories, Montgomery, TX). The immunoreaction was detected with Vectastain ABC kit (Vector Laboratory Inc., Burlingame, CA) according to the instructions of the manufacturer. Enzymatic development was performed in 0.05% diaminobenzene in PBS containing 0.003% hydrogen peroxide for 3-5 min. Colocalization of hypocretin mRNA and Bcl11b protein was performed as described previously (Thomas et al., 2001). Briefly, in situ hybridization for hypocretin mRNA was performed using an 35S-labeled, single-stranded antisense cRNA probe against hypocretin (nts 341- 459 of accession # NM_010410) at 107 cpm/ml, followed by immunostaining for Bcl11b protein.
Luciferase Assay
HEK293 cells were plated at 3x105 cells per well in six well tissue culture plates. The following day the cells were transfected with reporter plasmids containing the following mouse sequences: substance P/tachykinin (Tac1; -871 to +10 of translation start site), adenylate cyclase V (Adcy5; -454 to -13), neuronal guanine exchange factor (Ngef; -1110 to -650) and Bcl11b (-1002 to +9) cloned into pGL3-Basic (Promega, Madison, WI) in combination with an expression plasmid containing the cDNA of mouse Bcl11b (beta isoform; nts 272-2710 of accession #AB043553). In some experiments, cells were also co-transfected with exon 1 of the human HD gene containing 25 CAG (pQ25) or 72 CAG (pQ72) repeats. Transfections were carried out using Superfect transfection reagent according to the manufacturer's instructions (Qiagen Inc, Valencia, CA). Empty expression vector, pcDNA3.1, was used to ensure all wells received the same amount of total DNA. 48 hours after transfection, cells were analyzed for luciferase activity. For luciferase assays, cell monolayers were rinsed twice with wash buffer (40 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1 mM EDTA). Cells were lysed by the addition of 1 ml of lysis buffer (20 mM K2PO4, pH 7.8, 5 mM MgCl2, 0.5% Triton X-100). The lysate was transferred to a microfuge tube and centrifuged for 2 min to pellet cell debris. Luciferase assays were performed exactly as described previously (Archer et al., 1994). The protein concentration of each extract was determined by dye binding using a commercial kit (Bio-Rad).
Co-Immunoprecipitation experiments
For the co-immunoprecipitation of interacting proteins, striata from R6/1 transgenic mice and wild type littermate controls (n = 3 animals/condition, 6 months old) and whole brains from YAC72 transgenic mice (n=2 animals/condition, 8 months old) were homogenized with Teflon arrows in IP buffer (10 mM Tris/Hcl pH 7.8, 150 mM NaCl, 1 mM EDTA, 40 % v/v Nonidet P-40, 1 mM PMSF) at a ratio of 100 μl buffer/10 mg tissue. For in vitro assays, HEK293 cells were co-transfected with a Bcl11b construct (1 μg, pcDNA3.1: beta isoform; nts 272-2710 of accession # AB043553) in combination with HD exon 1 gene constructs containing 25 or 103 CAG repeats (1 μg each, pcDNA3.1; kindly provided by the Hereditary Disease Foundation) using Lipofectin Reagent (Invitrogen, Carlsbad, CA) according to the manufacturers instructions. After 3 passages of the lysate through a 1 ml syringe, the homogenates were spun at 10,000 x g 10 min at 4°C, and supernatants were collected. For immunoprecipitation, aliquots containing 650 μg of protein (determined by the bicinchoninic acid method) were brought to a final volume of 500 μl with NET-BSA buffer (50 mM Tris/HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1 % v/v Nonidet P-40, 0.25 % w/v BSA and 0.02 % sodium azide). Homogenates were incubated for 3 hrs with either, 1 μg of rabbit anti-Bcl11b (Bethyl Laboratories, Montgomery, TX), 1 μg mouse anti-htt/exon1 antibody, EM48 (MAB5374, Chemicon International, Temecula, CA), 1 μg mouse anti-htt/C-terminal antibody, 4C8 (MAB2166, Chemicon International, Temecula, CA) or 1 μg of a normal rabbit or mouse IgG (Santa Cruz Biotechnology, Santa Cruz, CA) as control, in a rocker at 4 °C. 40 μl of protein A-sepharose/PBS 50:50 v/v was added to the homogenates and incubated overnight on a rocker at 4 °C. Collection of the conjugates was done by spinning the samples at 12,000 x g for 30 sec and washing pellets for 10 min in a rocker at 4°C using 1 ml of NET-BSA buffer and finally with 1 ml of 10 mM Tris/HCl pH 7.5 – 0.1 % Nonidet P-40. Protein-sepharose A beads were resuspended in 50 mM TRIS/HCl, pH 6.5; 1 X Nupage loading buffer (Invitrogen, Carlsbad, CA) and 40 mM DTT and heated at 100 °C for 5 min before separation by SDS-PAGE and subsequent Western blotting. Western blotting was performed using the following antibodies: α-Bcl11b, 1:2,000 dilution (Bethyl Laboratories, Montgomery, TX), rabbit α-Actin, 1:200 dilution (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-htt, EM48, 1:1,000 (Chemicon International, Temecula, CA), anti-htt, 4C8 antibody, 1:5,000 dilution (MAB2166, Chemicon International, Temecula, CA) and anti-htt, 2E8 antibody, 1:5,000 dilution (MAB2168, Chemicon International, Temecula, CA). Although useful in immunoprecipitation and immunohistochemistry experiments on mouse brain, the EM48 antibody does not produce consistent results in mouse brain in Western blot analyses, hence was only used in HEK293 cells. Equal loading of Western blots was verified by Ponceau S stain.
Striatal Cell Culture
Conditionally immortalized wild-type STHdhQ7 striatal neuronal progenitor cells expressing endogenous normal huntingtin, with 7 glutamines and homozygous mutant STHdhQ111 striatal neuronal progenitor cell lines, expressing endogenous mutant huntingtin with 111-glutamines, were a kind donation from Dr. Marcy MacDonald (Trettel et al., 2000). The striatal cell lines were grown at 33°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% non-essential amino acids, 2mML-glutamine and 400 mg/ml G418 (Sigma-Aldrich, St Louis, MO). Cells were plated at 3x105 cells per well in six well tissue culture plates. The following day the cells were transfected with an expression plasmid containing the cDNA of mouse Bcl11b (beta isoform; nts 272-2710 of accession # AB043553) using Lipofectin transfection reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Empty expression vector, pcDNA3.1, was used to ensure all wells received the same amount of total DNA. 48 hours after transfection, cells were plated on 96-well ELISA plates and analyzed for mitochondrial activity by MTT assay. Transfection efficiency was assessed in duplicate sets of transfected cells by quantifying the percentage of green fluorescent protein (GFP)- positive cells using fluorescence microscopy. By this method, we observed transfection efficiency of ∼50%.
MTT assay
The metabolic activity of STHdhQ7 and STHdhQ111 cells was determined by using the MTT assay. This method is based on the reduction of the soluble yellow MTT tetrazolium salt to blue insoluble MTT formazan product by mitochondrial succinic dehydrogenase, an enzyme which is only active in cells with an intact metabolism and respiratory chain. 48 hours after transfection cells were treated with MTT reagents using CellQuanti-MTT kit (BioAssay Systems, Hayward, CA) as per manufacturer's instructions. After incubation for 6 hours at 33°C the reaction was stopped with acidified isopropyl alcohol to solubilize the colored formazan product. Absorbance was read at 550 nm on a multiwell spectrophotometer.
Results
Bcl11b Expression is decreased in HD
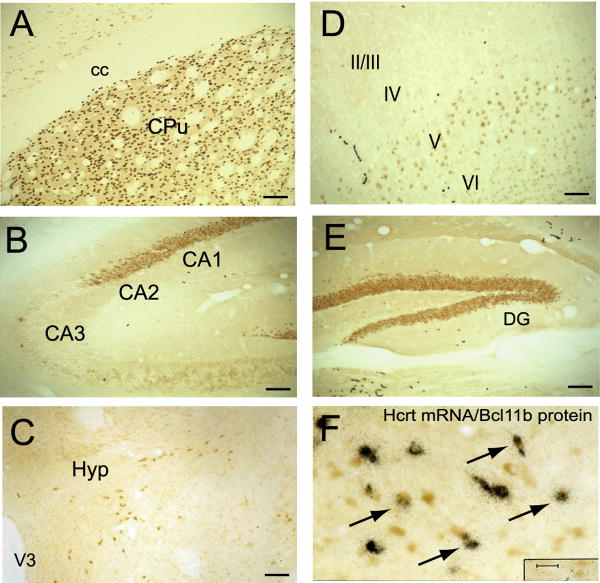
Our initial studies showed decreased expression of Bcl11b mRNA in the striata of symptomatic R6/1 transgenic mice (Desplats et al. 2006). In this study, we further investigated CNS-wide expression of Bcl11b protein and gene expression changes in R6/1 transgenic mice at different stages of disease, as well as in post-mortem caudate samples from human subjects with HD. In addition to showing high levels of expression in the striatum, Bcl11b mRNA is also expressed in other brain regions affected in HD, such as cortex, hippocampus and olfactory bulb from early embryonic stages of development (Leid et al., 2004; Arlotta et al. 2005; Desplats et al. 2006). Immunohistochemistry experiments on adult mouse brain (>2 months of age) demonstrate that Bcl11b protein expression is also expressed in these regions (Figure 1). Directly comparing the CNS-wide protein expression patterns of Bcl11b to its mRNA patterns revealed highly correlated expression profiles (Supplementary Table 2). Interestingly, we also detect expression of Bcl11b protein in the hypothalamus, specifically in a subset of hypocretin-containing neurons (Figure 1F), which are decreased in HD patients and mouse models (Petersen et al., 2005). Consistent with its role as a transcription factor, Bcl11b is localized to the nucleus of striatal neurons, as determined by precise colocalization with DAPI, which form fluorescent complexes with double-stranded DNA in the nucleus, and NeuN (Supplementary Figure 1).
Figure 1.
Localization of Bcl11b protein in adult mouse brain. Immunohistochemistry was performed using a polyclonal antibody directed against Bcl11b. Scale bar = 0.08 mm for panels A-E and 0.02 mm for panel F. Cortical layers are indicated in panel D. F, inset: co-localization of hypocretin (Hcrt) mRNA (black dots reflecting silver emulsion grains) and Bcl11b protein (brown stain) in the lateral hypothalamic area of the hypothalamus. Black arrows indicate neurons expressing both Hcrt mRNA and Bcl11b protein. CPu, caudate putamen; cc, corpus callosum; DG, dentate gyrus; Hyp, hypothalamus; V3, third ventricle.
Using real-time PCR analysis, we find that decreases in Bcl11b mRNA expression occur as early as 1 month of age in R6/1 transgenic mice and progressively decrease with increasing age (Figure 2A). These decreases detected at 1 and 2 months of age, precede the onset of disease symptoms, which typically do not begin until ∼4 months age (Mangiarini et al. 1996). Bcl11b transcript levels are also substantially reduced in the caudate of human subjects with HD (Figure 2A). Western blot analysis revealed decreases in Bcl11b protein expression in symptomatic (>6 months of age) R6/1 transgenic mice and in human caudate from HD subjects, in which an almost complete loss of protein is observed, consistent with the mRNA findings (Figure 2B).
Figure 2.
Bcl11b mRNA and protein levels are decreased in HD. (A): Real-time PCR analysis shows decrease in mRNA levels at different time points in HD R6/1 transgenic mice and in caudate samples from human subjects with HD pathology grade 3. Hprt was used as a housekeeping control for the mouse studies, while Bcl11b levels in human are shown relative to two housekeeping genes, beta-2-macroglobulin, B2M, and beta-actin (BA). Expression values are shown as fold-change. Asterisks denote significant differences from wt or control subjects as determined by student's t test (two-tailed; unpaired): *, p<0.05; **, p<0.01; ***, p<0.001 (B). Western blot analysis shows greatly reduced protein levels of Bcl11b (beta isoform) in human HD caudate and in the striatum of HD transgenic R6/1 mice. Caudate samples from three control human subjects (C1, C2 and C3) and three HD subjects (HD1, HD2 and HD3) are shown. Striata from three individual wt and three R6/1 transgenic mice are shown. Anti-actin immunoblotting was used for a loading control. Bar graphs show densitometry quantification of the Western blot signals, normalized to beta-actin.
Bcl11b binding sites are Overrepresented in Proximal Promoter Regions of Striatal-Expressed Genes
We hypothesized that, due to its specific localization in the brain, Bcl11b may play a role in the control of striatal gene expression, and hence, its decreased expression in HD may have pathological consequences. Because Bcl11b has known consensus DNA binding sites as determined from in vitro studies [GGCCG/AG/AAGG, or the core element GGCCG/AG/A (Avram et al. 2002)], we used in silico analysis to investigate the presence of these sites in different sets of genes. Most experimentally identified transcription factor binding sites lie in the proximal promoter region, which is within 1 kb upstream, and surrounding, the transcription start site (Zhang 1998). Therefore, we examined these promoter regions for 100 genes, 50 exhibiting “striatal-enriched” expression (Desplats et al. 2006) and 50 genes with low or undetectable levels of expression in the striatum (“non-striatal” genes) (Table 1), for consensus Bcl11b binding elements using SiteSeer, TFSEARCH and TESS search engines (Schug and Overton 1997; Boardman et al., 2003). We found a significant overrepresentation of Bcl11b binding sites in the proximal promoter regions of striatal-enriched genes compared to non-striatal-expressed genes using the Fisher's exact probability test (Long et al., 2004) (two-side probability threshold; p = 0.01) (Table 2). Scanning the putative promoter regions of these 100 genes for other transcription factors revealed the presence of many sites for known transcription factors, including consensus binding sites for the ubiquitous transcription factors, COUP-TF, which has been shown to interact with Bcl11b (Lin et al., 1999), Sp1, TBP and CREB. Transcriptional control by the latter factors is disrupted by the presence of mutant htt protein, with Sp1 and CBP specifically interacting with htt protein (McCampbell et al. 2000; Steffan et al., 2000; Dunah et al. 2002; Li et al. 2002). Nonetheless, binding sites for these and other transcription factors were equally represented in striatal and non-striatal gene groups (p values ranging from 0.16 to 1.0) (Table 2), suggesting that these transcription factors have less of a contribution to the specific control of striatal gene expression.
Table 1.
List of genes with striatal-enriched expression and representation of consensus binding sites for various transcription factors in their proximal promoter regions.
| Gene ID: |
Accession #: |
Bcl11b: |
CP-TF: |
Sp1: |
CREB1: |
TBP: |
TFIID: |
USF: |
AP-1: |
Oct-1: |
MASH1: |
Rxrg: |
Rarb: |
HD: |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Htr4 | NM_008313 | 0 | 8 | 0 | 0 | 1 | 0 | 4 | 6 | 3 | 0 | 0 | 0 | - |
| Htr6 | NM_021358 | 2 | 2 | >10 | 3 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | - |
| Actn2 | NM_033268 | 2 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 1 | 0 | 0 | + |
| Adora2a | NM_009630 | 1 | 1 | 0 | 1 | 1 | 1 | 3 | 3 | 0 | 1 | 1 | 0 | + |
| Adcy5 | XM_156060 | 1 | 1 | >10 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | + |
| Arpp19 | NM_021548 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 4 | 0 | 0 | 1 | + |
| Arpp21 | NM_033264 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | + |
| B3gnt1 | NM_016888 | 2 | 0 | >10 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | + |
| 4631426-5Rik | NM_029935 | 1 | 3 | >10 | 3 | 3 | 0 | 0 | 3 | 0 | 0 | 1 | 0 | - |
| Bcl11b | NM_021399 | 5 | 0 | >10 | 2 | 1 | 2 | 0 | 0 | 1 | 0 | 1 | 1 | + |
| Baiap2 | NM_130862 | 5 | 1 | 10 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | + |
| Klf9 | NM_010638 | 6 | 3 | 4 | 2 | 0 | 0 | 2 | 4 | 2 | 0 | 0 | 0 | + |
| Rasgrp2 | NM_011242 | 2 | 3 | >10 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | + |
| Cpne5 | NM_153166 | 1 | 0 | >10 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | + |
| Ppp1r1b | NM_144828 | 2 | 1 | 7 | 3 | 0 | 0 | 4 | 3 | 3 | 0 | 0 | 0 | + |
| Oprd1 | NM_013622 | 1 | 3 | 7 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | - |
| Dgke | NM_019505 | 3 | 7 | 6 | 2 | 0 | 1 | 2 | 0 | 3 | 1 | 1 | 1 | - |
| Drd1 | NM_010076 | 0 | 1 | 0 | 3 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | + |
| Drd2 | NM_010077 | 3 | 1 | >10 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 1 | 1 | + |
| Drd3 | NM_007877 | 1 | 3 | 0 | 2 | 1 | 0 | 0 | 1 | 3 | 1 | 0 | 0 | - |
| Klf16 | NM_078477 | 2 | 1 | 10 | 1 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | + |
| Penk1 | NM_001002927 | 0 | 6 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | + |
| Epha4 | NM_007936 | 3 | 4 | 4 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | + |
| FoxP1 | NM_053202 | 0 | 3 | 1 | 1 | 1 | 1 | 0 | 6 | 0 | 0 | 1 | 0 | + |
| FoxP2 | NM_053242 | 1 | 4 | 3 | 1 | 0 | 1 | 1 | 2 | 0 | 1 | 0 | 0 | - |
| Gna1 | NM_177137 | 2 | 4 | 2 | 6 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | + |
| Gng7 | NM_010319 | 1 | 2 | 5 | 3 | 0 | 1 | 4 | 1 | 3 | 0 | 1 | 0 | + |
| Gpr88 | NM_022427 | 1 | 4 | 2 | 0 | 0 | 1 | 0 | 2 | 1 | 0 | 1 | 1 | + |
| Hpca | NM_010471 | 2 | 3 | 1 | 4 | 0 | 0 | 3 | 0 | 0 | 1 | 1 | 1 | + |
| Isl1 | NM_021459 | 3 | 2 | 4 | 1 | 0 | 0 | 0 | 1 | 2 | 0 | 1 | 0 | + |
| Oprk1 | NM_011011 | 1 | 7 | 3 | 4 | 0 | 1 | 0 | 2 | 3 | 0 | 1 | 0 | + |
| Kcnip1/2 | NM_030716 | 2 | 1 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | + |
| PPP1-16B | NM_153089 | 1 | 3 | 2 | 3 | 0 | 1 | 0 | 3 | 0 | 0 | 1 | 1 | + |
| Oprm1 | NM_011013 | 0 | 8 | 2 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | - |
| Nts | NM_024435 | 0 | 4 | 0 | 2 | 2 | 0 | 0 | 1 | 4 | 0 | 0 | 0 | + |
| Ngef | NM_019867 | 2 | 2 | 3 | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | + |
| Zpf503 | NM_145459 | 1 | 3 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | + |
| Osbpl8 | NM_001003717 | 3 | 6 | 5 | 3 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | + |
| Pde10a | NM_011866 | 6 | 1 | 1 | 4 | 0 | 1 | 1 | 0 | 3 | 0 | 1 | 0 | + |
| Pde1b | NM_008800 | 5 | 2 | 5 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | + |
| Ppp3ca | NM_008913 | 4 | 1 | 6 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | + |
| Rarb | NM_011243 | 4 | 2 | 2 | 3 | 0 | 1 | 1 | 0 | 3 | 0 | 1 | 1 | + |
| Rgs9 | NM_011268 | 1 | 1 | 1 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | + |
| Rasd2 | XM_204287 | 0 | 4 | 0 | 1 | 0 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | + |
| Rxrg | NM_009107 | 1 | 3 | 1 | 1 | 0 | 0 | 0 | 2 | 2 | 0 | 2 | 0 | + |
| Scnb4 | BK001031 | 2 | 1 | 2 | 5 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | + |
| Ptpn5 | NM_013643 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | + |
| Strn4 | NM_133789 | 2 | 3 | 6 | 0 | 0 | 1 | 1 | 0 | 3 | 0 | 1 | 0 | + |
| Tac1 | NM_009311 | 0 | 2 | 5 | 5 | 1 | 0 | 2 | 2 | 0 | 2 | 0 | 0 | - |
| Synpr | NM_028052 | 0 | 5 | 3 | 5 | 1 | 0 | 0 | 2 | 1 | 0 | 2 | 0 | - |
| Tesc | NM_021344 | 4 | 0 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | + |
| Adcy2 | NM_153534 | 3 | 4 | >10 | 4 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | |
| Bcas1 | BC024553 | 0 | 5 | 3 | 3 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | |
| Cbln1 | NM_019626 | 5 | 2 | 7 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | |
| Cpne2 | BC023348 | 1 | 1 | 7 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | |
| Cpne4 | BC063081 | 0 | 0 | 4 | 0 | 1 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | |
| Cort | NM_007745 | 2 | 3 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 1 | |
| Fgf15 | NM_008003 | 4 | 1 | 5 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | |
| Gnai3 | NM_010306 | 1 | 0 | 2 | 4 | 1 | 0 | 2 | 5 | 2 | 0 | 0 | 0 | |
| Gnazx | NM_010311 | 2 | 4 | >10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Gng2 | NM_010315 | 0 | 4 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 0 | 2 | 0 | |
| Gbx2 | NM_010262 | 2 | 0 | 5 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| GFAP | NM_010277 | 0 | 5 | 2 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 3 | 1 | |
| Gpr51 | XM_143750 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Gpr85 | AF254416 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | |
| Hcrt | NM_010410 | 0 | 3 | 2 | 9 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | |
| IiiG9 | AY032664 | 3 | 6 | 2 | 6 | 0 | 1 | 2 | 2 | 0 | 1 | 1 | 0 | |
| Irs1 | L24563 2 | 1 | >10 | 2 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | |
| Irs2 | AK155277 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 3 | 0 | 1 | 0 | |
| Kctd4 | AK008741 | 0 | 2 | 0 | 0 | 1 | 0 | 2 | 3 | 2 | 0 | 0 | 0 | |
| Lix1 | AF351204 | 0 | 3 | 0 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 0 | 0 | |
| Pmch | NM_029971 | 0 | 2 | 0 | 1 | 7 | 2 | 0 | 2 | 2 | 0 | 0 | 0 | |
| Kcnh2 | NM_013569 | 9 | 9 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Mog | NM_010814 | 2 | 4 | 2 | 4 | 1 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | |
| Nelf | AF266508 | 1 | 1 | 1 | 5 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | |
| Nmur2 | NM_153079 | 1 | 2 | 2 | 1 | 0 | 0 | 0 | 2 | 3 | 0 | 0 | 0 | |
| Nnat | NM_010923 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 3 | 1 | 0 | 0 | 0 | |
| Chrn4 | AF225912 | 1 | 1 | >10 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| Npy | NM_023456 | 0 | 0 | 4 | 5 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | |
| Ncl | AF318184 | 0 | 2 | 7 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| OMP | U01213 0 | 5 | 4 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | |
| Pip5K1A | NM_008846 | 0 | 3 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | |
| PLP1 | M54927 1 | 0 | 3 | 2 | 3 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | |
| POMC | J00612 1 | 3 | 5 | 2 | 1 | 0 | 1 | 2 | 3 | 0 | 0 | 0 | 0 | |
| Pcp2 | NM_008790 | 0 | 7 | 3 | 2 | 0 | 1 | 2 | 4 | 1 | 1 | 2 | 2 | |
| Rgs1 | NM_015811 | 0 | 1 | 2 | 0 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| Rgs16 | NM_011267 | 3 | 4 | 5 | 0 | 0 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | |
| Rgs5 | NM_009063 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | |
| Rgs6 | NM_015812 | 1 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Rgs7 | NM_011880 | 2 | 0 | 3 | 1 | 0 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | |
| Rhcg | AF193810 | 1 | 7 | 6 | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | |
| RibS10 | BC003853 | 0 | 3 | 2 | 2 | 1 | 0 | 0 | 3 | 3 | 0 | 0 | 0 | |
| Scn1b | BC039140 | 1 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| Sla | AY217759 | 0 | 3 | 2 | 5 | 0 | 2 | 0 | 2 | 3 | 0 | 0 | 0 | |
| Scn5a | NM_021544 | 5 | 1 | 1 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 1 | |
| Sparc | NM_009242 | 1 | 5 | 5 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | |
| SPP | AF515662 | 1 | 8 | 1 | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | |
| Stra13 | NM_016665 | 0 | 2 | 4 | 5 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| SCP-1 | D16847 1 | 2 | 7 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 2 | 0 | 0 | |
| Galntl1 | AB045325 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 0 | 1 | 0 | |
| Uts2 | NM_011910 | 2 | 5 | 1 | 0 | 2 | 1 | 2 | 1 | 1 | 0 | 1 | 0 |
The list of striatal-enriched genes is taken from Desplats et al., 2006. The list of non-striatal-enriched genes was compiled from the literature and from our own unpublished studies examining differential gene expression in the CNS. When possible, family members of striatal-enriched genes showing no or low expression in the striatum were chosen in order to maintain a list of similar types of genes. Glial-expressed genes were also included. Unigene ID abbreviations for each gene and corresponding accession numbers are shown. Regions 1 Kb upstream from the transcription start site were searched for the indicatedtranscription factor binding sites using Transcription Element Search Software (TESS; http://www.cbil.upenn.edu/tess), TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) and Siteseer (http://rocky.bms.umist.ac.uk/SiteSeer/). User-defined sites were searched for the following factors: Bcl11b, 5′-GGCCG/AG/AAGG-3′, 5′-GGCCG/AG/A-3′ (Avram et al., 2002); COUP-TF (CP-TF), 5′-AGG/TTCA-3′ and 5′-TGTACT-3′ (Lin et al., 1999), USF, 5-CACGTG-3′ (Erno et al., 1996). The remaining sites were searched according to TESS and TFSEARCH defined sites at a probability threshold of 0.85. The numbers of binding sites for each of the indicated transcription factor are shown. USF, upstream stimulatory factor; AP-1, activator protein 1; Oct-1, octamer binding factor; MASH1, mammalian achaete-scute homolog 1; Rxrg, retinoid X receptor gamma; Rarb, retinoic acid receptor, beta. The last column “HD” (Huntington's disease) reflects those striatal-enriched genes that were found to be altered in their expression in mouse models of HD (indicated by a “+”) (Thomas, 2006).
Table 2.
P-values reflecting the significance of representation of consensus binding sites for various transcription factors in the 1 kb upstream regions of “striatal-enriched” versus “non-striatal-expressed” genes.
| Transcription Factor: |
# sites: |
p-value: |
# sites: |
p-value: |
|---|---|---|---|---|
| Bcl11b | 1 site | 0.01** | 2 sites | 0.03* |
| COUP-TF | 1 site | 0.48 | 2 sites | 0.66 |
| Sp1 | 1 site | 0.31 | 2 sites | 0.66 |
| CREB | 1 site | 0.31 | 2 sites | 0.54 |
| TBP | 1 site | 1.0 | 2 sites | 0.67 |
| USF | 1 site | 0.28 | 2 sites | 0.49 |
| AP-1 | 1 site | 0.29 | 2 sites | 0.67 |
| Oct-1 | 1 site | 0.16 | 2 sites | 0.36 |
| TFIID | 1 site | 0.54 | ||
| MASH-1 | 1 site | 0.81 | ||
| Rxrg | 1 site | 0.14 | ||
| Rarb | 1 site | 0.79 |
Fisher's exact probability test was used to determine the significance of representation of transcription factor binding sites in striatal-enriched versus non-striatal-expressed genes (see Table 1 for list of genes screened and presence of binding sites for individual factors). For a majority of genes containing sites for TFIID, MASH-1, Rxrg and Rarb, only 1 binding site was identified.
We further compared genes whose expression was co-regulated in HD mouse models, based on existing microarray profiling data in the literature [reviewed in (Thomas 2006)], for the presence of consensus binding sites for Bcl11b using the Fisher's exact test. For this analysis, we focused exclusively on those genes demonstrating enriched expression in the striatum. We find that 23 out of 41 genes shown to be regulated in HD mouse models contained ≥ 2 Bcl11b consensus binding sites in their proximal promoter regions, while only 2 genes that were not regulated in HD mouse models showed the same occurrence (see Table 1). These numbers indicate a significant enrichment of Bcl11b binding sites in the upstream sequences of genes regulated in HD mouse models (p<0.04).
Bcl11b Activates Promoter Regions of Genes with Striatal-Enriched Expression; Reversal by Mutant Htt
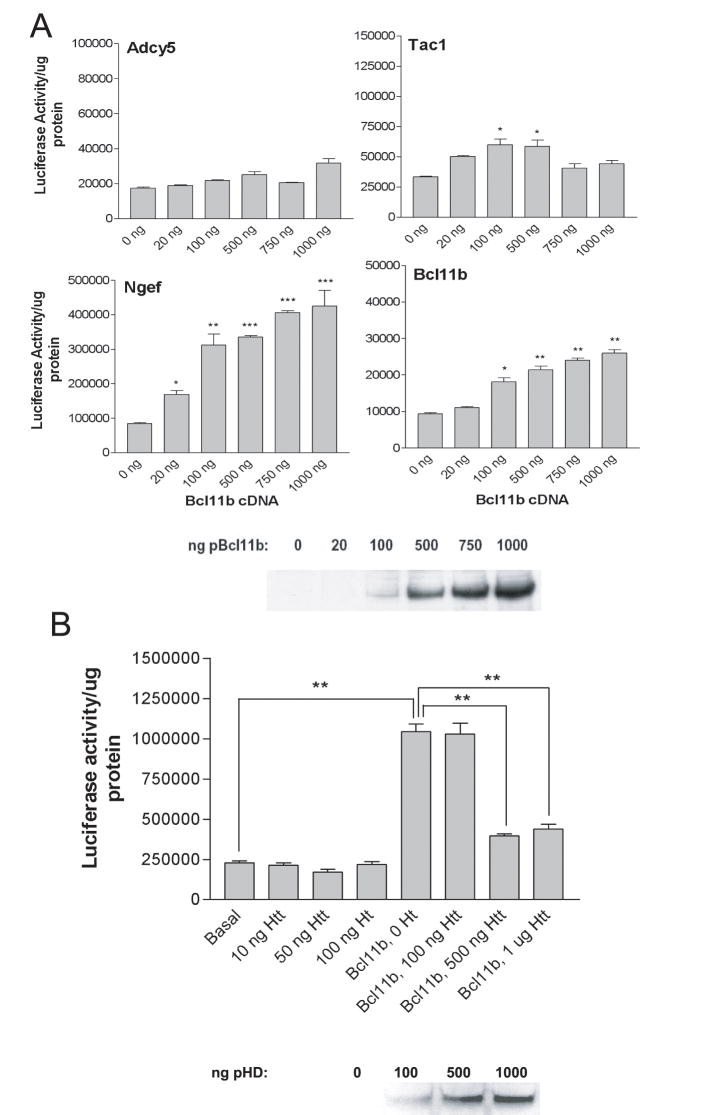
In order to test whether Bcl11b could directly regulate the basal expression of selected striatal genes, proximal promoter fragments of four striatal-enriched genes, Tac1, Adcy5, Ngef and Bcl11b itself, containing 0, 1, 2 and 5 consensus Bcl11b binding sites, respectively, were cloned into the pGL3 basic luciferase reporter vector. HEK293 cells were cotransfected with the different pGL3 constructs and increasing amounts of the full-length Bcl11b ORF (Figure 3). Overexpression of Bcl11b in the presence of the Adcy5 promoter construct had no significant effect on luciferase activity, while in the presence of Tac1, slight activation was observed at concentrations of 100 and 500 ng Bcl11b only. In contrast, overexpression of Bcl11b in the presence of the Ngef and Bcl11b promoters induced dose-dependent increases in luciferase activity with maximal effects of 5-fold and 2.7-fold at 1 μg Bcl11b/pcDNA3.1 (Figure 3A) (p=0.0001, one-way ANOVA). Bcl11b had no significant effects on basal luciferase activity (data not shown).
Figure 3.
Activation of proximal promoter regions of striatal-enriched genes by Bcl11b (A) and reversal of Ngef activation in the presence of mutant htt (B). Luciferase activity was measured in HEK293 cells transfected with proximal promoter regions for adenylate cyclase V (Adcy5), substance P/tachykinin 1 (Tac1), neuronal guanine nucleotide exchange factor (Ngef) and Bcl11b along with increasing concentrations of Bcl11b coding sequence (A). Panel B shows the effect of htt protein carrying 72 glutamines on promoter activity of Ngef in the absence and presence of Bcl11b. Western blots show representative expression levels for Bcl11b and htt protein (from pHD, Huntington's disease plasmid) in HEK293 cells. Significant increases in luciferase activity were determined by one-way ANOVA with Bonferroni's post tests: *, p<0.05; **, p<0.01; ***p<0.001.
To test the effect of mutant htt protein on Bcl11b-induced Ngef promoter activity, we cotransfected HEK293 cells with increasing concentrations of an HD/pCMV construct, expressing exon1 of human HD gene carrying 72 CAG repeats, in combination with the Ngef promoter construct and Bcl11b plasmid. The presence of htt had no effect on Ngef promoter activity alone, however, htt was able to almost completely reverse the activation of luciferase activity induced by Bcl11b (Figure 3B). We also compared the effects of transfecting HD exon1 carrying 25 CAG repeats (pQ25) vs. 72 CAG repeats (pQ72) on Bcl11b-induced Ngef promoter activity. Htt containing 25 glutamines was also able to inhibit Bcl11b-induced Ngef promoter activity. However, significantly greater inhibition was observed with pQ72 compared to pQ25 (47.3±0.13% vs. 55.8±0.072% inhibition for 500 ng pQ25 and pQ72, respectively; p<0.05, and 68.4±0.002% vs. 79.0±0.09% for 1 μg of pQ25 and pQ72, respectively; p<0.026). Previous studies, including our own unpublished studies, have shown that transient overexpression of mutant htt alone has no effect on cell death in HEK293 cells (Hackam et al., 2000; Zeron et al., 2001). Rather, htt fragments are only toxic in the presence of an additional stimulus, i.e. an apoptotic or excitotoxic stressor. Hence, it is very unlikely that the reduced effects of Bcl11b on promoter activity observed here are due to cytotoxic effects of mutant htt.
Bcl11b Interacts with Htt in the Brain
Because we found that the effects of Bcl11b on promoter activation were prevented in the presence of mutant htt, we investigated whether these two proteins could interact with one another. We first found that Bcl11b co-localized with intranuclear N-terminal htt aggregates in the R6/1 mouse model of HD, indicating proximal compartmental localization of these two proteins. Double immunofluorescence experiments revealed that 91.7±2.8% of Bcl11b-positive neurons in the striatum of symptomatic, 6-8 month-old R6/1 transgenic mice also contained htt aggregates (Supplementary Figure 1). To test for an interaction between Bcl11b and htt, we performed co-immunoprecipitation experiments both in vitro (HEK293 cells) and in vivo, using HD mouse models expressing truncated (R6/1 mice) and full-length (YAC72 mice) forms of the htt protein. Three isoforms of Bcl11b, alpha, beta and gamma (75, 89.4 and 97.1 kDa, respectively), generated by alternative splicing, are detected in mouse striatum, although the beta form is, by far, most abundant. HEK293 cells were first co-transfected with a construct containing the Bcl11b cDNA (beta isoform, Bcl1b-beta) in combination with constructs containing exon 1 of the HD gene carrying 25 or 103 CAG repeats. Immunoprecipitation of htt resulted in the co-precipitation of Bcl11b-beta (Figure 4), with a greater interaction being observed between Bcl11b-beta and htt containing 103 glutamines. Interactions between htt and Bcl11b protein were also observed in the brains of two different HD mouse models: R6/1 transgenic mice, which express exon 1 of the human HD gene carrying 115 CAG repeats (Mangiarini et al. 1996), and YAC72 transgenic mice, which express a full-length human HD gene carrying 72 CAG repeats (Hodgson et al. 1999). Immunoprecipitation of htt in striatal homogenates from wt and R6/1 transgenic mice using an antibody directed against the N-terminal region of htt, (antibody EM48), which is necessary due to the nature of the exon 1 model, resulted in the co-precipitation of all three Bcl11b isoforms (Figure 4B), although the gamma form was detected only at low levels. Greater interactions were detected between Bcl11b and mutant htt, with 1.4-fold and 2.6-fold increases in the amount of the alpha and beta isoforms, respectively, immunoprecipitated in the striata of R6/1 transgenic mice compared to wt mice (quantified relative to Ponceau S staining). We next tested whether Bcl11b could interact with the full-length htt protein by performing co-immunoprecipitation in whole brain of YAC72 transgenic mice. Immunoprecipitation of htt using an antibody directed against the C-terminal region of huntingtin, (antibody 4C8), also resulted in the co-precipitation of Bcl11b (Figure 4C). As we observed in the striata of R6/1 mice, the beta isoform of Bcl11b interacted with both wt and mutant htt protein, although a slightly greater interaction was observed in the transgenic mice. The alpha form was found to interact predominantly with the mutant htt protein, while the gamma isoform was only at detected very low levels in whole brain of wt or YAC72 mice (Figure 4C). It should be noted that the htt antibody used in this experiment, 4C8, can also recognize endogenous mouse htt protein, as well as htt proteolytic fragments. Therefore, we have used the human-specific anti-htt antibody, clone 2E8, to verify the immunoprecipitation of the mutant human transgenic protein in the reverse pull-down experiment using Bcl11b as the bait (Figure 4D).
Figure 4.
Interaction between Bcl11b and huntingtin protein. (A): Co-immunoprecipitation of Bcl11b and htt in HEK293 cells. HEK293 cells were transfected with Bcl11b, beta form (Bcl11b-beta), in combination with HD exon 1 constructs containing 25 CAG repeats (Q25) or 103 CAG repeats (Q103). (B): Co-immunoprecipitation of Bcl11b and htt in striatal homogenates from wt and R6/1 transgenic mice. The three isoforms of Bcl11b, alpha, beta and gamma, are indicated for the in vivo blot. (C): Co-immunoprecipitation of Bcl11b and full-length htt protein in whole brain of YAC72 transgenic mice. Note: the gamma form of Bcl11b is not detected in whole brain. (D): Reverse co-immunoprecipitation using Bcl11b for the immunoprecipitation (IP) in whole brain of YAC72 transgenic mice. Western blot of the immunoprecipitated material shows the Bcl11b-beta form, which is most abundant. For panels A and B, the IP was performed using EM48, while in C., the C-terminal antibody 4C8 was used. Immunoblotting was performed using three different anti-htt antibodies, as indicated. The expected band size for EM48 in the immunoprecipitate corresponds to the heavy chain immunoglobulin (∼50 kDa), which is recognized by our secondary antibody, hence Western blot using EM48 on immunoprecipitates in A and B are not shown. For the HEK293 cells, the input material is blotted with EM48 to demonstrate overexpression of htt in these cells. Equal loading of the lanes was verified by Ponceau S stain.
Bcl11b Expression Protects Against Mutant Htt-Induced Cellular Toxicity
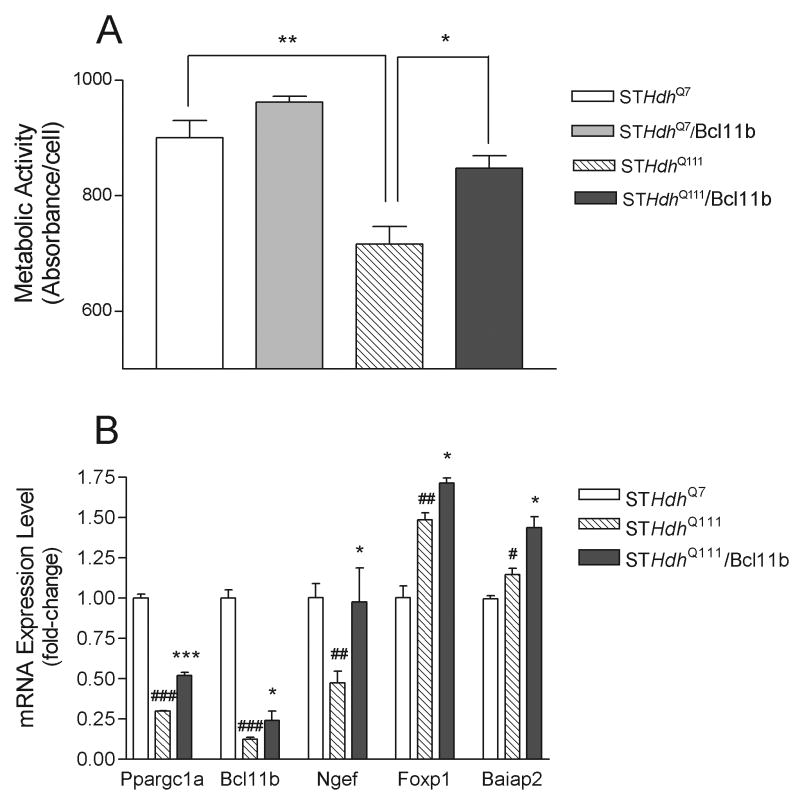
To test for a physiological role in HD pathology, we investigated the effects of Bcl11b in a well-established HD cell culture model, STHdhQ111 cells. These cells have been generated from the striatal tissue of HdhQ111 knock-in mice (Trettel et al. 2000), and express full-length endogenous htt protein containing 7 glutamines in the wt cell line, STHdhQ7 cells, and 111 glutamines in the mutant cell line, STHdhQ111 cells. STHdhQ111 cells display a mutant phenotype of decreased mitochondrial function and energy metabolism (Cui et al., 2006; Lee et al., 2007), which has also been implicated in HD pathogenesis (Browne and Beal 2006). We first tested whether these immortalized striatal cells express markers of in vivo mature striatal neurons, by performing PCR analysis on randomly selected striatal-enriched genes [see (Desplats et al. 2006)]. STHdhQ7 cells were found to express the dopamine and cAMP regulated phosphoprotein (DARPP-32), forkhead box P1 (Foxp1), adenosine A2a receptor (Adora2a), copine V (Cpne5), brain-specific angiogenesis inhibitor 1-associated protein 2 (Baiap2), regulator of G-protein signaling 9 (RGS9), Guanine nucleotide binding protein (G protein) gamma 7 subunit (Gng7), Ngef, Bcl11b and Adcy5; albeit, some of these were expressed at low levels (data not shown). As we observed in the brains of HD transgenic mice and human HD subjects, Bcl11b expression is greatly decreased in STHdhQ111 cells compared to wt cells (Figure 5B); this allowed us to examine the effects of Bcl11b overexpression in these cells. Using the MTT assay, we detected a defect in mitochondrial metabolism similar to previous reports in these cells (Trettel et al. 2000; Cui et al. 2006). Overexpression of Bcl11b in these cells significantly attenuated this deficit, suggesting that upregulation of Bcl11b can rescue the effects of mutant htt (Figure 5A).
Figure 5.
Physiological and molecular effects of Bcl11b overexpression in STHdhQ111 cells. (A): Overexpression of Bcl11b partially rescues the decrease in mitochondrial metabolic activity in STHdhQ111 cells. MTT activity was assayed by absorbance units measured at 550 nm and is reported as mean ± SEM on a per cell basis. Three independent experiments were performed. Expression of Bcl11b was verified by Western blot analysis. Asterisks denote significant differences: *, p<0.05; **, p<0.01. (B): Overexpression of Bcl11b causes an increase in the expression of selected genes, as indicated by their UniGene gene symbols. Cells were transfected with recombinant Bcl11b and the expression of the indicated genes was determined by real-time PCR analysis as described in the Experimental Procedures. Significant differences in gene expression were determined by one-way ANOVA. Asterisks denote significant differences between untransfected and Bcl11b-transfected STHdhQ111 cells: * p<0.05, *** p<0.001. Hash marks denote significant differences between wt cells (STHdhQ7 cells) and STHdhQ111 cells: # p<0.05, ## p<0.01; ### p<0.001.
Gene Expression Correlates to Bcl11b-Mediated Reversal of Phenotype in STHdhQ111 Cells
We next identified potential gene targets for Bcl11b in STHdhQ111 cells, focusing on striatal-enriched genes, as these are the putative targets for Bcl11b regulation. Consistent with its activation of the promoter region of Ngef, overexpression of Bcl11b resulted in a significant increase in transcription of Ngef in STHdhQ111 cells, completely reversing the deficit in Ngef expression exhibited by these cells (Figure 5B). Bcl11b elicited a small increase in the expression of itself, slightly reducing the deficit exhibited by these cells. Bcl11b also caused significant increases in the expression of striatal-enriched genes, Foxp1 and Baiap2 (Figure 5B), although STHdhQ111 cells did not show an underlying deficit in the expression of these genes. No effect of Bcl11b overexpression was observed for RGS9, Gng7 and Adcy5, whose promoter region was also not activated by Bcl11b (see Figure 3). We also examined the expression of peroxisome proliferative activated receptor, gamma, coactivator 1 alpha, (Ppargc1a), a mitochondrial respiration regulator (Puigserver and Spiegelman 2003) that has previously been shown to be important to mitochondrial dysfunction and neurodegeneration in these striatal HD cells (Cui et al. 2006). STHdhQ111 cells showed decreased expression of Ppargc1a, which was significantly attenuated by Bcl11b overexpression (Figure 5B). As we observed with many of the striatal-enriched genes, the putative promoter region for Ppargc1a contains two consensus binding sites for Bcl11b, suggesting that Bcl11b activates transcription via this site (data not shown).
Discussion
We have previously reported that the expression of 38 striatal-enriched genes is decreased in the brains of HD transgenic mice (Desplats et al. 2006); These differences are also present in caudate of human HD subjects (Hodges et al. 2006; Thomas 2006). These findings suggest that one or more transactivating factors may be responsible for such co-dyregulation of striatal-expressing genes. In this study, we present evidence indicating that Bcl11b is responsible for transcriptional control of important genes in the striatum and suggest that its dysregulation and/or aggregation caused by expanded htt protein may contribute to the striatal transcriptional deficits observed in HD.
The most significant neuropathological and neurodegenerative changes in HD are observed predominantly in the striatum. However, it is now appreciated that htt-induced deficits are more widespread, involving cortex, hippocampus, hypothalamus and olfactory function (Hedreen et al., 1991; Spargo et al., 1993; Murphy et al., 2000; Mitchell et al., 2005; Petersen et al. 2005; Wetter et al., 2005). These regions correspond to the specific localization of Bcl11b in the CNS, as described herein, and by others (Leid et al. 2004; Arlotta et al. 2005; Desplats et al. 2006). This led us to the hypothesis that Bcl11b may be an important transcription factor associated with striatal gene dysregulation in HD. Accordingly, we demonstrate significant decreases in expression of Bcl11b in HD striatal cells, striata from HD R6/1 transgenic mice and caudate samples from human subjects with HD. Importantly, we find that Bcl11b expression levels are decreased in R6/1 mice as early as 1 month of age, far prior to the onset of disease symptoms, suggesting that deficits in Bcl11b function may be pathogenic. We also find that Bcl11b expression deficits worsen with disease progression in mice and are dramatically reduced in the brains of human subjects with end-stage illness. It is possible some of the observed decrease in Bcl11b expression in human caudate could be due to neuronal loss, however this is not likely to explain the total decrease, as no changes in the mRNA or protein levels of beta-actin, which is commonly used to normalize neuronal levels of expression (Shim and Lubec 2002; Smart et al., 2003; Rinaldi et al., 2007), were observed.
From our in silico analyses, we predict that Bcl11b can potentially control the expression of up to 40 striatal-expressing genes; our luciferase and real-time PCR studies have confirmed this for a subset of striatal-expressing genes. In particular, we find that Bcl11b can activate its own promoter, which contains 5 consensus Bcl11b binding sites. Bcl11b also activated the Ngef promoter region, which contains two Bcl11b response elements. Previous studies have shown that Bcl11b, and a related isoform Bcl11a, repressed transcription of a reporter gene harboring multiple Bcl11b response elements in the context of the thymidine kinase promoter (Avram et al. 2002), hence we predict that at least two consensus binding sites are required for a full Bcl11b-induced effect. However, we did observe that Bcl11b caused a slight activation of the Tac1 proximal promoter region, although it did not contain a full Bcl11b response element in this region. A similar sequence, GGCCAAGG, is contained in the immediate upstream region of the translation start site, perhaps suggesting promiscuity in response element recognition by Bcl11b. Although Bcl11b was initially characterized as a transcriptional repressor (Avram et al. 2000; Avram et al. 2002), previous studies have shown activation of transcription of a target gene in the context of T-cell activation (Cismasiu et al., 2006), similar to our results.
Consistent with the promoter activation findings, we demonstrate that overexpression of Bcl11b in striatal STHdhQ111 cells resulted in increased mRNA expression levels of Ngef and Bcl11b, in addition to Foxp1, Ppargc1a and Baiap2, the latter two which contain Bcl11b binding sites in their proximal promoter regions. These expression increases were accompanied by Bcl11b-elicited attenuation of the metabolic deficits exhibited by these cells, indicating that Bcl11b can play a physiological role in htt-induced pathology in these cells.
In the context of transcriptional dysregulation in HD, it has been proposed that sequestration of transcription factors into htt nuclear aggregates plays an important role in pathogenesis (Cha 2000; Okazawa 2003; Sugars et al. 2003). In our studies, we demonstrate an interaction of Bcl11b with both truncated and full-length forms of htt protein, although the exact binding domain responsible for this interaction is not yet known. Such an interaction would likely result in loss-of-function of Bcl11b, which would lead to the observed deficits in striatal gene transcription. Because Bcl11b has the ability to regulate its own transcription, sequestration by htt protein would also result in decreased expression of Bcl11b, however decreases in Bcl11b expression could also be attributed to dysfunction of other transcription factors (see below). The downstream effects of either phenomenon are the same: altered expression of Bcl11b target genes. Bcl11b/htt interactions are not likely limited to insoluble aggregates, in light of the observed effects of Bcl11b on STHdhQ111 cells, which do not form of nuclear aggregates (Trettel et al. 2000). Hence we speculate that Bcl11b can also interact with soluble forms of htt, similar to findings suggesting that altered gene expression may result from the interactions of soluble mutant huntingtin with nuclear transcription factors, rather than from the depletion of transcription factors by nuclear inclusions (Yu et al., 2002).
Alterations in the functions of several other transcription factors have been reported in HD. Studies have shown that mutant htt interacts with, or can disrupt the function of, ubiquitous transcription factors such as CBP, Sp1, TBP and components of the basal transcriptional machinery, such as TFIID, TFIIF and TAF4 (McCampbell et al. 2000; Shimohata et al. 2000; Dunah et al. 2002; Li et al. 2002; van Roon-Mom et al. 2002; Zhai et al. 2005). While disruption of transcription via these transcription factors is likely to result in aberrant gene expression patterns, because these are ubiquitously expressed genes, they are unlikely to confer region specificity of transcription in the brain. In support of this hypothesis, we found that consensus binding sites for Sp1, CREB, TBP and TFIID were not enriched in the putative promoter regions of striatal-expressing genes, as we found for Bcl11b, suggesting that these transcription factors are not as pertinent to the control of global striatal gene expression, as Bcl11b. However, Bcl11b does contain several Sp1 and CREB consensus sites in its proximal promoter region, therefore, aberrant function of these factors may contribute to the decrease in expression of Bcl11b observed in HD patients and cells and mouse models, and therefore, may contribute indirectly to the control of striatal gene expression via regulation of Bcl11b. In addition to Bcl11b, we also detected greater than two-fold decreases in expression of other striatal transcription factors in the CNS of HD R6/1 mice, including Foxp1, kruppel-like factor 16 and the retinoid X receptor gamma (Desplats et al. 2006). Hence, it is possible that these transcription factors may also contribute to the dysregulation of striatal gene expression observed in HD.
The results from these studies demonstrate that Bcl11b can control transcriptional regulation of important genes in the striatum and suggest that its loss-of-function elicited by htt protein has detrimental consequences on the expression of genes important to striatal physiology. While it is likely that disruption of transcription due to the presence of an expanded mutant htt protein occurs via several mechanisms, transcriptional dysregulation involving Bcl11b may contribute to the specificity of regional pathology observed in this disease.
Supplementary Material
Acknowledgments
This study was funded by grants from the National Institutes of Health (NS44169 and MH069696 to E.A.T.). The authors wish to thank Kristi E. Kass for excellent technical assistance.
Abbreviations
- HD
Huntington's Disease
- HTT
human Huntington's disease gene
- htt
huntingtin
- CREB
cAMP response element-binding protein
- Sp1
specificity protein 1
- CBP
CREB-binding protein
- TBP
TATA binding box protein
- COUP-TF
chicken ovalbumin upstream promoter transcription factor
- TAF4
TATA box binding protein (TBP)-associated factor
- Tac1
substance P/tachykinin 1
- Adcy5
adenylate cyclase V
- Ngef
neuronal guanine exchange factor
- DARPP-32
dopamine and cAMP regulated phosphoprotein
- Foxp1
forkhead box P1
- Adora2a
adenosine A2a receptor
- Cpne5
copine V
- Rgs9
regulator of G protein signaling 9
- Gng7
guanine nucleotide binding protein (G protein), gamma 7 subunit
- Baiap2
brain-specific angiogenesis inhibitor 1-associated protein 2
- Ppargc1a
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Archer TK, Zaniewski E, Moyer ML, Nordeen SK. The differential capacity of glucocorticoids and progestins to alter chromatin structure and induce gene expression in human breast cancer cells. Mol Endocrinol. 1994;8:1154–1162. doi: 10.1210/mend.8.9.7838148. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram D, Fields A, Pretty On Top K, Nevrivy DJ, Ishmael JE, Leid M. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J Biol Chem. 2000;275:10315–10322. doi: 10.1074/jbc.275.14.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram D, Fields A, Senawong T, Topark-Ngarm A, Leid M. COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein. Biochem J. 2002;368:555–563. doi: 10.1042/BJ20020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman PE, Oliver SG, Hubbard SJ. SiteSeer: Visualisation and analysis of transcription factor binding sites in nucleotide sequences. Nucleic Acids Res. 2003;31:3572–3575. doi: 10.1093/nar/gkg511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne SE, Beal MF. Oxidative damage in Huntington's disease pathogenesis. Antioxid Redox Signal. 2006;8:2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- Cha JH. Transcriptional dysregulation in Huntington's disease. Trends Neurosci. 2000;23:387–392. doi: 10.1016/s0166-2236(00)01609-x. [DOI] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Sadri-Vakili G, Yohrling GJ, Braveman MW, Benn CL, Glajch KE, Dirocco DP, Farrell LA, Krainc D, Gines S, Macdonald ME, Cha JH. Decreased association of the transcription factor Sp1 with genes downregulated in Huntington's disease. Neurobiol Dis. 2006 doi: 10.1016/j.nbd.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Cismasiu VB, Ghanta S, Duque J, Albu DI, Chen HM, Kasturi R, Avram D. BCL11B participates in the activation of IL2 gene expression in CD4+ T lymphocytes. Blood. 2006;108:2695–2702. doi: 10.1182/blood-2006-05-021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Desplats PA, Kass KE, Gilmartin T, Stanwood GD, Woodward EL, Head SR, Sutcliffe JG, Thomas EA. Selective deficits in the expression of striatal-enriched mRNAs in Huntington's disease. J Neurochem. 2006;96:743–757. doi: 10.1111/j.1471-4159.2005.03588.x. [DOI] [PubMed] [Google Scholar]
- Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, Mouradian MM, Young AB, Tanese N, Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- Graybiel AM. The basal ganglia: learning new tricks and loving it. Curr Opin Neurobiol. 2005;15:638–644. doi: 10.1016/j.conb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Group, H.s.D.C.R. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Hackam AS, Yassa AS, Singaraja R, Metzler M, Gutekunst CA, Gan L, Warby S, Wellington CL, Vaillancourt J, Chen N, Gervais FG, Raymond L, Nicholson DW, Hayden MR. Huntingtin interacting protein 1 induces apoptosis via a novel caspase-dependent death effector domain. J Biol Chem. 2000;275:41299–41308. doi: 10.1074/jbc.M008408200. [DOI] [PubMed] [Google Scholar]
- Hedreen JC, Peyser CE, Folstein SE, Ross CA. Neuronal loss in layers V and VI of cerebral cortex in Huntington's disease. Neurosci Lett. 1991;133:257–261. doi: 10.1016/0304-3940(91)90583-f. [DOI] [PubMed] [Google Scholar]
- Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, Elliston LA, Hartog C, Goldstein DR, Thu D, Hollingsworth ZR, Collin F, Synek B, Holmans PA, Young AB, Wexler NS, Delorenzi M, Kooperberg C, Augood SJ, Faull RL, Olson JM, Jones L, Luthi-Carter R. Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- Hodgson JG, Agopyan N, Gutekunst CA, Leavitt BR, LePiane F, Singaraja R, Smith DJ, Bissada N, McCutcheon K, Nasir J, Jamot L, Li XJ, Stevens ME, Rosemond E, Roder JC, Phillips AG, Rubin EM, Hersch SM, Hayden MR. A YAC mouse model for Huntington's disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron. 1999;23:181–192. doi: 10.1016/s0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- Lee JM, Ivanova EV, Seong IS, Cashorali T, Kohane I, Gusella JF, MacDonald ME. Unbiased gene expression analysis implicates the huntingtin polyglutamine tract in extra-mitochondrial energy metabolism. PLoS Genet. 2007;3:e135. doi: 10.1371/journal.pgen.0030135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid M, Ishmael JE, Avram D, Shepherd D, Fraulob V, Dolle P. CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expr Patterns. 2004;4:733–739. doi: 10.1016/j.modgep.2004.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Cheng AL, Zhou H, Lam S, Rao M, Li H, Li XJ. Interaction of Huntington disease protein with transcriptional activator Sp1. Mol Cell Biol. 2002;22:1277–1287. doi: 10.1128/mcb.22.5.1277-1287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HB, Jurk M, Gulick T, Cooper GM. Identification of COUP-TF as a transcriptional repressor of the c-mos proto-oncogene. J Biol Chem. 1999;274:36796–36800. doi: 10.1074/jbc.274.51.36796. [DOI] [PubMed] [Google Scholar]
- Long F, Liu H, Hahn C, Sumazin P, Zhang MQ, Zilberstein A. Genome-wide prediction and analysis of function-specific transcription factor binding sites. Silico Biol. 2004;4:395–410. [PubMed] [Google Scholar]
- Luthi-Carter R, Strand A, Peters NL, Solano SM, Hollingsworth ZR, Menon AS, Frey AS, Spektor BS, Penney EB, Schilling G, Ross CA, Borchelt DR, Tapscott SJ, Young AB, Cha JH, Olson JM. Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum Mol Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- McCampbell A, Taylor JP, Taye AA, Robitschek J, Li M, Walcott J, Merry D, Chai Y, Paulson H, Sobue G, Fischbeck KH. CREB-binding protein sequestration by expanded polyglutamine. Hum Mol Genet. 2000;9:2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- Mitchell IJ, Heims H, Neville EA, Rickards H. Huntington's disease patients show impaired perception of disgust in the gustatory and olfactory modalities. J Neuropsychiatry Clin Neurosci. 2005;17:119–121. doi: 10.1176/jnp.17.1.119. [DOI] [PubMed] [Google Scholar]
- Murphy KP, Carter RJ, Lione LA, Mangiarini L, Mahal A, Bates GP, Dunnett SB, Morton AJ. Abnormal synaptic plasticity and impaired spatial cognition in mice transgenic for exon 1 of the human Huntington's disease mutation. J Neurosci. 2000;20:5115–5123. doi: 10.1523/JNEUROSCI.20-13-05115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazawa H. Polyglutamine diseases: a transcription disorder? Cell Mol Life Sci. 2003;60:1427–1439. doi: 10.1007/s00018-003-3013-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen A, Gil J, Maat-Schieman ML, Bjorkqvist M, Tanila H, Araujo IM, Smith R, Popovic N, Wierup N, Norlen P, Li JY, Roos RA, Sundler F, Mulder H, Brundin P. Orexin loss in Huntington's disease. Hum Mol Genet. 2005;14:39–47. doi: 10.1093/hmg/ddi004. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- Rinaldi T, Kulangara K, Antoniello K, Markram H. Elevated NMDA receptor levels and enhanced postsynaptic long-term potentiation induced by prenatal exposure to valproic acid. Proc Natl Acad Sci U S A. 2007;104:13501–13506. doi: 10.1073/pnas.0704391104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA. Huntington's disease: new paths to pathogenesis. Cell. 2004;118:4–7. doi: 10.1016/j.cell.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Sadri-Vakili G, Bouzou B, Benn CL, Kim MO, Chawla P, Overland RP, Glajch KE, Xia E, Qiu Z, Hersch SM, Clark TW, Yohrling GJ, Cha JH. Histones associated with downregulated genes are hypo-acetylated in Huntington's disease models. Hum Mol Genet. 2007;16:1293–1306. doi: 10.1093/hmg/ddm078. [DOI] [PubMed] [Google Scholar]
- Schug J, Overton GC. Technical Report CBIL. University of Pennsylvania; 1997. TESS: Transcription Element Search Software on the WWW. [Google Scholar]
- Shim KS, Lubec G. Drebrin, a dendritic spine protein, is manifold decreased in brains of patients with Alzheimer's disease and Down syndrome. Neurosci Lett. 2002;324:209–212. doi: 10.1016/s0304-3940(02)00210-0. [DOI] [PubMed] [Google Scholar]
- Shimohata T, Nakajima T, Yamada M, Uchida C, Onodera O, Naruse S, Kimura T, Koide R, Nozaki K, Sano Y, Ishiguro H, Sakoe K, Ooshima T, Sato A, Ikeuchi T, Oyake M, Sato T, Aoyagi Y, Hozumi I, Nagatsu T, Takiyama Y, Nishizawa M, Goto J, Kanazawa I, Davidson I, Tanese N, Takahashi H, Tsuji S. Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat Genet. 2000;26:29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- Smart FM, Edelman GM, Vanderklish PW. BDNF induces translocation of initiation factor 4E to mRNA granules: evidence for a role of synaptic microfilaments and integrins. Proc Natl Acad Sci U S A. 2003;100:14403–14408. doi: 10.1073/pnas.2436349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spargo E, Everall IP, Lantos PL. Neuronal loss in the hippocampus in Huntington's disease: a comparison with HIV infection. J Neurol Neurosurg Psychiatry. 1993;56:487–491. doi: 10.1136/jnnp.56.5.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, Wanker EE, Bates GP, Housman DE, Thompson LM. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci U S A. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugars KL, Rubinsztein DC. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003;19:233–238. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- Thomas EA. Striatal specificity of gene expression dysregulation in Huntington's disease. J Neurosci Res. 2006;84:1151–1164. doi: 10.1002/jnr.21046. [DOI] [PubMed] [Google Scholar]
- Thomas EA, Danielson PE, Nelson PA, Pribyl TM, Hilbush BS, Hasel KW, Sutcliffe JG. Clozapine increases apolipoprotein D expression in rodent brain: towards a mechanism for neuroleptic pharmacotherapy. J Neurochem. 2001;76:789–796. doi: 10.1046/j.1471-4159.2001.00027.x. [DOI] [PubMed] [Google Scholar]
- Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- van Roon-Mom WM, Reid SJ, Jones AL, MacDonald ME, Faull RL, Snell RG. Insoluble TATA-binding protein accumulation in Huntington's disease cortex. Brain Res Mol Brain Res. 2002;109:1–10. doi: 10.1016/s0169-328x(02)00450-3. [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y, Inoue J, Takahashi Y, Matsuki A, Kosugi-Okano H, Shinbo T, Mishima Y, Niwa O, Kominami R. Homozygous deletions and point mutations of the Rit1/Bcl11b gene in gamma-ray induced mouse thymic lymphomas. Biochem Biophys Res Commun. 2003;301:598–603. doi: 10.1016/s0006-291x(02)03069-3. [DOI] [PubMed] [Google Scholar]
- Wetter S, Peavy G, Jacobson M, Hamilton J, Salmon D, Murphy C. Olfactory and auditory event-related potentials in Huntington's disease. Neuropsychology. 2005;19:428–436. doi: 10.1037/0894-4105.19.4.428. [DOI] [PubMed] [Google Scholar]
- Yu ZX, Li SH, Nguyen HP, Li XJ. Huntingtin inclusions do not deplete polyglutamine-containing transcription factors in HD mice. Hum Mol Genet. 2002;11:905–914. doi: 10.1093/hmg/11.8.905. [DOI] [PubMed] [Google Scholar]
- Zeron MM, Chen N, Moshaver A, Lee AT, Wellington CL, Hayden MR, Raymond LA. Mutant huntingtin enhances excitotoxic cell death. Mol Cell Neurosci. 2001;17:41–53. doi: 10.1006/mcne.2000.0909. [DOI] [PubMed] [Google Scholar]
- Zhai W, Jeong H, Cui L, Krainc D, Tjian R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Zhang MQ. Identification of human gene core promoters in silico. Genome Res. 1998;8:319–326. doi: 10.1101/gr.8.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker B, Luthi-Carter R, Kama JA, Dunah AW, Stern EA, Fox JH, Standaert DG, Young AB, Augood SJ. Transcriptional dysregulation in striatal projection- and interneurons in a mouse model of Huntington's disease: neuronal selectivity and potential neuroprotective role of HAP1. Hum Mol Genet. 2005;14:179–189. doi: 10.1093/hmg/ddi014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.