Abstract
A key cellular response to DNA double-strand breaks (DSBs) is 5′-to-3′ DSB resection by nucleases to generate regions of ssDNA that then trigger cell cycle checkpoint signaling and DSB repair by homologous recombination (HR). Here, we reveal that in the absence of exonuclease Exo1 activity, deletion or mutation of the Saccharomyces cerevisiae RecQ-family helicase, Sgs1, causes pronounced hypersensitivity to DSB-inducing agents. Moreover, we establish that this reflects severely compromised DSB resection, deficient DNA damage signaling, and strongly impaired HR-mediated repair. Furthermore, we show that the mammalian Sgs1 ortholog, BLM—whose deficiency causes cancer predisposition and infertility in people—also functions in parallel with Exo1 to promote DSB resection, DSB signaling and resistance to DSB-generating agents. Collectively, these data establish evolutionarily conserved roles for the BLM and Sgs1 helicases in DSB processing, signaling, and repair.
Keywords: RecQ helicases, DNA resection, DNA repair, DNA damage checkpoint, Exonuclease 1, ATR
Results and Discussion
DNA double-strand breaks (DSBs) are highly cytotoxic lesions that are induced by ionizing radiation (IR) and certain anti-cancer chemotherapeutic agents. To survive DSB exposure and maximize genome stability, cells possess a complex machinery to detect DSBs, signal their presence, and mediate their repair (Zhou and Elledge 2000; Rouse and Jackson 2002; Lisby and Rothstein 2004; Wyman and Kanaar 2006). The importance of such responses is revealed by the fact that inherited defects in them lead to human pathologies, including cancer, infertility and neurodegeneration (Khanna and Jackson 2001; Kastan and Bartek 2004). In all eukaryotes, an early response to DSBs is their 5′-to-3′ resection to produce ssDNA (Lisby and Rothstein 2004; Wyman and Kanaar 2006). This is bound by replication protein A (RPA), which then recruits and activates the human protein kinase ATR (Mec1 in Saccharomyces cerevisiae) to phosphorylate downstream effector proteins, including the protein kinase CHK1 (the functional counterpart of S. cerevisiae Rad53; Zou and Elledge 2003). In addition to promoting signaling responses, ssDNA regions are also required for DNA repair by homologous recombination (HR), being bound by the HR proteins Rad51 and Rad52 (Wyman and Kanaar 2006; San Filippo et al. 2008). To understand the mechanisms and control of DSB signaling and HR repair, it is therefore crucial to define how DSBs are detected and processed into ssDNA.
While the S. cerevisiae nuclease Exo1 promotes DSB resection, its contribution to this is modest, with exo1Δ mutants exhibiting considerable residual resection and displaying little hypersensitivity to DNA damaging agents (Moreau et al. 2001; Maringele and Lydall 2002; Nakada et al. 2004; Tran et al. 2004; Cotta-Ramusino et al. 2005; Clerici et al. 2006; Bermejo et al. 2007). Furthermore, yeast cells deficient in the Mre11–Rad50–Xrs2 (MRX) complex also display impaired DSB processing (Lee et al. 1998; Nakada et al. 2004; Clerici et al. 2006), with the resection defect of mrxΔ exo1Δ double mutants being more severe than those of the single mutants (Nakada et al. 2004). Nevertheless, residual resection and HR still take place in mrxΔ exo1Δ mutant cells, indicating that additional, Exo1- and MRX-independent, pathways of DSB resection exist (Moreau et al. 2001; Nakada et al. 2004).
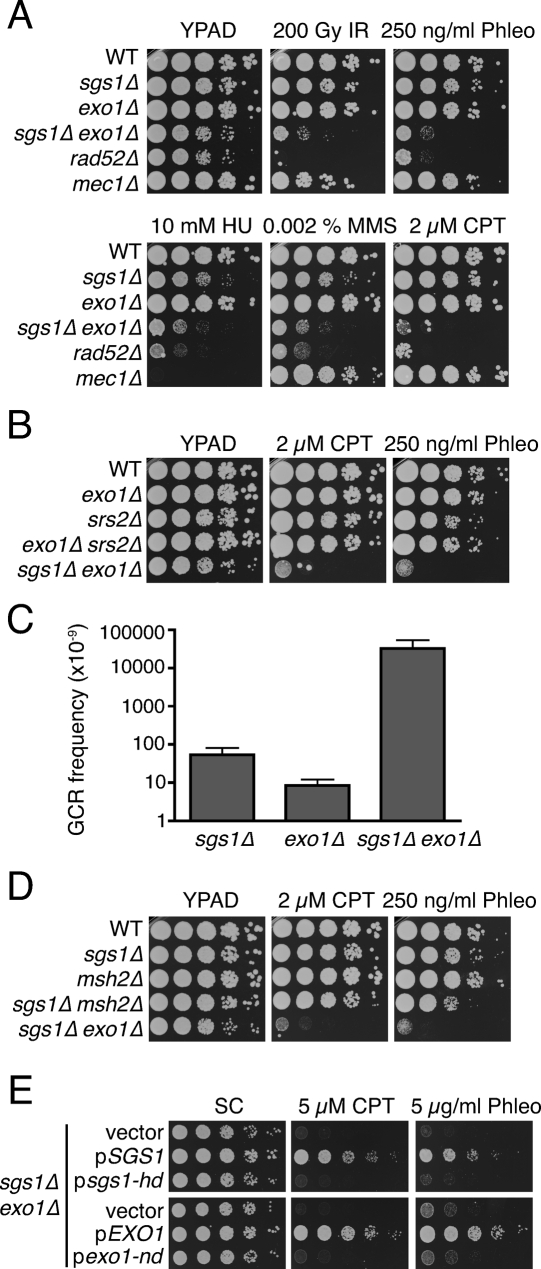
DNA helicases participate in multiple DNA transactions (Singleton et al. 2007; Lohman et al. 2008), but their possible involvement in DNA end resection has not been specifically addressed. While yeasts mutated in such helicases do not exhibit phenotypes suggestive of strong DSB resection defects, we reasoned that this might reflect them functioning in Exo1-independent resection pathways. Consequently, we tested the impact of disrupting genes for DNA helicases in an exo1Δ mutant background. Strikingly, this revealed that inactivation of the Sgs1 helicase in exo1Δ cells caused substantial hypersensitivity toward a wide range of DNA damaging agents, including IR, the IR-mimetic compound phleomycin, the DNA replication inhibitor hydroxyurea (HU), the DNA alkylating agent methyl methanesulphonate (MMS), and the topoisomerase I poison Camptothecin, which yields cytotoxicity primarily through it producing DSBs during S phase (Fig. 1A). Indeed, the sensitivities of the sgs1Δ exo1Δ mutant were comparable with those of rad52Δ cells, which are impaired in all HR pathways; and furthermore, in most cases sgs1Δ exo1Δ cells were considerably more sensitive than DNA damage checkpoint-deficient mec1Δ cells (Fig. 1A). In contrast, sgs1Δ or exo1Δ single mutants displayed little or no hypersensitivity toward the DNA damaging agents tested (Fig. 1A). Importantly, we found that the genetic interaction between SGS1 and EXO1 is specific, as no increased sensitivity to DNA damaging agents was observed when Srs2, another helicase with known roles in responding to DNA damage, was inactivated in the exo1Δ mutant background (Fig. 1B).
Figure 1.
SGS1 and EXO1 are components of parallel pathways promoting resistance to DNA-damaging agents. (A) sgs1Δ exo1Δ double mutants are hypersensitive to DNA damaging agents. Tenfold serial dilutions of the indicated strains were treated with IR or were plated on media containing the indicated drug, then were incubated for 3 d at 30°C. (B) Srs2 is not required for resistance to DNA damaging agents in the absence of Exo1. Strains were treated as in A. (C) GCR frequency was measured for the indicated strains; the mean and standard deviation of two fluctuation tests are shown. (D) Inactivation of MMR does not sensitize sgs1Δ cells to DNA damaging agents. Analyses were as in A. (E) The catalytic activities of Sgs1 and Exo1 promote resistance to DNA damaging agents. sgs1Δ exo1Δ mutants were transformed with an empty vector, with a vector expressing the wild-type (pSGS1) or a helicase-deficient (psgs1-hd) version of Sgs1, or with a vector expressing the wild-type (pEXO1) or a nuclease-deficient (pexo1-nd) version of Exo1. Analyses were as in A.
Earlier work has shown that sgs1Δ mutant cells suffer spontaneously arising gross chromosomal rearrangements (GCRs) at ∼20-fold higher rates than wild-type cells (Myung et al. 2001). As shown in Figure 1C, while we detected GCRs readily in sgs1Δ cells, exo1Δ mutant cells formed GCRs at low frequencies, as previously reported (Myung et al. 2001; Smith et al. 2005). In contrast, and in line with the DNA damage sensitivity data, deletion of both EXO1 and SGS1 had strikingly synergistic effects, with GCR frequencies in the double mutant being ∼600-fold and 3800-fold higher than those exhibited by the sgs1Δ and exo1Δ single mutants, respectively (Fig. 1C).
Because Exo1 functions in DNA mismatch repair (MMR), the phenotypes of sgs1Δ exo1Δ mutant cells might have reflected the combined inactivation of Sgs1 and MMR. However, when we inactivated the key MMR component Msh2 in an sgs1Δ mutant background, the resulting cells were nowhere near as sensitive as sgs1Δ exo1Δ cells, and had sensitivity profiles similar to those of the single mutants (Fig. 1D). These results therefore indicated that Exo1 imparts resistance to DSB-generating agents by mechanisms independent of its effects on MMR. Next, we addressed whether the helicase activity of Sgs1 and the nuclease activity of Exo1 were required for resistance to DNA damaging agents. Thus, sgs1Δ exo1Δ mutant cells were transformed with an empty plasmid, a plasmid encoding wild-type Sgs1 or Exo1, or a plasmid in which the coding sequence of SGS1 or EXO1 had been altered to produce a catalytically inactive protein. As shown in Figure 1E, the wild-type versions of Sgs1 and Exo1 rescued the DNA damage sensitivities of sgs1Δ exo1Δ mutant cells, whereas the helicase-defective Sgs1 derivative (sgs1-hd) and the nuclease-defective Exo1 derivative (exo1-nd) did not. These findings therefore indicated that the catalytic activities of both Sgs1 and Exo1 are required for resistance to DNA damaging agents.
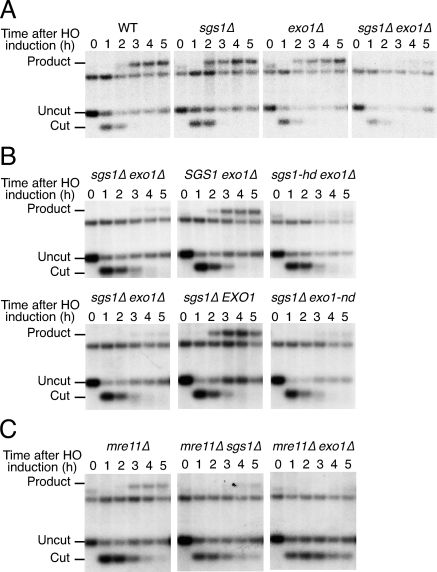
The observation that sgs1Δ exo1Δ double-mutant cells displayed DNA damage sensitivity profiles similar to those of HR-defective rad52Δ cells suggested that the combined loss of Sgs1 and Exo1 activity might prevent HR. To test this, we assessed the impact of SGS1 and EXO1 deletion on mating-type switching, an HR-mediated event that is initiated by the HO endonuclease and that replaces one MAT allele of a strain with DNA sequences encoding the opposite allele. Thus, we transformed wild-type and mutant cells of the MATa mating type with a plasmid that directed galactose-inducible HO expression. HO was then expressed in resulting strains by galactose addition, and glucose was provided 1 h afterward to inhibit HO production and cutting of the MAT locus, thereby allowing recombination to proceed. To assess such HR events, cells were recovered at various times before or after HO induction, then total genomic DNA was prepared and analyzed by Southern blotting (replacement of MATa by MATα generates restriction fragments of different sizes). As shown in Figure 2A, the MATα product was readily detected 2 h after HO induction in wild-type, sgs1Δ and exo1Δ cells. In contrast, MATα-product formation was dramatically delayed in sgs1Δ exo1Δ mutants and remained virtually undetectable even 5 h after HO induction (Fig. 2A). Moreover, while sgs1Δ exo1Δ mutants complemented with either wild-type Sgs1 or Exo1 had normal switching kinetics, the switching kinetics of cells expressing catalytically inactive versions of Sgs1 or Exo1 were similar to those of cells containing empty plasmids (Fig. 2B). These data therefore revealed that sgs1Δ exo1Δ double-mutant cells, but not the corresponding single mutants, are markedly impaired in HR-mediated events at the MAT locus. Furthermore, they established that the enzymatic functions of Sgs1 and Exo1 are required for normal kinetics of mating-type switching.
Figure 2.
sgs1Δ exo1Δ mutants have mating-type switching defects. (A) Time course of DSB repair at the MAT locus in the indicated strains. Galactose was added at time 0 to induce HO expression, and samples were removed at 1-h intervals for Southern blot analysis. Bands corresponding to uncut (MATa), HO endonuclease-cut and product (MATα) restriction fragments are indicated. The unlabeled band corresponds to the distal fragment and serves as a loading control. (B) The catalytic activities of Sgs1 and Exo1 promote mating-type switching. sgs1Δ exo1Δ cells transformed with an empty vector, or with a vector expressing the wild-type (pSGS1) or a helicase-deficient (psgs1-hd) version of Sgs1 (top panel), or with a vector expressing the wild-type (pEXO1) or a nuclease-deficient (pexo1-nd) version of Exo1 (bottom panel) were analyzed as in A. (C) Mre11 and Sgs1 promote mating-type switching by independent pathways. Analyses were as in A.
Previous work has shown that mating-type switching is greatly reduced when MRE11 is inactivated in an exo1Δ mutant background, indicating that Mre11 and Exo1 function in two independent pathways that promote HR (Moreau et al. 2001). Because we had observed similar synergistic effects when inactivating both SGS1 and EXO1, we wondered whether Sgs1 and Mre11 might promote HR through the same pathway. However, as was the case for EXO1 deletion, the deletion of SGS1 markedly reduced mating-type switching taking place in mre11Δ cells (Fig. 2C). These data therefore indicated that HR at the MAT locus is controlled by at least three distinct mechanisms involving Exo1, Mre11, and Sgs1, respectively. Consistent with these proteins controlling different functions, we found that the combined loss of MRX, EXO1, and SGS1 is lethal (Supplemental Fig. S1).
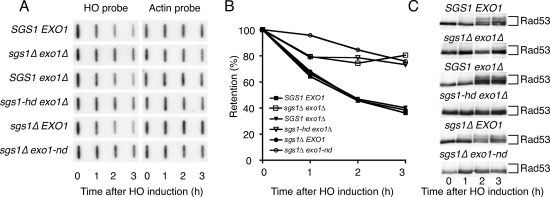
Although Sgs1 and Exo1 could potentially affect various steps of HR, we reasoned that they were most likely functioning at the level of DSB resection. To test this, we analyzed the DNA-degradation rate at HO-generated DSB ends by employing a slot blot hybridization procedure (Lee et al. 1998). When using a probe complementary to DNA sequences next to the HO recognition site, we detected a progressive, time-dependent decrease in signal in samples derived from wild-type, sgs1Δ, and exo1Δ strains. In contrast, the rate of degradation was markedly reduced in the sgs1Δ exo1Δ mutant (Fig. 3A,B). Furthermore, the helicase-defective SGS1 allele and the nuclease-defective EXO1 allele were incapable of complementing the resection defect of sgs1Δ exo1Δ mutant cells (Fig. 3A,B; as shown in Supplemental Fig. S2, the cell cycle profiles of the various strains were very similar at the time of HO induction, ruling out possible indirect, cell cycle-dependent effects of Exo1 and Sgs1 on resection). These findings therefore established that the catalytic activities of Exo1 and Sgs1 are needed for efficient DSB resection.
Figure 3.
Sgs1 and Exo1 act in parallel to promote DSB resection and checkpoint activation. (A) sgs1Δ exo1Δ mutants have resection defects. sgs1Δ exo1Δ cells containing an irreparable HO site were transformed with plasmids to generate the indicated strains. Galactose was added to cultures at time 0 and samples were removed at 1-h intervals for DNA analysis by denaturing slot blotting with a probe specific to sequences adjacent to the HO site. The blot was then stripped and reprobed with actin as a loading control. (B) Densitometric analysis of the data in A. For each time point, the signal detected by the HO probe was normalized to actin. (C) Extracts from samples taken during the experiment shown in A were analyzed by Western blotting with an anti-Rad53 antibody.
ssDNA generated by DSB processing provides the signal for Mec1-dependent DNA damage signaling (Zou and Elledge 2003; Ira et al. 2004). We therefore investigated whether the impaired resection in sgs1Δ exo1Δ cells affected Mec1-mediated Rad53 activation as monitored by phosphorylation-induced Rad53 mobility shifts on Western blots. Notably, Rad53 hyperphosphorylation was readily detectable 2 h after HO induction in wild-type cells but not in sgs1Δ exo1Δ double-mutant cells (Fig. 3C). Furthermore, while expression of either Sgs1 or Exo1 restored normal HO-induced Rad53 hyperphosphorylation in sgs1Δ exo1Δ mutant cells, the helicase-defective version of Sgs1 and the nuclease-defective version of Exo1 failed to complement this phenotype, indicating that the enzymatic activities of both proteins are required for effective Rad53 activation.
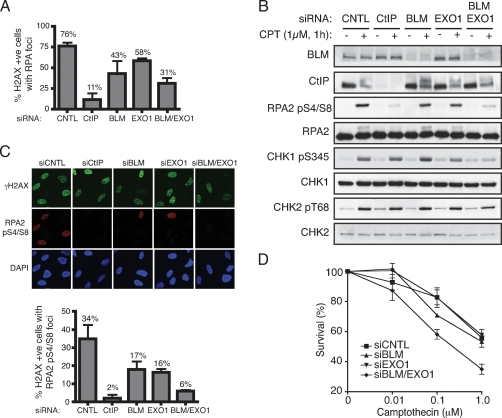
Given that many aspects of the DNA damage response have been conserved during evolution, we tested whether BLM, the mammalian counterpart of Sgs1 (Cobb et al. 2002; Bachrati and Hickson 2003), promotes DSB resection in human cells. Thus, we used siRNA treatment to deplete BLM, EXO1, or both BLM and EXO1 from U2OS cells, and then exposed depleted cells to Camptothecin for 1 h to induce DSB formation (Camptothecin was used because resection is needed for repair of Camptothecin-induced DSBs and because such DSBs occur in S phase, when CDK levels are sufficient to allow efficient resection to occur) (Pommier et al. 2003; Jazayeri et al. 2006; Sartori et al. 2007). Next, we measured ssDNA accumulation by using immunofluorescence microscopy to monitor the formation of RPA foci in cells that stained positive for phosphorylated histone H2AX (γH2AX), which is a marker of DNA DSBs (Rogakou et al. 1999). This revealed that BLM depletion reduced the proportion of cells with discernible RPA foci (Fig. 4A). Furthermore, and consistent with previous work implicating mouse EXO1 in DSB resection (Schaetzlein et al. 2007), we found that EXO1 depletion caused a mild but reproducible defect in Camptothecin-induced RPA focus formation in human cells (Fig. 4A). Moreover, codepletion of BLM and EXO1 had an additive effect, leading to a further reduction in the proportion of cells displaying RPA foci (Fig. 4A) (note that BLM and/or EXO1 depletion did not affect DSB formation per se because H2AX phosphorylation took place effectively in every case). These data therefore indicated that BLM and EXO1 function in distinct but complementary pathways of DSB resection in human cells. We also found that depletion of CtIP, a factor known to promote DSB resection (Sartori et al. 2007), had a greater impact on RPA-focus formation than codepletion of BLM and EXO1 (Fig. 4A,C). While this could reflect differences in depletion efficiencies, it is possible that CtIP performs a function particularly important for resection of Camptothecin-induced DSBs and/or provides a suitable environment for various resection pathways to operate efficiently.
Figure 4.
BLM and EXO1 promote DNA DSB resection and associated events in human cells. (A) BLM and EXO1 deficiency impairs Camptothecin-induced RPA focus formation. U2OS cells were transfected with siRNAs directed against Luciferase (siCNTL), CtIP, BLM, EXO1, or a combination of BLM and EXO1, then 72 h later were mock-treated or treated with 1 μM Camptothecin for 1 h. Cells were next detergent-extracted and fixed, then foci for phosphorylated histone H2AX (γH2AX) and RPA2 were visualized by indirect immunofluorescence. More than 100 cells were counted for each sample and the percentages of cells exhibiting both γH2AX and RPA foci was determined. Data represent the mean ± SEM from three independent experiments. All counting was done blind. (B) BLM and EXO1 promote DSB signaling. Extracts of mock-treated or Camptothecin-treated cells depleted for the indicated factors were analyzed by Western blotting with the indicated antibodies. Endogenous EXO1 levels were too low to allow detection with anti-EXO1 antisera and so verification of EXO1 siRNA depletion was done with cells stably expressing a GFP-Exo1 construct (see Supplemental Fig. S4). (C) RPA Ser-4 and Ser-8 phosphorylation (RPApS4/S8) is compromised by BLM and EXO1 depletion. CtIP, BLM, or EXO1 were depleted and cells were treated as in A, followed by analysis by indirect immunofluorescence with the indicated antibodies. More than 100 cells were counted for each sample, and data represent the mean ± SEM from three independent experiments. Counting was done blind. (D) Codepletion or BLM and EXO1 yields Camptothecin hypersensitivity. Seventy-two hours following transfection with the indicated siRNAs, U2OS cells were treated with Camptothecin for 1 h, and cell survival was determined by colony formation. Data represent the mean ± SEM from three independent experiments.
To explore the impact of BLM and EXO1 on DSB resection and signaling further, we used antibodies that recognize phosphorylation sites on downstream target proteins. The loading of RPA onto ssDNA promotes its hyperphosphorylation on various sites, including Ser-4 and Ser-8 of RPA2 (Shao et al. 1999; Anantha et al. 2007). Significantly, when we used phospho-specific antibodies against these sites in Western immunoblotting analyses, we found that while RPA2 Ser4/8 phosphorylation was only slightly reduced by BLM or EXO1 depletion, it was markedly diminished when BLM and EXO1 were codepleted (Fig. 4B; FACS analyses in Supplemental Fig. S3 show that depleting EXO1 and BLM alone or in combination had little effect on cell cycle distributions). Furthermore, BLM or EXO1 depletion reduced RPA phosphorylation when the phospho-specific antibodies were used in indirect immunofluorescence assays (Fig. 4C). Moreover, quantification revealed that BLM and EXO1 had additive effects, with doubly depleted cells exhibiting dramatically reduced focus formation (Fig. 4C, histogram). Phosphorylation of CHK1 on Ser-345, which is an established marker of ATR activation by ssDNA/RPA, was also markedly impaired by codepletion of BLM and EXO1 (Fig. 4B). In this case, however, BLM or EXO1 depletion alone had little impact, suggesting that in the presence of either factor resection was sufficient to promote ATR signaling (note that depletion of BLM and EXO1 alone or in combination had little effect on phosphorylation of CHK2 Thr-68, which can occur in the absence of extensive DSB resection) (Fig. 4B; Jazayeri et al. 2006). We also found that codepletion of BLM and EXO1 caused hypersensitivity toward Camptothecin, whereas the depletion of either factor alone had little or no effect (Fig. 4D).
In summary, we established that Exo1 and members of the RecQ-family of DNA helicases promote DSB resection in both yeast and human cells. Specifically, we showed that loss or mutation of the S. cerevisiae DNA helicase Sgs1 markedly impairs DSB resection in Exo1-deficient cells, leading to pronounced hypersensitivity toward DNA damaging agents, severely impaired HR and ineffective Mec1-mediated DNA damage signaling. Similarly, while depletion of either human BLM or EXO1 only mildly impairs DSB resection and ensuing ATR-mediated signaling, such responses are markedly defective when BLM and EXO1 are depleted simultaneously. Although other possibilities exist, the simplest explanation for our findings is that Sgs1/BLM and Exo1/EXO1 constitute parts of two complementary resection pathways. Furthermore, data in yeast cells suggest that Exo1 and Sgs1 facilitate DSB resection by mechanisms that are at least partly distinct from resection events promoted by the MRX complex. One key question remaining is why cells possess multiple DSB resection mechanisms. While this could reflect redundancy—possibly to provide cells with a buffer against genotoxic challenges—we speculate that the different pathways will turn out to be more complementary than redundant, with each pathway being best suited for different physiological situations. Thus, it will be interesting to compare the abilities of the Sgs1/BLM-, MRX/MRN-, and Exo1/EXO1-dependent DSB resection pathways to operate in different phases of the cell cycle and in different cell types. It will also be of interest to assess their relative abilities to process stalled replication forks and promote replication restart, and to compare their activities on different classes of DSB lesions, which include clean DNA ends and DSBs containing complex base damage and/or difficult-to-process structures such as protein–DNA cross-links. Other challenges for future research will be to identify other protein players functioning in the various DSB-processing pathways and define how their activities are controlled. Finally, it is tempting to speculate that defective DSB processing and ensuing ssDNA signaling might underlie the cancer predisposition and/or infertility phenotypes of human Bloom’s syndrome patients, and that defective EXO1 function might also be associated with certain cancers.
Materials and methods
Plasmids and yeast strains
Plasmids encoding wild-type Sgs1 (pJL31: LEU2 CEN6 ARS4) and a helicase-dead version of Sgs1 (pJL37: LEU2 CEN6 ARSH4) are described in Lu et al. (1996), and were kindly provided by S. Brill. The EXO1 ORF plus 250 base pairs (bp) of upstream sequences were amplified by PCR and cloned in pRS414 (TRP1 CEN6 ARSH4). A C-terminal myc epitope tag was added to Exo1 at the same time to generate pExo1-myc. To generate a catalytically inactive version of Exo1, a 533-bp NsiI/BamHI fragment from pJAS-exo1-D173A-Flag (a kind gift of M. Liskay; described in Tran et al. 2002) was used to replace the wild-type sequence of pExo1-myc. Yeast strains used in Figures 1 and 2 and Supplemental Figure S1 are W303 derivatives (with the exception of Fig. 1C, in which RDKY3615 derivatives were used; Myung et al. 2001). To generate the sgs1Δ exo1Δ hmlΔ hmrΔ GAL-HO strain used in Figure 3, W303 sgs1Δ exo1Δ were crossed to JKM139 (hmlΔ hmrΔ GAL-HO) (Lee et al. 1998). The resulting diploids were sporulated and microdissected, and relevant genetic elements were identified by marker segregation or PCR.
GCRs measurements
GCRs were determined as described previously (Myung et al. 2001). Experiments were repeated twice using five independent cultures in each case.
Physical analysis of mating type switching
Cells to be tested were transformed with a plasmid permitting the galactose-dependent induction of HO endonuclease and with plasmids encoding wild-type and catalytically inactive versions of SGS1 and EXO1 when indicated. Cells were grown in raffinose-containing media to an OD600 of 0.3–0.4, then 2% galactose was added. One hour after addition of galactose, cells were harvested and resuspended in glucose-containing media. Samples were removed prior to HO induction (t = 0), and at 1-h intervals after induction for DNA analysis. Total genomic DNA was prepared, digested with StyI, separated on 0.8 % agarose gels, and then transferred to nylon membrane and hybridized with a radioactively labeled probe overlapping the first StyI site downstream from the Z region of MAT.
DNA end resection assay
sgs1Δ exo1Δ hmlΔ hmrΔ GAL-HO cells were transformed with plasmids to generate strains of the genotype indicated in Figure 3. These cells were grown in media containing raffinose to an OD600 of 0.3–0.4, then 2% galactose was added. Samples were removed prior to HO induction (t = 0), and at 1-h intervals after induction for DNA analysis. Total genomic DNA was prepared, denatured, and blotted onto a nylon membrane using a slot blot manifold. DNA was hybridized with a radiolabeled probe complementary to DNA sequences adjacent to the HO recognition site, then stripped and reprobed with an actin probe. Signals were quantified with a FLA-5000 instrument (Fuji), and values obtained with the HO probe were normalized to those obtained with the actin probe.
Cell culture and transfection
Human U2OS osteosarcoma cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and standard antibiotics. All siRNA duplexes were purchased from MWG-Biotech, with the exception of the BLM siGENOME Smartpool (Dharmacon). siRNA sequences are as follow: CNTL (luciferase: 5′-CGUACGC GGAAUACUUCGATT-3′), CtIP (CtIP-1) (Sartori et al. 2007), EXO1 (5′-CAAGCCUAUUCUCGUAUUUTT-3′), BLM (siGENOME sequences: 5′-GAGCACAUCUGUAAAUUAA-3′; 5′-GAGAAACUCACUUCAAU AA-3′; 5′-CAGGAUGGCUGUCAGGUUA-3′; 5′-CUAAAUCUGUGG AGGGUUA-3′). For optimal depletion of protein factors, siRNA was transfected into cells using Oligofectamine reagent (Invitrogen) in two consecutive rounds in which siRNA was added to cells to a final concentration of 80 nM as follows: CNTL (80 nM luciferase siRNA), CtIP (40 nM CtIP siRNA + 40 nM CNTL siRNA), BLM (40 nM BLM siRNA + 40 nM CNTL siRNA), EXO1 (40 nM EXO1 siRNA + 40 nM CNTL siRNA), or combined BLM and EXO1 (40 nM BLM siRNA + 40 nM EXO1 siRNA). Experiments were typically performed 72 h following first siRNA transfection. Data for survival curves were generated by colony formation assays. In brief, 48 h following the second siRNA transfection, cells were treated with Camptothecin or DMSO (CNTL). After 1 h, the drug was removed and cells were left for 10–14 d at 37°C to allow colonies to form. Colonies were stained with 0.5% crystal violet/20% ethanol and counted.
Antibodies
The antibodies used in this study were rabbit α-Rad53 (a gift from Noel Lowndes), goat α-BLM (C18, Santa Cruz Biotechnologies), mouse α-RPA2 (Ab1, 9HD, Lab Vision), rabbit α-RPA pS4/S8 (Bethyl), rabbit α-γH2AX (Cell Signaling), rabbit α-Chk2 pT68 (Cell Signaling), rabbit α-Chk2 (Abcam), rabbit α-Chk1 pS345 mAb (133D3, Cell Signaling), mouse α-Chk1 (G4, Santa Cruz Biotechnologies), and a mouse monoclonal antibody to CtIP (provided by R. Baer) (Yu and Baer 2000).
Immunofluorescence microscopy
Cells grown on glass coverslips were pre-extracted for 5 min on ice with cytoskeletal buffer (10mM PIPES at pH 6.8, 300 mM sucrose, 50 mM NaCl, 3 mM EDTA, 0.5% Triton X-100, Protease Inhibitor Cocktail [Complete, EDTA-free; Roche]) before fixation in 2% paraformaldehyde for 15 min at room temperature. After fixation, cells were washed with PBS and then blocked with 5% BSA and 0.1% Triton X-100 in PBS. Cells were stained with primary antibodies in block for 1 h, washed in PBS + 0.1% Triton X-100, and then stained with Alexa Fluor 488 goat anti-mouse/rabbit, and Alexa Fluor 598 goat anti-mouse/rabbit (Molecular Probes) for 1 h at room temperature in block. DNA was counterstained with DAPI (0.2 μg/mL) in Vectorshield mounting agent (Vector Laboratories). Images were acquired using a Bio-Rad Radiance 2100 (on a Nikon Eclipse E800 upright) microscope using Lasersharp 2000 software (Zeiss).
Acknowledgments
We thank all members of the S.P.J. laboratory for their advice and encouragement. We are grateful to Steve Brill, Mike Liskay, James Haber, and Richard Kolodner for providing plasmids and strains, and to Noel Lowndes and Richard Baer for the Rad53 and CtIP antibodies, respectively. Research in the S.P.J. laboratory is supported by grants from Cancer Research UK and the European Union.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.503108.
References
- Anantha R.W., Vassin V.M., Borowiec J.A. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J. Biol. Chem. 2007;282:35910–35923. doi: 10.1074/jbc.M704645200. [DOI] [PubMed] [Google Scholar]
- Bachrati C.Z., Hickson I.D. RecQ helicases: Suppressors of tumorigenesis and premature aging. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R., Doksani Y., Capra T., Katou Y.M., Tanaka H., Shirahige K., Foiani M. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes & Dev. 2007;21:1921–1936. doi: 10.1101/gad.432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M., Mantiero D., Lucchini G., Longhese M.P. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 2006;7:212–218. doi: 10.1038/sj.embor.7400593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J.A., Bjergbaek L., Gasser S.M. RecQ helicases: At the heart of genetic stability. FEBS Lett. 2002;529:43–48. doi: 10.1016/s0014-5793(02)03269-6. [DOI] [PubMed] [Google Scholar]
- Cotta-Ramusino C., Fachinetti D., Lucca C., Doksani Y., Lopes M., Sogo J., Foiani M. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol. Cell. 2005;17:153–159. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N.M., et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A., Falck J., Lukas C., Bartek J., Smith G.C., Lukas J., Jackson S.P. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Kastan M.B., Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Khanna K.K., Jackson S.P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- Lee S.E., Moore J.K., Holmes A., Umezu K., Kolodner R.D., Haber J.E. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Lisby M., Rothstein R. DNA damage checkpoint and repair centers. Curr. Opin. Cell Biol. 2004;16:328–334. doi: 10.1016/j.ceb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Lohman T.M., Tomko E.J., Wu C.G. Non-hexameric DNA helicases and translocases: Mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- Lu J., Mullen J.R., Brill S.J., Kleff S., Romeo A.M., Sternglanz R. Human homologues of yeast helicase. Nature. 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- Maringele L., Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes & Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S., Morgan E.A., Symington L.S. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Datta A., Chen C., Kolodner R.D. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- Nakada D., Hirano Y., Sugimoto K. Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol. Cell. Biol. 2004;24:10016–10025. doi: 10.1128/MCB.24.22.10016-10025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y., Redon C., Rao V.A., Seiler J.A., Sordet O., Takemura H., Antony S., Meng L., Liao Z., Kohlhagen G., et al. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat. Res. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Boon C., Redon C., Bonner W.M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J., Jackson S.P. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaetzlein S., Kodandaramireddy N.R., Ju Z., Lechel A., Stepczynska A., Lilli D.R., Clark A.B., Rudolph C., Kuhnel F., Wei K., et al. Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell. 2007;130:863–877. doi: 10.1016/j.cell.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao R.G., Cao C.X., Zhang H., Kohn K.W., Wold M.S., Pommier Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Smith S., Gupta A., Kolodner R.D., Myung K. Suppression of gross chromosomal rearrangements by the multiple functions of the Mre11–Rad50–Xrs2 complex in Saccharomyces cerevisiae. DNA Repair (Amst.) 2005;4:606–617. doi: 10.1016/j.dnarep.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Tran P.T., Erdeniz N., Dudley S., Liskay R.M. Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair (Amst.) 2002;1:895–912. doi: 10.1016/s1568-7864(02)00114-3. [DOI] [PubMed] [Google Scholar]
- Tran P.T., Erdeniz N., Symington L.S., Liskay R.M. EXO1—A multi-tasking eukaryotic nuclease. DNA Repair (Amst.) 2004;3:1549–1559. doi: 10.1016/j.dnarep.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Wyman C., Kanaar R. DNA double-strand break repair: All’s well that ends well. Annu. Rev. Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- Yu X., Baer R. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J. Biol. Chem. 2000;275:18541–18549. doi: 10.1074/jbc.M909494199. [DOI] [PubMed] [Google Scholar]
- Zhou B.B., Elledge S.J. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]