Abstract
Caenorhabditis elegans SIR-2.1, a member of the sirtuin family related to Saccharomyces cerevisiae Sir2p, has previously been implicated in aging. The mammalian homolog SIRT1 plays important roles in multiple cellular processes including transcriptional repression and stress response. We show that sir-2.1 is essential for the execution of apoptosis in response to DNA damage, and that sir-2.1 genetically acts in parallel to the worm p53-like gene cep-1. This novel cep-1-independent proapoptotic pathway does not require the daf-16 FOXO transcription factor. Cytological analysis of SIR-2.1 suggests a novel mechanism of apoptosis induction. During apoptosis SIR-2.1 changes its subcellular localization from the nucleus to the cytoplasm and transiently colocalizes with the C. elegans Apaf-1 homolog CED-4 at the nuclear periphery. SIR-2.1 translocation is an early event in germ cell apoptosis and is independent of apoptosis execution and cep-1, raising the possibility that SIR-2.1 translocation is linked to the induction of DNA damage-induced apoptosis.
Keywords: sir-2.1, SIR2, sirtuin, C. elegans, DNA damage response, apoptosis
Apoptosis during worm development generally requires the transcriptional induction of egl-1 encoding for a proapoptotic BH3-only domain protein (Conradt and Horvitz 1998, 1999). EGL-1 then interacts with the anti-apoptotic Bcl2 family member CED-9 that is localized at the outer mitochondrial membrane and forms a complex with CED-4, a protein related to mammalian Apaf-1 (Chen et al. 2000). EGL-1 binding to CED-9 leads to the disruption of the CED-9/CED-4 complex. CED-4 is released, accumulates at the nuclear periphery by binding to SUN-1, oligomerizes, and induces the autoactivation of the caspase CED-3, leading to apoptosis (Yang et al. 1998; Horvitz 1999; Chen et al. 2000; Yan et al. 2004, 2005; Fairlie et al. 2006; Tzur et al. 2006). Within somatic tissues apoptosis only occurs during development.
In the proliferative germline of adult worms multiple pathways are able to trigger apoptosis. Interestingly, only late pachytene stage meiotic germ cells have the potential to undergo apoptosis (Fig. 1D). While physiological germ cell apoptosis occurs independently of exogenous stimuli and is thought to be necessary for maintaining tissue homeostasis (Gumienny et al. 1999), genotoxic stress can also elicit an apoptotic response in germ cells (Gartner et al. 2000). In DNA damage-induced apoptosis, egl-1 transcription is induced by a pathway that includes ATL-1, a worm ATR-like PI-3 type protein kinase (Garcia-Muse and Boulton 2005); Caenorhabditis elegans MRT-2 and HUS-1, which are part of the so-called 9–1–1 DNA sliding clamp complex (Hofmann et al. 2002); and CLK-2, which acts in a pathway parallel to the 9–1–1 complex (Gartner et al. 2000; Ahmed et al. 2001). While upstream sensors and transducers affect all DNA damage responses including DNA repair, cell cycle arrest, and apoptosis, downstream effectors like cep-1, which encodes a primordial worm p53-like protein, are only needed for a subset of responses. cep-1 is required for ionizing radiation (IR)-induced apoptosis, and for cell cycle arrest and apoptosis in response to UV radiation (Fig. 2D; Derry et al. 2001, 2007; Schumacher et al. 2001; Stergiou et al. 2007).
Figure 1.
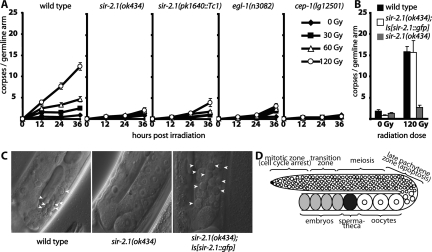
sir-2.1 is required for DNA damage-induced germ cell apoptosis. (A) Worms were irradiated with the indicated doses of ionizing radiation at the late L4 larval stage, and apoptotic corpses were scored by DIC optics after 12, 24, and 36 h. (B) Worms were treated as in A and corpses were scored 24 h after irradiation (C) Representative pictures of germlines of worms scored in B. Arrowheads indicate apoptotic corpses. (D) Schematic drawing of a C. elegans germline arm. Germ cell proliferation occurs in the mitotic zone and cells progress through meiosis as they are moved along the germline. In response to DNA damage cell cycle arrest occurs only in the mitotic zone, while only late pachytene cells are able to undergo apoptosis.
Figure 2.
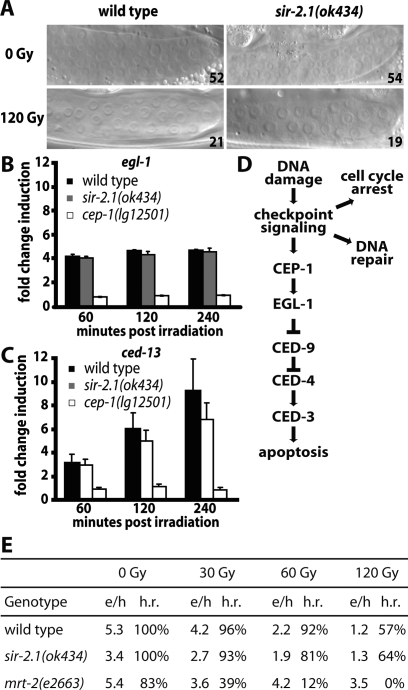
sir-2.1 does not affect the DNA damage response pathway upstream of cep-1. (A) Cell cycle arrest following DNA damage is not affected by a sir-2.1 deletion. The number of nuclei is shown in the bottom right corner of each picture. (B,C) sir-2.1 is not required for cep-1-dependent transcriptional induction of egl-1 and ced-13. Worms were irradiated with 120 Gy 24 h after the L4 stage. RNA was extracted 60, 120, and 240 min after irradiation and egl-1 and ced-13 transcript levels were assayed by qRT–PCR as described previously (Schumacher et al. 2005a, b). (D) Genetic pathway connecting DNA damage to apoptosis and cell cycle arrest/DNA repair. (E) sir-2.1 loss of function does not increase radiation sensitivity. Eggs laid per hour (e/h) and the percentage of surviving embryos (hatch rate, h.r.) with and without IR treatment are indicated.
Interestingly, as in mammals where Apaf-1 and Bcl-2 do not directly interact, the regulation of CED-4 seems to be more complex than previously thought (Meier and Vousden 2007), especially in germ cell apoptosis (see below). Indeed, it has been shown that CED-4 translocates to the nuclear periphery in irradiated mitotic germ cells without concomitantly inducing germ cell apoptosis (Zermati et al. 2007). Furthermore, we show here that CED-4 is predominantly localized at the nuclear periphery in healthy nonapoptotic late pachytene cells. These results, together with the notion that physiological germ cell death occurs independently of egl-1 (Gumienny et al. 1999), raise the possibility that additional factors are needed to transmit a proapoptotic signal from mitochondrial CED-9 to perinuclear CED-4 to trigger apoptosis.
C. elegans SIR-2.1 is a member of the Sirtuins, a ubiquitous family of NAD+-dependent protein deacetylases with members present in virtually every species from archaea to mammals (Brachmann et al. 1995). The founding member, budding yeast Sir2p, is involved in the transcriptional silencing of the mating-type loci (Rine and Herskowitz 1987), telomeres (Gottschling et al. 1990), and rDNA repeats (Bryk et al. 1997; Smith and Boeke 1997). Apart from its role in epigenetic regulation of gene expression, Sir2p has been implicated in the nonhomologous end-joining DNA repair pathway (Martin et al. 1999; Hegde and Klein 2000), as well as in double-strand break repair by homologous recombination (Tamburini and Tyler 2005).
In mammals, seven sirtuins exist, the most widely studied of which, SIRT1, is most closely related to budding yeast Sir2p and C. elegans SIR-2.1. Interestingly, while most SIR2 mutant phenotypes in budding yeast are related to transcriptional silencing, mammalian SIRT1 interacts with and deacetylates many nonhistone substrates. In tissue culture-based experiments SIRT1 has been reported to have an anti-apoptotic role in response to genotoxic stress. SIRT1 can deacetylate the p53 tumor suppressor protein, which leads to its inactivation and destabilization (Luo et al. 2001; Vaziri et al. 2001; Langley et al. 2002; Cheng et al. 2003). SIRT1 binds and deacetylates FOXO transcription factors, resulting in differential target gene expression. FOXO3 deacetylation by SIRT1 leads to the transcriptional repression of proapoptotic Bim, but also to the up-regulation of the stress resistance gene GADD45 (Brunet et al. 2004). SIRT1-mediated deacetylation of FOXO1 represses its proapoptotic activity in prostate cancer cells (Yang et al. 2005). In addition, SIRT1 has been reported to deacetylate Ku70, which in the deacetlylated form sequesters the proapoptotic factor Bax from mitochondria, thereby inhibiting apoptosis (Cohen et al. 2004). However, recent studies in knockout mice revealed that the in vivo role of SIRT1 in stress-induced apoptosis is less clear (Kamel et al. 2006).
In C. elegans, sir-2.1 overexpression is reported to significantly extend post-mitotic life span, whereas deleting sir-2.1 results in a modestly shortened life span (Tissenbaum and Guarente 2001; Berdichevsky et al. 2006; Wang and Tissenbaum 2006). Extended longevity mediated by sir-2.1 overexpression requires DAF-16 a forkhead family transcription factor related to the mammalian FOXOs. DAF-16 is negatively regulated by the conserved C. elegans insulin/IGF-1 pathway, and is repressed by the DAF-2 insulin receptor (Tissenbaum and Guarente 2001). The life span extension by sir-2.1 overexpression is mediated by the C. elegans PAR-5 and FTT-2 14–3–3 proteins that interact with both DAF-16 and SIR-2.1 (Berdichevsky and Guarente 2006; Wang et al. 2006). sir-2.1 is also required for life span extension of eat-2 worms that are long lived, likely due to caloric restriction (Wang and Tissenbaum 2006). In addition, sir-2.1 is required for germline silencing of multicopy transgenic arrays (Jedrusik and Schulze 2003) and seems to influence the subcellular localization of the linker histone HIS-24 (H1.1) in the germline (Jedrusik and Schulze 2007).
As part of an ongoing program to screen for novel genes involved in DNA repair and/or damage response signaling we screened through C. elegans homologs of genes reported to be involved in these processes and included sir-2.1. Here we examine the role of C. elegans SIR-2.1 in germline DNA damage response pathways. We show that sir-2.1 specifically affects DNA damage-induced apoptosis in parallel to or downstream from cep-1, while not impinging on developmental apoptosis or physiological germ cell apoptosis. Finally, we show that SIR-2.1 is located in germ cell nuclei, but is lost from nuclei undergoing apoptosis. SIR-2.1 nuclear loss is an early event in germ cell apoptosis and occurs independent of apoptosis execution. During apoptosis, SIR-2.1 colocalizes with CED-4, suggesting a functional connection between these proteins.
Results
sir-2.1 is required for DNA damage-induced apoptosis
To address whether sir-2.1 has a role in DNA damage-induced apoptosis, we γ-irradiated late L4 worms and scored apoptosis 12, 24, and 36 h post-treatment. Apoptosis was nearly completely abolished in sir-2.1(ok434) mutants (Fig. 1 A,C). The degree of reduction of apoptosis was close to that observed in egl-1 or cep-1 mutants (Fig. 1A). To determine if the apoptotic response might just be delayed, rather than absent, we also assayed worms 48 h after irradiation. At this time point we observed a significantly higher number of corpses in the germlines of egl-1(n3082) animals as compared with sir-2.1(ok434), while germlines of wild-type worms essentially disintegrated due to excessive apoptosis under those conditions, confirming that SIR-2.1 is indeed necessary for DNA damage-induced apoptosis (Supplemental Fig. 1). To further corroborate these results, we also introduced DNA double-strand breaks by X-ray treatment confirming the absence of apoptosis under those conditions (Supplemental Fig. 2B). We next wanted to verify that the defect in DNA damage-induced apoptosis is due to sir-2.1(ok434) rather than due to a secondary, possibly genetically linked mutation. As to this we tested for DNA damage-induced apoptosis with a second sir-2.1 loss of function allele sir-2.1(pk1640∷Tc1) that disrupts the sir-2.1 catalytic domain and confirmed that DNA damage-induced apoptosis is dramatically reduced (Fig. 1A). SIR-2.1 protein was absent in both mutant alleles of sir-2.1 (Supplemental Fig. 3). To further confirm a role of sir-2.1 in DNA damage-induced apoptosis we rescued the sir-2.1(ok434) deletion with a construct containing the entire sir-2.1 expressing operon with GFP fused to the C terminus of sir-2.1. We found only one line that rescued the sir-2.1 defect (Fig. 1B), but this became silenced within three to four generations. While the level of DNA damage-induced apoptosis when averaged was comparable with wild type in the rescue line, we noted higher than normal fluctuations in DNA damage-induced apoptosis when scoring individual germlines, with only ∼30% of worms showing evidence of DNA damage-induced apoptosis, which then appeared to occur at higher than wild-type levels (data not shown). Thus, these results suggest that overexpressing sir-2.1 might lead to enhanced apoptosis, which could explain the difficulty of establishing a stable line.
Given that life span extension by sir-2.1 overexpression is mediated by daf-16 (Tissenbaum and Guarente 2001), we wanted to see whether the function of SIR-2.1 in DNA damage-induced apoptosis might also depend on DAF-16. It has previously been reported that DAF-16 may be required for DNA damage-induced apoptosis (Pinkston et al. 2006). We therefore assayed the apoptotic response to DNA damage in three daf-16 mutants: daf-16(mu27), daf-16(mu86), and daf-16(mgDf50). We found only a slight reduction in DNA damage-induced apoptosis in daf-16(mu86) and daf-16(mgDf50) at early time points and at intermediate doses of irradiation throughout the time course experiment, whereas apoptosis appeared close to wild-type levels in the latest time points (Supplemental Fig. 4). These results indicate that, unlike SIR-2.1, DAF-16 is not an integral part of the pathway leading to DNA damage-induced apoptosis. Our data differ from those reported by Pinkston et al. (2006), but are in line with results reported by Quevedo et al. (2007). In summary, our results suggest that sir-2.1 mutants are defective in DNA damage-induced apoptosis, and that the proapoptotic function of sir-2.1 is independent of daf-16.
sir-2.1 does not overtly affect DNA repair and acts in a pathway parallel to or downstream from cep-1 to effect DNA damage-induced germ cell apoptosis
Some C. elegans mutants that are defective for DNA damage-induced apoptosis also play roles in orchestrating DNA repair and Sarccharomyces cerevisiae SIR2 mutants are compromised in the nonhomologous end-joining DNA double-strand break repair pathway (Martin et al. 1999; Hegde and Klein 2000). To ask if sir-2.1 is required for double-strand break repair in C. elegans we tested whether repair by homologous recombination or DNA end joining is defective in sir-2.1. We first examined the potential role of sir-2.1 in nonhomologous end joining, a DNA repair pathway that plays a major role in late-stage C. elegans embryos, where most somatic cells are arrested in the G1 phase of the cell cycle (Clejan et al. 2006). We confirmed that upon ionizing irradiation of late-stage embryos, progression to the L4 developmental stage is dramatically retarded in end-joining defective cku-80 mutants. In contrast, we could not find such an effect in wild-type or sir-2.1(ok434) mutant worms (Supplemental Fig. 2A). Thus, sir-2.1 is dispensable for nonhomologous end-joining in C. elegans. We also found that sir-2.1 has no overt role in repair by homologous recombination, which is the predominant form of repair in the germline (Clejan et al. 2006). In contrast to the known checkpoint mutants mrt-2(e2663) and hus-1(op241) (Ahmed et al. 2001) we found that sir-2.1 mutants do not have an obvious DNA repair defect upon ionizing irradiation (Fig. 2E). Even though we found that sir-2.1 worms are not overtly radiation-sensitive, we decided to look more closely at whether sir-2.1 might act as an upstream checkpoint gene by assessing DNA damage-induced cell cycle arrest, which we found to be indistinguishable from that of wild type (Fig. 2A). Defective DNA damage-induced apoptosis in the presence of IR-dependent cell cycle arrest and DNA repair was also found in cep-1 mutants (Derry et al. 2001; Schumacher et al. 2001). cep-1 encodes for the C. elegans p53-like transcription factor, which we and others have shown to be required for DNA damage-dependent germ cell apoptosis and the transcriptional induction of the BH3-only domain encoding gene egl-1 (Derry et al. 2001; Schumacher et al. 2001, 2005a, b). Thus sir-2.1 might, like cep-1, specifically affect DNA damage-induced apoptosis.
Given the phenotypic similarity between cep-1 and sir-2.1, we tested whether sir-2.1 might be required for cep-1 activation by comparing the transcriptional level of the cep-1 targets egl-1 and ced-13 in wild-type, cep-1, and sir-2.1 strains after irradiation. These experiments show that cep-1 activity is not affected by sir-2.1 (Fig. 2B,C), suggesting that sir-2.1 acts in a pathway leading to DNA damage-induced germ cell apoptosis in parallel to or downstream from cep-1. Alternatively, sir-2.1 might affect the core apoptosis pathway or only affect germ cell apoptosis without being specific to DNA damage-induced apoptosis.
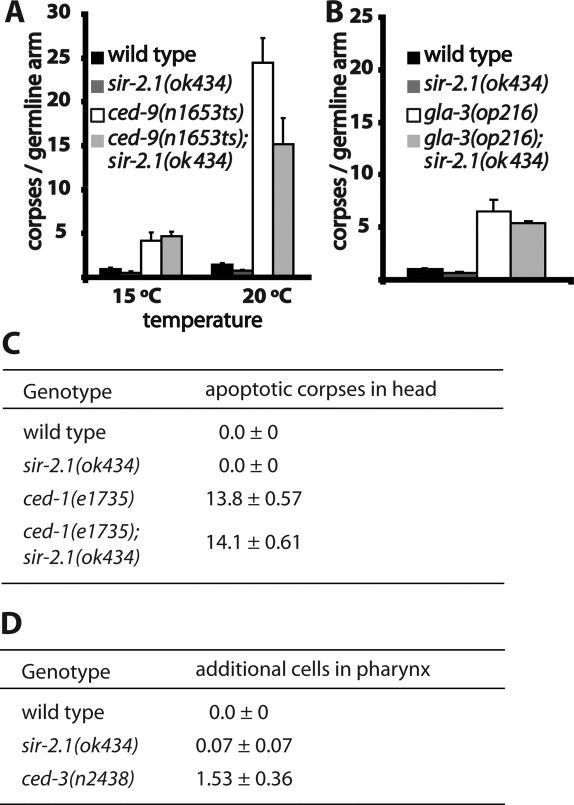
To assess whether sir-2.1 affects the core apoptosis pathway used during somatic development we scored developmental apoptosis in early L1 larvae by taking advantage of the ced-1(e1735) apoptotic corpse engulfment mutant, which allows for visualization of persisting corpses generated during embryonic development in L1 larvae. We found no difference in the number of apoptotic corpses between ced-1(e1735) and ced-1(e1735); sir-2.1(ok434) animals, indicating that sir-2.1 does not affect general apoptosis (Fig. 3C). Consistent with this interpretation, we found no extra (undead) cells in the anterior part of the pharynx in late L3 early L4 sir-2.1(ok434) animals as would be expected if apoptosis had failed (Fig. 3D).
Figure 3.
sir-2.1 specifically affects only DNA damage-induced germ cell apoptosis. (A) Germ cell apoptosis induced by the ced-9(n1653) temperature-sensitive loss of function mutation is largely unaffected by loss of sir-2.1. Worms were grown at 15°C until the late L4 stage and then shifted to 20°C. Apoptosis was scored 24 h later. The slight reduction in sir-2.1(ok434); ced-9(n1653) worms is likely due to slightly reduced germ cell proliferation of sir-2.1(ok434) worms. (B) sir-2.1(ok434) has no effect on excessive physiological germ cell apoptosis conferred by the gla-3(op216) mutation. Corpses were scored at 20°C 24 after the L4 stage. (C,D) Developmental apoptosis is not affected by sir-2.1. (C) Apoptotic corpses that persist until the early L1 stage in ced-1(e1735) animals were scored as described (Ellis et al. 1991). (D) Defects in developmental apoptosis were assayed by counting extra cells in the pharynx of old L3/young L4 larvae as described (Ledwich et al. 2000).
To exclude the possibility of the core apoptotic pathway being differentially regulated in the germline, or being differentially affected by sir-2.1 in the germline, we constructed a double mutant with ced-9(n1653ts) (Hengartner et al. 1992; Hengartner and Horvitz 1994). This allele of ced-9 encodes a protein that is unable to repress apoptosis at temperatures above 15°C. The increase in apoptosis observed in ced-9(n1653ts) worms was not suppressed by sir-2.1(ok434) in comparison with apoptosis suppression upon IR (Fig. 3A). These results indicate that sir-2.1 does not largely affect the core apoptotic pathway in the germline. The slight apoptosis reduction in ced-9(n1653ts); sir-2.1(ok434) worms is likely due to the weak reduction in germ cell proliferation of sir-2.1(ok434) worms (Fig. 2E). We next considered a potential role for sir-2.1 in DNA damage-independent germ cell apoptosis. First, we noted that the basal, radiation, and cep-1-independent level of germ cell apoptosis referred to as physiological germ cell apoptosis was not obviously diminished in sir-2.1 mutants (Fig. 1A). We next asked whether sir-2.1 affects apoptosis in the gla-3(op216) mutant background, which shows increased physiological germ cell apoptosis (Kritikou et al. 2006). Figure 3D indicates that there is no overall reduction of apoptosis in a gla-3(op216); sir-2.1(ok434) background, indicating that physiological, egl-1-independent apoptosis is unlikely to be affected by sir-2.1 (Fig. 3B). Taken together, our results suggest that the proapoptotic activity of sir-2.1 is confined to apoptosis occurring in response to DNA damage.
SIR-2.1 is expressed in germline nuclei and is lost in dying germ cells
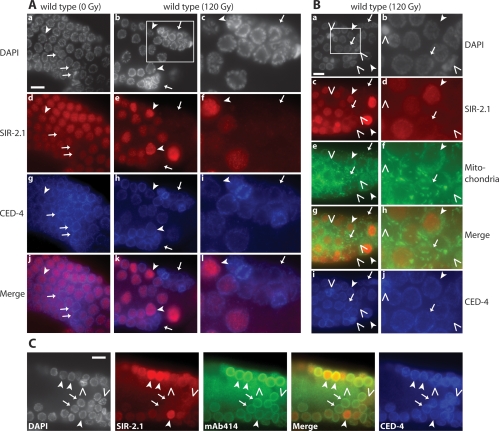
To investigate the localization of SIR-2.1 we expressed recombinant full-length SIR-2.1 protein and generated rabbit and goat anti-SIR-2.1 antibodies (Supplemental Fig. 5). Using these antibodies, we found that SIR-2.1 was present in the nuclei of almost all germ cells (Fig. 4; data not shown). This is consistent with previous reports suggesting that SIR-2.1 is nuclear in nongerm cells (Berdichevsky et al. 2006; Wang and Tissenbaum 2006). However, upon irradiation, SIR-2.1 disappeared from the nuclei of many, but not all late-stage pachytene germ cells, while in corresponding nonirradiated samples only very few cells lost SIR-2.1 nuclear staining (Fig. 4 A, arrows for exemplary cells; Supplemental Movie 1 for scanning through the depicted irradiated germline). The above staining, done with goat anti-SIR-2.1, was confirmed using the rabbit anti-SIR-2.1 antibody (Supplemental Fig. 6; Supplemental Movie 6).
Figure 4.
SIR-2.1 translocates from the nucleus to the cytoplasm of dying cells. Worms were irradiated at the late L4 larval stage and germlines were extracted and fixed 24 h after treatment. Arrows indicate nuclei of dying cells that have lost SIR-2.1 but where the nucleus is still intact. Arrowheads indicate surviving nuclei with strong SIR-2.1 staining. Empty arrowheads indicate cells in the late stages of apoptosis where the nucleus has disintegrated. (A) Worms were stained with anti-SIR-2.1 and anti-CED-4 antibodies and with DAPI. Pictures in the third column are an enlargement of the region indicated by the white rectangle in b. (B) Worms were treated as described in A and stained with anti-SIR-2.1, anti-CED-4, DAPI, and with a mix of five monoclonal antibodies recognizing different mitochondrial proteins. Pictures taken are a projection of a single cell layer. The right column is an enlargement of the region indicated by the white rectangle in panel a. (C) Worms were irradiated as in A and B and stained as indicated. MAb414 was used as a nuclear envelope marker (Lee et al. 2000). Bars, 10 μm. Germlines were stained with goat anti-SIR-2.1 (126.3) and rabbit anti-CED-4 (9103.1) antibodies.
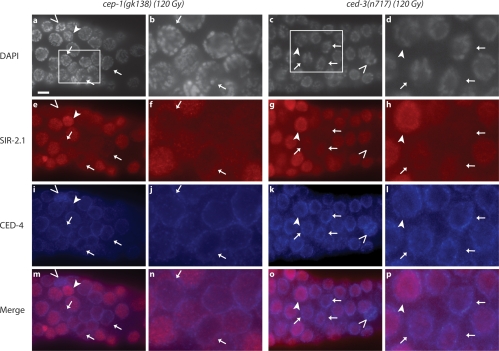
Given that only late-stage pachytene cells die by apoptosis, we next asked whether SIR-2.1 disappearance occurs in dying cells or surviving cells. We aimed at using CED-4 localization as a marker for apoptotic cells. Previous studies of developmental apoptosis have shown that during developmental apoptosis CED-4 accumulates at the nuclear periphery in dying somatic cells (Chen et al. 2000). We generated specific rabbit and goat anti-CED-4 antibodies (Supplemental Fig. 5) and using both antibodies (data not shown) found that CED-4 accumulates at the nuclear periphery of dying germ cells (identified by their intense DAPI staining of condensed chromatin) (Fig. 4A, arrows, panels b,e,h,k). Surprisingly, CED-4 also localized around the nuclei of apparently healthy cells in irradiated germlines or in untreated germlines albeit with lower intensity (Fig. 4A, arrowhead in panels a,b). In irradiated sir-2.1 and cep-1 mutants CED-4 did not accumulate around germ cell nuclei as strongly as in irradiated wild-type worms, consistent with the absence of DNA damage-induced apoptosis in these mutants (Supplemental Fig. 7). Thus, although CED-4 is presumably activated through cep-1-dependent egl-1 transcriptional induction in sir-2.1 mutants, DNA damage-induced perinuclear hyperaccumulation of CED-4 is disrupted, suggesting that execution of apoptosis depends on the SIR-2.1-dependent recruitment or retention of CED-4 near the nucleus.
Since we observed fewer nuclei showing CED-4 hyperaccumulation as compared with the number of corpses under DIC optics and given that SIR-2.1 was lost from many nuclei without CED-4 hyperaccumulation (Fig. 4B, panels c,i), we suspected that CED-4 perinuclear hyperaccumulation might be a late-stage germ cell apoptosis marker, consistent with the condensed chromatin of those cells (Fig. 4A, panels b,h). We therefore decided to employ a further apoptosis marker. In apoptotic cells mitochondria fragment, leading to the loss of the fine-meshed mitochondrial network and the accumulation of condensed morphologically distinct organelles (Jagasia et al. 2005). We confirmed these changes in mitochondrial morphology by staining irradiated germ cells with a cocktail of commercially available monoclonal antibodies against conserved mitochondrial proteins (Fig. 4B; Materials and Methods). The disappearance of the fine-meshed mitochondrial network and the concomitant appearance of condensed punctiform mitochondria correlated with the loss of nuclear SIR-2.1 (Fig. 4B, arrows; Supplemental Movies 2–5 for scanning through the depicted germline) while the fine-meshed mitochondrial network typical for healthy cells was present in cells with nuclear SIR-2.1 (Fig. 4B, arrowheads; Supplemental Fig. 8). Scanning through irradiated germlines indicates that a fine-meshed mitochondrial network persists in the nucleus free rachis located at the center of the germline, and that the absence of the fine-meshed mitochondrial network best correlates with SIR-2.1 nuclear loss (Supplemental Movies 2–5 for scanning through the depicted germline). In the outer rim of the germline, mitochondria appeared as condensed (and not fine-meshed) elongated structures in nonapoptotic cells (Supplemental Movies 2–5). We next assessed if SIR-2.1 nuclear disappearance is due to nuclear degradation. We scanned many apoptotic nuclei and found that some cells without nucleoplasmic SIR-2.1 contained SIR-2.1 halos around the nucleus, indicating that SIR-2.1 is likely to be translocated from the nuclei of dying cells rather than being degraded within the nucleus (Figs. 4B [empty arrowheads, panels c,d], 5 [panels e,f]; Supplemental Fig. 6; Supplemental Movies 4–6). These halos also indicated that the nuclear envelope was still intact in those cells, as SIR-2.1 was excluded from the nucleoplasm.
We next wished to address whether SIR-2.1 nuclear loss is an early or late apoptotic event by comparing SIR-2.1 loss with the loss of nuclear envelope integrity that occurs at a late stage of apoptosis, by using the MAb414 antibody recognizing the nuclear pore complex (Lee et al. 2000). We found that the majority of SIR-2.1-negative nuclei (Fig. 4C, arrows) had an intact nuclear membrane (Fig. 4C, middle panel). Nuclei that lacked both SIR-2.1 and MAb414 staining tended to occur more proximal in the germline, indicating that these nuclei represented cells in the very late stages of apoptosis, a notion confirmed by the condensed appearance of chromatin and accumulation of CED-4. We thus conclude that SIR-2.1 nuclear disappearance is an early apoptotic event.
We next asked whether SIR-2.1 translocation might be an active regulatory event or a mere consequence of early apoptosis progression. If SIR-2.1 export is regulatory and sir-2.1 acts genetically in parallel to cep-1, SIR-2.1 export should also occur in cep-1 mutants. We therefore examined SIR-2.1 and CED-4 localization as well as chromatin morphology in late stage pachytene cells in a cep-1 mutant background where the basal level of germ cell apoptosis, termed physiological germ cell apoptosis, is not affected, whereas irradiation-induced apoptosis is almost completely blocked. Under these conditions many late pachytene cell nuclei had largely reduced levels of SIR-2.1 even in the absence of CED-4 perinuclear accumulation (Fig. 5, arrows) or changes in mitochondrial morphology (data not shown) while SIR-2.1 was lost from only very few nuclei of unirradiated germ cells (Supplemental Fig. 8). Similar to wild-type dying cells SIR-2.1 accumulated in the cytoplasm around some nuclei that had lost SIR-2.1 staining (Fig. 5, arrows, panels e,f). Interestingly, we occasionally also found apoptotic cells with strong CED-4 staining and condensed chromatin, which we consider to be apoptotic (Fig. 5, empty arrowhead, left panel). These cells may be dying by the cep-1-independent physiological germ cell apoptosis pathway or due to damage-induced apoptosis triggered by spontaneous recombination failure or residual cep-1 activity or endogenous DNA damage (Gumienny et al. 1999; Gartner et al. 2000). Consistent with the idea that SIR-2.1 translocation might be linked to an early apoptotic event independent of apoptosis execution, we also observed that SIR-2.1 exits from the nucleus of germlines of irradiated ced-3(n717) caspase defective worms that rarely show a single apoptotic corpse (Fig. 5). In contrast, SIR-2.1 remains nuclear in unirradiated germlines (Supplemental Fig. 8).
Figure 5.
SIR-2.1 translocation is independent of cep-1 and ced-3. Worms were treated and germlines were stained as described in Figure 4. Arrows indicate nuclei that have lost SIR-2.1 but where the nucleus is still intact. Arrowheads indicate nuclei with strong SIR-2.1 staining. Empty arrowheads indicate cells in the late stages of apoptosis. Germlines were stained with goat anti-SIR-2.1 (126.3) and rabbit anti-CED-4 (9103.1) antibodies.
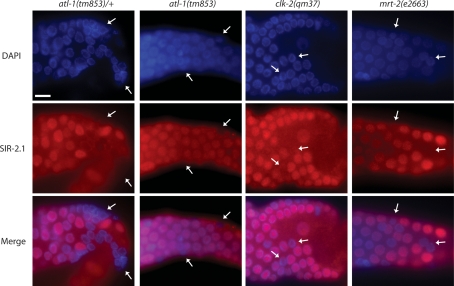
We next tested if the upstream DNA damage checkpoint mutants atl-1, clk-2, and mrt-2 affect SIR-2.1 translocation. We therefore treated atl-1(tm853), clk-2(qm37), and mrt-2(e2663) worms with 120 Gy of ionizing irradiation. While we see apoptosis induction and SIR-2.1 nuclear loss in the majority of late pachytene cells in wild-type (Fig. 4; Supplemental Fig. 6; Supplemental Movies 1,2,4,5,6) and in atl-1/+ heterozygotes (Fig. 6), SIR-2.1 is only lost from a small minority of late pachytene nuclei in clk-2 and atl-1 checkpoint mutants, while the reduction in mrt-2 appears as less dramatic (Fig. 6). We think that there are several reasons why SIR-2.1 translocation is not completely blocked in these strains. We recently showed that DNA damage checkpoint mutants do not fully block egl-1 induction (Greiss et al. 2008), indicating that a residual level of checkpoint signaling still occurs in those mutants. Furthermore, as checkpoint responses are defective in mrt-2, clk-2, and atl-1 germlines (Gartner et al. 2000; Ahmed et al. 2001; Garcia-Muse and Boulton 2005), there are more late pachytene cells in these germlines as compared with wild type due to the lack of apoptosis and cell cycle arrest. In summary, these data suggest that SIR-2.1 translocation largely depends on the DNA damage checkpoint pathway.
Figure 6.
SIR-2.1 translocation depends on the DNA damage checkpoint pathway. Worms were irradiated as described in Figure 4, and germlines stained with anti-SIR-2.1 antibodies and DAPI. Arrows indicate nuclei that have lost SIR-2.1. Bar, 10 μm.
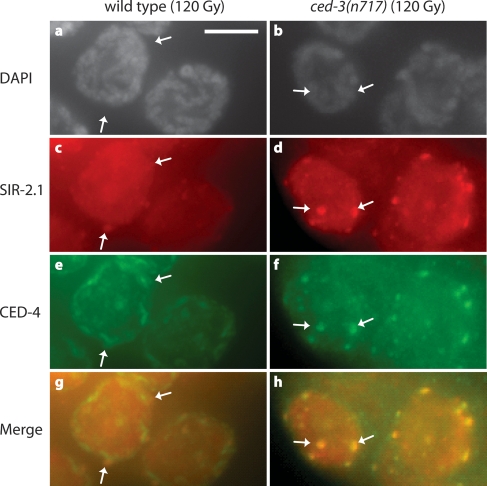
The loss of SIR-2.1 from dying cells raises the possibility that there might be a direct link between SIR-2.1 and apoptosis proteins. However, we could not observe a direct interaction between SIR-2.1 and the C. elegans cell death proteins CED-9, CED-4, or EGL-1 by coimmunoprecipitation experiments (data not shown). This may be a consequence of only a small minority of worm cells being in the process of dying at any given time. However, close examination of wild-type irradiated early pachytene nuclei revealed perinuclear dots of accumulated SIR-2.1 that often also showed increased CED-4 staining, most apparent when using goat anti-CED-4 and rabbit anti-SIR-2.1 antibodies (Fig. 7, left panel). We next examined if these structures, which may reflect a very transient colocalization between SIR-2-1 and CED-4 before cells apoptose, might accumulate in apoptosis defective ced-3(n717) germ cells. SIR-2.1 and CED-4 were indeed more extensively colocalized in ced-3(n717) mutants (Fig. 7, right panel). In summary, our cytological data are consistent with a model that SIR-2.1 translocation and a possible functional interaction with CED-4 might be an integral part of DNA damage-induced apoptosis.
Figure 7.
SIR-2.1 colocalizes with CED-4 in germ cells after irradiation. The apparent weak intranuclear CED-4 staining is nonspecific. Bar, 5 μm. Germlines were stained with rabbit anti-SIR-2.1 (1434.3) and goat anti-CED-4 (10147.1).
Discussion
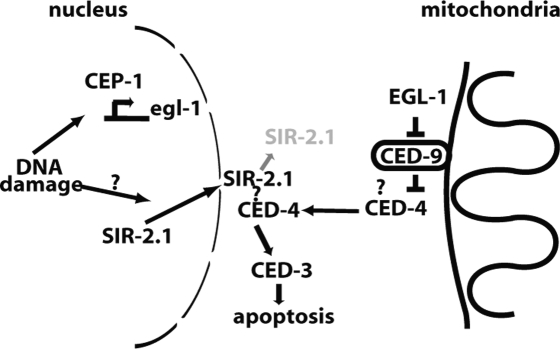
Our studies identified a novel function of sir-2.1 in promoting DNA damage-induced germ cell apoptosis. This proapoptotic function of sir-2.1 is restricted to DNA damage-induced apoptosis, as neither developmental apoptosis nor physiological germ apoptosis nor germ cell apoptosis in gla-3 and ced-9 loss-of-function mutants is overtly affected by sir-2.1. sir-2.1 seems to be specifically needed for DNA damage-induced apoptosis, as other DNA damage responses such as transient germ cell cycle arrest and DNA repair are not affected in the mutant. DNA damage signaling, which induces cep-1-dependent egl-1 transcription (Schumacher et al. 2005a, b), is not compromised in sir-2.1 mutants, suggesting that sir-2.1 genetically acts in parallel to, or downstream from, cep-1-dependent transcription to affect DNA damage-induced apoptosis (Fig. 8).
Figure 8.
Model. SIR-2.1 is exported from germ cell nuclei upon DNA damage and impinges on the apoptotic pathway at the genetic level of EGL-1/CED-9/CED-4.
The finding that sir-2.1 is required for DNA damage-induced apoptosis is surprising in light of in vitro and tissue culture-based studies that indicate that mammalian SIRT1 suppresses stress-induced apoptosis by deacetylating p53 (Luo et al. 2001; Vaziri et al. 2001). The hypothesis that SIRT1 is a negative regulator of p53 was supported by observation of increased apoptosis in thymocytes from irradiated SIRT1 knockout mice (Cheng et al. 2003). However, recent studies indicate that thymocyte apoptosis is not modulated by SIRT1 and that SIRT1 does not affect transcription of p53 targets even though SIRT1 and p53 can physically interact (Kamel et al. 2006). Our results in C. elegans may suggest proapoptotic in vivo function(s) in response to DNA damage for mammalian sirtuins.
Our observations that SIR-2.1 functions independently of cep-1-induced transcription of the egl-1 BH3-only gene, and that DNA damage-induced translocation of SIR-2.1 occurs independently of the core apoptotic machinery are supported by previous studies that apoptosis induction can be affected downstream from or in parallel to egl-1 and or ced-9. For example, pal-1-dependent transcriptional induction of ced-3 occurs during the death of the worm tail spike cell, where egl-1 and ced-9 only play minor roles (Maurer et al. 2007). Similarly, the ceh-30 transcription factor, which regulates the sex-specific death of the CEM neurons, acts downstream from or in parallel to egl-1 and ced-9 (Peden et al. 2007; Schwartz and Horvitz 2007). In germline apoptosis it has been reported that the C. elegans retinoblastoma gene homolog lin-35 and the E2F like transcription factor components efl-2 and dpl-1 are required for IR-induced germ cell apoptosis independent of egl-1 regulation, likely through transcriptional regulation downstream from and/or in parallel to cep-1, or through transcriptional regulation of ced-9, ced-4, and ced-3 (Schertel and Conradt 2007). In contrast to lin-35 or efl-2 and dpl-1 mutations, we did not find any effect on ced-9, ced-4, and ced-3 transcription in sir-2.1 mutants (data not shown).
How could SIR-2.1 impinge on germ cell apoptosis independent of transcriptional regulation? Our data indicate that it is unlikely that sir-2.1 affects apoptosis through transcriptional regulation. If the loss of SIR-2.1 from the nucleus was needed for the activation of genes promoting germ cell apoptosis, deletion of sir-2.1 should phenocopy the effect of SIR-2.1 translocation and lead to excessive rather than blocked apoptosis. Our results indicate that at least in germ cells, apoptosis execution might be more complex than previously thought based on studies on developmental apoptosis. In this system, mitochondrial-bound CED-9 is thought to be complexed to CED-4 to keep CED-4 at bay in order to prevent CED-3 caspase activation. Once egl-1 is transcriptionally induced EGL-1 releases CED-4, which then translocates to a perinuclear location and triggers CED-3 activation, likely through oligomerization (Yang et al. 1998; Horvitz 1999; Chen et al. 2000). In contrast, we found that CED-4 constitutively localizes around the nuclear membrane in late pachytene stage cells. CED-4 perinuclear hyperaccumulation, which in developmental apoptosis is considered an early event of apoptosis activation (Chen et al. 2000), only occurs in few rather late-stage corpses after SIR-2.1 nuclear exit. Thus, it is unlikely that the direct displacement of CED-4 from the mitochondrial-bound CED-9/CED-4 complex occurs solely by direct EGL-1 binding. The worm germline system might thus be more analogous to mammalian Apaf-1 regulation than previously thought, and may lack a direct physical link between mitochondrially located Bcl-2-like CED-9 and the Apaf-1-like CED-4 to regulate apoptosis (Danial and Korsmeyer 2004). We speculate that one of the factors regulating CED-4 during DNA damage-induced germ cell apoptosis might be SIR-2.1, which appears to transiently colocalize with CED-4, once its translocation from the nucleus to the cytoplasm commences. This regulation is likely not a direct consequence of CED-4 deacetylation, as we could not detect a direct interaction between recombinant CED-4 and SIR-2.1, and coimmunoprecipitation from irradiated whole worm extracts failed. Nevertheless, aside from our genetic data, the weak perinuclear colocalization of CED-4 and SIR-2.1, observed in wild-type germlines, which is enhanced in apoptosis execution-defective backgrounds, suggests a functional relationship between these proteins. Future studies will address exactly how DNA damage-induced germ cell apoptosis is triggered and if these mechanisms are conserved in mammals.
Materials and methods
C. elegans strains and maintenance
Worms were maintained at 20°C on NGM agar plates according to standard protocols, unless otherwise indicated. Alleles used were LG I: cep-1(lg12501), cep-1(gk138) gla-3(op216), ced-1(e1935), daf-16(mu26), daf-16(mu86), daf-16(mgDf50); LG III: mrt-2(e2663), clk-2(qm37), ced-9(n1653), ced-4(n1162), lig-4(ok716), cku-80(ok861), unc-119(ed3); LG IV: sir-2.1(ok434), sir-2.1(pk1640∷Tc1), ced-3(n717), ced-3(n2438); LG V: atl-1(tm853), egl-1(n1084n3082). sir-2.1(pk1640∷Tc1) carries a Tc1 transposon insertion between the nucleotides corresponding to nucleotides 17,144 and 17,145 of cosmid R11A8.
C. elegans apoptosis and DNA damage response assays
DNA damage-induced apoptosis and radiation sensitivity (rad) assays were performed as described (Gartner et al. 2000). For γ-irradiation a Cs137 source, 2.9 Gy/min (IBL 437C, CIS Bio International) and for X-ray treatment a Stabilipan (Siemens) source was used (11.25 Gy/min). For rad assays, late L4 stage worms were γ-irradiated, transferred to fresh plates 24 h later, and removed after 12 h. Defects in NHEJ repair were scored as described (Clejan et al. 2006). Developmental apoptosis was scored as described previously (Ellis et al. 1991). egl-1 and ced-13 quantitative RT–PCR (qRT–PCR) was performed using ∼1000 age-synchronized worms using tbg-1 as an internal normalization control as described previously (Schumacher et al. 2005b). The following primers were used: tbg-1: GA1760 (5′-AAGATCTATTGTTCTACCAGGC-3′) and GA1761 (5′-CTT GAACTTCTTGTCCTTGAC-3′); egl-1: GA1762 (5′-CCTCA ACCTCTTCGGATCTT-3′) and GA1763 (5′-TGCTGATCT CAGAGTCATCAA-3′); ced-13: GA1764 (5′-GCTCCCTGTT TATCACTTCTC-3′) and GA1765 (5′-CTGGCATACGTCTT GAATCC-3′).
The SIR-2.1∷GFP fusion plasmid (pGA291) was constructed by cloning the C. elegans operon CEOP4372 including the endogenous promoter and 3′UTR into a vector containing the C. elegans unc-119 gene as a transformation marker (Praitis et al. 2001). The sequence encoding for GFP and containing artificial introns was amplified from Addgene plasmid 1587 and cloned 5′ of the sir-2.1 stop codon, resulting in a C-terminal GFP fusion. Primers used were GA1263 (5′-GCGTGGGCGCGCCATGC GAAAGTTAGACACTAGGCG-3′), GA1351 (5′-AGATGCG GCCGCAGATACGCATTTCTTCACACAAA-3′), GA1350 (5′-TCAGTGCGGCCGCTGAATCTCATGTTAAAAAATTTCAA-3′), and GA1299 (5′-CGCAGGCCGGCCCTACCAGCCATGA TACTCTACGC-3′). Transgenic lines were created by Biolistic bombardment using a PDS-100/He Biolistic Particle Delivery System (Bio-Rad).
Protein expression and antibody production
6xHis-tagged full-length SIR-2.1 (pGA225) and CED-4 (pGA333), and MBP-tagged SIR-2.1(pGA226) and CED-4 (pGA334) were amplified from cDNA derived from adult worms using the same protocol as used for the qRT–PCR (Schumacher et al. 2005b) using primers GA1346 (5′-TTCAGGCCGGCCTGATACGCA TTTCTTCACACAAA-3′) and GA1348 (5′-AACGTGGCGC GCCATGTCACGTGATAGTGGCAAC-3′) for sir-2.1, and GA1911 (5′-TAACGGCGCGCCATGCTCTGCGAAATCGAA TGC-3′) and GA1912 (5′-ATCAGGCCGGCCCACAGCATG CAAAATTTTTGAGG-3′) for ced-4 to introduce AscI at the 5′ of the start codon and FseI at the 3′ of the stop codon and cloned into appropriately modified pQE-80L (6xHis) and pMAL-c2 (MBP). Protein expression was done in BL21(DE3) CodonPlus grown at 37°C to an OD600 = 0.6 before shifting to 20°C and adding IPTG (1 mM). Bacteria were harvested after incubation for 3 h (SIR-2.1) or overnight (CED-4). 6xHis-tagged SIR-2.1 was soluble and purification was carried out with Ni-NTA (Qiagen). 6xHis-tagged CED-4 was recovered from inclusion bodies using BugBuster (Novagen), solubilized with 10 mM Tris, 300 mM NaCl, 10 mM imidazole, 8 M urea, and purified with Ni-NTA (Qiagen). After elution in the presence of 8 M urea the protein was refolded by stepwise dialysis at 4°C in PBS + 4 M urea (overnight), PBS + 2 M urea (2 h), PBS + 1 M urea (2 h), PBS (2 h) ×2, PBS (overnight). Maltose-tagged proteins were purified according to standard protocols on amylose resin (New England Biolabs). For affinity purification, proteins were covalently linked to AffiGel 15 (Bio-Rad). 6xHis-tagged proteins were used to immunize rabbits and goats. Antibodies were then affinity-purified from the final bleeds using MBP-tagged protein.
Immunostaining of isolated C. elegans germlines
Worms were dissected on poly lysine coated slides in egg buffer (Edgar 1995) supplemented with 0.1% Tween 20 and 0.2 mM levamisol. Germlines were then fixed in 1.8% formaldehyde for 5 min at room temperature followed by freeze cracking by submersion in liquid nitrogen. Post-fixation was done in a 1:1 mixture of methanol:acetone at −20°C, followed by permeabilization with PBS + 1% Triton X-100 (three times for 10 min, room temperature). Blocking was performed by incubating the samples with Image-iT FX signal enhancer (Invitrogen) for 20 min, followed by 15 min of incubation in PBS + 0.1% Tween 20 + 1% BSA (PBSTB). Primary antibodies were diluted in PBSTB and allowed to bind at 4°C overnight in a humid chamber. Samples were washed three times for 10 min in PBS + 0.1% Tween 20 (PBST). Binding of secondary antibodies was performed for 2 h at room temperature with antibodies diluted in PBSTB supplemented with 1 μg/μL DAPI. After washing three times for 10 min in PBST, the samples were mounted in mounting medium (90% glycerol, 20 mM Tris at pH 8.0, 1 mg/mL p-phenylenediamine). Pictures were taken with a Leica LMF Spectris using SoftWorX software (Applied Precision).
The following primary antibodies were used: goat anti-SIR-2.1 (126.3), 100× dilution; goat anti-CED-4 (10147.1), 250× dilution; rabbit anti-SIR-2.1 (1434.3), 250× dilution; and rabbit anti-CED-4 (9103.1), 100× dilution.
Mitochondria were stained with a 250× dilution (final concentration 0.08 μg/mL for each antibody) of a mixture containing equal amounts of mouse monoclonal antibodies (MitoSciences): MS404 (anti-Complex IV subunit I), MS503 (anti-ATP synthase [Complex V] subunit β), MS507 (anti-ATP synthase [Complex V] subunit α), MSP07 (anti-PDH subunit E1 α), and MSA06 (anti-Cytochrome c). Mouse monoclonal antibody MAb414 (Covance Research Products) was used at a 250× dilution (final concentration 4 μg/mL).
The following secondary antibodies were used for detection: Cy3 labeled donkey anti-rabbit (Jackson Immunochemicals); 1000× dilution, 1.4 μg/mL; FITC labeled donkey anti-goat (Jackson Immunochemicals); 100× dilution, 15 μg/mL; FITC labeled donkey anti-mouse (Jackson Immunochemicals); 50× dilution, 28 μg/mL; Alexa 488-labeled donkey anti-goat (Molecular Probes); 200× dilution, 10 μg/mL; Alexa 647-labeled donkey anti-mouse (Molecular Probes); 200× dilution, 10 μg/mL; and Alexa 647 labeled donkey anti-goat (Molecular Probes); 200× dilution, 10 μg/mL.
Western blots
Worms were lysed in 4× LDS sample buffer (Invitrogen) by boiling for 15 min. After gel electrophoresis the protein was transferred to a nitrocellulose membrane, blocked with PBST supplemented with 5% milk powder, and probed with antibody 1434.3 (1000× dilution) overnight at 4°C. Horseradish peroxidase-conjugated goat anti-rabbit antibodies (Jackson Immunochemicals) were used as secondary antibodies.
Acknowledgments
We are grateful to Julian Blow, Arno Mueller, and Bjoern Schumacher for critical comments on the manuscript; Ehsan Pourkarimi, Caroline Fleischer, and Jean-Luc Tison for help with generating antibodies and immunostaining; and Ronald Plasterk and Marcel Tijsterman for sir-2.1(pk1640::Tc1). A.G. and S.G. were funded by AICR, CRUK (CDA award to A.G.), the DFG, and the Max Planck Society (Erich Nigg). S.A. and J.H. were supported by an Ellison Medical Foundation New Scholar in Aging Award
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.482608.
References
- Ahmed S., Alpi A., Hengartner M.O., Gartner A. C. elegans RAD-5/CLK-2 defines a new DNA damage checkpoint protein. Curr. Biol. 2001;11:1934–1944. doi: 10.1016/s0960-9822(01)00604-2. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A., Guarente L. A stress response pathway involving sirtuins, forkheads and 14–3–3 proteins. Cell Cycle. 2006;5:2588–2591. doi: 10.4161/cc.5.22.3513. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A., Viswanathan M., Horvitz H.R., Guarente L. C. elegans SIR-2.1 interacts with 14–3–3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Sherman J.M., Devine S.E., Cameron E.E., Pillus L., Boeke J.D. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes & Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Bryk M., Banerjee M., Murphy M., Knudsen K.E., Garfinkel D.J., Curcio M.J. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes & Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- Chen F., Hersh B.M., Conradt B., Zhou Z., Riemer D., Gruenbaum Y., Horvitz H.R. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science. 2000;287:1485–1489. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- Cheng H.L., Mostoslavsky R., Saito S., Manis J.P., Gu Y., Patel P., Bronson R., Appella E., Alt F.W., Chua K.F. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clejan I., Boerckel J., Ahmed S. Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics. 2006;173:1301–1317. doi: 10.1534/genetics.106.058628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen H.Y., Miller C., Bitterman K.J., Wall N.R., Hekking B., Kessler B., Howitz K.T., Gorospe M., de Cabo R., Sinclair D.A. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Conradt B., Horvitz H.R. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- Conradt B., Horvitz H.R. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell. 1999;98:317–327. doi: 10.1016/s0092-8674(00)81961-3. [DOI] [PubMed] [Google Scholar]
- Danial N.N., Korsmeyer S.J. Cell death: Critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Derry W.B., Putzke A.P., Rothman J.H.2001Caenorhabditis elegans p53: Role in apoptosis, meiosis, and stress resistance Science 294591 -595 [DOI] [PubMed] [Google Scholar]
- Derry W.B., Bierings R., van Iersel M., Satkunendran T., Reinke V., Rothman J.H. Regulation of developmental rate and germ cell proliferation in Caenorhabditis elegans by the p53 gene network. Cell Death Differ. 2007;14:662–670. doi: 10.1038/sj.cdd.4402075. [DOI] [PubMed] [Google Scholar]
- Edgar L.G. Blastomere culture and analysis. Methods Cell Biol. 1995;48:303–321. doi: 10.1016/s0091-679x(08)61393-x. [DOI] [PubMed] [Google Scholar]
- Ellis R.E., Jacobson D.M., Horvitz H.R. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics. 1991;129:79–94. doi: 10.1093/genetics/129.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairlie W.D., Perugini M.A., Kvansakul M., Chen L., Huang D.C., Colman P.M. CED-4 forms a 2:2 heterotetrameric complex with CED-9 until specifically displaced by EGL-1 or CED-13. Cell Death Differ. 2006;13:426–434. doi: 10.1038/sj.cdd.4401762. [DOI] [PubMed] [Google Scholar]
- Garcia-Muse T., Boulton S.J. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J. 2005;24:4345–4355. doi: 10.1038/sj.emboj.7600896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A., Milstein S., Ahmed S., Hodgkin J., Hengartner M.O. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- Gottschling D.E., Aparicio O.M., Billington B.L., Zakian V.A. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Greiss S., Schumacher B., Grandien K., Rothblatt J., Gartner A. Transcriptional profiling in C. elegans suggests DNA damage dependent apoptosis as an ancient function of the p53 family. BMC Genomics. 2008;9:334. doi: 10.1186/1471-2164-9-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny T.L., Lambie E., Hartwieg E., Horvitz H.R., Hengartner M.O. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- Hegde V., Klein H. Requirement for the SRS2 DNA helicase gene in non-homologous end joining in yeast. Nucleic Acids Res. 2000;28:2779–2783. doi: 10.1093/nar/28.14.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner M.O., Horvitz H.R. C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. Cell. 1994;76:665–676. doi: 10.1016/0092-8674(94)90506-1. [DOI] [PubMed] [Google Scholar]
- Hengartner M.O., Ellis R.E., Horvitz H.R. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature. 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- Hofmann E.R., Milstein S., Boulton S.J., Ye M., Hofmann J.J., Stergiou L., Gartner A., Vidal M., Hengartner M.O.2002Caenorhabditis elegansHUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis Curr. Biol. 121908 -1918 [DOI] [PubMed] [Google Scholar]
- Horvitz H.R. Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res. 1999;59:1701s–1706s. [PubMed] [Google Scholar]
- Jagasia R., Grote P., Westermann B., Conradt B. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- Jedrusik M.A., Schulze E. Telomeric position effect variegation in Saccharomyces cerevisiae by Caenorhabditis elegans linker histones suggests a mechanistic connection between germ line and telomeric silencing. Mol. Cell. Biol. 2003;23:3681–3691. doi: 10.1128/MCB.23.10.3681-3691.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik M.A., Schulze E. Linker histone HIS-24 (H1.1) cytoplasmic retention promotes germ line development and influences histone H3 methylation in Caenorhabditis elegans. Mol. Cell. Biol. 2007;27:2229–2239. doi: 10.1128/MCB.01713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel C., Abrol M., Jardine K., He X., McBurney M.W. SirT1 fails to affect p53-mediated biological functions. Aging Cell. 2006;5:81–88. doi: 10.1111/j.1474-9726.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- Kritikou E.A., Milstein S., Vidalain P.O., Lettre G., Bogan E., Doukoumetzidis K., Gray P., Chappell T.G., Vidal M., Hengartner M.O. C. elegans GLA-3 is a novel component of the MAP kinase MPK-1 signaling pathway required for germ cell survival. Genes & Dev. 2006;20:2279–2292. doi: 10.1101/gad.384506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley E., Pearson M., Faretta M., Bauer U.M., Frye R.A., Minucci S., Pelicci P.G., Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledwich D., Wu Y.C., Driscoll M., Xue D. Analysis of programmed cell death in the nematode Caenorhabditis elegans. Methods Enzymol. 2000;322:76–88. doi: 10.1016/s0076-6879(00)22009-0. [DOI] [PubMed] [Google Scholar]
- Lee K.K., Gruenbaum Y., Spann P., Liu J., Wilson K.L. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol. Biol. Cell. 2000;11:3089–3099. doi: 10.1091/mbc.11.9.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Nikolaev A.Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W.2001Negative control of p53 by Sir2α promotes cell survival under stress Cell 107137 -148 [DOI] [PubMed] [Google Scholar]
- Martin S.G., Laroche T., Suka N., Grunstein M., Gasser S.M. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- Maurer C.W., Chiorazzi M., Shaham S. Timing of the onset of a developmental cell death is controlled by transcriptional induction of the C. elegans ced-3 caspase-encoding gene. Development. 2007;134:1357–1368. doi: 10.1242/dev.02818. [DOI] [PubMed] [Google Scholar]
- Meier P., Vousden K.H. Lucifer’s labyrinth—Ten years of path finding in cell death. Mol. Cell. 2007;28:746–754. doi: 10.1016/j.molcel.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Peden E., Kimberly E., Gengyo-Ando K., Mitani S., Xue D. Control of sex-specific apoptosis in C. elegans by the BarH homeodomain protein CEH-30 and the transcriptional repressor UNC-37/Groucho. Genes & Dev. 2007;21:3195–3207. doi: 10.1101/gad.1607807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkston J.M., Garigan D., Hansen M., Kenyon C. Mutations that increase the life span of C. elegans inhibit tumor growth. Science. 2006;313:971–975. doi: 10.1126/science.1121908. [DOI] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevedo C., Kaplan D.R., Derry W.B. AKT-1 regulates DNA-damage-induced germline apoptosis in C. elegans. Curr. Biol. 2007;17:286–292. doi: 10.1016/j.cub.2006.12.038. [DOI] [PubMed] [Google Scholar]
- Rine J., Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertel C., Conradt B. C. elegans orthologs of components of the RB tumor suppressor complex have distinct pro-apoptotic functions. Development. 2007;134:3691–3701. doi: 10.1242/dev.004606. [DOI] [PubMed] [Google Scholar]
- Schumacher B., Hofmann K., Boulton S., Gartner A. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 2001;11:1722–1727. doi: 10.1016/s0960-9822(01)00534-6. [DOI] [PubMed] [Google Scholar]
- Schumacher B., Hanazawa M., Lee M.H., Nayak S., Volkmann K., Hofmann E.R., Hengartner M., Schedl T., Gartner A. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005a;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Schumacher B., Schertel C., Wittenburg N., Tuck S., Mitani S., Gartner A., Conradt B., Shaham S. C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage. Cell Death Differ. 2005b;12:153–161. doi: 10.1038/sj.cdd.4401539. [DOI] [PubMed] [Google Scholar]
- Schwartz H.T., Horvitz H.R. The C. elegans protein CEH-30 protects male-specific neurons from apoptosis independently of the Bcl-2 homolog CED-9. Genes & Dev. 2007;21:3181–3194. doi: 10.1101/gad.1607007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.S., Boeke J.D. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes & Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Stergiou L., Doukoumetzidis K., Sendoel A., Hengartner M.O. The nucleotide excision repair pathway is required for UV-C-induced apoptosis in Caenorhabditis elegans. Cell Death Differ. 2007;14:1129–1138. doi: 10.1038/sj.cdd.4402115. [DOI] [PubMed] [Google Scholar]
- Tamburini B.A., Tyler J.K. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell. Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum H.A., Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tzur Y.B., Margalit A., Melamed-Book N., Gruenbaum Y. Matefin/SUN-1 is a nuclear envelope receptor for CED-4 during Caenorhabditis elegans apoptosis. Proc. Natl. Acad. Sci. 2006;103:13397–13402. doi: 10.1073/pnas.0604224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H., Dessain S.K., Ng E.E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A.2001hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase Cell 107149 -159 [DOI] [PubMed] [Google Scholar]
- Wang Y., Tissenbaum H.A. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech. Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Wang Y., Oh S.W., Deplancke B., Luo J., Walhout A.J., Tissenbaum H.A. C. elegans 14–3–3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech. Ageing Dev. 2006;127:741–747. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Yan N., Gu L., Kokel D., Chai J., Li W., Han A., Chen L., Xue D., Shi Y. Structural, biochemical, and functional analyses of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4. Mol. Cell. 2004;15:999–1006. doi: 10.1016/j.molcel.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Yan N., Chai J., Lee E.S., Gu L., Liu Q., He J., Wu J.W., Kokel D., Li H., Hao Q., et al. Structure of the CED-4–CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature. 2005;437:831–837. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- Yang X., Chang H.Y., Baltimore D. Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science. 1998;281:1355–1357. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- Yang Y., Hou H., Haller E.M., Nicosia S.V., Bai W. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J. 2005;24:1021–1032. doi: 10.1038/sj.emboj.7600570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zermati Y., Mouhamad S., Stergiou L., Besse B., Galluzzi L., Boehrer S., Pauleau A.L., Rosselli F., D'Amelio M., Amendola R., et al. Nonapoptotic role for Apaf-1 in the DNA damage checkpoint. Mol. Cell. 2007;28:624–637. doi: 10.1016/j.molcel.2007.09.030. [DOI] [PubMed] [Google Scholar]