Abstract
The Shugoshin/Aurora circuitry that controls the timely release of cohesins from sister chromatids in meiosis and mitosis is widely conserved among eukaryotes, although little is known about its function in organisms whose chromosomes lack a localized centromere. Here we show that Caenorhabditis elegans chromosomes rely on an alternative mechanism to protect meiotic cohesin that is shugoshin-independent and instead involves the activity of a new chromosome-associated protein named LAB-1 (Long Arm of the Bivalent). LAB-1 preserves meiotic sister chromatid cohesion by restricting the localization of the C. elegans Aurora B kinase, AIR-2, to the interface between homologs via the activity of the PP1/Glc7 phosphatase GSP-2. The localization of LAB-1 to chromosomes of dividing embryos and the suppression of mitotic-specific defects in air-2 mutant embryos with reduced LAB-1 activity support a global role of LAB-1 in antagonizing AIR-2 in both meiosis and mitosis. Although the localization of a GFP fusion and the analysis of mutants and RNAi-mediated knockdowns downplay a role for the C. elegans shugoshin protein in cohesin protection, shugoshin nevertheless helps to ensure the high fidelity of chromosome segregation at metaphase I. We propose that, in C. elegans, a LAB-1-mediated mechanism evolved to offset the challenges of providing protection against separase activity throughout a larger chromosome area.
Keywords: LAB-1, meiosis, Aurora B kinase, cohesin, Shugoshin, Caenorhabditis elegans
Faithful chromosome segregation is vital to the maintenance of genomic integrity. Failure to tightly regulate the sorting of sister chromatids in either mitotically or meiotically dividing cells results in aneuploidy with significant deleterious consequences such as tumorigenesis and congenital defects (Hassold and Hunt 2001; Kops et al. 2005). Central to accurate chromosome segregation during both mitosis and meiosis is the formation of sister chromatid cohesion during DNA replication, ensuring the stable association between newly replicated DNA strands (for review, see Cohen-Fix 2001). Sister chromatid cohesion is established by components of cohesin, a highly conserved protein complex constituted by the association of two SMC (structural maintenance of chromosomes) core proteins (Smc1 and Smc3) and at least two other non-SMC proteins (Scc1/Rad21 kleisin and the Scc3 accessory protein) (Michaelis et al. 1997). The antiparallel ends of the Smc1/Smc3 heterodimers are linked by the kleisin subunit into a stable ring-like structure thought to join both DNA strands (Nasmyth and Haering 2005). During meiosis in many organisms, Scc1/Rad21 is specifically replaced by its paralog, Rec8 (Watanabe and Nurse 1999). In preparation for chromosome segregation at metaphase, the stable association between sister chromatids is progressively lost by the active removal of cohesins (for review, see Cohen-Fix 2001). This is accomplished by controlling the structural integrity of the cohesin ring via selective targeting and degradation of the kleisin subunit from subsets of cohesin complexes at particular timepoints during cell division (Ciosk et al. 1998; Uhlmann et al. 1999; Buonomo et al. 2000).
In most eukaryotes, the stepwise loss of sister chromatid cohesion during mitosis is accomplished, in part, by distinct phosphorylation events. Specifically, chromosome arm cohesin is released by the activity of a POLO-like kinase (Plk1/Cdc5) prior to entry into metaphase (Sumara et al. 2002; Hauf et al. 2005). As kinetochores are captured by the spindle, sister chromatid association relies entirely on the cohesin left around the centromeres. At the metaphase to anaphase transition, phosphorylation of this residual Scc1 potentiates cohesin removal by the protease separase, ultimately triggering chromosome segregation (Alexandru et al. 2001; Hornig and Uhlmann 2004). An extra layer of complexity is added to this process during meiosis, when a single round of DNA replication is followed by two consecutive rounds of chromosome segregation (meiosis I and II) to generate haploid gametes. While homologs dissociate at the end of meiosis I, sister chromatid association has to be maintained until segregation at meiosis II. To accomplish this, chromosomes undergo a series of unique steps during meiosis I. These include the formation of a proteinaceous structure (the synaptonemal complex or SC) that connects the axes of paired homologous chromosomes until late prophase, and the completion of crossover recombination between homologs. These crossover events, underpinned by flanking cohesion, afford physical connections (chiasmata) between homologs that persist after the disassembly of the SC and promote proper homolog alignment at the metaphase I plate where sister kinetochores co-orient to face the same spindle pole. Moreover, following the completion of homologous recombination, sister chromatids undergo a process referred to as chromosome remodeling, when amidst rapid DNA decondensation and recondensation, chromosomes are restructured around the crossover site (see below) and stripped from structural proteins that provided support for synapsis in early prophase I, while being imprinted with a new set of factors thought to mediate segregation competency (Chan et al. 2004; Nabeshima et al. 2005). Finally, at anaphase I, the subset of Rec8 mediating interhomolog association is selectively removed while Rec8 near the centromere is preserved to secure sister chromatid cohesion until anaphase II (Clyne et al. 2003; Lee and Amon 2003).
The selective removal of mitotic and meiotic cohesin in monocentric organisms, such as yeast, flies, and vertebrates, is mediated through the protective activity of the MEI-S332/Shugoshin family of proteins (Kerrebrock et al. 1995; Katis et al. 2004; Kitajima et al. 2004; Marston et al. 2004; Rabitsch et al. 2004; Hamant et al. 2005; Watanabe 2005). Research on both yeast and human shugoshin (Sgo) indicates that it specifically associates with the centromeric regions of metaphasic chromosomes and transiently prevents the degradation of cohesin until segregation at anaphase, in part by antagonizing the phosphorylation activity of mitotic and meiotic kinases via recruitment of the protein phosphatase 2A (PP2A) to the centromere (Kitajima et al. 2006; Riedel et al. 2006; Tang et al. 2006). This is critical given that centromeres represent the last point of contact between sister chromatids in early anaphase and act as the sites for kinetochore assembly and attachment of spindle microtubules. In contrast, far less is known about the mechanisms mediating protection during the removal of cohesins in organisms lacking a localized centromere such as Caenorhabditis elegans.
Mitotic chromosome segregation in C. elegans involves the assembly of a diffuse kinetochore and microtubule attachments along the whole length of chromosomes (Albertson and Thomson 1982). However, while a presumptive kinetochore surrounds each bivalent during meiosis, it is still unclear where microtubules attach on meiotic chromosomes (Howe et al. 2001; for review, see Dernburg 2001). Interestingly, during meiosis in C. elegans, autosome pairs undergo a single crossover preferentially within the terminal thirds of chromosomes (Albertson et al. 1997). Moreover, it has been suggested that the bivalents then remodel themselves around the off-center crossover (or crossover precursor), which would explain in part how the bivalents adopt a cross-shaped configuration composed of two perpendicular chromosomal axes (one long and one short) intersecting at the chiasma (Nabeshima et al. 2005). Upon congression, bivalents align such that the short arms occupy an equatorial position on the metaphase plate whereas the longer arms are positioned perpendicular to the spindle poles (Albertson et al. 1997; Maddox et al. 2004). The ends of the long arms in the aligned bivalents have, therefore, been suggested to lead the way poleward at anaphase I (Albertson and Thomson 1993). In preparation for homolog segregation, it has been proposed that REC-8 located along the short arms of the bivalent is phosphorylated by the C. elegans Aurora B kinase AIR-2, leading to its removal via separase activity in this region (Kaitna et al. 2002; Rogers et al. 2002). In contrast, REC-8 localized along the long arms is spared from AIR-2-mediated phosphorylation due to the activity of the C. elegans PP1 phosphatases GSP-1 (GLC-7α) and GSP-2 (GLC-7β) (Rogers et al. 2002). Thus, protection of cohesin during meiosis I in C. elegans is similar to monocentric species in that it relies on phosphatase activity to counterbalance the phosphorylation state of a subset of cohesins. Remarkably different from the monocentric paradigm, however, is the fact that in C. elegans the chromosomal domain requiring cohesin protection during meiosis I spans approximately two thirds of the entire bivalent.
Here we report that the protection of cohesin in C. elegans meiosis I is independent of SGO-1, the predicted C. elegans shugoshin homolog (Rabitsch et al. 2004), and instead relies on the activity of LAB-1 (Long Arms of the Bivalent protein) to counteract AIR-2-mediated phosphorylation on the long arms of the bivalent. LAB-1 localizes to meiotic chromosomes beginning in early prophase I. Upon initiation of chromosome remodeling in mid to late prophase, LAB-1 localization is progressively restricted to the long arms of the asymmetric bivalent where it remains until completion of homolog segregation at anaphase I. The kinetics of LAB-1 localization and its presence exclusively on the AIR-2-free domain of late prophase bivalents support a role for LAB-1 in the protection of sister chromatid cohesion. As in gsp-2/glc-7β mutants, lab-1 depletion results in the mislocalization of AIR-2 to the long arms of the bivalent and subsequently the premature loss of sister chromatid cohesion at metaphase I. Importantly, LAB-1-mediated antagonism of AIR-2 localization depends on the activity of GSP-2. In addition, LAB-1 is also present on mitotic chromosomes where it likely antagonizes AIR-2 activity as well. Finally, LAB-1 is conserved in at least three other Caernorhabditis species. Therefore, we propose a model in which LAB-1-mediated protection represents a mechanism evolved in the context of a chromosome architecture where protection of sister chromatid integrity from separase-mediated removal is required specifically at the long arms of meiosis I bivalents.
Results
Decrease in LAB-1 activity results in a weakening of sister chromatid association during prophase I
An RNAi-based screen performed as in Colaiacovo et al. (2002), to identify meiotic genes from among new candidates with germline-enriched expression in C. elegans (Reinke et al. 2004; M. Colaiacovo, unpubl.), led us to the identification of lab-1 (ORF C03D6.6) (Fig. 1A). Depletion of lab-1 by RNAi yields phenotypes suggestive of errors in meiotic chromosome segregation such as increased embryonic lethality (Emb = 57%; n = 428) and a high incidence of males (Him = 6%) among the surviving progeny (Fig. 1B). Cytological analysis of DAPI-stained germlines from lab-1(RNAi) worms revealed the presence of univalents in late diakinesis oocytes (up to 12 DAPI-stained bodies instead of the six bivalents observed in wild type), indicating a lack of chiasmata (Fig. 1C; Supplemental Fig. 1A,C; Dernburg et al. 1998). In addition, the presence of >12 DAPI-stained bodies in 1% (n = 109) of lab-1(RNAi) oocytes at diakinesis suggests a further defect in sister chromatid cohesion (Fig. 1C; Pasierbek et al. 2001). Studies that will be described in detail elsewhere indicate that these results likely reflect an AIR-2 and GSP-2-independent role for LAB-1 in early prophase I events. Specifically, fluorescence in situ hybridization (FISH) analysis and immunostaining of lab-1(RNAi) gonads revealed that LAB-1 is required for proper chromosome pairing, progression of meiotic double-strand break repair resulting in chiasmata, and maintenance of sister chromatid cohesion (data not shown). Accordingly, the chromosome association defect observed in the oocyte at the end of diakinesis (referred to hereafter as the −1 oocyte, based on its position in the gonad relative to the spermatheca; Fig. 2A) in lab-1 gonads is independent of separase activity, since univalents were still detected in worms with impaired separase activity (Supplemental Fig. 1B).
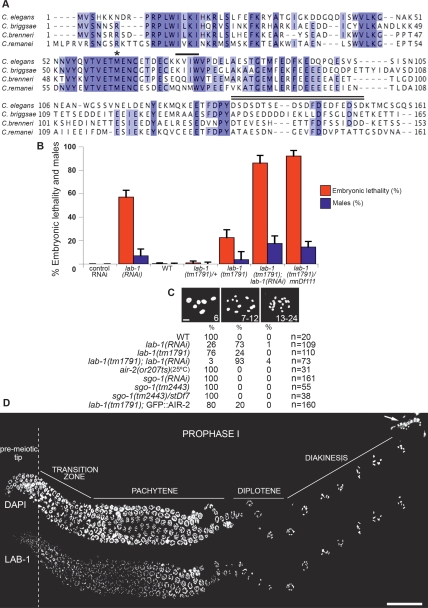
Figure 1.
LAB-1 protein conservation and phenotypes. (A) Sequence alignment between LAB-1 and its homologs in other Caenorhabditis species (C. elegans: C03D6.6; Caenorhabditis briggsae: CBP06688; Caenorhabditis remanei: CR03512). A cosmid (1003.1) carrying the putative Caenorhabditis brenneri (former PB2801) lab-1 homolog was identified by a BLAST database search of the C.elegans LAB-1 sequence against a C. brenneri cosmid library (Genome Sequencing Center, Washington University School of Medicine). Prediction of the coding sequence in the respective region in 1003.1 and conceptual translation were performed using FGENESH+ (http://www.softberry.com/berry.phtml). Residue conservation is shown in different shades of blue (light, −75% conserved; dark, −100% conserved). The location of the putative first methionine in lab-1(tm1791) is indicated by an asterisk. The [(R/K) (V/I) X (F/W)] core motif associated with binding and regulation of PP1/Glc7 proteins is shown below a solid line. The C terminus peptide region used for antibody production is marked by a double line. (B) Percentage of embryonic lethality and incidence of males observed among the progeny of worms of the indicated genotypes. Error bars represent standard deviation of the mean. (C) Number of DAPI-stained bodies present in −1 oocytes at diakinesis in the indicated genotypes. Bar, 2 μm. (D) Immunolocalization of LAB-1 in meiotic prophase I. No signal is detected on mature sperm (arrow in DAPI panel). Bar, 20 μm.
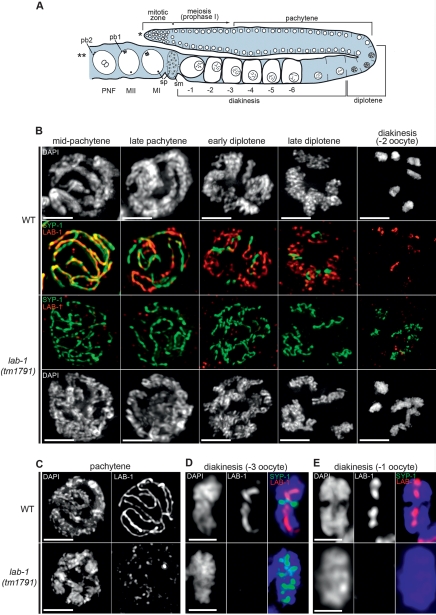
Figure 2.
Chromosome remodeling establishes a LAB-1-specific domain on bivalents. (A) Schematic representation of one arm of the C. elegans gonad. Germline nuclei undergoing mitotic proliferation in the premeiotic region (distal tip is marked by a single asterisk) enter meiosis as they move proximally toward the spermatheca (sm) and the uterus (double asterisk). A chromosome remodeling process initiated in late pachytene culminates in the formation of six discrete bivalents at diakinesis, where cellularization individualizes single oocytes. A sperm-associated signal then promotes the maturation of the oocyte closest to the spermatheca (−1), which in turn signals its own ovulation. Fertilization than ensues with sperm (sp) entry in the oocyte triggering completion of the meiotic program with subsequent segregation of homologs (anaphase I) and sister chromatids (anaphase II). The presence of two polar bodies (pb1 and pb2) at the surface of the fertilized oocyte marks the end of meiosis. Finally, the female and male pronuclei migrate toward each other and fuse to initiate embryo development. MI, metaphase I; MII, metaphase II; PNF, pronuclei fusion. (B) Immunolocalization of LAB-1 and SYP-1 in mid to late prophase I nuclei of wild-type and lab-1(tm1791) gonads. Bar, 2 μm. (C) Punctate and weak expression of LAB-1 in pachytene nuclei of lab-1(tm1791) mutants. Bar, 2 μm. (D) LAB-1 and SYP-1 immunolocalization on a single bivalent from a −3 oocyte at diakinesis in wild-type and lab-1(tm1791) gonads. Bar, 1 μm. (E) LAB-1 and SYP-1 immunolocalization on a single bivalent from a −1 oocyte at diakinesis in wild-type and lab-1(tm1791) gonads. Bar, 1 μm.
lab-1 encodes a 161-amino acid protein containing the highly degenerate [(R/K) (V/I) X (F/W)] motif, which has been associated with binding, localization, and function of PP1/Glc7 phosphatases (Fig. 1A; Egloff et al. 1997; Zhao and Lee 1997; Wakula et al. 2003). Position-specific iterated (PSI) BLAST searches revealed the presence of LAB-1 homologs in other Caenorhabditis species, but not among any other eukaryote (Fig. 1A).
A mutant carrying a 421-base-pair (bp) deletion (tm1791) encompassing most of the promoter, exon 1, and part of intron 1 of lab-1 was used to further characterize the meiotic role of this gene (Supplemental Fig. 2; see the Materials and Methods). lab-1(tm1791) worms are homozygous viable at 20°C and show only low Emb (22%; n = 2770) and Him (4%) phenotypes when compared with RNAi-depleted worms (Fig. 1B). Accordingly, only 24% of late diakinesis oocytes carry univalents in lab-1(tm1791) mutants (Fig. 1C; Supplemental Fig. 1C).
Three main observations suggest that tm1791 is a reduction-of-function (rf) allele of lab-1. First, the meiotic phenotypes of tm1791 worms are enhanced both by further depletion of lab-1 via RNAi and in trans-heterozygotes for a deficiency (mnDf111) encompassing the lab-1 locus (Fig. 1B,C). Second, a shorter transcript is detected, by RT–PCR in lab-1(tm1791) worms, with primers downstream from the deletion region (Supplemental Fig. 2A–C). An alternative in frame ATG is present in the coding sequence downstream from the tm1791 deletion (exon 3) and could potentially be used for translation initiation in these mutants (Fig. 1A). Furthermore, an expressed sequence tag lacking the predicted start codon in exon 1, but carrying the alternative ATG in exon 3, has been reported in wild-type worms (EC037036; WormBase release WS189), supporting a potential physiological role for this shorter transcript. Finally, although an antibody against LAB-1 (Fig. 1A) fails to detect a LAB-1 signal on bivalents at diakinesis in either lab-1(RNAi) or lab-1(tm1791) worms (Fig. 2B,D,E; Supplemental Fig. 1C), a weak signal is detectable on chromosomes in mid-pachytene nuclei of the latter (Fig. 2C), presumably representing the translation product of the short tm1791 transcript. We therefore concluded that lab-1(tm1791) worms display only a moderate reduction in LAB-1 activity by virtue of expressing a shorter C-terminal protein that can localize to early prophase I chromosomes and partially substitute for the wild-type protein function. Under these conditions, most lab-1(tm1791) oocytes present at diakinesis carry structurally intact bivalents. We took advantage of this scenario to investigate the role of LAB-1 both at late diakinesis and after fertilization, when oocytes complete meiosis and separase-mediated degradation of cohesin is critical to drive chromosome segregation.
LAB-1 localizes to chromosomes in prophase I and is necessary for the establishment of a molecular asymmetry in the bivalent
Immunolocalization of LAB-1 in wild-type gonads, using an antibody generated against its C-terminal region, revealed that it associates with chromosomes from entry into prophase I until the end of meiosis I (Figs. 1D, 2A,B,D,E, 4, below). During pachytene, the LAB-1 signal intensifies and is observed as continuous tracks at the interface between homologous chromosomes, in a similar localization to that detected at this stage for the SC component SYP-1 (Figs. 1D, 2B,C; MacQueen et al. 2002). Strikingly, during late pachytene, LAB-1 and SYP-1 progressively acquire a reciprocal localization. Specifically, the LAB-1 signal is observed on chromosome regions where SYP-1 is lost, and in a reciprocal fashion, SYP-1 remains on regions where the LAB-1 signal is no longer apparent (Fig. 2B). Upon chromosome resolution, SYP-1 localization is restricted to the short arms of the bivalents at diakinesis (Nabeshima et al. 2005), whereas LAB-1 is observed exclusively on the long arms (Fig. 2D). Moreover, while the SYP-1 signal is lost prior to the end of diakinesis, LAB-1 remains localized to chromosomes until homolog segregation at anaphase I (Figs. 2E, 4, below). Closer examination of the long arm domain revealed a distinct ring-like structure formed by LAB-1 and REC-8 that presumably delineates the interface between sister chromatids (Supplemental Fig. 3A,B). Furthermore, coimmunostaining of LAB-1 and SYP-1 revealed that SC disassembly in lab-1(tm1791) mutants, although completed by the end of diakinesis (−1 oocyte), is considerably delayed when compared with wild type (Fig. 2B,D,E). Thus, LAB-1 associates with the interchromatid domain harboring the cohesin complex on the bivalent and undergoes an unprecedented chromatin reorganization process in late prophase I nuclei that is essential for the correct kinetics of SC disassembly and the adoption of a long arm identity.
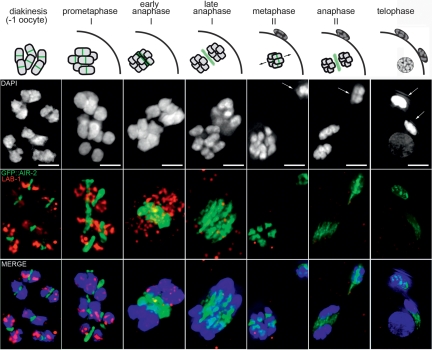
Figure 4.
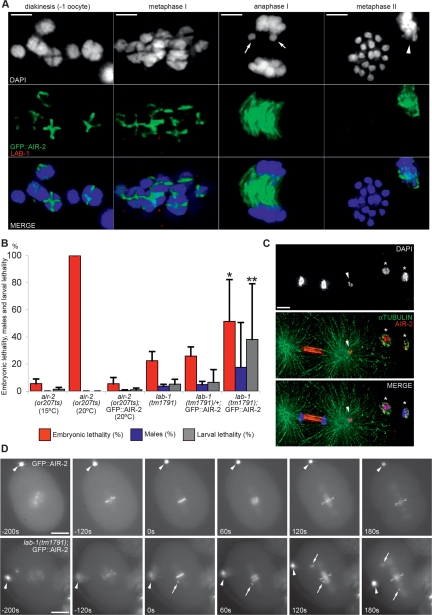
LAB-1 is present specifically in the interchromatid domain of the bivalent where cohesin protection is required during homolog segregation. LAB-1 and GFP∷AIR-2 localization during meiosis. The top row is a schematic representation of the localization of AIR-2 at different stages of meiosis (green). AIR-2 activity is necessary for the dissociation of subsets of cohesins in the bivalent (first meiotic division) and sister chromatids (second meiotic division and mitosis). As chromosomes separate in anaphase, AIR-2 shuttles to the spindle midzone where it participates in the completion of cytokinesis. During meiosis I, AIR-2-chromosomal localization is restricted to the short arms of the bivalents, in striking contrast to the localization of LAB-1. Bar, 2 μm. Arrows indicate polar bodies.
LAB-1 acts through GSP-2 to antagonize AIR-2 and prevent the untimely removal of REC-8 during meiosis I
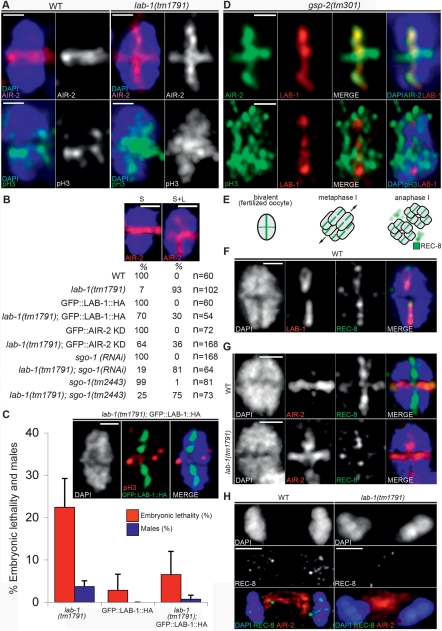
The restricted localization of AIR-2 to the short arms of the bivalents at late diakinesis preserves sister chromatid cohesion on the long arms (Fig. 3A; Kaitna et al. 2002; Rogers et al. 2002). Because the targeting of AIR-2 to the short arms relies on the previous establishment of distinct domains on the bivalent, we examined whether a decrease in LAB-1 function would disturb this distinct chromosomal localization of AIR-2. Indeed, in a lab-1(tm1791) background, 93% (n = 102) of the bivalents at late diakinesis showed AIR-2 mislocalized to both chromosome arms, whereas in wild type (n = 60), AIR-2 is only observed occupying the short arms of the bivalents at that stage (Fig. 3A,B). This phenotype is reproducible by RNAi experiments and consistent with the idea that partial depletion of LAB-1 activity in lab-1(tm1791) worms, while insufficient to disrupt homolog association in all oocytes, is enough to alter the molecular blueprint of most bivalents (Supplemental Fig. 1A,D). This is further supported by the observation that CSC-1, the C. elegans Borealin homolog and another member of the chromosomal passenger complex (CPC) required for targeting AIR-2 to the short arms (Romano et al. 2003), is also mislocalized to both arms of lab-1(tm1791) bivalents in oocytes at late diakinesis (Supplemental Fig. 4). Moreover, the abnormal localization of AIR-2 is biologically relevant since the phosphorylation of Histone H3, a canonical chromosomal substrate of AIR-2 (Hsu et al. 2000; Kaitna et al. 2002), is also widespread along both the short and long arms of lab-1(tm1791) bivalents at late diakinesis (Fig. 3A). The mislocalization of AIR-2 also correlates with a reduction of the REC-8 signal along the long arms of metaphase I bivalents, and with an absence of detectable REC-8 signal in 60% (n = 5/8) of anaphase I oocytes in lab-1(tm1791) gonads, in contrast to wild type where REC-8 is always present on both sets of segregating homologs (n = 0/6 oocytes without REC-8 signal) at anaphase I (Fig. 3E–H). Finally, a GFP∷LAB-1 transgene, whose localization reiterates the expression of the native LAB-1 protein (Supplemental Fig. 5), rescues the mislocalization defects of AIR-2 on late diakinesis bivalents and partially alleviates the embryonic lethality of lab-1(tm1791) (P < 0.0001 by the two-tailed Mann-Whitney test, 95% C.I.) (Fig. 3B,C). Taken together, these results suggest a global deregulation of AIR-2-mediated phosphorylation on lab-1(tm1791) bivalents and point to a role for LAB-1 in the proper localization of AIR-2 on bivalents at late diakinesis.
Figure 3.
LAB-1 localizes to the AIR-2-free domain of the bivalent in late prophase I. (A) AIR-2 and phospho Histone H3 (pH3) localization in wild-type and lab-1(tm1791) diakinesis bivalents (−1 oocytes). Bar, 0.5 μm. (B) AIR-2 localization on the short (S) or both short and long (S + L) arms of bivalents in −1 oocytes at diakinesis of different genotypes. Bar, 1 μm. (C) Expression of a GFP∷LAB-1∷HA transgene, detected with an anti-GFP antibody, partially rescues the lab-1(tm1791) phenotypes. Error bars represent standard deviation of the mean. Bar, 1 μm. (D) LAB-1 and AIR-2 (top) or LAB-1 and phospho Histone H3 (bottom) localization on bivalents in −1 oocytes at diakinesis in gsp-2(tm301) gonads. Bar, 0.5 μm. (E) Schematic representation of chromosome-associated REC-8 during meiosis I segregation. Upon fertilization, bivalents align at the metaphase plate and rotate to assume a position perpendicular to the cell cortex prior to homolog segregation. REC-8 in the region distal to the chiasma (short arms) progressively weakens until it is completely removed by separase in the metaphase to anaphase transition. The subset of REC-8 present on the long arms of the bivalent is spared from separase activity and sustains sister chromatid cohesion as chromatids enter meiosis II. (F) REC-8 and LAB-1 localization on a bivalent from a fertilized oocyte in wild type. Bar, 0.5 μm. (G) AIR-2 and REC-8 localization on metaphase I bivalents of wild-type and lab-1(tm1791) worms. Bar, 0.5 μm. (H) AIR-2 and REC-8 localization on anaphase I chromosomes of wild-type and lab-1(tm1791) worms. Bar, 2 μm.
AIR-2 phosphorylation is also antagonized by the phosphatase activity of the C. elegans GLC-7-like protein GSP-2 (Hsu et al. 2000; Kaitna et al. 2002). Similarly to what is observed in lab-1 mutants, AIR-2 is mislocalized to the long arms of the bivalents at late diakinesis in gsp-2-depleted mutants (Fig. 3D; Kaitna et al. 2002; Rogers et al. 2002). Considering the segregated domains occupied by AIR-2 and LAB-1 on bivalents at late diakinesis in wild type, we were surprised to find these proteins colocalizing on the long arms of the bivalents in this stage in the gsp-2 mutant (Fig. 3D). This observation strongly argues against a scenario where LAB-1 passively deters AIR-2 docking on the long arms, and instead suggests that LAB-1 antagonizes AIR-2 via GSP-2.
If the role of LAB-1 following chromosome remodeling and bivalent resolution is solely to mediate the protection of cohesin along the long arms, it should be absent from chromosomes during the second meiotic division when phosphorylation of the remaining REC-8 by AIR-2 is essential to drive sister chromatid dissociation. Indeed, LAB-1 loses chromosome association as homologs segregate and is not detected on chromosomes in meiosis II (Fig. 4). Surprisingly, a weak LAB-1 signal is observed again on chromosomes during prophase of the first embryonic division and is subsequently present on mitotic chromatin from late prophase to anaphase during early embryonic development (up to the ∼100-cell stage embryo). Reminiscent of the segregated localization observed on bivalents in late meiosis I, LAB-1 and AIR-2 are seen occupying distinct regions of the separating mitotic chromosome at the onset of anaphase (Supplemental Fig. 6).
Increase in AIR-2 activity mediates chromosome segregation defects in lab-1(tm1791) worms
Despite the mislocalization of AIR-2 on bivalents in −1 oocytes and the premature reduction of REC-8 signal detected on both arms of the bivalents in metaphase I in lab-1(tm1791), levels of embryonic lethality among lab-1(tm1791) progeny are relatively low when compared with the 98% reported Emb for gsp-2 mutants (Fig. 1B; Sassa et al. 2003). One possible explanation for this difference predicts that the kinase activity of the mislocalized AIR-2 in lab-1(tm1791) bivalents is either poor or insufficient to drive chromosome dissociation. To test whether hyperactivation of AIR-2 in a lab-1 mutant background could further compromise sister chromatid cohesion, we introduced exogenous AIR-2 activity into lab-1(tm1791) gonads via expression of a functional GFP∷AIR-2 transgene. GFP∷AIR-2 reproduces the complex localization pattern of the native AIR-2 protein (Fig. 4) and is able to rescue the embryonic lethality phenotype of or207, a mitosis-specific allele of air-2 (Fig. 5B; Severson et al. 2000; Kaitna et al. 2002). Moreover, we detected no overt meiotic or mitotic defects due to the expression of GFP∷AIR-2 per se, despite its kinase activity (Supplemental Fig. 7B; data not shown). As in wild-type gonads, six bivalents were observed aligned at the metaphase I plate in GFP∷AIR-2 cells. During homolog segregation, at anaphase I, two distinct masses of chromosomes (each consisting of six pairs of sister chromatids as attested by the presence of REC-8) (data not shown) were observed migrating toward opposite poles. Upon polar body extrusion, a single mass of chromosomes (consisting of six distinct pairs of sister chromatids) realigned at the metaphase II plate in preparation for meiosis II segregation. In contrast, a higher embryonic lethality is observed in the progeny of lab-1(tm1791); GFP∷AIR-2 worms compared with lab-1(tm1791) (P = 0.0008 by the two-tailed Mann-Whitney test, 95% C.I.) (Fig. 5B). This cannot be explained as an enhancement of the early role of LAB-1 in promoting cohesion, since the frequency of oocytes at diakinesis with >12 DAPI-stained bodies (representing premature sister chromatid dissociation) observed in lab-1(tm1791) worms was not altered by the introduction of the GFP∷AIR-2 transgene (Fig. 1C). However, premature sister chromatid separation at the metaphase to anaphase I transition increased even further, as seen by the presence of >12 DAPI-stained bodies in 33% (n = 4/12) of lab-1(tm1791); GFP∷AIR-2 oocytes at metaphase II when compared with 6% (n = 1/15) of lab-1(tm1791) oocytes at this stage (Fig. 5A).
Figure 5.
AIR-2 activity mediates some of lab-1(tm1791) meiotic defects. (A) LAB-1 and GFP∷AIR-2 localization during meiosis in lab-1(tm1791) mutants. Arrows indicate lagging chromosomes during anaphase I. Arrowhead indicates polar body I. Bar, 2 μm. (B) Embryonic lethality, larval lethality, and incidence of males among the progeny of hermaphrodites of the indicated genotypes. Error bars represent standard deviation of the mean. (C) Defect in chromosome segregation in lab-1(tm1791); GFP∷AIR-2 embryos. Anaphase in the one-cell embryo. Arrowhead indicates chromosome unattached to the spindle. Asterisks indicate both meiotic polar bodies. Bar, 5 μm. (D) Time-lapse analysis of the first mitotic division in GFP∷AIR-2 and lab-1(tm1791); GFP∷AIR-2 embryos (n = 10, respectively). Arrows show unattached chromosomes. Filled arrowheads indicate the polar body. Open arrowheads point to the spindle midzone where AIR-2 migrates during anaphase. Bar, 10 μm.
Surprisingly, the expression of GFP∷AIR-2 in lab-1(tm1791) embryos can result in missegregation during mitotic anaphase, which presumably contributes to both the increased embryonic and larval lethality observed (Fig. 5B,C). In these cases, segregation ensues before completion of chromosome congression as evidenced by chromosomes that fail to be properly captured by the spindle [observed in n = 2/10 of the lab-1(tm1791); GFP∷AIR-2 embryos compared with n = 0/10 in GFP∷AIR-2 embryos] (Fig. 5D).
Taken together, our data suggest that the chromosome association defects detected in lab-1 mutant gonads can be enhanced by increasing AIR-2 activity. Furthermore, LAB-1 may also play a role either in promoting proper kinetochore/microtubule attachments or in the activation of the spindle checkpoint during mitosis perhaps by regulating AIR-2 activity with respect to the kinetochore/microtubule interaction.
LAB-1 depletion can suppress defects associated with a kinase-deficient AIR-2 protein during embryogenesis
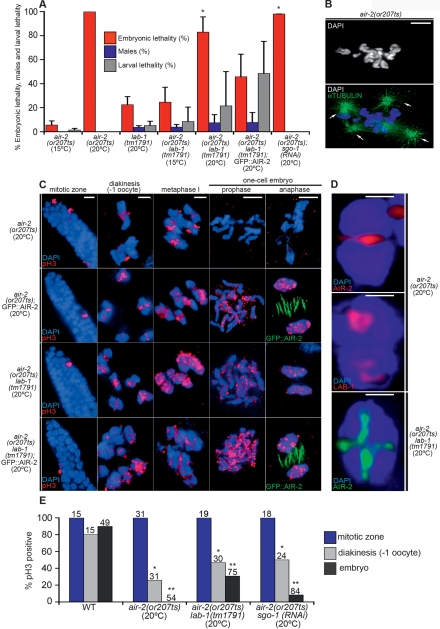
or207 is a mitosis-specific temperature-sensitive allele of air-2 carrying a mutation in the predicted kinase domain (Severson et al. 2000). Most air-2(or207) worms show no obvious meiotic or mitotic defects when grown at 15°C. At the restrictive temperature (20°C or 25°C), air-2(or207) mutants undergo normal meiosis and pronuclear fusion, but eggs fail to successfully complete the first embryonic division due to defects in chromosome segregation (albeit not due to premature sister chromatid dissociation) and cytokinesis (Fig. 6A,B; Severson et al. 2000; Kaitna et al. 2002). The mitotic specificity of defects observed in air-2(or207) worms is further supported by the observation that Histone H3 phosphorylation is completely abrogated in the embryos, but not in germline nuclei (Fig. 6C,E). Since the existence of a redundant kinase function resulting in meiotic Histone H3 phosphorylation in the gonad has been previously ruled out (Hsu et al. 2000), we reasoned that the mitosis-specific defects of air-2(or207) are a result of a real physiological difference in the contribution of the mutant kinase (AIR-2or207) to meiotic and mitotic chromosome segregation. Of note, the reduced phosphorylation ability of AIR-2or207 is not a product of disrupted protein localization, since AIR-2or207 loading on meiotic and mitotic chromosomes is similar to the wild-type kinase (Fig. 6D; data not shown). This is further confirmed by the mislocalization to the long arms of lab-1 mutant bivalents observed both for AIR-2or207 and a mutant GFP∷AIR-2 transgenic protein that lacks kinase activity (GFP∷AIR-2-Kinase Dead) (Figs. 3B, 6D; Supplemental Fig. 7A). Therefore, our analysis suggests that the levels of AIR-2 kinase activity on chromosomes of air-2(or207) worms are sufficient to mediate homolog and sister chromatid dissociation during meiosis, but inadequate to drive mitotic segregation.
Figure 6.
lab-1(tm1791) partially suppresses the mitotic defects of air-2(or207ts). (A) Embryonic lethality, larval lethality, and incidence of males among the progeny of hermaphrodites of the indicated genotypes. Error bars represent standard deviation of the mean. Asterisks indicate statistically significant reduction in embryonic lethality in air-2(or207); lab-1(tm1791) (P < 0.0001) and air-2(or207); sgo-1(RNAi) (P = 0.0005 by the two-tailed Mann-Whitney test, 95% C.I.) when compared with air-2(or207). (B) air-2(or207ts) mitotic defects at 25°C. Failure in chromosome segregation and cytokinesis in air-2(or207ts) embryos leading to polyploid cells. Bar, 5 μm. (C) Histone H3 phosphorylation is abolished in air-2(or207) embryos, but not in the germline. A GFP∷AIR-2 transgene or lab-1(tm1791) can independently restore embryonic phosphorylation of Histone H3 in air-2(or207) mutants. Bars: premeiotic tip, 5 μm; diakinesis and metaphase I, 2 μm; late prophase and anaphase, 5 μm. (D) AIR-2 and LAB-1 localization on prometaphase I bivalents in air-2(or207ts) worms grown at the restrictive temperature. Bar, 0.5 μm. (E) Quantification of Histone H3 phosphorylation in the gonad and embryo. The sample sizes are shown above each bar. Phosphorylation is significantly reduced in −1 diakinesis oocytes and embryos (P < 0.0001, respectively, by the two-sided Fisher’s Exact Test, 95% C.I.), but not at the premeiotic tip (histone H3 phosphorylation was observed in 100% of the examined premeiotic tips) of air-2(or207) worms when compared with the wild type. Phosphorylation is significantly improved in −1 oocytes (one asterisk) and embryos (two asterisks) of air-2(or207) lab-1(tm1791) (P < 0.0019 and P < 0.0001, respectively) and air-2; sgo-1(RNAi) mutants (P < 0.0004 and P < 0.0001, by the two-sided Fisher’s Exact Test, 95% C.I.) when compared with air-2(or207).
The ability of LAB-1 to counteract AIR-2 poses the possibility that a reduction in LAB-1 activity might suppress defects associated with insufficient AIR-2-mediated phosphorylation. To assess whether lab-1 genetically interacts with air-2, we therefore examined if lab-1(tm1791) could rescue the defects observed in air-2(or207) embryos. Indeed, a significant increase in embryonic viability is observed in air-2(or207) lab-1(tm1791) double mutants compared with air-2(or207) worms grown at 20°C (P < 0.0001 by the two-tailed Mann-Whitney test, 95% C.I.) (Fig. 6A). This rescue from embryonic lethality is not as strong as observed with the introduction of GFP∷AIR-2 (Fig. 6A). Nevertheless, whereas virtually all air-2(or207) embryos laid at the restrictive temperature fail to hatch, 17% (n = 3027) of air-2(or207) lab-1(tm1791) embryos successfully complete embryogenesis and reach adulthood as fertile worms (Fig. 6A,C). Moreover, consistent with an improvement in the phosphorylation mediated by AIR-2or207, 30% (n = 75) of air-2(or207) lab-1(tm1791) embryos show H3 phosphorylation signal on mitotic chromosomes in contrast to 0% of air-2(or207) embryos (n = 54) (Fig. 6E). Therefore, our genetic analysis suggests that, as in meiosis, LAB-1 also plays a role in antagonizing the AIR-2 kinase activity during mitosis.
C. elegans shugoshin is not required for AIR-2 localization on the bivalent in meiosis I
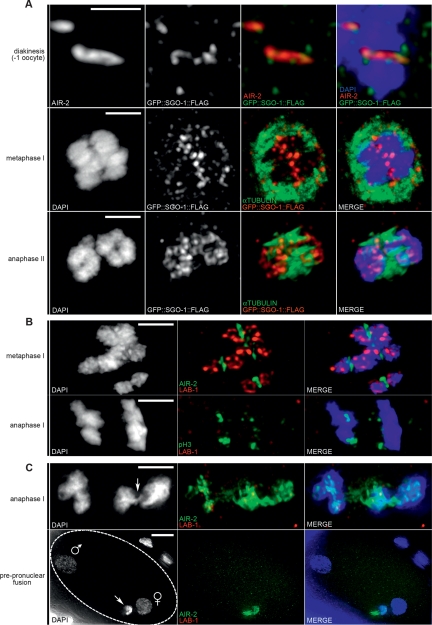
The extraordinary role of the MEI-S332/Shugoshin proteins in preserving centromeric cohesin from premature release is based on their ability to spatially regulate the phosphorylation effects of meiotic and mitotic kinases on chromosomes (Kitajima et al. 2006; Riedel et al. 2006). The fact that the chromosomes in C. elegans do not contain discrete centromeres prompted us to investigate whether the predicted C. elegans shugoshin, SGO-1 (Rabitsch et al. 2004), shared a role in AIR-2 regulation with LAB-1 and GSP-2. Expression of an integrated GFP∷SGO-1∷Flag transgene was used to assess the localization of SGO-1 in C. elegans gonads and embryos. In the gonads, GFP∷SGO-1∷Flag localized to the short arms of bivalents in late diakinesis and remained chromosome-associated until anaphase II (Fig. 7A). Unlike LAB-1, GFP∷SGO-1∷Flag signal was never detected along the long arms of the diakinesis bivalents (Fig. 7A), raising the possibility that, in C. elegans, SGO-1 may not directly protect cohesin during meiosis I. Indeed, homolog association was normal at diakinesis in sgo-1(RNAi), two sgo-1 deletion mutants (tm2443 and tm2344) and sgo-1(tm2443) trans-heterozygotes for a deficiency (stDf7) encompassing the sgo-1 locus (Fig. 1C; see Materials and Methods). Moreover, no defects in homolog association or premature loss of sister chromatid cohesion were detected at metaphase I [sgo-1(tm2443) = 0/12; sgo-1(tm2443);sgo-1(RNAi) = 0/6; sgo-1(tm2443)/stDf7 = 0/9]. In support of these observations, AIR-2 localization remained restricted to the short arms of the bivalents in sgo-1(RNAi) and sgo-1(tm2443) worms during late prophase I (Fig. 3B). Furthermore, Histone H3 phosphorylation signal and the localization of AIR-2 and LAB-1 on metaphase I bivalents of sgo-1(RNAi) and sgo-1(tm2443) worms were indistinguishable from wild type, suggesting that SGO-1 does not affect the access nor the activity of these proteins in chromosomes at the exit of prophase I (Fig. 7B; Supplemental Fig. 8C). Finally, sgo-1 depletion failed to increase premature loss of sister chromatid cohesion during metaphase I in lab-1(tm1791) oocytes [n = 3/14 for lab-1(tm1791); sgo-1(tm2443) compared with n = 2/15 for lab-1(tm1791)], arguing against a role of shugoshin in mediating cohesin protection during this critical period. Nevertheless, SGO-1 activity appears to play some role in mediating proper chromosome segregation during meiosis I and II. This is supported, first, by a low penetrance of segregation defects such as chromosome bridges observed at anaphase I for sgo-1(tm2443) (n = 1/25), sgo-1(RNAi) (n = 2/20), sgo-1(tm2443); sgo1(RNAi) (n = 3/19), and lab-1(tm1791); sgo-1(tm2443) (n = 1/14) compared with lab-1(tm1791) (n = 0/23) (Fig. 7C). Second, by the observation during meiosis II of chromosome bridges, lagging chromosomes and failure in polar body extrusion in a few sgo-1(RNAi) (n = 2/20), sgo-1(tm2443); sgo-1(RNAi) (n = 2/8), lab-1(tm1791); sgo-1(tm2443) (n = 1/3), and lab-1(tm1791); sgo-1(RNAi) oocytes (n = 3/6) compared with lab-1(tm1791) (n = 0/4). These defects likely account for the slight, but significant increase in embryonic lethality observed by sgo-1 depletion in lab-1(tm1791) worms [P = 0.0080 in lab-1(tm1791); sgo-1(tm2443) by the two-tailed Mann-Whitney test, 95% C.I] (Supplemental Fig. 8B).
Figure 7.
Expression of a GFP∷SGO-1 transgene and sgo-1(RNAi) phenotype. (A) GFP∷SGO-1∷Flag (anti-Flag) colocalizes with AIR-2 on the short arms of bivalents in −1 oocytes, is present along the interface of homologs setting up to segregate away from each other at metaphase I, and on segregating chromatids at anaphase II. Bars: −1 diakinesis, 0.5 μm; metaphase I and anaphase II, 2 μm. (B) Normal homolog segregation, as well as AIR-2, phospho H3, and LAB-1 localization on meiosis I chromosomes, in most sgo-1-depleted oocytes. (C) The small number of homolog segregation defects observed in sgo-1-depleted cells include chromosomal bridges (arrow in anaphase I) and failure of polar body extrusion (arrow in prepronuclear fusion) (n = 2/20, respectively). Male and female pronuclei as seen in the end of meiosis II (encircled oocyte) are indicated. Bars: metaphase I and anaphase I, 2 μm; prepronuclear fusion, 10 μm.
During embryogenesis, the GFP∷SGO-1∷Flag signal is detected on metaphase chromosomes as well as on the centrosomes, partially intersecting with the dynamic shuttling of the Aurora B kinase between chromatin and the cytoskeleton (Supplemental Fig. 9). This observation is particularly relevant in light of the fact that sgo-1(RNAi) and sgo-1(tm2443) weakly suppress the mitotic defects of air-2(or207) embryos, [2% viability in the air-2(or207); sgo-1(RNAi) progeny, P = 0.0005 by the two-tailed Mann-Whitney test, 95% C.I.] (Fig. 6A,E; data not shown). Taken together, these results are consistent with a potential role of SGO-1 in the regulation of AIR-2 during mitosis.
Discussion
LAB-1 counteracts AIR-2 phosphorylation effects during homolog segregation
As bivalents exit diakinesis, long and short arms must carry different molecular cues to guide the stepwise release of cohesin and to regulate chromosome orientation and capture by the spindle (Paliulis and Nicklas 2000). By the time of nuclear envelope breakdown (NEBD), the localization of kinetochore components, such as the histone H3 variant HCP-3 (CENP-A), HCP-4 (CENP-C), and KNL-1, is coincident with chromosomal DNA (Buchwitz et al. 1999; Maddox et al. 2004; Monen et al. 2005). In contrast, the C. elegans CPC proteins (AIR-2, BIR-1, ICP-1, and CSC-1) specifically occupy the short arms of the bivalents (Kaitna et al. 2000; Speliotes et al. 2000; Bishop and Schumacher 2002; Rogers et al. 2002; Romano et al. 2003; Nabeshima et al. 2005). This restricted localization of the CPC is correlated with separase-dependent REC-8 degradation and homolog dissociation in meiosis I. Although the CPC has the intrinsic ability to also load on the long arms of the bivalents, it is normally excluded from this subdomain by the activity of the Glc7-like phosphatases GSP-1 and GSP-2 (Kaitna et al. 2002; Rogers et al. 2002). However, exactly how these enzymes counteract AIR-2 activity specifically on the long arm domain remains to be determined.
Here we show that the long arm domain of bivalents in −1 oocytes is also specified before NEBD as indicated by the exclusive localization of LAB-1 on the region proximal to the chiasma. Like GSP-2/GLC-7β, LAB-1 is needed to constrain the localization of AIR-2 to the short arms of the bivalent until homologs separate at anaphase I. Furthermore, LAB-1 itself does not constitute a physical obstacle to AIR-2 loading but instead depends on GSP-2 to antagonize AIR-2. Although the precise subcellular localization of GSP-2 on C. elegans gonads has not been characterized, we propose a model in which LAB-1 acts as a platform to recruit and/or spread GSP-2 protective activity throughout the interchromatid domain of the long arms of the bivalents (Fig. 8). Precedents for such a mechanism have been reported previously in yeast, where antagonism of Ipl1/Aurora is achieved by specifically redistributing Glc7 (Pinsky et al. 2006). Targeting of PP1/Glc7 to different subcellular compartments and the modulation of its activity on specific substrates has been linked to binding of the PP1/Glc7 catalytic subunit to the degenerate [(R/K) (V/I) X (F/W)] consensus motif present in a collection of regulatory proteins in yeast and mammals (Egloff et al. 1997; Zhao and Lee 1997; Aggen et al. 2000; Bollen 2001; Wakula et al. 2003). These specificity factors comprise a diverse set of otherwise unrelated proteins that share the [(R/K) (V/I) X (F/W)] motif and are essential for different cellular processes, including the regulation of mitotic and meiotic progression (Zhao and Lee 1997). The presence of a putative [(R/K) (V/I) X (F/W)] motif in the C. elegans LAB-1 sequence (Fig. 1A) suggests that LAB-1 may antagonize AIR-2-mediated phosphorylation by regulating the accessibility of GSP-1/GSP-2 to the long arms of the bivalent. Under this interpretation, the reconfiguration of the localization of LAB-1 during chromosome remodeling is essential to prevent the early dissociation of sister chromatids at metaphase I, while also permitting the destruction of the subset of cohesin holding homologs together after SC disassembly.
Figure 8.
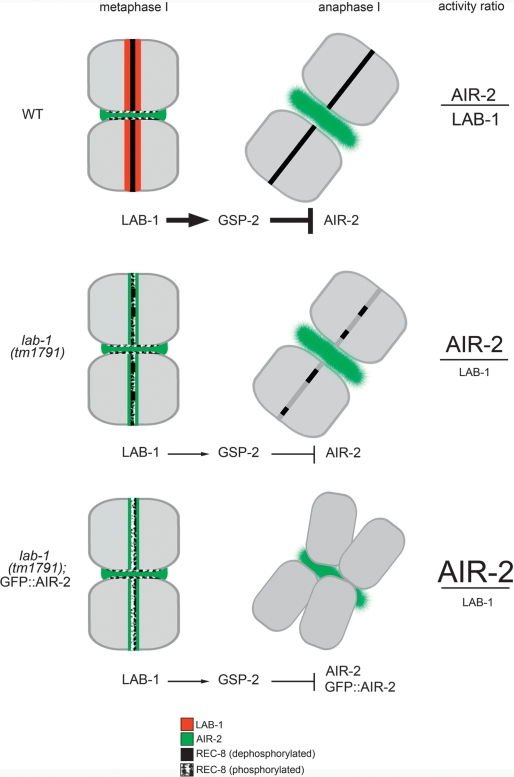
A model for LAB-1-mediated protection of sister chromatid cohesion. LAB-1 participates in the protection of cohesin by specifically counteracting AIR-2 activity on the long arm domains of the bivalent. This presumably involves the facilitation of GSP-2 activity in this domain. LAB-1 could serve as docking sites for GSP-2, actively recruiting the phosphatase in a shugoshin-like role or it could directly participate in dephosphorylating REC-8. Defects observed due to a decrease in LAB-1 activity, such as premature sister chromatid separation, are proportional to increased doses of AIR-2-mediated phosphorylation. Progressively harsher phenotypes are thus observed when worms carrying a reduced-loss-of-function allele of lab-1(tm1791) are further exposed to ectopic AIR-2 activity from a transgene (GFP∷AIR-2).
Consistent with a role of AIR-2 in mediating the defects observed in lab-1 mutants, ectopically increasing AIR-2 activity can enhance the defects observed in late meiosis I of lab-1(tm1791). This is reminiscent of gsp-1/2(RNAi) and gsp-2(tm301) mutant phenotypes where unbalanced AIR-2 activity causes hyperphosphorylation of its chromosomal targets and weakening of sister chromatid cohesion (Kaitna et al. 2002; Sassa et al. 2003). The milder sister chromatid dissociation phenotype observed in lab-1(tm1791) bivalents at the end of meiosis I probably reflects the presence of sufficient LAB-1 activity in tm1791 worms to engage GSP-2 and preserve chromosome association. Whether the GSP-2 paralog, GSP-1, equally participates in the LAB-1-mediated protection of cohesin on the bivalent is not known at the moment. Because GSP-1 and GSP-2 have partially overlapping functions during meiosis and mitosis in C. elegans (Hsu et al. 2000), it is possible that GSP-1 activity in lab-1(tm1791) oocytes compensates for the reduced antagonism of AIR-2 via GSP-2, attenuating the effects of LAB-1 depletion on the long arms of the bivalent.
The opposing roles of PP1/Glc7 and Aurora/Ipl1 in controlling the reversible phosphorylation of cohesins predict that the defects derived from decreased kinase activity should be compensated in part by a simultaneous loss of phosphatase activity. Indeed, knockdown of yeast Glc7 and C. elegans gsp-1/gsp-2 partially suppress the defects in Ipl1 and air-2-depleted cells, respectively (Francisco et al. 1994; Hsu et al. 2000; Pinsky et al. 2006). Suppression of ipl1 mutant phenotypes can also be achieved by depleting the regulatory subunits that promote Glc7 activity. For instance, binding of the regulatory protein Ypi1 to cytoplasmic Glc7 through a [(R/K) (V/I) X (F/W)] motif is required to translocate the phosphatase to the nucleus (Garcia-Gimeno et al. 2003; Pedelini et al. 2007; Bharucha et al. 2008). Loss of Ypi1 phenocopies the defects observed in a Glc7 mutant and can be explained by unchallenged nuclear Ipl1 activity (Bharucha et al. 2008). Here we demonstrate that a decrease in LAB-1 function, which likely affects GSP-2 activity, results in the disruption of sister chromatid association during meiosis I due to mislocalization of AIR-2 and can ameliorate the defects of air-2-deficient embryos possibly by partially restoring AIR-2-mediated phosphorylation during mitosis. Taken together, our analysis is consistent with a role for LAB-1 as a positive regulator of GSP-2 phosphatase activity. Moreover, it suggests that the stability of cohesin along the long arms of the bivalents during meiosis as well as the regulation of mitotic segregation in the embryo respond to a relative AIR-2/LAB-1 activity ratio (Fig. 8).
Protection of sister chromatid cohesion is shugoshin-independent in C. elegans meiosis I
Shugoshin proteins have two main functions during eukaryotic cell division: the protection of sister chromatid cohesion via PP2A phosphatase recruitment and the activation of the spindle checkpoint by surveying kinetochore/microtubule attachments (Kitajima et al. 2004; Kawashima et al. 2007). Although shugoshin mostly antagonizes the phosphorylation effects of mitotic and meiotic kinases to preserve centromeric cohesin, the Drosophila MEI-S332/Shugoshin protein promotes meiotic cohesion by cooperating with the Aurora B kinase (Resnick et al. 2006). In addition, tension-generating attachment of kinetochores has been reported to require the reciprocal activities of both shugoshin and Aurora B in the pericentromeric region (Kawashima et al. 2007; Pouwels et al. 2007). This complexity in shugoshin function is underscored by the marginal sequence identity of shugoshin homologs across phyla as well as the evolution of paralogs in mammals and fission yeast that have only partially overlapping roles (Kitajima et al. 2004). The C. elegans sole predicted shugoshin protein (product of ORF C33H5.15) helps to ensure the high fidelity of meiotic chromosome segregation, as seen by the occasional chromosome bridges and defects in polar body extrusion in sgo-1-depleted oocytes. However, we found no evidence of the involvement of SGO-1 in directly protecting meiotic cohesin; sgo-1 depletion does not compromise sister chromatid association in late prophase I or metaphase I and a GFP∷SGO-1 protein failed to localize to the long arms of the bivalents where protection against a premature release of sister chromatid cohesion is required. Although it is possible that SGO-1 activity protects meiotic cohesin on the long arms either redundantly and/or indirectly, we favor the idea that its main role in C. elegans meiosis lies in ensuring bipolar attachment of homologs to the metaphase I spindle, perhaps via regulation of AIR-2 activity. This is consistent with the notion that surveillance of kinetochore–microtubule association likely represents the ancestral role of shugoshin proteins with cohesin protection evolving at a later stage to facilitate this process (Kawashima et al. 2007). Accordingly, we observed GFP∷SGO-1 colocalizing with chromosomal AIR-2 both in meiosis as well as in mitosis. How homologs aligned at the metaphase I plate avoid merotelic attachments during C. elegans meiosis is not well understood. It is possible that AIR-2 and SGO-1 indirectly promote the ends of the long arms in the aligned bivalents to lead the way poleward at anaphase I, perhaps by destabilizing microtubule interactions along the short arms. This is supported by the observation that defects in homolog segregation in air-2-depleted oocytes cannot be solely explained by the failure to degrade cohesin on the short arms, as well as new evidence in mammalian oocytes linking lack of kinetochore tension with shugoshin-dependent protection of centromeric cohesin (Rogers et al. 2002; Gomez et al. 2007; Lee et al. 2007). Finally, the suppression of air-2(or207) defects in sgo-1-depleted embryos identifies a role for SGO-1 in counterbalancing AIR-2-mediated phosphorylation during mitosis. Recent evidence has emerged describing a major splice variant of Sgo1 (sSgo1) that localizes to centrosomes of murine fibroblasts where it regulates centriole cohesion in a Plk-dependent manner (Wang et al. 2008). Therefore, the presence of GFP∷SGO-1∷Flag on mitotic chromosomes and centrosomes of C. elegans embryos might reflect a localized need for shugoshin activity in two subcellular domains with distinct AIR-2 substrates. The characterization of the native localization pattern of the C. elegans shugoshin protein will be necessary to clarify the relevance of these results.
In conclusion, our observations indicate that, in C. elegans, a shugoshin-independent mechanism exists in meiosis to allow cohesin proximal to the chiasma to bypass separase-mediated degradation and support the integrity of sister chromatid cohesion until anaphase II. We hypothesize that the evolution of LAB-1 proteins in Caenorhabditis represents, at least in part, the response to the challenge of protecting meiotic cohesin along an extensive domain of the bivalent.
Materials and methods
Strains and alleles
The N2 Bristol strain was used as the wild-type background. C. elegans strains were cultured at 20°C under standard conditions as described in Brenner (1974). For temperature sensitivity analysis of air-2(or207ts) mutants, worms were incubated at 15°C as the permissive temperature and transferred at the L4 stage to 20°C or 25°C as restrictive conditions. The embryonic lethality observed among air-2(or207) progeny is 100% at either 20°C or 25°C. However, the partial suppression of the Emb phenotype observed in air-2(or207) worms after either lab-1 or sgo-1 depletion was only evident at 20°C, presumably because of a less severe constraint for AIR-2or207 activity at 20°C compared with 25°C. The following mutations and chromosome rearrangements were used: LGI: air-2(or207ts), unc-13(e1091), lab-1(tm1791, tm3002), mnDf111; LGIII: gsp-2(tm301); LGIV: sgo-1(tm2443, tm2344), stDf7 (Maruyama and Brenner 1991; Labouesse 1997; Severson et al. 2000; Sassa et al. 2003; this work).
The lab-1 and sgo-1 alleles were generated by the Japanese National BioResource Project (NBP) for C. elegans. The tm1791 allele of lab-1 carries an N-terminal 421-bp deletion that removes most of its promoter sequence along with exon 1 and part of intron 1 (Supplemental Fig. 2A). The tm3002 allele of lab-1 carries an N-terminal 327-bp deletion that removes exons 1 and 2 without affecting a putative alternative start site at exon 3 (Supplemental Fig. 2A). Analysis of chromosome morphology throughout DAPI-stained germlines, as well as monitoring of brood size, embryonic lethality, and frequency of male progeny, indicated that tm3002 is a weaker reduction-of-function allele of lab-1 compared with tm1791 and therefore our studies focused on the latter (data not shown).
The available sgo-1 alleles, tm2344 and tm2443, are not predicted to affect the conserved coiled-coil domain in the N-terminal region of sgo-1 required for centromeric localization, although the equally conserved basic carboxy domain is ablated by the deletion in tm2443 (Watanabe 2005). Indeed, because tm2443 is also able to partially suppress air-2(or207) (P = 0.0020 by the two-tailed Mann-Whitney test, 95% C.I.), as observed with sgo-1 depletion via RNAi (P = 0.0005 by the two-tailed Mann-Whitney test, 95% C.I.) (Fig. 6A), it may be a weak loss-of-function allele of sgo-1. However, as suggested by the comparison of embryonic lethality among sgo-1(tm2443) (Emb = 0.9%; n = 2481), sgo-1(RNAi) (Emb = 2%; n = 1319), or sgo-1(tm2443);sgo-1(RNAi) (Emb = 0%; n = 1622) worms (Supplemental Fig. 8B), and the lack of premature sister chromatid separation in sgo-1(tm2443)/stDf7 (see the Results), the analysis of sgo-1(tm2443) may constitute the full extent of the defects observed upon depletion of SGO-1 function. Therefore, for completion, our analysis of SGO-1 function relied on RNAi-depleted gonads and analysis of sgo-1(tm2443).
RT–PCR
cDNA was produced from single-worm RNA extracts using the SuperScript III One-step RT–PCR System (Invitrogen) as in Stothard et al. (2002). The effectiveness of RNAi was determined by assaying the expression of the transcript being depleted in at least four individual animals subjected to RNAi. Expression of either myo-3 (K12F2.1) or fbxa-192 (F57G4.8) transcripts was used as a control.
RNAi
Feeding RNAi experiments were performed at either 20°C or 25°C as described in Timmons et al. (2001). The entire coding sequence of lab-1 was originally cloned into pGEMT (Promega) to generate pCEC03. lab-1 cDNA from pCEC03 was subsequently subcloned into the pL4440 feeding vector to generate pCEC11 used in RNAi experiments. HT115 bacterial clones from a C. elegans feeding RNAi library (Geneservice) were used for sgo-1 (C33H5.15) RNAi experiments. Control RNAi was performed by feeding HT115 bacteria carrying the empty pL4440 vector. RNAi depletion by gonad microinjection was also performed for sgo-1. The full sgo-1 genomic sequence in pCEC49 (pGEMT-sgo-1) was used as template to generate sgo-1 dsRNA as in Colaiacovo et al. (2002).
GFP constructs and integrative transformation by microparticle bombardment
To create an air-2 GFP construct, the full-length air-2 cDNA was PCR-amplified and cloned into the Gateway donor plasmid pDONR201 (Invitrogen). The pDONR201 air-2 construct was recombined with the pID3.01B destination vector to generate an in-frame N-terminal GFP fusion protein (Stitzel et al. 2006). The kinase-dead (KD) air-2 GFP transgene was made by introducing the point mutation K65M into the pDONR201 air-2 construct using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The genomic sequences of lab-1 and sgo-1 were used for making N-terminal GFP fusions. Specifically, a PCR-amplified lab-1 genomic fragment carrying a 3′ end HA tag was cloned into pGEMT (pCEC13). The genomic lab-1 insert in pCEC13 was next subcloned in frame with the GFP cassette in pJH4.52 using its SpeI site (Pellettieri et al. 2003). Finally, a rescue unc-119(+) fragment was inserted in the SacII site of pJH4.52 (pCEC20). To obtain the sgo-1 GFP construct, a genomic sgo-1 fragment carrying a 3′ end Flag tag was PCR-amplified and cloned into pTOPO (Invitrogen) (pCEC32). The sgo-1∷Flag insert and a unc-119(+) rescue fragment were subsequently subcloned into pJH4.52 (pCEC31). The constructs were introduced into unc-119(ed3) animals by microparticle bombardment (Praitis et al. 2001). The isolated integrated lines used in this work were GFP∷AIR-2 (WH371); GFP∷AIR-2-KD (JS533); GFP∷LAB-1∷HA (CV40) and GFP∷SGO-1∷Flag (CV62).
Antibody production and immunofluorescence
Rabbit polyclonal antibodies against C-terminal peptides of C. elegans LAB-1 (DSDSDTSEDSDFDEDFEDSD) and AIR-2 (KI RAEKQQKIEKEASLRNH) were generated by Sigma-Genosys. Antisera were affinity-purified against either a bacterially expressed HIS tag fusion protein (LAB-1) or using the original peptide-antigen (AIR-2) as described in Chase et al. (2001). To express the HIS-tagged LAB-1 in bacteria (pCEC09), lab-1 cDNA was subcloned from pCEC03 into pRSETa (Invitrogen). Whole mount preparation of dissected gonads, fixation, and immunostaining procedures were carried out as in Colaiacovo et al. (2003) and Rogers et al. (2002). α-AIR-2 and α-LAB-1 affinity-purified antibodies were used at 1:100 and 1:300 dilutions, respectively. Other antibodies and dilutions used in this work were: α-REC-8 (1:50; Abcam), α-GFP (1:100; Abcam), α-Flag (1:100; Sigma), α-phospho Ser10 (Histone H3) (1:400; Upstate Biotechnologies), α-tubulin (1:100; Sigma), and α-CSC-1 (1:100; Romano et al. 2003). For AIR-2 and LAB-1 coimmunostaining, 5 μg of affinity-purified α-LAB-1 was Cy3-labeled using the Cy3 Ab labeling kit (Amersham) according to the manufacturer’s instructions. To avoid cross-reactivity, dissected gonads were first stained with α-AIR-2, followed by a 20-min treatment with whole rabbit serum (1:25 dilution), before being incubated with α-LAB-1Cy3 antibody (1:20 dilution).
Imaging and time-lapse microscopy
Immunofluorescence images were collected at 0.2-μm intervals with an IX-70 microscope (Olympus) and a cooled CCD camera (model CH350; Roper Scientific) controlled by the DeltaVision system (Applied Precision). Images were subjected to deconvolution analysis using the SoftWorx 3.0 program (Applied Precision) as in Nabeshima et al. (2005). For time-lapse microscopy, embryos were released by cutting open gravid hermaphrodites in M9 and transferred to a 3% agarose pad on a slide. Images were captured on a Leica DM5000 B microscope (60× objective) every 30 or 40 sec.
Acknowledgments
Some strains were kindly provided by the Japanese National Bioresource Project for C. elegans and by the Caenorhabditis Genetics Center. We acknowledge B. DuBoff for help in the initial characterization of tm1791, as well as T. Furuta and K. Verbrugghe for generating the GFP::AIR-2 strain. We thank M. Glotzer for the CSC-1 antibody and N. Svrzipaka for technical help with microparticle bombardment. We are grateful to A. Amon and members of the Colaiácovo laboratory for critical reading of this manuscript. This work was supported by National Institutes of Health grant R01GM072551 to M.P.C. and R01GM62181 to J.M.S.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1691208.
References
- Aggen J.B., Nairn A.C., Chamberlin R. Regulation of protein phosphatase-1. Chem. Biol. 2000;7:R13–R23. doi: 10.1016/S1074-5521(00)00069-7. [DOI] [PubMed] [Google Scholar]
- Albertson D.G., Thomson J.N. The kinetochores of Caenorhabditis elegans. Chromosoma. 1982;86:409–428. doi: 10.1007/BF00292267. [DOI] [PubMed] [Google Scholar]
- Albertson D.G., Thomson J.N. Segregation of holocentric chromosomes at meiosis in the nematode Caenorhabditis elegans. Chromosome Res. 1993;1:15–26. doi: 10.1007/BF00710603. [DOI] [PubMed] [Google Scholar]
- Albertson D.G., Rose A.M., Villeneuve A.M. Chromosome organization, mitosis and meiosis. In: Riddle D.L., et al., editors. C. elegans II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 47–48. [PubMed] [Google Scholar]
- Alexandru G., Uhlmann F., Mechtler K., Poupart M., Nasmyth K. Phosphorylation of the cohesin subunits Scc1 by Polo/Dcd5 kinases regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- Bharucha J.P., Larson J.R., Gao L., Daves L.K., Tatchell K. Ypi1, a positive regulator of nuclear protein phosphatase type 1 activity in Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:1032–1045. doi: 10.1091/mbc.E07-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J.D., Schumacher J.M. Phosphorylation of the carboxy terminus of inner centromere protein (INCENP) by the Aurora B kinase stimulates Aurora B kinase activity. J. Biol. Chem. 2002;277:27577–27580. doi: 10.1074/jbc.C200307200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen M. Combinatorial control of protein phosphatase-1. Trends Biochem. Sci. 2001;26:426–431. doi: 10.1016/s0968-0004(01)01836-9. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz B.J., Ahmad K., Moore L.L., Roth M.B., Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- Buonomo S.B., Clyne R.K., Fuchs J., Loidl J., Uhlmann F., Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Chan R.C., Severson A.F., Meyer B.J. Condensin restructures chromosomes in preparation for meiotic divisions. J. Cell Biol. 2004;167:613–625. doi: 10.1083/jcb.200408061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase D.L., Patikoglou G.A., Koelle M.R. Two RGS proteins that inhibit Gα0 and Gαq signaling in C. elegans neurons require a Gβ5-like subunit for function. Curr. Biol. 2001;11:222–231. doi: 10.1016/s0960-9822(01)00071-9. [DOI] [PubMed] [Google Scholar]
- Ciosk R., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Clyne R.K., Katis V.L., Jessop L., Benjamin K.R., Herskowitz I., Lichten M., Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O. The making and breaking of sister chromatid cohesion. Cell. 2001;106:137–140. doi: 10.1016/s0092-8674(01)00439-1. [DOI] [PubMed] [Google Scholar]
- Colaiacovo M.P., Stanfield G.M., Reddy K.C., Reinke V., Kim S.K., Villeneuve A.M. A targeted RNAi screen for genes involved in chromosome morphogenesis and nuclear organization in the Caenorhabditis elegans germline. Genetics. 2002;162:113–128. doi: 10.1093/genetics/162.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo M.P., MacQueen A.J., Martinez-Perez E., McDonald K., Adamo A., La Volpe A., Villeneuve A.M. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Dernburg A.F. Here, there, and everywhere: Kinetochore function on holocentric chromosomes. J. Cell Biol. 2001;153:F33–F38. doi: 10.1083/jcb.153.6.F33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A.F., McDonald K., Moulder G., Dresser M., Villeneuve A.M. Meiotic recombination in C.elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Egloff M.P., Johnson D.F., Moorhead G., Cohen P.T.W., Cohen P., Barford D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997;16:1876–1887. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco L., Wang W., Chan C.S. Type 1 protein phosphatase acts in opposition to Ipl1 protein kinase in regulating yeast chromosome segregation. Mol. Cell. Biol. 1994;14:4731–4740. doi: 10.1128/mcb.14.7.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gimeno M.A., Munoz I., Arino J., Sanz P. Molecular characterization of Ypi1, a novel Saccharomyces cerevisiae type1 protein phosphatase inhibitor. J. Biol. Chem. 2003;278:47744–47752. doi: 10.1074/jbc.M306157200. [DOI] [PubMed] [Google Scholar]
- Gomez R., Valdeolmillos A., Parra M.T., Viera A., Carreiro C., Roncal F., Rufas J.S., Barbero J.L., Suja J.A. Mammalian SGO2 appears at the inner centromere domain and redistributes depending on tension across centromeres during meiosis II and mitosis. EMBO Rep. 2007;8:173–180. doi: 10.1038/sj.embor.7400877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O., Golubovskaya I., Meeley R., Fiume E., Timofejeva L., Schleiffer A., Nasmyth K., Cande W.Z. A REC8-dependent plant shugoshin is required for maintenance of centromeric cohesion during meiosis and has no mitotic functions. Curr. Biol. 2005;15:948–954. doi: 10.1016/j.cub.2005.04.049. [DOI] [PubMed] [Google Scholar]
- Hassold T., Hunt P. To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Hauf S., Roitinger E., Koch B., Dittrich C.M., Mechtler K., Peters J.M. Dissociation of cohesin from chromosomes arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig N.C., Uhlmann F. Preferential cleavage of chromatin-bound cohesin after targeted phosphorylation by Polo-like kinase. EMBO J. 2004;23:3144–3153. doi: 10.1038/sj.emboj.7600303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M., McDonald K.L., Albertson D.G., Meyer B.J. HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J. Cell Biol. 2001;153:1227–1238. doi: 10.1083/jcb.153.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.Y., Sun Z.W., Li X., Reuben M., Tatchell K., Bishop D.K., Grushcow J.M., Brame C.J., Caldwell J.A., Hunt D.F., et al. Mitotic phosphorylation of histone H3 is governed by Ilp1/Aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Kaitna S., Mendoza M., Jantsch-Plunger V., Glotzer M. Incenp and an Aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol. 2000;10:1172–1181. doi: 10.1016/s0960-9822(00)00721-1. [DOI] [PubMed] [Google Scholar]
- Kaitna S., Pasierbek P., Jantsch M., Loidl J., Glotzer M. The Aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr. Biol. 2002;12:798–812. doi: 10.1016/s0960-9822(02)00820-5. [DOI] [PubMed] [Google Scholar]
- Katis V.L., Galova M., Rabitsch K.P., Gregan J., Nasmyth K. Maintenance of cohesin at centromere after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr. Biol. 2004;14:560–572. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Kawashima S.A., Tsukahara T., Langegger M., Hauf S., Kitajima T.S., Watanabe Y. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes & Dev. 2007;21:420–435. doi: 10.1101/gad.1497307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrebrock A.W., Moore D.P., Wu J.S., Orr-Weaver T.L. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–256. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Kawashima S.A., Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Sakuno T., Ishiguro K.I., Iemura S.I., Natsume T., Kawashima S.A., Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- Kops G.J., Weaver B.A., Cleveland D.W. On the road to cancer: Aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- Labouesse M. Deficiency screen based on the monoclonal antibody MH27 to identify genetic loci required for morphogenesis of the Caenorhabditis elegans embryo. Dev. Dyn. 1997;210:19–32. doi: 10.1002/(SICI)1097-0177(199709)210:1<19::AID-AJA3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Amon A. Role of polo-like kinase CDC5 in programming meiosis I segregation. Science. 2003;300:482–486. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
- Lee J., Kitajima T.S., Yanno Y., Yoshida K., Morita T., Miyano T., Miyake M., Watanabe Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat. Cell Biol. 2007;10:42–52. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
- MacQueen A.J., Colaiacovo M.P., McDonald K., Villeneuve A.M. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes & Dev. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P.S., Oegema K., Desai D., Cheeseman I.M. ‘Holo’er than thou: Chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 2004;12:641–653. doi: 10.1023/B:CHRO.0000036588.42225.2f. [DOI] [PubMed] [Google Scholar]
- Marston A.L., Tham W.H., Shah H., Amon A. A genome-wide screen identified genes required for centromeric cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- Maruyama I.N., Brenner S. A phorbol ester/diacylglycerol-binding protein encoded by the unc-13 gene of Caenorhabditis elegans. Proc. Natl. Acad. Sci. 1991;88:5729–5733. doi: 10.1073/pnas.88.13.5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Monen J., Maddox P.S., Hyndman F., Oegema K., Desai A. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat. Cell Biol. 2005;12:1248–1255. doi: 10.1038/ncb1331. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve A.M., Colaiacovo M.P. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J. Cell Biol. 2005;168:683–689. doi: 10.1083/jcb.200410144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Paliulis L.V., Nicklas R.B. The reduction of chromosome number in meiosis is determined by properties built on the chromosomes. J. Cell Biol. 2000;150:1223–1232. doi: 10.1083/jcb.150.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D., Loidl J. A Caenorhabditis elegans cohesin protein with functions in meiotic chromosome pairing and disjunction. Genes & Dev. 2001;15:1349–1360. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedelini L., Marquina M., Arino J., Casamayor A., Sanz S., Bollen M., Sanz P., Garcia-Gimeno M.A. YPI1 and SDS22 protein regulate the nuclear localization and function of yeast type 1 phosphatase Glc7. J. Biol. Chem. 2007;282:3282–3292. doi: 10.1074/jbc.M607171200. [DOI] [PubMed] [Google Scholar]
- Pellettieri J., Reinke V., Kim S.K., Seydoux G. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Dev. Cell. 2003;5:451–462. doi: 10.1016/s1534-5807(03)00231-4. [DOI] [PubMed] [Google Scholar]
- Pinsky B.A., Kotwaliwale C.V., Tatsutani S.Y., Breed C.A., Biggins S. Glc7/Protein phosphatase 1 regulatory subunits can oppose the Ipl1/Aurora protein kinase by redistributing Glc7. Mol. Cell. Biol. 2006;26:2648–2660. doi: 10.1128/MCB.26.7.2648-2660.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouwels J., Kukkonen A.M., Lan W., Daum J.R., Gorbsky G.J., Stukenberg T., Kallio M.J. Shugoshin 1 plays a central role in kinetochore assembly and is required for kinetochore targeting of Plk1. Cell Cycle. 2007;6:1579–1585. doi: 10.4161/cc.6.13.4442. [DOI] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch K.P., Gregan J., Schleiffer A., Javerzat J.P., Eisenhaber F., Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr. Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- Reinke V., Gil I.S., Ward S., Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- Resnick T.D., Satinover D.L., MacIsaac F., Stukenberg P.T., Earnshaw W.C., Orr-Weaver T.L., Carmena M. INCENP and Aurora B promote meiotic chromatid cohesion through localization of the shugoshin MEI-S332 in Drosophila. Dev. Cell. 2006;11:57–68. doi: 10.1016/j.devcel.2006.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel C.G., Katis V.L., Katou Y., Mori S., Itoh T., Helmhart W., Galova M., Petronczki M., Gregan J., Cetin B., et al. Protein phosphatase 2A protects centromeric sister cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- Rogers E., Bishop J.D., Waddle J.A., Schumacher J.M., Lin R. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J. Cell Biol. 2002;157:219–229. doi: 10.1083/jcb.200110045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano A., Guse A., Krascenicova I., Schnabel H., Schnabel R., Glotzer M. CSC-1: A subunit of the Aurora B kinase complex that binds to the Survivin-like protein BIR-1 and the Incenp-like protein ICP-1. J. Cell Biol. 2003;161:229–236. doi: 10.1083/jcb.200207117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa T., Ueda-Ohba H., Kitamura K.I., Harada S.I., Hosono R. Role of Caenorhabditis elegans protein phosphatase type 1, CeGLC-7β, in metaphase to anaphase transition during embryonic development. Exp. Cell Res. 2003;287:350–360. doi: 10.1016/s0014-4827(03)00157-5. [DOI] [PubMed] [Google Scholar]
- Severson A.F., Hamill D.R., Carter J.C., Schumacher J., Bowerman B. The Aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr. Biol. 2000;10:1162–1171. doi: 10.1016/s0960-9822(00)00715-6. [DOI] [PubMed] [Google Scholar]
- Speliotes E.K., Uren A., Vaux D., Horvitz H.R. The Surviving-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol. Cell. 2000;6:211–223. doi: 10.1016/s1097-2765(00)00023-x. [DOI] [PubMed] [Google Scholar]
- Stitzel M.L., Pellettieri J., Seydoux G. The C. elegans DYRK kinase MBK-2 marks oocyte proteins for degradation in response to meiotic maturation. Curr. Biol. 2006;16:56–62. doi: 10.1016/j.cub.2005.11.063. [DOI] [PubMed] [Google Scholar]
- Stothard P., Hansen D., Pilgrim D. Evolution of the PP2C family in Caenorhabditis: Rapid divergence of the sex-determining protein FEM-2. J. Mol. Evol. 2002;54:267–282. doi: 10.1007/s0023901-0008-y. [DOI] [PubMed] [Google Scholar]
- Sumara I., Vorlaufer E., Stukenberg P.T., Kelm O., Redemann N., Nigg E.A., Peters J.M. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol. Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- Tang Z., Shu H., Qi W., Mahmood N., Mumby M.C., Yu H. PP2A is required for centromere localization of Sgo1 and proper chromosome segregation. Dev. Cell. 2006;10:1–11. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Timmons L., Court D.L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Lottspeich F., Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Wakula P., Beullens M., Ceulemans H., Stalmans W., Bollen M. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J. Biol. Chem. 2003;278:18817–18823. doi: 10.1074/jbc.M300175200. [DOI] [PubMed] [Google Scholar]
- Wang X., Yang T., Duan Q., Huang Y., Darzynkiewicz Z., Dai W. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Pkl1. Dev. Cell. 2008;14:331–341. doi: 10.1016/j.devcel.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y. Shugoshin: Guardian spirit at the centromere. Curr. Opin. Cell Biol. 2005;17:590–595. doi: 10.1016/j.ceb.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Zhao S., Lee E.Y.C. A protein phosphatase-1 motif identified by the panning of a random peptide display library. J. Biol. Chem. 1997;272:28368–28372. doi: 10.1074/jbc.272.45.28368. [DOI] [PubMed] [Google Scholar]