Abstract
Bloom Syndrome is an autosomal recessive cancer-prone disorder caused by mutations in the BLM gene. BLM encodes a DNA helicase of the RECQ family, and associates with Topo IIIα and BLAP75/RMI1 (BLAP for BLM-associated polypeptide/RecQ-mediated genome instability) to form the BTB (BLM–Topo IIIα–BLAP75/RMI1) complex. This complex can resolve the double Holliday junction (dHJ), a DNA intermediate generated during homologous recombination, to yield noncrossover recombinants exclusively. This attribute of the BTB complex likely serves to prevent chromosomal aberrations and rearrangements. Here we report the isolation and characterization of a novel member of the BTB complex termed BLAP18/RMI2. BLAP18/RMI2 contains a putative OB-fold domain, and several lines of evidence suggest that it is essential for BTB complex function. First, the majority of BLAP18/RMI2 exists in complex with Topo IIIα and BLAP75/RMI1. Second, depletion of BLAP18/RMI2 results in the destabilization of the BTB complex. Third, BLAP18/RMI2-depleted cells show spontaneous chromosomal breaks and are sensitive to methyl methanesulfonate treatment. Fourth, BLAP18/RMI2 is required to target BLM to chromatin and for the assembly of BLM foci upon hydroxyurea treatment. Finally, BLAP18/RMI2 stimulates the dHJ resolution capability of the BTB complex. Together, these results establish BLAP18/RMI2 as an essential member of the BTB dHJ dissolvasome that is required for the maintenance of a stable genome.
Keywords: RECQ, double Holliday junction, Chromatin, BLAP18/RMI2, BLM
The autosomal recessive disorder Bloom syndrome (BS) is characterized by severe growth retardation, immunodeficiency, anemia, reduced fertility, and cancer predisposition (German 1969; German and Ellis 2002). BS patients show different malignancies that appear early in life and within various tissues (German 1969; German and Ellis 2002). Cells derived from BS patients show cytogenetic abnormalities, such as chromosomal breaks and an elevated rate (10-fold) of sister chromatid exchanges (SCEs). These cytogenetic abnormalities are used as a molecular diagnostic test for the disease (German 1969; German et al. 1977c).
BS stems from mutations in the BLM gene (Ellis et al. 1995). BLM protein belongs to the RecQ helicase family, which also includes RECQ1, WRN, RECQ4/RTS, and RECQ5, all of which play a unique role in the maintenance of genomic stability. WRN and RECQ4/RTS are also needed for the suppression of cancer and premature aging in humans (Ellis et al. 1995; Hanada and Hickson 2007), while the ablation of RECQ5 in mice engenders a late-onset tumor susceptibility phenotype (Hu et al. 2007).
BLM is a structure-specific helicase that can unwind 3′-tailed duplexes, bubble structures, forked duplexes, G-quadruplex structures, DNA displacement loops (D-loops), and four-way junctions that model Holliday junction (HJ) recombination intermediates (for review, see Hanada and Hickson 2007). The BLM–Topo IIIα complex has been shown to resolve double Holliday junction (dHJ) in vitro in a noncrossover fashion, and the recently discovered BLAP75/RMI1 (BLAP for BLM-associated polypeptide/RecQ-mediated genome instability) strongly stimulates this reaction (Raynard et al. 2006; Wu et al. 2006). The BLM–Topo IIIα–BLAP75/RMI1 ensemble has been termed the BTB (or RecQ–Topo IIIα–RMI1) complex (Raynard et al. 2006; Wu et al. 2006). BLM and Topo IIIα interact with the OB-fold-containing N-terminal region of BLAP75/RMI1 (Raynard et al. 2008).The ability of the BTB complex to dissolve dHJs to yield noncrossovers is thought to play a crucial role in the avoidance of chromosomal rearrangements, such as translocations, during the homology-directed repair of chromosomal lesions and injured replication forks (Sung and Klein 2006; Wu and Hickson 2006).
BLM has been shown to localize to promyelocytic leukemia (PML) bodies in the absence of DNA damage (Bischof et al. 2001). Upon the occurrence of DNA damage or inhibition of DNA replication, however, BLM dissociates from PML bodies to form nuclear foci, where it colocalizes with other DNA repair proteins, such as RAD51, BRCA1, and the MRE11–RAD50–NBS1 complex (Bischof et al. 2001). Consistent with these observations, BLM is recruited to laser-induced DNA double-strand breaks (DSBs) (Dutertre et al. 2000; Karmakar et al. 2006).
Here, to better understand the function of BLM in DNA damage repair and response, we sought to determine whether the BTB complex harbors other protein components and, if so, to ascertain the function of these novel BTB components. Earlier immunopurification approaches utilizing BLM antibody had a disadvantage in that the IgG light chain of the antibody might have masked BTB-associated proteins (Meetei et al. 2003). We therefore used a recently developed two-step affinity purification approach by expressing BLM fused with a double tag containing (His)6 and Flag. This new approach has led to the identification of BLAP18/RMI2 as a novel component of the BTB complex. We find that BLAP18/RMI2 forms a core complex with Topo IIIα and BLAP75/RMI1. We also find that BLAP18/RMI2 is required for the recruitment of BLM to chromatin and replication stress-induced nuclear foci. Depletion of BLAP18/RMI2 yields a profile of chromosome instability and sensitivity to DNA damage similar to that observed in BS cells. These results thus help define the nature of the BLM-associated protein complex in cells and reveal a critical role of BLAP18/RMI2 in the promotion of BLM-dependent genome maintenance pathway.
Since the Saccharomyces cerevisiae BLAP75 ortholog is called Rmi1, we will henceforth refer to BLAP75 as RMI1 and BLAP18 as RMI2 to be consistent with the yeast literature.
Results
RMI2 is a novel component of BLM-containing complexes
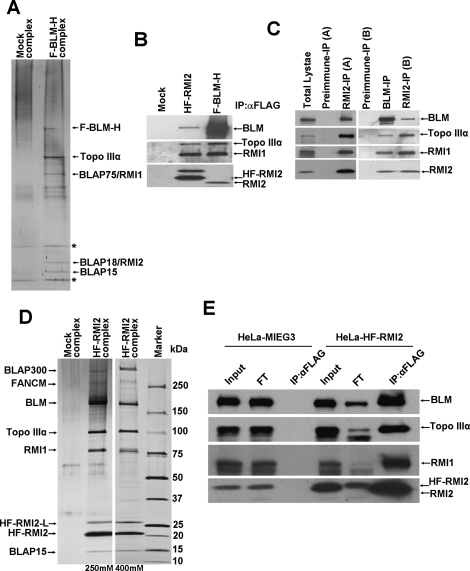
In order to gain more insight into the cellular function of BLM-containing complexes, we used a two-step affinity purification coupled mass spectrometry (MS) approach to isolate and identify novel BLM-associated polypeptides. BLM that harbors an N-terminal Flag tag and a C-terminal (His)6-tagged (F-BLM-H) was stably expressed in HT1080 cells by retroviral-mediated gene transfer, and BLM and its associated proteins were purified by a two-step affinity chromatographic procedure as described in the Materials and Methods. MS analysis of the polypeptides in the purified fraction identified several proteins that are known to interact with BLM, including the other components of the BTB complex, Topo IIIα, and RMI1. In addition, we also identified novel polypeptides of 18 kDa and 15 kDa molecular mass. Here we focus on the 18-kDa polypeptide, which we named as a BLM-associated polypeptide of 18-kDa mass, or RMI2 (Fig. 1A). The 15-kDa polypeptide, BLAP15, will not be considered further here.
Figure 1.
RMI2 is a novel component of the BTB complex. (A) Silver-stained gel showing the polypeptide bands purified from the nuclear extract of HT1080 cells transduced with vector alone (Mock) and cells expressing F-BLM-H, as described in Materials and Methods. The polypeptides identified by MS analysis are indicated by an arrow, and the asterisk (*) marks polypeptides also found in the mock purification. (B) Immunoblot of complexes purified from HeLa or HT1080 cells using anti-Flag M2 agarose. HeLa cells were transduced with vector alone or with vector that contained HF-RMI2. HT1080 cells stably expressed F-BLM-H. Immunoblots were probed with the antibodies indicated on the right. (C) Immunoblot of endogenous RMI2 complex immuno-isolated using anti-RMI2 antibodies raised against either MBP-RMI2 (RMI2-A) or GST-RMI2 (RMI2-B) fusions and probed with antibodies against known BTB members. RMI2 was detected with RMI2-A antibody on these immunoblots. (D) Silver-stained gel showing polypeptide bands purified from the nuclear extract of HeLa S3 cells transduced with vector alone (Mock) and cells stably expressing HF-RMI2. The RMI2 complex was purified using two different NaCl concentrations (250 and 400 mM, as indicated). Note that the unique polypeptide with molecular mass ∼27 kDa (denoted as HF-RMI2-L) was identified as a longer version of RMI2 that resulted from a leaky start site in the retroviral construct used. (E) Immunoblot of RMI2 complex purified using anti-Flag M2 agarose from HeLa cells stably expressing either the vector alone or HF-RMI2. Immunoblots were probed with antibodies against BTB members. The flow-through (FT) fraction is shown as a measure of depletion of BTB proteins along with HF-RMI2. Note that there is a substantial amount of BLM not depleted along with HF-RMI2, Topo IIIα, and RMI1.
RMI2 is clearly one of the prominent polypeptide components of the purified BLM protein complex (Fig. 1A). RMI2 corresponds to an uncharacterized hypothetical protein named C16orf75 (chromosome 16 ORF 75), otherwise also known as MGC24665 or LOC116028. RMI2 has no known function and appears to harbor an OB-fold domain (Supplemental Fig. 1).
We generated two different antibodies against RMI2 (A and B), both of which proved suitable for immunoblot analysis of RMI2 tagged with the (His)6 and Flag epitopes at its N terminus (HF-RMI2) and expressed in HeLa cells (Fig. 1B), and of endogenous RMI2 present in the BLM complex (Fig. 1B,C). When we immunoprecipitated RMI2 using the anti-RMI2 antibodies (Fig. 1C), we found by immunoblot analysis the presence of BLM, Topo IIIα, and RMI1 (Fig. 1C).
Likewise, two-step affinity purification of HF-RMI2 from extracts of HeLa S3 cells under high stringency (400 mM salt) coupled with MS revealed its association with BLM, Topo IIIα, and RMI1. BLAP15 was also found in the purified fraction (Fig. 1D). We established that the polypeptide denoted as HF-RMI2-L stems from a leaky upstream start site in the retroviral construct used for tagged RMI2 expression.
Immunoprecipitation of HF-RMI2 using anti-Flag antibody significantly depleted both Topo IIIα and RMI1, and to a large extent BLM, from the flow-through fraction (Fig. 1E). Taken together, the results help identify RMI2 as a novel and integral component of the BTB complex.
RMI2 contains an OB-fold domain and is conserved among higher eukaryotes
BLAST analysis of human RMI2 identified likely orthologs in vertebrate and invertebrate species that show a high degree of amino acid sequence identity to the human protein (Supplemental Fig. 1). Sequence analysis using InterProScan revealed a putative OB-fold domain. The OB-fold domain is a compact structural motif involved in both nucleic acid-binding and protein–protein interactions (Theobald et al. 2003; Raynard et al. 2008). Proteins with OB-fold domains have been implicated in DNA repair, DNA replication, DNA recombination, transcription, translation, cold shock response, and telomere maintenance. The recently discovered BLM-interacting protein, RMI1, also contains OB-fold domain that apparently mediates the interaction of RMI1 with Topo IIIα and BLM (Raynard et al. 2008). Additional examples of OB-fold-containing proteins include the single-strand-binding protein RPA and the DNA repair protein and tumor suppressor BRCA2. OB-fold domains from different proteins share the characteristic feature of five β-sheets (Theobald et al. 2003). All five β-sheets are present in the putative OB-fold domain of RMI2 as predicted using the program PSIPRED (Supplemental Fig. 1). We did not find any RMI2 ortholog in lower organisms like Caenorhabiditis elegans or yeast based on protein sequence alignment analysis.
RMI2 is required for the stability of Topo IIIα and RMI1
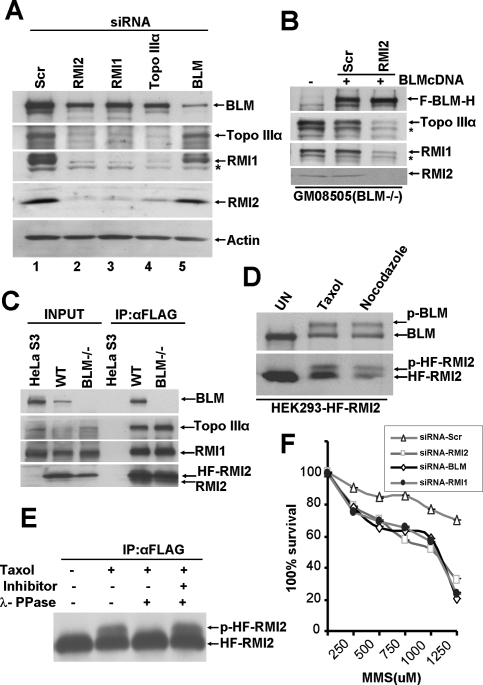
The absence of a critical subunit of a multicomponent protein complex often destabilizes the complex (Yin et al. 2005). As shown in Figure 2A, upon depletion of RMI2 in HeLa cells by siRNA treatment, a decrease in Topo IIIα and RMI1 protein level was observed (Fig. 2A, lane 2), although the level of BLM protein remained little affected. Conversely, depletion of Topo IIIα or RMI1 by siRNA treatment reduced the level of RMI2 (Fig. 2A, lanes 3,4). The results thus reveal an interdependence of RMI2, RMI1, and Topo IIIα for their stability in cells. On the other hand, depletion of BLM by siRNA resulted in no significant change in RMI2, Topo IIIα, or RMI1 protein level (Fig. 2A, lane 5).
Figure 2.
RMI2 is essential for the integrity of the BTB complex. (A) Immunoblot showing levels of BTB members, RMI2, and actin in lysates from HeLa cells that were transfected with the indicated siRNA oligos for 72 h. HeLa cells transfected with a scrambled siRNA oligo were used a control. Asterisk (*) denotes a nonspecific crossreactive band detected by anti-RMI1 antibodies. (B) Immunoblot showing levels of BTB members and RMI2 in lysates from GM08505 (BLM−/−) cells (lane 1) and the same cells corrected with BLM cDNA and transfected with either the control siRNA (lane 2) or RMI2 siRNA (lane 3). (C) Immunoblot showing input (lanes 1–3) and anti-Flag M2 agarose immunoprecipitated complex (lanes 4–6) from HeLa S3 cells transduced with vector alone or GM0067 (wild-type) and GM08505 (BLM−/−) fibroblast cells transduced with HF-RMI2. (D) Immunoblot showing the emergence of a slower mobility species of BLM and RMI2 (lanes 2–3) after HEK293 cells stably expressing HF-RMI2 were treated with either taxol or nocodazole for 16 h, as compared with untreated cells. (E) Immunoblot showing the effect of λ-protein phosphatase treatment on the slower migrating form of RMI2. (F) Graph showing MMS survival curve of RMI2, BLM. and RMI1 knockdown cells. HEK293 cells were transfected with either scramble or siRNA oligos targeting either BLM, RMI1, or RMI2, and were subsequently treated with the indicated concentration of MMS. Visible colonies from 200 cells were counted after 10 d. The data represent the percent survival, as compared with untreated cells. Each experiment was independently repeated three times and representative data are shown. Each experiment was performed in triplicate and mean values are shown with standard deviations.
We also investigated the level of RMI2, Topo IIIα, and RMI1 in BLM-deficient cells (GM08505) derived from a BS patient and the same cells reconstituted with functional BLM (Fig. 2B, lanes 1,2). We observed a comparable level of RMI2, Topo IIIα, and RMI1 in both cell lines (Fig. 2B, lanes 1,2). SiRNA-mediated depletion of RMI2 in BLM-reconstituted cells also led to the destabilization of Topo IIIα and RMI1 (Fig. 2B, lane 3). These results are consistent with earlier reports suggesting that the stability of Topo IIIα is dependent on RMI1 but that Topo IIIα and RMI1 are stable without BLM (Meetei et al. 2003; Yin et al. 2005).
Since RMI2, Topo IIIα, and RMI1 levels were unchanged in BLM-depleted cells, we next investigated the role of BLM in maintaining the integrity of RMI2, Topo IIIα, and the RMI1 complex. We stably expressed HF-RMI2 in GM0067 (wild type) and GM08505 (BLM-null cells) and purified the RMI2 complex from these cells using anti-Flag M2 agarose. HF-RMI2 was able to interact with, and form a stable complex with Topo IIIα and RMI1 in these BLM-null cells (Fig. 2C). Thus, the formation of a stable RMI2, RMI1, and Topo IIIα complex can occur in the absence of BLM.
RMI2 is phosphorylated during mitosis
BLM is phosphorylated during mitosis and becomes hyperphosphorylated in the presence of microtubule destabilizing agents, such as demecolcine and nocodazole, or microtubule stabilizing agents, such as taxol (Dutertre et al. 2000; Leng et al. 2006). We found that HF-RMI2 overexpressed in HeLa cells displayed a slower gel mobility when cells were grown in the presence of taxol or nocodazole (Fig. 2D). Determination of DNA content by flow cytometry of cells treated with demecolcine and nocodazole confirmed cell arrest at the G2/M phase (data not shown). We also observed a slower moving form of endogenous RMI2 upon treatment of cells with taxol and nocodazole (Supplemental Fig. 2A). These results suggest that RMI2, like BLM, is post-translationally modified during mitosis (Fig. 2D). To determine whether the mobility shift is due to phosphorylation, HF-RMI2 was immunoprecipitated with anti-Flag M2 agarose, and the immunoprecipitate was treated with λ-phosphatase. As shown in Figure 2E, the slower migrating form of HF-RMI2 disappeared upon phosphatase treatment. Blocking phosphatase activity with phosphatase inhibitors prevented the disappearance of the slower migrating HF-RMI2 form, indicating that RMI2 is indeed phosphorylated during mitosis. Immunoprecipitation of HF-RMI2 from taxol-treated HeLa cells expressing HF-RMI2, using anti-Flag M2 agarose, brought down the phosphorylated form of BLM (Supplemental Fig. 2B), suggesting that the BLM–RMI2 complex is not affected by BLM phosphorylation.
RMI2-depleted cells show methyl methanesulfonate (MMS) sensitivity and increased chromosome breaks
BS cells and BLM−/− chicken DT40 cells show hypersensitivity to MMS (So et al. 2004). Since RMI2 is an integral component of the BLM complex, we examined the sensitivity of RMI2- or RMI1-depleted HEK293 cells to MMS (Fig. 2F). Both RMI2- and RMI1-depleted cells showed sensitivity to MMS that was comparable with BLM-depleted cells (Fig. 2F).
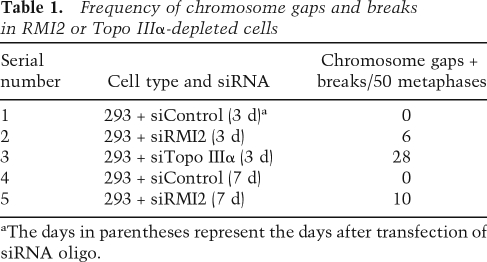
We found that depletion of either RMI2 or Topo IIIα engenders an increase in spontaneous chromosome gaps and breaks in HEK293 cells (Table 1), a result that is in congruence with the recent observation that Topo IIIα-depleted DT40 cells accumulate the same chromosome aberrations (Seki et al. 2006). Taken together, the above results further strengthen the conclusion that RMI2 is essential for BLM complex function.
Table 1.
Frequency of chromosome gaps and breaks in RMI2 or Topo IIIα-depleted cells
aThe days in parentheses represent the days after transfection of siRNA oligo.
RMI2 is required for chromatin targeting of BLM in response to replication block
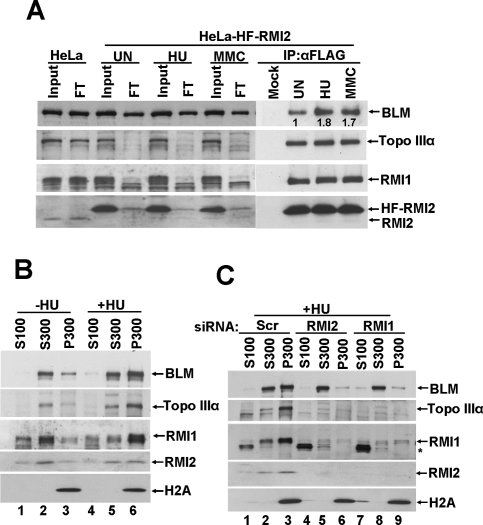
Having established that RMI2 forms a stable complex with BLM, Topo IIIα, and RMI1 in untreated cells, we investigated the interaction of RMI2 with BLM, Topo IIIα, and RMI1 after DNA damage or blockage of DNA replication. HeLa cells stably expressing HF-RMI2 were treated with either hydroxyurea (HU) or MMC for 16 h, cells were lysed, and HF-RMI2 was immunoprecipitated. Figure 3A shows that the levels of BLM, Topo IIIα, RMI1, and HF-RMI2 were not affected by DNA damage or replication stress. While a majority of the Topo IIIα and RMI1 are depleted from the flow-through fraction before and after DNA damage, there is a slight decrease of BLM in the flow-through in extracts from treated cells (Fig. 3A). These data suggest that BLM may exist in cells apart from the BTB–RMI2 complex. Importantly, there is a concomitant increase of BLM in the IP fraction using extracts derived from treated cells (Fig. 3A). These results suggest that there is an increased interaction of BLM with HF-RMI2 after DNA damage or replication block.
Figure 3.
RMI2 is essential for the chromatin targeting of BLM in response to replication fork blockage. (A) Immunoblot showing that RMI2 and BLM form a tighter complex in response to DNA damage or replication blockage. HeLa cells stably expressing HF-RMI2 were left untreated (UN), or were treated with HU (1.5 mM) or MMC (100 ng/mL) for 16 h. Cells were lysed and HF-RMI2 was immunodepleted with anti-Flag M2 agarose. Immunoblot showing the levels of BTB members and RMI2 in input and flow-through (FT) fractions and in immunoprecipitates. Cell lysate from HeLa cells transduced with vector alone was used as a control. (B,C). Immunoblot showing the levels of BTB members and RMI2 in the following cellular fractions: cyto-nucleoplasmic (S100), nucleoplasmic (S300), and chromatin (P300) (see the Material and Methods for details). (B) Comparison between untreated (lanes 1–3) and HU treated (lanes 4–6) cellular fractions. (C) Comparison of the effect of a control siRNA (scramble) (lanes 1–3), RMI2 siRNA (lanes 4–6), or RMI1 siRNA (lanes 7–9) on the distribution of BLM, Topo IIIα, RMI1, and RMI2 to various fractions. Cells were treated with HU prior to fractionation, and H2A serves as a marker for the chromatin fraction. Asterisk (*) denotes a nonspecific crossreactive band detected by anti-RMI1 antibodies.
Since RMI1 is required for BLM foci formation at the site of DNA damage or replication block in chromatin (Yin et al. 2005), we hypothesized that RMI2 is also required for the localization of BLM to chromatin. We first compared the distribution of BLM, Topo IIIα, RMI1, and RMI2 in untreated HeLa cells or following exposure to HU. Cells were lysed in low-salt-containing (100 mM) and detergent-containing buffer, yielding a soluble fraction (S100) and an insoluble fraction. The S100 fraction contained cytoplasmic and nucleoplasmic proteins, and the insoluble fraction contained proteins stably bound to nuclear structures (mainly chromatin and nuclear matrix proteins). The insoluble fraction was further extracted with a higher salt buffer (300 mM) to give a soluble fraction (S300: proteins loosely bound to nuclear structures) and an insoluble fraction (P300: proteins tightly bound to nuclear structures). Immunoblotting revealed that in untreated HeLa cells, most of the BLM and Topo IIIα was in the S300 and P300 fractions (Fig. 3B), while RMI2 and RMI1 were distributed in all fractions (S100, S300 and P300) (Fig. 3B). Interestingly, in the HU-treated samples, BLM, Topo IIIα, RMI1, and RMI2 became preferentially associated with the P300 fraction.
We next investigated the role of RMI2 in targeting the BTB complex to chromatin. We knocked down RMI2 in HeLa cells by siRNA and treated the cells with HU for 16 h The cellular fractionation was carried out as described above. As shown in Figure 3C, RMI2 depletion resulted in abrogation of chromatin targeting of BLM in response to DNA replication stress. We also established that RMI1 is similarly critical for the targeting of BLM to chromatin.
RMI2 is required for HU-induced BLM foci formation
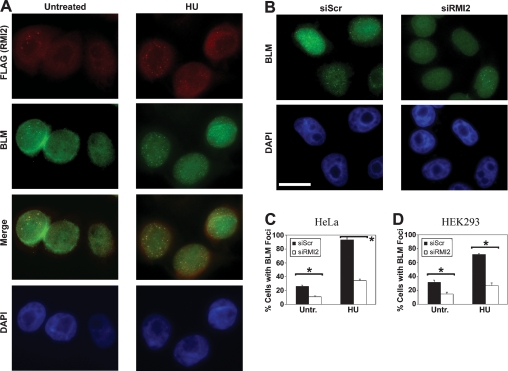
The subcellular localization of HF-RMI2 was studied with an antibody directed against the Flag epitope tag. While only a subset of untreated cells displayed RMI2 foci, the percentage of cells with RMI2 foci increased following treatment with HU for 24 h. It has been reported that BLM foci localize to stalled replication forks following treatment with HU (Davalos and Campisi 2003). As shown in Figure 4A, RMI2 foci strongly colocalized with BLM foci, both in untreated cells and following HU treatment. Since RMI2 is required for the efficient targeting of BLM to chromatin in response to replication stress (Fig. 3C), we sought to determine whether RMI2 has a role in the assembly of BLM foci. Examples of HeLa cells transfected with a siRNA directed against RMI2 or a scrambled siRNA (control) are shown in Figure 4B. In the control population, most cells displayed BLM foci following HU exposure, whereas the assembly of these foci was attenuated in most HeLa cells following RMI2 depletion (Fig. 4B,C). As summarized in Figure 4D, the assembly of HU-induced BLM foci in HEK293 cells is similarly dependent on RMI2.
Figure 4.
RMI2 regulates the assembly of BLM nuclear foci. (A) HF-RMI2 was expressed in HeLa cells. Foci assembled by this fusion protein and by BLM following exposure to 2 mM HU for 24 h were detected by immunofluorescence microscopy using anti-Flag and anti-BLM antibodies, respectively. HF-RMI2 foci were detected in red, while BLM foci were detected in green. A merged image shows colocalizing foci. The position of nuclei is indicated by a counterstain with DAPI (blue). Examples are shown for an untreated population of cells (top panels) and for cells treated with 1 mM HU for 24 h (bottom panels). (B) Examples of BLM foci in HeLa cells transfected with a scrambled control siRNA or with a siRNA that targeted RMI2 are shown. Cells were treated with 2 mM HU for 24 h prior to fixation and preparation for immunofluorescence microscopy. Bar, 10 μm. (C,D) Quantification of the assembly of BLM foci in HeLa (C) and HEK293 (D) cells. Cells were transfected with a scrambled control siRNA or with a siRNA that targeted RMI2 and were fixed for immunofluorescence microscopy at 96 h after transfection. Cells were either left untreated or were exposed to 2 mM HU for 24 h. The mean percentage of cells with five or more BLM foci from three counts of 150 or more cells each is shown with the standard deviation. Statistical significance (P < 0.01) using a Z-test for two sample proportions is indicated by an asterisk (*).
Purification of the RMI2/RMI1 complex
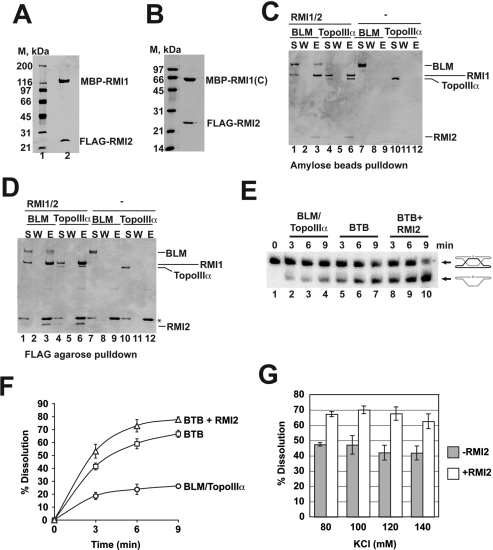
In order to elucidate the biochemical function of RMI2, we attempted to purify N-terminally Flag-tagged RMI2 from bacterial cells. However, the resulting protein appeared aggregated when analyzed by gel filtration. It also did not interact with BLM, Topo IIIα, or RMI1 in our in vitro pull-down assays (data not shown). Importantly, upon coexpression of Flag-tagged RMI2 and MPB-tagged RMI1 in bacteria, a stable complex of the two proteins could be obtained. We could purify the RMI2/RMI1 complex to a nearly homogeneous state using affinity chromatographic steps on anti-Flag and amylose matrices, as described in the Materials and Methods. The purified preparation consisted of a monodispersed, nonaggregated species, as determined by sizing in a Sephracryl S200 gel filtration column (data not shown). The resulting protein complex (Fig. 5A) appears to harbor stoichiometric amounts of RMI1 and RMI2.
Figure 5.
RMI2 stimulates dHJ dissolution activity of the BTB complex. (A) Purified RMI2/RMI1 complex, 3 μg, was analyzed by Coomassie Blue-stained SDS-PAGE. (B) Purified RMI2/RMI1-(C) complex, 3 μg, was resolved by SDS-PAGE and stained with Coomassie Blue. (C) BLM (5 μg) or Topo IIIα (5 μg) was incubated with or without RMI2/RMI1 complex (5 μg), and protein complexes were captured on amylose resin, which was washed and treated with SDS to elute bound proteins. The supernatant (S) that contained unbound proteins, wash (W), and SDS eluate (E) were analyzed by SDS-PAGE. (D) Experiment performed as in C, but protein complexes were captured on anti-Flag agarose. (*) IgG light chain dissociated from anti-Flag M2 agarose. (E) Time course of dHJ dissolution by combinations of BLM–Topo IIIα, RMI1, and RMI2. (BTB) BLM, Topo IIIα, and RMI1 combined. (F) Time course of dHJ dissolution by combinations of BLM–Topo IIIα, RMI1, and RMI2 from five independent experiments presented in the graph. The average values ± SEM. (G) dHJ dissolution by the BTB complex with or without RMI2 as a function of KCl concentration. The histogram shows the average levels of dissolution ± SEM. from four independent experiments.
We previously found that the N-terminal portion of RMI1 (residues 1–211) was responsible for binding both BLM and TopoIIIα. Moreover, the RMI1-N fragment stimulates the dHJ dissolution reaction as well as the full-length protein (Raynard et al. 2008). In order to determine which portion of RMI1 harbors the RMI2 interaction domain, we expressed the RMI1-N, RMI1-M (residues 212–424) and RMI1-C (residues 425–625) fragments (Raynard et al. 2008) in Escherichia coli, either alone or with Flag-tagged RMI2. The cell lysates containing these proteins were incubated with anti-Flag agarose and the eluted proteins analyzed by SDS-PAGE and Coomassie Blue staining. As expected, none of the RMI1 fragments was retained on the anti-Flag beads when expressed alone (Supplemental Fig. 3A). Upon coexpression with Flag-RMI2, a near stoichiometric amount of the RMI1 C-terminal fragment, but little if any of the RMI1 N or M fragment, became associated with RMI2 on the anti-Flag beads (Supplemental Fig. 3B). We could purify the RMI1-C/RMI2 complexwith the procedure developed for the RMI1/RMI2 complex (Fig. 5B). This finding indicates that RMI1 interacts with BLM, TopoIIIα, and RMI2 through different epitopes.
RMI2/RMI1 complex interacts with BLM and Topo IIIα in vitro
We used affinity pull-down assays to confirm that our purified RMI2/RMI1 complex interacts with BLM and Topo IIIα. When amylose resin was used to pull down the RMI2–RMI1 complex through the maltose-binding protein (MBP) tag on RMI1, purified BLM and Topo IIIα were retained on the affinity matrix (Fig. 5C, lanes 3,6). Similar results were obtained when anti-Flag agarose was used to pull down the RMI2–RMI1 complex through the Flag tag on RMI2 (Fig. 5D, lanes 3,6). Neither BLM nor Topo IIIα bound to amylose resin or anti-Flag agarose in the absence of the RMI2–RMI1 complex (Fig. 5C,D, lanes 9,12).
Enhancement of dHJ dissolution by RMI2
We wished to determine whether RMI2 affects the dHJ dissolution reaction mediated by the BTB complex. For this, a radiolabeled dHJ substrate, constructed as described (Fu et al. 1994; Wu and Hickson 2003; Raynard et al. 2006), was incubated with the combination of BLM, Topo IIIα, and either RMI1 or RMI2–RMI1, and the dissolution products were resolved in a polyacrylamide gel under denaturing conditions and then visualized by phosphorimaging analysis (Wu and Hickson 2003; Raynard et al. 2006). As expected, time course experiments showed an enhancement of dHJ dissolution by RMI1, with ∼40% and 65% of dissolution in 3 and 9 min, respectively, compared with 18% and 24% by BLM–Topo IIIα in the same time frame (Fig. 5E,F). Importantly, inclusion of RMI2 caused a further increase in dHJ dissolution catalyzed by the BTB complex, with around ∼52% and >75% of the dHJ substrate resolved at 3 and 9 min, respectively (Fig. 5E,F). The same level of stimulation of dHJ dissolution by RMI2 was observed at higher KCl concentrations (Fig. 5G).
RMI2 mutants deficient in BTB complex assembly
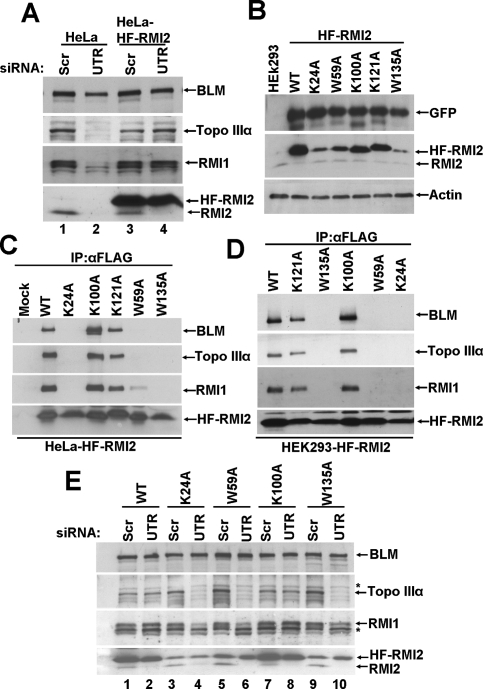
A highly conserved lysine residue of RMI1 (K166A) has been shown to be required for the interaction of RMI1 with Topo IIIα (Raynard et al. 2008). In order to map the residues that are important for RMI2 function and interaction with BTB complex, we used site-directed mutagenesis to change conserved residues in RMI2 (Supplemental Fig. 1). Specifically, we substituted the following highly conserved residues in HF-RMI2 with alanine: lys-24 (K24A), trp-59 (W59A), lys-100 (K100A), lys-121 (K121A), and trp-135 (W135A). Since a RMI2 knockout cell line is not available, we established a siRNA-based complementation system using HeLa cells that specifically knock down endogenous RMI2 but permit the expression of exogenous RMI2. HeLa cells were transduced with retroviruses expressing wild type or one of the RMI2 variants and sorted for EGFP-positive cells by FACS. Then, the cells were transfected with siRNA oligos targeting the 3′ untranslated region of RMI2 (siRMI2-3′UTR). Since the exogenous RMI2 construct lacks the 3′UTR, this siRNA should deplete only the endogenous RMI2. As shown in Figure 6A, transfection of HeLa cells with siRMI2-3′UTR knocked down endogenous RMI2, thereby destabilizing Topo IIIα and RMI1. As expected, siRMI2-3′UTR did not affect the level of ectopically expressed HF-RMI2, and both Topo IIIα and RMI1 were protected from degradation by HF-RMI2.
Figure 6.
Point mutations in the conserved residues of RMI2 that bind inefficiently to the BTB complex are unstable. (A, lanes 2,4) Immunoblot showing that siRMI2-3′UTR specifically depletes endogenous RMI2 and not the exogenously expressed HF-RMI2. (Lanes 1,3) Scrambled oligos were used as a control. (B) Immunoblot showing that RMI2 mutants K24A, W59A, and W135A were unstable, as compared with wild-type (WT) RMI2 or the K100A or K121A mutants. GFP was used as an internal control to show that mRNA expression is comparable in all the cells stably expressing HF-RMI2 variants. Immunoprecipitation coupled with immunoblotting to assay for the binding of the various variants to the BTB complex in HeLa cells (C) and HEK293 cells (D). (E) Immunoblot showing the levels of BTB members in HeLa cells depleted of the endogenous RMI2. Note that when the endogenous RMI2 was knocked down using siRMI2-3′UTR, HF-RMI2 mutants (K24A, W59A, and W135A), which could not bind to BTB complex, were unable to protect Topo IIIα and RMI1 from degradation (lanes 4,6,10), while wild type or the K100A could rescue Topo IIIα and RMI1 from degradation (lanes 2,8). Asterisk (*) denotes nonspecific crossreactive bands detected by anti-Topo IIIα and anti-RMI1 antibodies.
Immunoblot analysis confirmed the expression of the RMI2 mutants, with the K24A, W59A, and W135A, being present at a lower level (Fig. 6B). Since the amount of EGFP coexpressed with the RMI2 mutants from the same bicistronic transcript appeared to be the same in each case, we attributed the lower steady level of some of the RMI2 mutants to reduced protein stability (Fig. 6B). We then tested the RMI2 variants for their ability to associate with BLM, Topo IIIα, and RMI1 in HeLa or HEK293 cells by immunoprecipitation. As shown in Figure 6, C and D, the RMI2-K100A and RMI2-K121A variants were proficient in interaction with BLM, Topo IIIα, and RMI1. Interestingly, RMI2-K24A, RMI2-W59A and RMI2-W135A variants were unable to interact with the other BTB complex components. Next, we examined whether variants that were deficient in binding the other BTB components could protect RMI1 and Topo IIIα against proteolysis in the absence of endogenous RMI2. We used the siRNA-based complementation system described above to address this question. As shown in Figure 6E, unlike wild-type RMI2 and the K100A variant, the K24A, W59A, and W135A mutants did not prevent the degradation of RMI1 and Topo IIIα. However, we cannot rule out the possibility that the lack of association of the K24A, W59A, and W135A mutants with the BTB complex stems from their reduced steady state level in cells rather than an inability to interact with the BTB complex.
Discussion
Cancer predisposition is a major characteristic of BS, which arises likely because of the elevated genomic instability in the cells of patients. The BLM protein has been shown to interact with many other proteins involved in DNA damage repair and response (Brosh et al. 2000; Wang et al. 2000, 2001; Langland et al. 2001; Meetei et al. 2003; Yin et al. 2005), and consistent with this, BS cells are hypersensitive to DNA damaging agents, such as MMS (German and Ellis 2002). This phenotype and the increased rate of SCEs in BS cells (German 1969; German et al. 1977b) are consistent with the notion that BLM mediates its genome preservation function via influencing DNA damage repair. In this regard, a thorough understanding of the partner proteins of BLM is essential for deciphering the mechanism of the BLM-dependent genome maintenance pathway.
In this study, we used an affinity purification approach (Ling et al. 2007) to isolate a BLM-containing protein complex that harbors a novel protein of 18 kDa molecular mass, which we refer to as RMI2. Previous work has shown that BLM forms a complex, termed the BTB complex, with Topo IIIα and RMI1 (Yin et al. 2005; Raynard et al. 2006; Wu et al. 2006). Here we presented extensive biochemical and cytological evidence that RMI2 is also a critical component of the BTB complex and, as discussed further below, influences the function of the BTB complex in various ways.
RMI2 is a novel component of the BTB complex
Our studies revealed remarkable interdependence among Topo IIIα, RMI2, and RMI1, such that depletion of any one of these proteins leads to the destabilization of the others. Interestingly, the cellular level of these proteins is not affected by the BLM status, and their association occurs in the absence of BLM. Furtheremore, we found that RMI2 colocalizes with BLM in nuclear foci. The results from our cytological and cell extract fraction experiments showed that the complex of Topo IIIα, RMI2, and RMI1 is required for the efficient targeting of BLM to chromatin and to nuclear foci. These nuclear foci form both spontaneously and in response to replication stress induced by HU. The observation that there is an increased interaction of BLM with the Topo IIIα–RMI1–RMI2 complex in chromatin in cells treated with a replication blocker or DNA interstrand cross-linker strengthens the hypothesis that BLM function depends upon the Topo IIIα–RMI1–RMI2 complex.
The other proteins that were isolated with the RMI2 complex as determined by MS analysis and clearly visible on the silver-stained SDS-PAGE gel are BLAP300, FANCM, and BLAP15. These proteins may regulate the function of the BTB–RMI2 complex. Some other proteins were not visible as a clear band on the gel, but they were identified in the MS analysis of the purified RMI2-associated complex. These proteins included MSH2, MSH6, and RFC, which are part of the BASC complex (Wang et al. 2000), as well as all the three subunits of RPA. Understanding the functional interaction of these proteins with the BTB–RMI2 complex will be an important focus of future studies. The presence of FANCM and its partner FAAP24 in the RMI2 complex suggests that RMI2 is also a component of the BRAFT complex that contains BLM and components of the Fanconi anemia core complex (Meetei et al. 2003).
Biochemical functions of the RMI2 protein
RMI2 is unique to higher eukaryotes and contains a putative OB-fold domain. Even though OB-folds are generally assumed to be DNA-binding modules, we previously provided evidence that the OB-fold located within the N-terminal region of the RMI1 protein is not responsible for DNA binding but and may in fact be involved in mediating the interaction of RMI1 with BLM and Topo IIIα. In this study, by mutating highly conserved residues within the putative OB-fold of RMI2, three mutants—K24A, W59A and W135A—that fail to associate with the BTB complex, have been identified. It thus seems likely that the RMI2 OB-fold is involved in protein–protein interactions with RMI1 and possibly Topo IIIα. Future studies will address whether RMI2 has DNA-binding activity and the relevance of its putative OB-fold in such an activity.
We provided evidence that RMI2 enhances the dHJ dissolvase activity of the BTB complex. Based on our previous finding that complex formation with Topo IIIα as being indispensable for the ability of RMI1 to function as a stimulatory factor of the dHJ dissolution reaction, it seems reasonable to suggest that the dHJ dissolvase stimulatory attribute of RMI2 is likewise reliant on its interactions with RMI1 and possibly with Topo IIIα as well.
Like BLM, RMI2 also undergoes mitotic phosphorylation. It will be interesting to determine whether phosphorylation of BLM and RMI2 is mediated by the same protein kinase(s) and to ascertain the functional significance of phosphorylation. In this regard, since it has been demonstrated previously that ATM, ATR, and MPS1 phosphorylate BLM (Davies et al. 2004; Leng et al. 2006), it will be important to test the relevance of these kinases in RMI2 phosphorylation.
The RMI2-binding domain in RMI1
We showed previously that RMI1 interacts with BLM and Topo IIIα through its N-terminal region, likely its OB-fold (Raynard et al. 2008). In contrast, we demonstrated that RMI1 interacts with RMI2 through its C-terminal region. Comparison of the RMI1 protein and its presumptive Saccharomyces cerevisiae ortholog Rmi1 protein shows that the middle and C-terminal of RMI1 are absent from Rmi1 (Raynard et al. 2008). It has been suggested that this region has evolved in higher eukaryotes to perform specific cellular functions and may mediate interaction with other DNA repair proteins (Raynard et al. 2008). The observations that RMI2 is absent in yeast and that the RMI1 C terminus mediates RMI2 interaction is certainly in congruence with this particular premise.
Closing remarks
There is compelling evidence linking the BLM protein to tumor suppression. First, BS patients exhibit a marked predisposition to various cancers (German et al. 1977a; German 1997). Second, mutation of BLM is found in a variety of tumors in the general population (Gruber et al. 2002). Finally, heterozygosity for BLM increases the incidence of cancer in mice (Goss et al. 2002). Given that depletion of RMI2 results in DNA damage sensitivity and chromosomal instability in the form of elevated levels of chromosomal gaps and breaks, it will be particularly interesting to determine whether RMI2 and the other BTB components, including RMI1 and Topo IIIα, also function as tumor suppressors.
Materials and methods
Cell culture
HeLa, HT1080, HEK293, and U2OS cells were cultured in DMEM medium (Invitrogen) supplemented with 10% FBS (Atlanta Biologicals) and 1× penicillin and streptomycin solution (Invitrogen) in a humidified atmosphere of 5% CO2 at 37°C.
Chemicals
MMS was purchased from Sigma. A stock solution was diluted to 2000 μM with DMSO. Nocodazole and taxol (Sigma) were resuspended in DMSO to a stock concentration of 1 mg/mL and 10 mM, respectively. HU (Sigma) was resuspended in water, to a stock concentration of 1 M. Mitomycin C (MMC; Sigma) was dissolved in 70% ethanol (Sigma) to a stock concentration of 250 ng/μL.
Protein knockdown by siRNA treatment
All siRNA oligos were purchased from Dharmacon. For the knockdown of RMI2, we designed an siRNA in the 3′UTR (3′UTR1C: 5′-UGUUGGAACUGUCGUUAAAUU-3′), and we also used ON-TARGETplus SMARTpool siRNA (catalog no. L-015684-01, human C16orf75, NM_152308). The sequence of the siRNA for RMI1 is described in Yin et al (2005). ON-TARGETplus SMARTpool siRNAs were also used for the knockdown of BLM (catalog no. L-007287-00, human BLM, NM_000057) and Topo IIIα (catalog no. L-005279-00, human Topo IIIα, NM_004618). A nonspecific control siRNA (catalog no. D-001210-01) was used in all experiments. Cells were transfected with siRNA using lipofectamine2000 for 5 h in reduced serum OptiMEM medium, as recommended by the manufacturer (Invitrogen). After 5 h, OptiMEM was removed and replaced by complete DMEM medium. Cells were harvested 4 d post-transfection and analyzed by immunoblotting.
Computational analysis
Different orthologs of RMI2 were identified by searching protein sequence databases available at NCBI using the BLAST program. Multiple sequence analysis of the various orthologs of RMI2 was carried out using the multiple sequence alignment program ClustalW 1.8 available at the BCM search launcher (http://searchlauncher.bcm.tmc.edu). Shading of the multiple alignments was carried out using the BOXSHADE 3.21 program (EMBnet server; http://www.ch.embnet.org/software/BOX_form.html). Putative domains were identified by searching the InterPro database using the InterProScan tool (http://www.ebi.ac.uk/Tools/InterProScan). Secondary structure (β-sheets) characteristic of OB-fold domains, was identified using PSIPRED, a highly accurate method for protein secondary structure prediction (http://bioinf.cs.ucl.ac.uk/psipred).
Antibodies
Two rabbit RMI2 polyclonal antibodies were raised against fusion proteins containing full-length RMI2 with either MBP (Antibody A) or glutathione-S-transferase (GST) (Antibody B) and affinity-purified. These fusion proteins were expressed and purified from E. coli in accordance with the manufacturer’s protocols. A polyclonal antibody against BLM (69D) has been described elsewhere (Meetei et al. 2003), and Topo IIIα and RMI1 antibodies were described elsewhere (Yin et al. 2005). A goat polyclonal anti-β-actin antibody and a rabbit anti-GFP antibody were from Santa Cruz Biotechnologies. Anti-histone H2A antibody was purchased from Millipore.
Construction of double-tagged BLM and RMI2 plasmids and retroviruses
The pMIEG3 retroviral vector described in this study is a kind gift from Dr. David Williams (Cincinnati Children’s Hospital, Cincinnati, OH), and has been described earlier (Chandra et al. 2005; Ling et al. 2007). In order to generate a double-tagged BLM construct, we obtained a BLM cDNA tagged with a (His)6 affinity epitope at its C terminus (Raynard et al. 2006). N-terminal Flag was introduced by site-directed mutagenesis using PCR and cloned into pMIEG3 vector to generate pMIEG3-F-BLM-H. Human RMI2 cDNA was obtained from Open Biosystems. RMI2 ORF was PCR-amplified with high-fidelity pfu Polymerase (Stratagene) with an N-terminal (His)6-Flag tag. This product was cloned into EcoR1 and Xho1 sites of the pMIEG3 vector to generate pMIEG3-HF-RMI2. Next, amphotropic retroviruses were created and used to infect the target cells. To prepare transient virus stocks, 1.5 × 106 293T cells were plated in 10-cm dishes. The next day, cells were cotransfected using lipofectamine2000 with the retroviral expression vectors as described above, together with the appropriate helper plasmid (gag-pol and RD114). The medium was changed 5 h post-transfection, and retrovirus-containing medium was harvested in 12-h increments at 24 h post-transfection.
Generation of stable cell lines
Cells were seeded in six-well plates at the density of 5 × 104 cells per well in 3 mL of complete medium. The next day, cells were transduced in the presence of 8 ng/mL polybrene (Sigma Aldrich). The plates were then spun at 1000g for 1 h, transferred to a humidified incubator (5% CO2), and cultured for 17 hat 37°C. Cells were washed twice to remove polybrene and resuspended in complete DMEM medium (GIBCO-BRL). After culture for 48–72 h, the EGFP-positive cells were sorted using a Becton Dickinson FACS Vantage SE instrument (Becton Dickinson Immunocytometry Systems).
Purification of BLM and RMI2 complexes
BLM and RMI2 complexes were isolated from HT1080-F-BLM-H and HeLa S3-HF-RMI2 nuclear extracts, respectively, by using a modified two-step affinity chromatography protocol described previously (Ling et al. 2007). Briefly, 50 150-mm-diameter plates of cells expressing F-BLM-H or HF-RMI2 were grown to 85%–95% confluency. Cells were washed with phosphate-buffered saline and collected as a pellet. Nuclear extracts were prepared as described (Meetei et al. 2003). The first purification step was performed with anti-Flag M2 agarose beads (Sigma). Multiple tubes containing 1.5 mL each of the nuclear extracts were incubated with 25 μL anti-Flag M2 agarose beads overnight, washed four times with 250-lysis buffer (20 mM HEPES at pH 7.9, 250 mM NaCl, 0.2 DTT, 0.5 mM sodium orthovanadate, 50 mM sodium fluoride, protease inhibitor cocktail, 2 mM PMSF, 1% Triton X-100, 10% glycerol) with 10 min rotation at 4°C, and eluted with 3XFlag peptide (Sigma) for 1 h on ice. The second purification step was performed by incubating the eluate of the first step with 30 μL Talon metal affinity Resin (BD) for 3 h at 4°C in the presence of 3 mM imidazole. Then the beads were washed four times with 250-lysis buffer containing 10 mM imidazole, and eluted with 50 mM EDTA. The purified protein complex was resolved on an 8%–16% SDS-PAGE gel and stained by either SilverQuest silver or colloidal blue staining kit according to the manufacturer’s instructions (Invitrogen). Protein identification by MS was carried out by using either excised gel pieces or the entire gel, as described previously (Meetei et al. 2003). Actual MS data are not shown but are available upon request.
For immunodepletion, the total cell lysate of HeLa S3 cells stably expressing HF-RMI2 was incubated with 50 μL of anti-Flag M2 agarose beads overnight, then washed four times with 250-lysis buffer and eluted with 3XFlag peptide (Sigma) for 1 h on ice. CoIP experiments using anti BLAP-18 and anti-BLM antibodies were carried out as described (Meetei et al. 2003).
Protein phosphatase treatment
We treated HeLa cells expressing HF-RMI2 with taxol or nocodazol for 16 h or left the cells untreated. Cells were prepared with lysis buffer, then HF-RMI2 was immunoprecipitated with anti-M2 agarose. Flag immunoprecipitates were incubated at 30°C with 400 U of λ-protein phosphatase, either in the presence or absence of phosphatase inhibitors, for 60 min prior to immunoblot analysis according to the manufacturer's instructions (New England Biolabs). For phosphatase inhibition, a combination of 10 mM sodium orthovanadate and 50 mM sodium fluoride was added to the protein samples prior to the addition of the λ-protein phosphatase.
Preparation of cellular fractions
Cells were treated with HU (1.5 mM) for 16 h Then the cells were trypsinized and washed with cold phosphate-buffered saline (PBS). The pellets were resuspended in cold buffer A (10 mM PIPES at pH 7.0, 100 mM NaCl, 1 mM EGTA, 300 mM sucrose, 0.5 mM sodium orthovanadate, 50 mM sodium fluoride, protease inhibitor cocktail, 1 mM PMSF, 0.5% Triton X-100) at five times the volume of the cell pellet and were incubated for 2 min at room temperature to permeabilize the cells. The suspension was then centrifuged at 200g for 3 min, and the supernatant (S100; detergent-soluble nuclear proteins) was collected. Pellets were washed with cold buffer A. The pellet was then extracted with buffer B (buffer A with the NaCl concentration increased to 300 mM) for 5 min. The supernatant was collected (S300). The pellet was washed once with buffer A and then boiled with 2× SDS buffer (P300).
Purification of the RMI2/RMI1 complex
All purification steps were conducted at 0 to 4°C, and the elution of the RMI2/RMI1 protein complex was monitored by 12% SDS-PAGE and Coomassie Blue staining. For purification, E. coli BL21:DE3 Rosetta cells were transformed with pRSF-Duet vector (Invitrogen) carrying Flag-RMI2 cDNA and pMAL-p2X vector (New England Biolabs) carrying MBP-RMI1-(His)6 cDNA. Cells were grown at 37°C to OD600 = 0.8, and the expression of proteins was induced by the addition of 0.1 mM IPTG at 16°C for 16 h. Bacterial pellets (30 g) were suspended in 100 mL of Buffer K (20 mM KH2PO4, 10% glycerol, 0.5 mM EDTA, 0.01% IGEPAL, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM benzamidine, and 5 μg/mL each of aprotinin, chymostatin, leupeptin, pepstatin) containing 150 mM KCl. The cells were disrupted by sonication, and the crude lysate was subjected to ultracentrifugation (100,000g for 90 min). The clarified lysate was incubated with 5 mL of anti-Flag M2 agarose (Sigma) for 4 h with constant agitation. The matrix was poured into a column with an internal diameter of 1 cm and was washed with 100 mL of Buffer K with 150 mM KCl. The bound proteins were eluted in 5 mL of PBS containing 250 ng/μL of Flag peptide (Sigma). The elutions were repeated five times, 30 min each with constant agitation. Fractions containing RMI2/RMI1 were incubated with 3 mL Amylose beads (New England Biolabs) for 3 h with constant agitation. The matrix was washed as above, and proteins were eluted with 15 mL of 10 mM maltose in Buffer K with 150 mM KCl. Fractions containing RMI2/RMI1 complexes were combined and concentrated in an Amicon Ultra microconcentrator (Millipore) to 600 μL and then subjected to gel filtration in a 35 mL Sephacryl S200 column in Buffer K with 150 mM KCl. The size of collected fractions was 1 mL. Fractions containing the RMI2/RMI1 peak were combined and concentrated as before to 6 mg/mL (RMI1) and stored in small portions at −80°C. The concentration of the proteins was determined by densitometric comparison of multiple loadings of the purified proteins against known amounts of bovine serum albumin in a Coomassie Blue-stained polyacrylamide gel. The identities of the proteins were determined by Western blotting with α-Flag (Sigma) and α-MBP (New England Biolabs) antibodies.
Purification of RMI2/RMI1(C) complex
MBP-RMI1(N)-(His)6, MBP-RMI1(M)-(His)6, and MBP-RMI1(C)-(His)6 (Raynard et al. 2008) were each expressed alone or coexpressed with Flag-RMI2 in the E. coli cells and the cell lysates were prepared and incubated with anti-Flag M2 agarose as above. A small portion of anti-Flag M2 agarose (25 μL bead bed) was taken from each sample and bound proteins were eluted with 2% SDS and analyzed by 12% SDS-PAGE and Coomassie Blue staining. Only MBP-RMI1(C)-(His)6 was found in the complex with RMI2 and the complex was purified as above.
Purification of RMI1 and other proteins
MBP-RMI1-(His)6 was expressed from a modified pMAL-p2X vector (New England Biolabs), as described above, and purified using our published procedure devised for GST-RMI1-(His)6 (Raynard et al. 2006) with the following modifications: amylose resin (New England Biolabs) was substituted for glutathione-Sepharose (GE Healthcare) and elution of proteins bound to the resin was done with 10 mM maltose in K buffer containing 150 mM KCl. Recombinant (His)6-tagged BLM and (His)6-tagged Topo IIIα were expressed in yeast and E. coli, respectively, and purified following our published procedures (Bussen et al. 2007). The concentration of these proteins was determined as above.
DNA substrates
The dHJ substrate was prepared by hybridizing and ligating two partially complementary oligonucleotides, as described (Fu et al. 1994; Wu and Hickson 2003; Raynard et al. 2006).
dHJ dissolution assay
BLM (7.5 nM) was incubated with Topo IIIα (120 nM) and RMI1 or RMI1/RMI2 (50 nM) for 10 min on ice in 24 μL of reaction buffer (50 mM Tris-HCl at pH 7.8, 1 mM DTT, 0.8 mM MgCl2, 200 μg/mL bovine serum albumin, 2 mM ATP, 80 mM KCl [or amount as indicated], an ATP regenerating system consisting of 10 mM creatine phosphate and 50 μg/mL creatine kinase), followed by the addition of dHJ substrate (1.2 nM) in 1 μL of water. The reactions were incubated at 37°C, and after 5 min, or time as indicated, 8-μL aliquots of the reaction mixtures were removed and mixed with 2 μL of 1% SDS and 0.5 μL of proteinase K (10 μg/μL stock), followed by a 3-min incubation at 37°C. The deproteinized reactions were mixed with an equal volume of sample loading buffer (20 mM Tris-HCl at pH 7.5, 50% glycerol, and 0.08% Orange G) containing 50% urea, incubated at 95°C for 3 min, and resolved in an 8% polyacrylamide gel containing 20% formamide and 8 M urea in TAE buffer at 55°C.
Affinity pull-down assays
For pull-down assays, 5 μg of BLM or Topo IIIα were incubated in 30 μL of Buffer K containing 200 mM KCl with or without RMI2/RMI1 complex (5 μg of RMI1) for 30 min at 4°C. The reactions were mixed with 15 μL of Amylose beads (which recognize the MBP tag at the N terminus of RMI1) (Fig 5B) or 20 μL Flag agarose (which binds Flag epitope at the N terminus of RMI2) (Fig 5C) for 30 min at 4°C. After washing the beads twice with 200 μL of the same buffer, bound proteins were eluted with 25 μL of 2% SDS. Fifteen percent of total supernatant (S) and elution (E) fractions, and 2% of total wash (W) fraction were analyzed by 4%–15% gradient SDS-PAGE and Coomassie Blue staining.
Immunofluorescence microscopy
Adherent cells were grown on 12-mm-diameter glass coverslips, which were coated with poly-D-lysine, for a minimum of 24 h prior to treatment or fixation. Cells were left untreated or were exposed to 2 mM HU for 24 h. For depletion, RMI2 cells were transfected with siRMI2 or the scrambled control and treated with HU beginning at 72 h after transfection.
Cells were fixed with 2% paraformaldelyde for 20 min at 37°C. Cells were then washed with PBS, permeabilized 3 min with 0.2% Triton X-100 in PBS, and washed again with PBS. Cells were incubated 1 h at 37°C with primary antibodies diluted in antibody buffer (PBS/3% bovine serum albumin/0.05% Tween 20/0.04% sodium azide). Primary antibodies included mouse anti-Flag (M2; 1:100; Sigma) and goat anti-BLM (1:200; Santa Cruz Biotechnologies) antibodies. After incubation with primary antibodies, cells were washed with PBS (3×, 5 min each) and then incubated with the appropriate secondary antibody (Jackson Immunoresearch; 1:500) for 30 min at 37°C. Secondary antibodies included FITC-conjugated donkey anti-goat IgG and Rhodamine B-conjugated donkey anti-mouse IgG diluted in antibody buffer. Cells were washed three times with PBS and mounted over Vectashield containing DAPI (Vector Laboratories) to stain DNA, sealed with nail polish, and stored at −20°C pending analysis.
Labeled cells were observed with a Zeiss Axiovert 200M microscope, and images were collected with a Hamamatsu Camera using Openlab software (Improvision). Images were processed into figures using Photoshop (Adobe).
MMS sensitivity assay
HEK293 cells were transfected with siRNA targeting RMI2, RMI1, BLM, or control oligos, as described above. At 3 d post-transfection, 200 cells were seeded per 10-cm dish containing the indicated concentration of MMS in DMEM medium. After 10 d cells were fixed, stained, and visible colonies were counted.
Detection of chromosome gaps/breaks
Analysis of chromosome gaps/breaks was carried out at the cytogenetic facility at Cincinnati Children’s Hospital, using a standard protocol. Three days post-transfection with siRNA, HEK 293 cells were cultured in colcemid for 2.5 h. The cells were harvested and treated with hypotonic solution containing 0.35% Sodium citrate and 0.28% KCl for 30 min at room temperature. Cells were then fixed with methanol/acetic acid (3:1) for 60 min. The cell suspension was dropped onto ice-cold wet glass slides and air dried. The cells were stained with Giemsa solution for 1.5 min and examined by light microscopy.
Acknowledgments
We thank the Mass Spectrometry and Proteomics Facility at Ohio State University and the Taplin Biological Mass Spectrometry Facility at Harvard Medical School for MS analysis. We thank Drs. Weidong Wang and David A Williams for reagents. We thank Dr. Teresa Smolarek, Audra Birri, and Jenny Coffman of the Cytogenetics Laboratory at Cincinnati Children’s Hospital Medical Center for chromosome gaps/breakage analyses. We also thank Wahengbam Kebola Devi for technical assistance. This work was supported by National Institutes of Health Research Grants RO1 HL085587 (to P.R.A.), ES015632 (to P.S.), and R01 HL084082 (to A.R.M.), and an American Society of Hematology Junior Faculty Award (to A.R.M). V.B. was supported by Ruth L. Kirschtein post-doctoral fellowship F32 GM079816.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1725108.
References
- Bischof O., Galande S., Farzaneh F., Kohwi-Shigematsu T., Campisi J. Selective cleavage of BLM, the Bloom syndrome protein, during apoptotic cell death. J. Biol. Chem. 2001;276:12068–12075. doi: 10.1074/jbc.M006462200. [DOI] [PubMed] [Google Scholar]
- Brosh R.M., Li J.L., Kenny M.K., Karow J.K., Cooper M.P., Kureekattil R.P., Hickson I.D., Bohr V.A. Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J. Biol. Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- Bussen W., Raynard S., Busygina V., Singh A.K., Sung P. Holliday junction processing activity of the BLM–Topo IIIα–BLAP75 complex. J. Biol. Chem. 2007;282:31484–31492. doi: 10.1074/jbc.M706116200. [DOI] [PubMed] [Google Scholar]
- Chandra S., Levran O., Jurickova I., Maas C., Kapur R., Schindler D., Henry R., Milton K., Batish S.D., Cancelas J.A., et al. A rapid method for retrovirus-mediated identification of complementation groups in Fanconi anemia patients. Mol. Ther. 2005;12:976–984. doi: 10.1016/j.ymthe.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Davalos A.R., Campisi J. Bloom syndrome cells undergo p53-dependent apoptosis and delayed assembly of BRCA1 and NBS1 repair complexes at stalled replication forks. J. Cell Biol. 2003;162:1197–1209. doi: 10.1083/jcb.200304016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies S.L., North P.S., Dart A., Lakin N.D., Hickson I.D. Phosphorylation of the Bloom’s syndrome helicase and its role in recovery from S-phase arrest. Mol. Cell. Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S., Ababou M., Onclercq R., Delic J., Chatton B., Jaulin C., Amor-Gueret M. Cell cycle regulation of the endogenous wild type Bloom’s syndrome DNA helicase. Oncogene. 2000;19:2731–2738. doi: 10.1038/sj.onc.1203595. [DOI] [PubMed] [Google Scholar]
- Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Fu T.J., Tse-Dinh Y.C., Seeman N.C. Holliday junction crossover topology. J. Mol. Biol. 1994;236:91–105. doi: 10.1006/jmbi.1994.1121. [DOI] [PubMed] [Google Scholar]
- German J. Bloom’s syndrome. I. Genetical and clinical observations in the first twenty-seven patients. Am. J. Hum. Genet. 1969;21:196–227. [PMC free article] [PubMed] [Google Scholar]
- German J. Bloom’s syndrome. XX. The first 100 cancers. Cancer Genet. Cytogenet. 1997;93:100–106. doi: 10.1016/s0165-4608(96)00336-6. [DOI] [PubMed] [Google Scholar]
- German J., Ellis N.A. Bloom syndrome. In: Vogelstein B., Kinzler K.W., et al., editors. The genetic basis of human cancer. McGraw-Hill; New York: 2002. pp. 267–288. [Google Scholar]
- German J., Bloom D., Passarge E. Bloom’s syndrome. V. Surveillance for cancer in affected families. Clin. Genet. 1977a;12:162–168. doi: 10.1111/j.1399-0004.1977.tb00919.x. [DOI] [PubMed] [Google Scholar]
- German J., Bloom D., Passarge E., Fried K., Goodman R.M., Katzenellenbogen I., Laron Z., Legum C., Levin S., Wahrman J. Bloom’s syndrome. VI. The disorder in Israel and an estimation of the gene frequency in the Ashkenazim. Am. J. Hum. Genet. 1977b;29:553–562. [PMC free article] [PubMed] [Google Scholar]
- German J., Schonberg S., Louie E., Chaganti R.S. Bloom’s syndrome. IV. Sister-chromatid exchanges in lymphocytes. Am. J. Hum. Genet. 1977c;29:248–255. [PMC free article] [PubMed] [Google Scholar]
- Goss K.H., Risinger M.A., Kordich J.J., Sanz M.M., Straughen J.E., Slovek L.E., Capobianco A.J., German J., Boivin G.P., Groden J. Enhanced tumor formation in mice heterozygous for Blm mutation. Science. 2002;297:2051–2053. doi: 10.1126/science.1074340. [DOI] [PubMed] [Google Scholar]
- Gruber S.B., Ellis N.A., Scott K.K., Almog R., Kolachana P., Bonner J.D., Kirchhoff T., Tomsho L.P., Nafa K., Pierce H., et al. BLM heterozygosity and the risk of colorectal cancer. Science. 2002;297:2013. doi: 10.1126/science.1074399. [DOI] [PubMed] [Google Scholar]
- Hanada K., Hickson I.D. Molecular genetics of RecQ helicase disorders. Cell. Mol. Life Sci. 2007;64:2306–2322. doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Raynard S., Sehorn M.G., Lu X., Bussen W., Zheng L., Stark J.M., Barnes E.L., Chi P., Janscak P., et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes & Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmakar P., Seki M., Kanamori M., Hashiguchi K., Ohtsuki M., Murata E., Inoue E., Tada S., Lan L., Yasui A., et al. BLM is an early responder to DNA double-strand breaks. Biochem. Biophys. Res. Commun. 2006;348:62–69. doi: 10.1016/j.bbrc.2006.07.037. [DOI] [PubMed] [Google Scholar]
- Langland G., Kordich J., Creaney J., Goss K.H., Lillard-Wetherell K., Bebenek K., Kunkel T.A., Groden J. The Bloom’s syndrome protein (BLM) interacts with MLH1 but is not required for DNA mismatch repair. J. Biol. Chem. 2001;276:30031–30035. doi: 10.1074/jbc.M009664200. [DOI] [PubMed] [Google Scholar]
- Leng M., Chan D.W., Luo H., Zhu C., Qin J., Wang Y. MPS1-dependent mitotic BLM phosphorylation is important for chromosome stability. Proc. Natl. Acad. Sci. 2006;103:11485–11490. doi: 10.1073/pnas.0601828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C., Ishiai M., Ali A.M., Medhurst A.L., Neveling K., Kalb R., Yan Z., Xue Y., Oostra A.B., Auerbach A.D., et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 2007;26:2104–2114. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei A.R., Sechi S., Wallisch M., Yang D., Young M.K., Joenje H., Hoatlin M.E., Wang W. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 2003;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynard S., Bussen W., Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIα, and BLAP75. J. Biol. Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- Raynard S., Zhao W., Bussen W., Lu L., Ding Y.Y., Busygina V., Meetei A.R., Sung P. Functional role of BLAP75 in BLM–topoisomerase IIIα-dependent holliday junction processing. J. Biol. Chem. 2008;283:15701–15708. doi: 10.1074/jbc.M802127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Nakagawa T., Seki T., Kato G., Tada S., Takahashi Y., Yoshimura A., Kobayashi T., Aoki A., Otsuki M., et al. Bloom helicase and DNA topoisomerase IIIα are involved in the dissolution of sister chromatids. Mol. Cell. Biol. 2006;26:6299–6307. doi: 10.1128/MCB.00702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So S., Adachi N., Lieber M.R., Koyama H. Genetic interactions between BLM and DNA ligase IV in human cells. J. Biol. Chem. 2004;279:55433–55442. doi: 10.1074/jbc.M409827200. [DOI] [PubMed] [Google Scholar]
- Sung P., Klein H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- Theobald D.L., Mitton-Fry R.M., Wuttke D.S. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cortez D., Yazdi P., Neff N., Elledge S.J., Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes & Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Tseng A., Ellis N.A., Spillare E.A., Linke S.P., Robles A.I., Seker H., Yang Q., Hu P., Beresten S., et al. Functional interaction of p53 and BLM DNA helicase in apoptosis. J. Biol. Chem. 2001;276:32948–32955. doi: 10.1074/jbc.M103298200. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I.D. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I.D. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- Wu L., Bachrati C.Z., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G.W., Hickson I.D. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl. Acad. Sci. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Sobeck A., Xu C., Meetei A.R., Hoatlin M., Li L., Wang W. BLAP75, an essential component of Bloom’s syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24:1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]