Abstract
Canonical microRNAs (miRNAs) require two processing steps: the first by the Microprocessor, a complex of DGCR8 and Drosha, and the second by a complex of TRBP and Dicer. dgcr8Δ/Δ mouse embryonic stem cells (mESCs) have less severe phenotypes than dicer1Δ/Δ mESCs, suggesting a physiological role for Microprocessor-independent, Dicer-dependent small RNAs. To identify these small RNAs with unusual biogenesis, we performed high-throughput sequencing from wild-type, dgcr8Δ/Δ, and dicer1Δ/Δ mESCs. Several of the resulting DGCR8-independent, Dicer-dependent RNAs were noncanonical miRNAs. These derived from mirtrons and a newly identified subclass of miRNA precursors, which appears to be the endogenous counterpart of shRNAs. Our analyses also revealed endogenous siRNAs resulting from Dicer cleavage of long hairpins, the vast majority of which originated from one genomic locus with tandem, inverted short interspersed nuclear elements (SINEs). Our results extend the known diversity of mammalian small RNA-generating pathways and show that mammalian siRNAs exist in cell types other than oocytes.
Keywords: ES cells, small RNAs, high throughput sequencing
Small RNAs that mediate RNAi-related processes are classified by their biogenesis pathways. Central to the processing of most small RNAs is the RNase III-containing enzyme, Dicer, which forms a complex with TRBP in mammals and cleaves dsRNA precursors into the characteristic ∼22-nucleotide (nt) final product (Hammond 2005; Maniataki and Mourelatos 2005). Dicer products can be classified into two main categories: siRNAs and microRNAs (miRNAs). siRNAs are generated from multiple Dicer cleavages along a long precursor dsRNA, whereas miRNAs are generated from a single Dicer cleavage of a short hairpin pre-miRNA (Bartel 2004). miRNAs require additional upstream processing to convert a longer pol II expressed pri-miRNA transcript to the short pre-miRNA hairpin. For canonical miRNAs, this processing event is performed by the Microprocessor complex, which consists of the RNase III enzyme Drosha (Lee et al. 2003) and the dsRNA-binding protein DGCR8 (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004, 2006; Landthaler et al. 2004). A subclass of pre-miRNAs, the mirtrons, bypass the Microprocessor; for these noncanonical miRNAs, the upstream processing is performed by the spliceosome and debranching enzyme, which produce a short hairpin directly suitable for Dicer cleavage without further processing (Okamura et al. 2007; Ruby et al. 2007a).
A central role for miRNAs in metazoan development is well established (Bartel 2004). Endogenous siRNAs play important roles in plants (Poethig et al. 2006), Schizosaccharomyces pombe (Verdel and Moazed 2005) and Tetrayhymena (Lee and Collins 2006). They also have been observed in metazoa including Caenorhabditis elegans (Ambros et al. 2003), Drosophila melanogaster (Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008), and mouse oocytes (Watanabe et al. 2006, 2008; Tam et al. 2008). Roles for siRNAs in metazoan development have not been clearly delineated. However, exogenous siRNAs are used regularly in the experimental setting to knock down gene function (Elbashir et al. 2001; Hannon and Rossi 2004; Dykxhoorn and Lieberman 2005). Exogenous siRNAs are often introduced using vectors expressing shRNAs driven from a pol III promoter (Lee et al. 2002; Paul et al. 2002). These shRNAs are structurally similar to pre-miRNAs and, therefore, bypass the Microprocessor and are processed directly by Dicer.
Small RNAs play a central role in embryonic stem (ES) cells (Stadler and Ruohola-Baker 2008). Deletion of Dicer in mouse ES cells (mESCs) results in severe proliferation and differentiation defects (Kanellopoulou et al. 2005; Murchison et al. 2005). Sequencing of small RNAs from both mouse and human ES cells has revealed a miRNA population distinct from those of other cells and tissues (Houbaviy et al. 2003; Suh et al. 2004; Calabrese et al. 2007; Morin et al. 2008). Notably, the mir-290 cluster is highly expressed in mESCs and is rapidly down-regulated upon differentiation (Houbaviy et al. 2003). An important role for ES cell miRNAs has been confirmed by the deletion of Dgcr8, which specifically affects the processing of canonical miRNAs (Wang et al. 2007). The Dgcr8 knockout cells show a proliferation defect with the accumulation of cells in the G1 phase as well as a failure to silence self-renewal upon differentiation. However, these defects are less severe than those reported in Dicer1 knockouts. In particular, dicer1Δ/Δ ES cells are more difficult to isolate and are thought to require a secondary genetic or epigenetic event to grow (Murchison et al. 2005). Also, Dicer1 knockout ES cells have a more complete block in differentiation (Kanellopoulou et al. 2005). Previous sequencing of small RNAs from dicer1Δ/Δ cells has suggested that such defects result primarily from the loss of canonical miRNAs (Calabrese et al. 2007), but the differences between the dgcr8Δ/Δ and dicer1Δ/Δ phenotypes suggest a physiological role for additional small RNAs. To begin to explore this possibility, we sequenced small RNA populations from wild-type, Dgcr8, and Dicer knockout mESCs, reasoning that noncanonical miRNAs and/or endogenous siRNAs would be enriched in dgcr8Δ/Δ and depleted in dicer1Δ/Δ cells. This approach uncovered a diverse population of DGCR8-independent, Dicer-dependent small RNAs in mESCs, including mirtron- and endogenous shRNA-derived miRNAs, as well as endogenous hairpin-derived siRNAs (hp-siRNAs).
Results
Classification of small RNAs based on processing dependencies
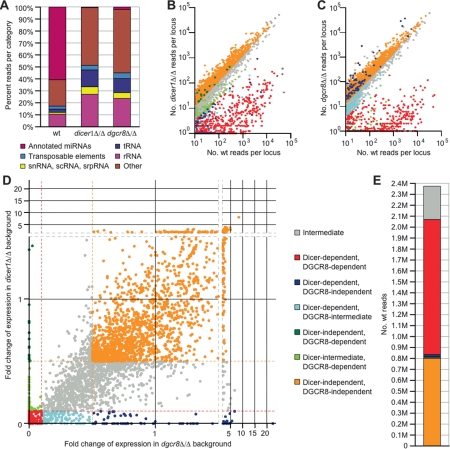
Small RNAs collected from wild-type, dgcr8Δ/Δ, and dicer1Δ/Δ mESCs were sequenced using the Illumina and 454 high-throughput sequencing technologies (Seo et al. 2004; Margulies et al. 2005). A total of 2,570,847 genome-matching sequence reads were identified from the wild-type sample, 2,925,826 from the dgcr8Δ/Δ sample, and 2,211,458 from the dicer1Δ/Δ sample. The majority of the wild-type small RNAs derived from annotated miRNA loci, and as expected, the fraction of miRNA-derived RNAs, was drastically diminished in the dgcr8Δ/Δ and dicer1Δ/Δ samples (Fig. 1A; Supplemental Table 1). Among the other noncoding RNA ncRNA) gene products, such as tRNAs, snRNAs, scRNAs, and srpRNAs, the relative proportions of sequenced degradation fragments were consistent between the three samples (Fig. 1A; Supplemental Fig. S1A). These were used for normalization between the samples (see the Materials and Methods).
Figure 1.
DGCR8 and Dicer dependence of mouse small RNAs. (A) Classification of small RNAs based on UCSC mouse genome annotations (mm8). See Supplemental Table 1 for a tabular representation of the data. (B) Read counts from the wild-type versus dicer1Δ/Δ libraries for genomic loci with at least 10 reads from the wild-type library, colored as in D. (C) Read counts from the wild-type versus dgcr8Δ/Δ libraries for genomic loci defined in B, colored as in D. (D) Dependencies of the small RNAs derived from genomic loci defined in B. Each axis indicates the following quotient: read count from mutant library/read count from wild-type library, with read counts normalized to the number of t/sn/sc/srpRNA-derived reads from that library. Ratios <0.1 defined enzyme dependence (red hashed line) and ratios >0.5 defined enzyme independence (gold hashed line). Loci were categorized in consideration of their dependencies on both Dicer and DGCR8, and are color coded according to the key. (E) The number of reads from each dependency category from D in the wild-type library. See Supplemental Table 3 for tabular representation.
To determine the enzyme dependencies for the small RNAs, 5230 independent genomic loci, to which at least 10 reads from the wild-type data set mapped, were identified and evaluated according to read number from wild-type versus mutant cells (see the Materials and Methods; Fig. 1B,C; Supplemental Table 2). These analyses revealed a large number of loci with a diminished number of reads from either one or both mutant cells. The DGCR8 and Dicer dependencies of small RNAs from each locus were then considered concurrently by plotting the read frequency fold change between each mutant versus wild type (Fig. 1D). We required at least a 10-fold decrease in the number of reads in either mutant background to classify the locus as DGCR8- or Dicer-dependent and less than a twofold decrease to classify the locus as DGCR8- or Dicer-independent. Using this approach, all loci were classified into seven categories based on their dependencies for Dicer and/or DGCR8 (Fig. 1D,E; Supplemental Table 3).
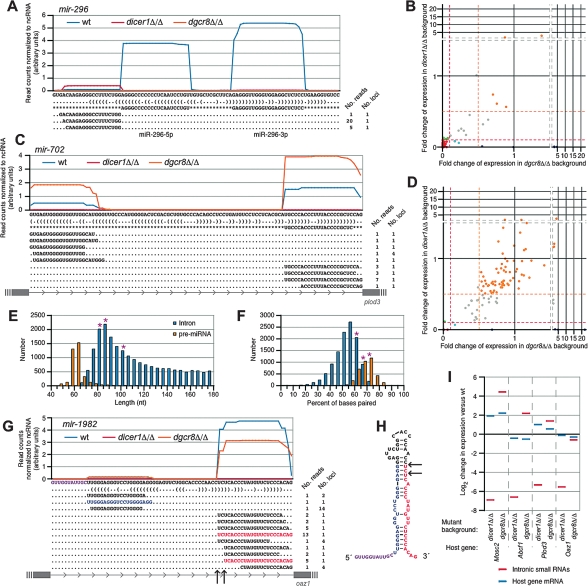
The majority of small RNA reads were both DGCR8- and Dicer-dependent (Fig. 1E; Supplemental Table 3), as expected for canonical miRNAs. Indeed, the majority of reads from this category (68%) derived from annotated miRNA hairpins (miRBase version 10.0; Griffiths-Jones et al. 2008). The strict dependency of canonical miRNA loci on Dicer and DGCR8 was illustrated by the mir-296 locus (Fig. 2A). This locus produced a large number of reads from both arms of the hairpin in wild-type, but not in either mutant background. Interestingly, additional small RNA reads mapped to the region immediately 5′ of the mir-296 pre-miRNA (Fig. 2A). These reads were enriched in the Dicer but absent in the Dgcr8 knockout background and had heterogeneous 5′ ends but homogenous 3′ ends that corresponded precisely to the Drosha cleavage site. Analogous reads observed among Drosophila high-throughput data are attributed to Drosha cleavage followed by 5′ → 3′ degradation of the 5′ leader segment (Ruby et al. 2007b), a hypothesis consistent with the DGCR8-dependent and Dicer-independent biogenesis inferred from our mutant mESC analysis. Our whole-genome analysis revealed 17 loci with sequence reads having similar enzymatic dependencies (Fig. 1D, green; Supplemental Table 2): 15 corresponding to segments immediately flanking annotated pre-miRNAs, one corresponding to a segment immediately flanking a previously unannotated pre-miRNA (mir-6691) (Supplemental Fig. S2A), and one unclear locus (Supplemental Fig. S2B).
Figure 2.
Canonical and mirtronic miRNAs. (A) The distribution of reads across the mir-296 hairpin. Y-axis indicates the read count at each nucleotide position from the indicated library, normalized to the number of t/sn/sc/srpRNA-derived reads from that library. The predicted secondary structure is shown below, along with the 5′ pre-miRNA-flanking reads from the wild-type library (see the text). For each RNA species, the number of reads that were obtained with that sequence and the number of genomic loci to which the sequence maps are indicated at the right. See Supplemental Figure S9 for PhastCons scores. (B) The Dicer and DGCR8 dependencies of annotated miRNA genes (miRBase 10.0), plotted as in Figure 1D. (C) The normalized read frequency from each library, predicted secondary structure, and wild-type reads mapping to the mirtron mir-702, presented as in A. The boundaries of host gene exons are indicated at the bottom. See Supplemental Figure S9 for PhastCons scores. (D) The DGCR8 and Dicer dependencies of all introns <500 bp, plotted as in Figure 1D. (E) The length distribution of intron (blue) and canonical pre-miRNAs (orange). The lengths of the three mirtrons described here are indicated by pink asterisks. (F) The distribution is shown for the percentage of nucleotides that are base paired in the predicted secondary structures of 60- to 120-nt introns (blue) and canonical pre-miRNAs (orange). The value for the three mirtrons described here are indicated by pink asterisks. (G) mir-1982, a tailed-mirtron, was processed from Oaz1. Shown in magenta is the 5′ 11-bp tail that must be removed prior to Dicer processing. Plotted as in C. See Supplemental Figure S9 for PhastCons scores. (H) The secondary structure of mir-1982. Arrows indicate the 5′ ends of the two most abundant Dicer products. (I) A comparison of the levels of the mirtron small RNAs (red) to those of their parent mRNAs (blue). Changes in small RNA levels were determined as the ratio of t/sn/sc/srpRNA-normalized read counts between the wild-type and mutant libraries with a single pseudocount added. Changes in mRNA expression were determined by microarrays.
The second most abundant class consisted of small RNA loci that were independent of both DGCR8 and Dicer (Fig. 1B,C; Supplemental Table 2). Many of these loci corresponded to the tRNA, snRNA, scRNA, and srpRNA genes, which were used for normalization. Analysis of the 20 loci with the most reads revealed that a majority (18 out of 20) overlapped with rRNA repeats. Reads that mapped to these loci were extremely heterogeneous, often spanning the entire locus, as expected for degradation fragments of abundant cellular RNAs. Because our study was designed to identify loci dependent on either DGCR8 or Dicer, these doubly independent loci were not considered further.
Identification of novel mouse mirtrons
Analysis of the enzymatic dependencies for annotated miRNA hairpins confirmed the requirements for DGCR8 and Dicer for most but not all expressed miRNAs (Fig. 2B; Supplemental Table 4). Reads from five annotated miRNA loci were DGCR8-independent and Dicer-dependent: mir-320, mir-344, mir-484, mir-668, and mir-702 (Fig. 2B, blue). One of these, mir-702, had miRNA and miRNA* reads flanking the edges of its host intron in a pattern reminiscent of mirtron miRNA precursors (Fig. 2C). The dependence of these reads on Dicer but not DGCR8 extended demonstration of enzymatic requirements for mirtron biogenesis beyond flies to mammals (Okamura et al. 2007; Ruby et al. 2007a), thereby confirming the proposal that mammals have mirtrons (Berezikov et al. 2007). Additional mouse mirtrons were sought by examining the dependencies of small RNAs from all annotated introns <500 nt. Those intronic reads were generally independent of both Dicer and DGCR8, but four short introns were identified with DGCR8-independent but Dicer-dependent reads (Fig. 2D; Supplemental Table 5). The host genes of these introns included Plod3, host of mir-702 (Fig. 2C), Abcf1, host of a recently proposed mirtron mir-877 (Supplemental Fig. S3A; Berezikov et al. 2007), and Mosc2, host of a newly identified mirtron mir-1981 (Supplemental Fig. S3B).
Closer inspection of the hairpin structure of these mirtrons provided additional insights into the evolutionary constraints placed on the originating loci. The lengths of the three mirtrons were statistically more similar to those of short introns than to canonical pre-miRNAs (KS test vs. short introns, P = 0.954; vs. pre-miRNAs, P = 0.008) (Fig. 2E). Despite their longer-than-average pre-miRNA length, the fractions of nucleotides predicted to be paired in each of the mirtronic hairpins were statistically more akin to those of canonical pre-miRNAs than they were to those of short introns (KS test vs. short introns, P = 0.053; vs. pre-miRNAs, P = 0.886) (Fig. 2F). Thus, the larger size of the mammalian mirtrons seemed to impose a larger informational load in the form of extended secondary structure than is imposed on the shorter mirtrons of other species (Ruby et al. 2007a).
Beyond the three mirtrons described above, a fourth short intron gave rise to small RNAs in a Dicer-dependent, DGCR8-independent manner (Fig. 2G). This intron of host gene Oaz1 was predicted to fold into a hairpin structure, with miRNA species arising from the 3′ arm of the hairpin and miRNA* species arising from the 5′ arm (Fig. 2G,H). Two abundant miRNA 5′ ends were observed, one of which generated a 2-nt 3′ overhang when paired to the miRNA* species (Fig. 2G,H). Like mirtrons, the 3′ end of the miRNA was defined by the 3′ splice site, but unlike mirtrons, there was an 11-nt tail at the 5′ end. In Drosophila, the biogenesis of a miRNA whose gene has a similar tailed-mirtron structure, mir-1017, was attributed to the nucleolytic cleavage of a large overhang from a debranched intron to generate a pre-miRNA-like hairpin (Ruby et al. 2007a). Such tailed mirtrons were also suggested recently based on small RNA sequences from developing chicks, although their biogenesis was not evaluated (Glazov et al. 2008). The observed Dicer dependence of small RNAs from the Oaz1 tailed-mirtron confirmed the ability of such a structure to enter the miRNA biogenesis pathway, and the DGCR8 independence indicated that Drosha was not responsible for cleaving off the overhanging sequence.
An alternative explanation for the loss of mirtron and tailed-mirtron expression in the dicer1Δ/Δ mESCs could have been an indirect reduction of host gene expression. However, microarray expression analysis of these genes (Plod3, Abcf1, Mosc2, and Oaz1) indicated that their mRNA expression profiles in the dgcr8Δ/Δ and dicer1Δ/Δ mutant backgrounds did not decrease relative to wild type (Fig. 2I). Therefore, the dramatic Dicer-dependent nature of the intronic small RNAs was attributed to a defect in processing, not precursor expression.
Identification of short hairpin-derived miRNAs
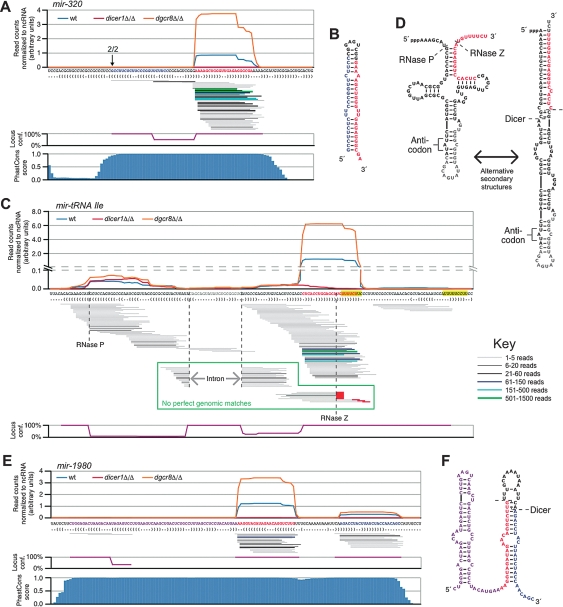
Of the nonmirtronic genomic loci producing Dicer-dependent and DGCR8-independent small RNA reads (Fig. 1D), four were also annotated as miRNA genes: mir-320 (2925 wild-type reads), mir-484 (2285 wild-type reads), mir-668 (55 wild-type reads), and mir-344 (36 wild-type reads). The most prolific of these loci, mir-320, was highly conserved. High PhastCons scores, which reflect high probabilities of selective maintenance through evolution (Siepel et al. 2005), extended to the edges of the pre-miRNA hairpin (Fig. 3A). This pattern differed from that of most conserved miRNA loci, which display conservation of the positions flanking the pre-miRNA because these flanking residues are under selective pressure to maintain the pairing needed for proper Microprocessor recognition (Lim et al. 2003; Han et al. 2006). Moreover, a vast majority of the small RNAs from the mir-320 locus mapped to the 3′ arm of the hairpin (>1000-fold excess). This finding might be expected if the small RNAs from the 5′ arm did not possess the 5′ monophosphate characteristic of Drosha cleavage and required for inclusion in the sequencing libraries. 5′ RACE in the dicer1Δ/Δ mESCs confirmed the predicted 5′ end of the mir-320 Dicer substrate (Fig. 3A, arrow). Importantly, the 5′ RACE method, unlike the library-construction method, was unbiased by the type of 5′ RNA modification. An additional locus, mir-484, had features resembling those of mir-320, including abundant reads, strong pre-miRNA conservation, and reads deriving primarily from the 3′ arm of the hairpin (Supplemental Fig. S4). As commonly observed for other DGCR8-independent, Dicer-dependent small RNAs (Fig. 1C), the normalized small RNA frequencies from both of these loci were elevated in the dgcr8Δ/Δ background (Fig. 3A; data not shown). This elevation may be because, in the absence of canonical miRNAs, there was an increase in available silencing complexes, which were able to protect the remaining small RNAs from nucleases. Moreover, in the absence of the canonical pre-miRNAs, Dicer may have more efficiently cleaved the remaining Dicer substrates into mature small RNAs.
Figure 3.
Small RNAs that are Dicer-dependent and DGCR8-independent and derive from highly conserved shRNA. (A) Small RNAs from mir-320 were DGCR8-independent, Dicer-dependent. (Top) The normalized read count from each library, plotted above the genomic sequence and predicted secondary structure, as in Figure 2A. RNA species are represented below as lines, with thickness/color representing the number of reads (see the key). Below that is the mean locus confidence for small RNA reads at each position; 100% represents a unique match to the genome; percentages are then divided by the mean number of genomic loci to which the sequences map. (Bottom) PhastCons score at each nucleotide position. The arrow represents the 5′ end as determined by RACE in dicer1Δ/Δ and the number above represents the number of clones observed. (B) Predicted secondary structure of the mir-320 pre-miRNA. (C) Small RNAs mapping to a tRNA-Ile gene, shown as in A. Green boxed area represents reads that do not match the genome due to tRNA processing, and red lines represent untemplated CCA addition. mir-tRNA Ile has been annotated as mir-1983. (D) Manually predicted secondary structures for the tRNA cloverleaf structure and alternative shRNA fold. (E) Small RNAs mapping to mir-1980, presented as in A. (F) Predicted secondary structure of the presumed mir-1980 precursor.
The enzymatic dependencies of these loci suggested that they were either transcribed directly as short hairpins or preprocessed by an unknown Microprocessor-independent mechanism. To gain insight into these alternative hypotheses for the Microprocessor independence of mir-320 and mir-484, we analyzed the genome for clear examples of each of these potential biogenic mechanisms. This analysis identified two compelling loci. The first was a locus giving rise to Dicer-dependent, DGCR8-independent reads that was also annotated as an isoleucine tRNA gene (Fig. 3C,D). This locus gave rise to 1954 wild-type reads, the majority of which were clustered at the 3′ end of the tRNA. Unlike other tRNA genes, the primary transcript from this locus was predicted to have the potential to form a long hairpin as an alternative to the tRNA cloverleaf secondary structure, with the acceptor stem extended at its base by 7 base pairs (bp) (Fig. 3D). The cluster of abundant, Dicer-dependent small RNAs mapped to the 3′ end of this extended hairpin, with their 3′ termini extending into a poly-U stretch characteristic of a terminator pol III, the polymerase responsible for transcribing tRNAs (Dieci et al. 2007). The pol III transcription, with termination producing a 2-nt 3′ overhang at the end of a hairpin product had striking resemblance to the design of engineered shRNAs expressed from exogenous vectors for gene knockdown experiments (Paddison and Hannon 2002).
In addition to its processing as an endogenous shRNA, several data suggested that this gene had retained the ability to be processed into a mature tRNA. Reads were scattered across the locus that were generated in both mutant as well as the wild-type samples, many of which mapped uniquely to this locus (Fig. 3C). Reads diagnostic of the mature tRNA accumulated, including a cluster with 5′ ends matching the RNase P cleavage site (Altman 2000), a set that spanned the splice junction at the anti-codon loop (Abelson et al. 1998), and a set that did not match the genome because of untemplated CCA addition at their 3′ ends (Fig. 3C,D; Weiner 2004). Most of these could also have derived from other tRNA genes, but five of 28 CCA-appended reads mapped uniquely to this locus. The 3′ termini of these uniquely mapping reads clustered at the first of two pol III termination signals rather than at the RNase Z cleavage site (Fig. 3C), suggesting CCA addition in the absence of RNase Z cleavage. The second pol III termination signal was detected further downstream, concurrent with the 3′ ends of a small cohort of reads (Fig. 3C), and may have generated an alternative primary transcript more suited to tRNA processing than was the putative shRNA primary transcript.
The second locus that provided insight into potential mechanisms for mir-320 and mir-484 biogenesis, mir-1980, had not been annotated previously but was highly conserved (Fig. 3E). In contrast to the mir-320, mir-484, and tRNA loci, more reads were generated from the 5′ arm than the 3′ arm of the mir-1980 predicted pre-miRNA hairpin, an observation that was inconsistent with the initiation of transcription defining the 5′ end of these reads. The region upstream of the pre-miRNA hairpin was also highly conserved and had predicted capacity to form a second hairpin. A small number of reads was detected at the 5′ edge of the conserved block, consistent with its transcription. However, no reads were generated from the 3′ arm of this upstream hairpin, suggesting that it might only serve to prevent competitive base pairing of this portion of the primary transcript with the pre-miRNA. This tailed shRNA locus appears to be processed in a fashion analogous to that of the tailed mirtron, with an unidentified, non-Microprocessor nuclease removing nucleotides up to the 5′ end of the pre-miRNA hairpin.
Endogenous hp-siRNAs derived from tandem, inverted SINE elements
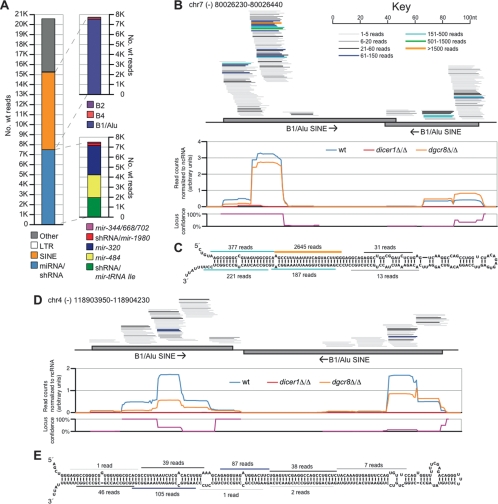
A large population (38%) of DGCR8-independent, Dicer-dependent small RNAs mapped to SINE elements (Fig. 4A; Supplemental Table 6). Rather than being evenly distributed across the subclasses of SINE elements, almost all of the SINE-derived reads (98%) mapped to the B1/Alu subclass (Vassetzky et al. 2003). Because many of these reads mapped to a multitude of SINE elements across the genome, the Dicer-dependent, DGCR8-independent, SINE-annotated locus with the highest normalized wild-type read count was examined as a representative of these loci. Notably, many Dicer-dependent reads mapped uniquely to this locus on chromosome 7 (Fig. 4C), which contained two annotated B1/Alu SINE elements located adjacent to one another, convergently oriented (Fig. 4B). Transcription across SINE elements so arranged would yield a long hairpin RNA (hp-RNA) that could be a dsRNA substrate for successive Dicer cleavage to yield endogenous siRNAs (Fig. 4C). Consistent with this model of hp-siRNA biogenesis, all of the reads that mapped uniquely to this locus mapped to one strand of the genome; the Dicer-dependent reads were phased at ∼21-nt intervals (only a small cohort of Dicer-independent reads mapping to ∼15 other loci disrupted this phasing; Fig. 4B,C), and the reads from the 5′ arm of the predicted hairpin precursor shared 2-nt 3′overhangs with those from the 3′ arm (Fig. 4C). qRT–PCR confirmed that the expression of this locus was similar between wild type, dgcr8Δ/Δ, and dicer1Δ/Δ (Supplemental Fig. S5). Therefore, the absence of the small RNA reads in dicer1Δ/Δ must be predominantly due to a loss in processing, rather than a loss in expression of the precursor.
Figure 4.
SINE-derived hp-siRNAs. (A) Sources of Dicer-dependent small RNAs. See Supplemental Table 6 for tabular representation. (B) Small RNAs from the wild-type library that derived from the indicated genomic locus, which contains two inverted B1/Alu SINE elements. Presented as in Figure 3A. (C) Predicted secondary structure of a transcript corresponding to the locus from B, with the most abundantly sequenced small RNA from each register indicated. (D) A second hp-siRNA SINE locus, presented as in B. (E) Predicted secondary structure of a transcript corresponding to the locus from D, with the most abundantly sequenced small RNA from each register indicated.
The arrangement of reads across the hp-siRNA locus suggested that the majority of reads mapping to that locus could be ascribed to it despite additional perfect matches to the genome. So considered, the phased small RNAs of this single hp-siRNA locus accounted for 87% of the B1/Alu-derived, DGCR8-independent small RNAs (85% when excluding the out-of-phase, Dicer-independent reads described above). Excluding those reads, the SINE-annotated locus with the second-most Dicer-dependent reads, present on chromosome 4, also had two B1/Alu elements in a tandem, inverted arrangement (Fig. 4D). The predicted hairpin structure at this hp-siRNA precursor was longer then that from the chromosome-7 locus, and phased reads sharing ∼2-nt 3′ overhangs with those from the opposing arm of the hairpin spanned a greater length, presenting an even more compelling case for successive Dicer cleavage through this dsRNA precursor (Fig. 4E). Together, these two hp-siRNA loci accounted for 91% (89% excluding the out-of-phase reads from above) of the B1/Alu-derived Dicer-dependent, DGCR8-independent endogenous siRNAs. hp-siRNAs have also been described deriving from the inverted repeats flanking LTR elements in mouse oocytes (Tam et al. 2008; Watanabe et al. 2008), but LTR-derived RNAs represented a negligible fraction of the transposon-derived siRNAs in mESCs (Fig. 4A). The 26% of Dicer-dependent, DGCR8-independent reads remaining unclassified were mostly short reads that derived from a small number of loci (examples in Supplemental Fig. S6). Despite their short length, these sequences generally mapped uniquely to the genome, and were found clustered with other short sequences exhibiting heterogeneity at both their 5′ and 3′ ends. The biogenesis and possible roles for these small RNAs was unclear.
Discussion
Here we report a global analysis of the enzymatic requirements for the production of small RNAs in mESCs. The experimental identification of the enzymatic dependencies of the small RNAs allowed for categorization based on their biogenesis pathways. This approach allowed canonical miRNAs to be distinguished from mirtron- and shRNA-derived miRNAs (Fig. 5A). It also allowed unambiguous identification of genomically clustered small RNAs as endogenous siRNAs, which in mESCs were predominantly hp-siRNAs. This observation is in contrast to mouse oocytes, where endo-siRNAs included hp-siRNAs, cis-nat-siRNAs, and trans-natsiRNAs (Fig. 5B; Tam et al. 2008; Watanabe et al. 2008).
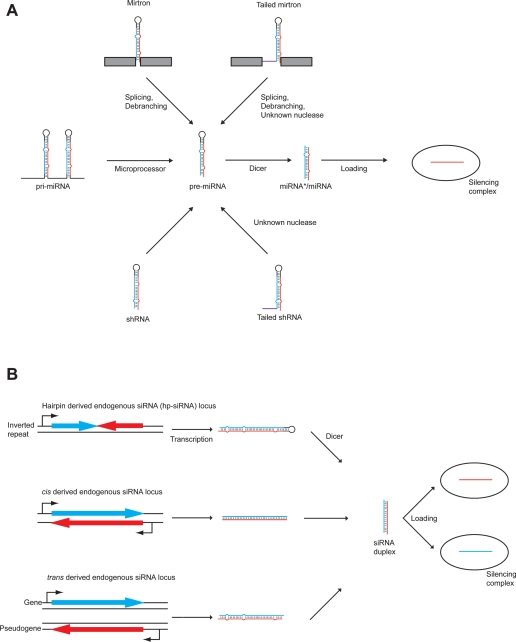
Figure 5.
Alternative biogenesis pathways of Dicer-dependent small RNAs. (A) Canonical and noncanonical pathways for miRNA biogenesis. DGCR8-independent, Dicer-dependent (noncanonical) miRNAs arise from splicing (mirtrons) or direct transcription (shRNA). Some mirtrons and shRNAs have an additional tail that is removed by an unknown nuclease. (B) Endogenous siRNA biogenesis. Inverted repeats can form long hairpins, which are processed by Dicer into siRNAs (hp-siRNAs). dsRNA duplexes can also arise from convergent transcription, or gene/pseudogene pairs. Note that convergent transcription and gene/pseudogene pairs have only been identified in mouse oocytes and were not detected in ES cells.
Most annotated miRNAs were observed to be canonical, as loss of either DGCR8 or Dicer resulted in a loss of their expression. However, several annotated miRNAs were present in the absence of DGCR8, but lost in the absence of Dicer. Further analysis of the enzymatic dependencies of all small RNA generating loci in the mESCs identified more DGCR8-independent, Dicer-dependent small RNAs. These loci included novel murine mirtrons, analogous to those of mirtrons in flies and worms (Okamura et al. 2007; Ruby et al. 2007a). A previous study had mapped small RNAs from human and macaque to the ends of short introns and used this mapping data to propose that mammals have mirtrons (Berezikov et al. 2007). The DGCR8 independence and Dicer dependence observed in our study confirmed the mirtronic identity of one proposed locus, mir-877, which is conserved between mouse, humans, and chimps. We also identified two other mirtrons, including the reclassified mir-702. In addition, a tailed mirtron was identified that had a short extension 5′ of the pre-miRNA hairpin. A similar miRNA precursor structure is found in mir-1017 of Drosophila, but with the tail at the 3′ end of the putative pre-miRNA (Ruby et al. 2007a). These tailed mirtrons expand the repertoire of introns that can give rise to miRNAs (Fig. 5A).
Overall, the mirtrons made up a small fraction of the DGCR8-independent, Dicer-dependent small RNAs. A much larger fraction of reads derived from endogenously expressed short hairpins (Fig. 5A). Although exogenous shRNAs are common laboratory reagents designed to knock down genes of interest, this type of small RNA precursor had not been reported to produce endogenous small RNAs in any species. Several lines of evidence support the annotation of these loci as endogenous shRNA genes: First, the small RNAs mapping to shRNAs were Dicer-dependent, linking them to RNA silencing. Second, these loci did not map to introns and lacked the canonical splicing signals, suggesting a pathway for bypassing Microprocessor biogenesis distinct from that of mirtrons. Third, analysis of the sequences aligning to these hairpins revealed a paucity of reads mapping to the 5′ arms of the hairpins. Inspection of data from recent sequencing efforts in human ES cells (Morin et al. 2008) revealed that human mir-320 and mir-484 have a similar lack of 5′ reads (Supplemental Fig. S7). Because the protocols for constructing small RNA libraries required a 5′ monophosphate for adapter ligation, the lack of reads matching the 5′ arms could be explained by an alternative modification at the 5′ end of the hairpin. Indeed, the pol III-expressed tRNA-IIe transcript, expected to have a 5′ triphosphate, showed a similar paucity of 5′ reads. Taken together, these three lines of evidence suggest that mir-320, mir-484, and mir-tRNA Ile loci were expressed as shRNAs. We note that one difference between endogenous and engineered shRNAs is that engineered hairpins are typically designed to have more perfectly paired stems, whereas endogenous hairpins resemble most pre-miRNAs in having more mismatches in the stem.
An interesting potential variation on shRNA biogenesis was exemplified by mir-1980, which was predicted to form two hairpins with only the second hairpin producing a substantial number of sequenced small RNAs. The Microprocessor independence of this locus suggests that an unknown nuclease removes the 5′ tail up to the 5′ end of the second hairpin, creating a parallel between the biogenesis of tailed mirtrons and that of this tailed shRNA.
Dicer-dependent small RNAs deriving from SINE elements have been a curiosity since they were found in mESCs (Calabrese et al. 2007). We found that these small RNAs were DGCR8-independent and derived almost exclusively from B1/Alu SINE elements (Fig. 5B). Moreover, the bulk of these small RNAs appeared to derive from only one locus containing tandem, inverted SINE B1 elements with the potential to produce an hp-RNA transcript. Phased reads with 2-nt 3′ overhangs when paired to reads from the other arm of the hairpin strongly implied that this hairpin is produced from transcriptional read-through of the inverted SINE elements and then successively cleaved by Dicer to generate endogenous hp-siRNAs. A similar hp-siRNA biogenesis pathway has been reported in mouse oocytes (Tam et al. 2008; Watanabe et al. 2008) and in Drosophila (Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008). In oocytes, siRNAs can also arise from long dsRNAs formed in trans by pseudogene/gene pairing (trans-nat-siRNAs) or in cis by antisense transcription (cis-nat-siRNAs), but our mESCs data sets revealed no such siRNAs. Conversely, analysis of the oocyte small RNA data revealed few, if any reads from the hairpin-forming SINE elements we uncovered in ES cells (Supplemental Fig. S8). When considered together with the recent results from oocytes, our data showed that endo-siRNAs are produced in mammalian cell types other than oocytes and suggested that endo-siRNAs are tightly regulated during the transition from oocytes and blastocyst-stage embryos.
It is unclear how commonly these different subclasses of noncanonical miRNAs and siRNAs are expressed in mammalian somatic tissues. Until very recently, endo-siRNAs were presumed largely absent from mammalian cells, a notion bolstered by the assumption that long dsRNA would trigger the interferon response (Stark et al. 1998). Indeed, the two cell types where a large number of mammalian siRNAs have been unambiguously identified, oocytes (Tam et al. 2008; Watanabe et al. 2008) and mESCs (this study) lack an interferon response (Wianny and Zernicka-Goetz 2000; Yang et al. 2001). The mirtron and shRNA biogenesis pathways would not face this limitation, and are likely to have a more broad distribution of expression. ES cells have a less complex miRNA expression profile than do differentiated cells (Strauss et al. 2006), with a small pool of miRNAs making up vast majority of the small RNA population (Calabrese et al. 2007). Therefore, we postulate that many more examples of small RNAs deriving from the alternative biogenesis pathways described here will be uncovered in other cell types.
Materials and methods
ES cell culture
mESCs were grown on gelatin-coated plates in standard, feeder-free conditions, using mouse embryonic fibroblast (MEF)-conditioned media as described previously (Wang et al. 2007). DGCR8 knockout ES cells were described previously (Wang et al. 2007). Dicer knockout cells were produced by first deriving ES cells (Blelloch et al. 2006) from Dicer flox/flox mice (Harfe et al. 2005). Resulting cells were treated with cre recombinase to loop out the floxed Dicer alleles. Homozygous loop out was confirmed by genotyping (Harfe et al. 2005).
Small RNA cloning and sequencing
Total RNA from wild type, dgcr8Δ/Δ, and dicer1Δ/Δ was extracted using Trizol (Invitrogen). Small RNA libraries were constructed essentially as described elsewhere (Grimson et al. 2008). Small RNAs ranging from 18 to 32 nt were gel-purified and ligated to the 3′ adaptor and 5′ adaptor oligonucleotides. Products from the second ligation were purified, reverse transcribed, and amplified using corresponding primers. Sequences of the adaptors and primers are as published (Illumina, 454 Life Sciences). Samples were sequenced on an Illumina 1G Genome Analyzer and 454 Genome Sequencer.
Bioinformatic analysis of small RNA libraries
5′ and 3′ adapter sequences were removed from 454-generated reads as described (Ruby et al. 2006). For Illumina-derived reads, the 5′ end of the read was treated as the small RNA 5′ nucleotide, and the 3′ end of the small RNA was determined by the 3′-most perfect match to the first 6 nt of the 3′ adaptor. Reads without a perfect 6-nt adapter match were discarded. All adapter-extracted reads 16–27 nt in length were mapped to the University of California at Santa Cruz mm8 assembly of the mouse genome (Waterston et al. 2002; Karolchik et al. 2003) by searching for exact matches over the entire sequence. All reads mapping perfectly to the genome at <500 loci were considered in downstream analysis. Read counts for any locus were normalized to the number of genomic matches as described (Ruby et al. 2006), unless otherwise noted. RNA secondary structures were predicted using RNAfold (Hofacker et al. 1994), unless otherwise noted. miRNA annotations were taken from miRBase version 10.0 (Griffiths-Jones et al. 2008), and mm8 genome coordinates were obtained using BLAT (Kent 2002), unless otherwise noted. Repeat element and NCBI refSeq gene annotations were obtained from UCSC (Jurka 2000; Karolchik et al. 2003; Pruitt et al. 2005). When considering reads overlapping the border of an annotation, counts were multiplied by the fraction of the read length that overlapped. Antisense reads were considered nonoverlapping, unless otherwise noted.
Small RNA dependencies were determined by sliding a 100-nt window along each strand of each chromosome at 1-nt increments. Any such window with ≥10 reads from the wild-type data set was placed into one of five categories: (1) Dicer-dependent, DGCR8-dependent; (2) Dicer-dependent, DGCR8-independent; (3) Dicer-independent, DGCR8-dependent; (4) Dicer-independent, DGCR8-independent; or (5) intermediate. A window’s dependence on each enzyme was calculated by dividing the number of reads in that enzyme’s knockout data set by the number of wild-type data set reads after normalizing the read count of each data set to that data set’s count of tRNA-, snRNA-, scRNA-, and srpRNA-derived reads; bioinformatic analysis confirmed that members of these species were relatively constant between wild type, dgcr8Δ/Δ, and dicer1Δ/Δ (Supplemental Fig. S1). For each window, quotients ≤0.1 indicated dependence, while quotients ≥0.5 indicated independence. An intermediate value with respect to either enzyme placed the window in the “intermediate” category. After classification, overlapping windows from each category were combined into nonredundant loci, and the dependencies of each locus were re-evaluated according to the criteria above, but with the “intermediate” category split into three subcategories: “Dicer-dependent, DGCR8-intermediate,” “Dicer-intermediate, DGCR8-dependent,” or “intermediate.” In analysis of miRNA genes and short introns, the same requirements were applied to those annotations as to the windows/loci above.
Microarrays
Wild-type, dgcr8Δ/Δ, and dicer1Δ/Δ ES cells were plated on 60-mm plates in triplicate. Total RNA was extracted using Trizol (Invitrogen). cRNAs were synthesized and hybridized to the Affymetrix Mouse Gene 1.0 ST arrays at the Genomics Core Facility, Gladstone Institute, University of California at San Francisco. Data were normalized with GCRMA algorithm.
5′ RACE
Total RNA from dicer1Δ/Δ was extracted using Trizol reagent (Invitrogen). 5′ RACE was performed per manufacturer’s instructions (Invitrogen 5′ RACE System for Rapid Amplification of cDNA Ends, Version 2.0 kit). Nested PCR was carried out to enrich for the mir-320 pre-miRNA hairpin. Primer sequences were as follows: mir-320 GSP1, CGCCCTCTCAACCCAGC; mir-320 GSP2, CCCGACTCCGGGAAGAACC; mir-320 GSP3, CCGGGAAGAACCGGGAAGAG; 5′ RACE Abridged Anchor Primer, GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG; Abridged Universal Amplification Primer, GGCCACGCGTC GACTAGTAC. The PCR product was TA-cloned into pCR2.1-TOPO (Invitrogen) and sequenced. Two clones gave unambiguous reads, while an additional three clones were indeterminate due to the ambiguity of a single cytosine residue adjacent to the adapter/insert boundary following a long stretch of guanines.
Accession numbers
All small RNA reads are available at the GEO database with accession number GSE12521. Data from 454 sequencing are deposited in GPL7196, and data from Illumina sequencing are in GPL7195.
Acknowledgments
We thank the Vincent J. Coates Genomics Sequencing Laboratory at University of California at Berkeley and the Whitehead Center for Advanced Genome Technology for sequencing services, Michael McManus for providing Dicer conditional mice, and Susan Fisher, Marco Conti, Monica Venere, and Deepa Subramanyam for critical reading of the manuscript. This work was supported by funds to R.B. from NIH grants R01 NS057221 and K08 NS048118 and by funds to D.B. from NIH grant R01 GM067031. J.B. is supported by NIH training fellowship T32 HD007263. Y.W. is supported by a California Institute of Regenerative Medicine post-doctoral fellowship. D.B. is an HHMI Investigator, and R.B. is a Pew Scholar. Y.W. developed the small RNA libraries for deep sequencing and RNA for microarrays. J.B. and J.G.R. performed the bioinformatic analyses. J.B. performed 5′ RACE, Northerns, and qRT–PCR experiments. J.B., J.G.R., D.B., and R.B. analyzed the data and wrote the manuscript.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1705308.
References
- Abelson J., Trotta C.R., Li H. tRNA splicing. J. Biol. Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- Altman S. The road to RNase P. Nat. Struct. Biol. 2000;7:827–828. doi: 10.1038/79566. [DOI] [PubMed] [Google Scholar]
- Ambros V., Lee R.C., Lavanway A., Williams P.T., Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Chung W.J., Willis J., Cuppen E., Lai E.C. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch R., Wang Z., Meissner A., Pollard S., Smith A., Jaenisch R. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells. 2006;24:2007–2013. doi: 10.1634/stemcells.2006-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese J.M., Seila A.C., Yeo G.W., Sharp P.A. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc. Natl. Acad. Sci. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B., Malone C.D., Zhou R., Stark A., Schlingeheyde C., Dus M., Perrimon N., Kellis M., Wohlschlegel J.A., Sachidanandam R., et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Dieci G., Fiorino G., Castelnuovo M., Teichmann M., Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn D.M., Lieberman J. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Annu. Rev. Med. 2005;56:401–423. doi: 10.1146/annurev.med.56.082103.104606. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M., Seitz H., Horwich M.D., Li C., Du T., Lee S., Xu J., Kittler E.L., Zapp M.L., Weng Z., et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov E.A., McWilliam S., Barris W.C., Dalrymple B.P. Origin, evolution, and biological role of miRNA cluster in DLK–DIO3 genomic region in placental mammals. Mol. Biol. Evol. 2008;25:939–948. doi: 10.1093/molbev/msn045. [DOI] [PubMed] [Google Scholar]
- Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A., Srivastava M., Fahey B., Woodcroft B.J., Chiang H.R., King N., Degnan B.M., Rokshar D.S., Bartel D.P.2008The early origins and evolution of microRNAs and piRNAs in animals Nature (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.M. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha–DGCR8 complex in primary microRNA processing. Genes & Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hannon G.J., Rossi J.J. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- Harfe B.D., McManus M.T., Mansfield J.H., Hornstein E., Tabin C.J. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl. Acad. Sci. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer L.S., Tacker M., Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- Houbaviy H.B., Murray M.F., Sharp P.A. Embryonic stem cell-specific MicroRNAs. Dev. Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Jurka J. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes & Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y., Saito K., Kin T., Ono Y., Asai K., Sunohara T., Okada T.N., Siomi M.C., Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M., Yalcin A., Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lee S.R., Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes & Dev. 2006;20:28–33. doi: 10.1101/gad.1377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N.S., Dohjima T., Bauer G., Li H., Li M.J., Ehsani A., Salvaterra P., Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P. The microRNAs of Caenorhabditis elegans. Genes & Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E., Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes & Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin R.D., O’Connor M.D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison E.P., Partridge J.F., Tam O.H., Cheloufi S., Hannon G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Hagen J.W., Duan H., Tyler D.M., Lai E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Chung W.J., Ruby J.G., Guo H., Bartel D.P., Lai E.C. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison P.J., Hannon G.J. RNA interference: The new somatic cell genetics? Cancer Cell. 2002;2:17–23. doi: 10.1016/s1535-6108(02)00092-2. [DOI] [PubMed] [Google Scholar]
- Paul C.P., Good P.D., Winer I., Engelke D.R. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- Poethig R.S., Peragine A., Yoshikawa M., Hunter C., Willmann M., Wu G. The function of RNAi in plant development. Cold Spring Harb. Symp. Quant. Biol. 2006;71:165–170. doi: 10.1101/sqb.2006.71.030. [DOI] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Jan C., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Ruby J.G., Jan C.H., Bartel D.P. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007a;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Stark A., Johnston W.K., Kellis M., Bartel D.P., Lai E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007b;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo T.S., Bai X., Ruparel H., Li Z., Turro N.J., Ju J. Photocleavable fluorescent nucleotides for DNA sequencing on a chip constructed by site-specific coupling chemistry. Proc. Natl. Acad. Sci. 2004;101:5488–5493. doi: 10.1073/pnas.0401138101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler B.M., Ruohola-Baker H. Small RNAs: Keeping stem cells in line. Cell. 2008;132:563–566. doi: 10.1016/j.cell.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G.R., Kerr I.M., Williams B.R., Silverman R.H., Schreiber R.D. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Strauss W.M., Chen C., Lee C.T., Ridzon D. Nonrestrictive developmental regulation of microRNA gene expression. Mamm. Genome. 2006;17:833–840. doi: 10.1007/s00335-006-0025-7. [DOI] [PubMed] [Google Scholar]
- Suh M.R., Lee Y., Kim J.Y., Kim S.K., Moon S.H., Lee J.Y., Cha K.Y., Chung H.M., Yoon H.S., Moon S.Y., et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Tam O.H., Aravin A.A., Stein P., Girard A., Murchison E.P., Cheloufi S., Hodges E., Anger M., Sachidanandam R., Schultz R.M., et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassetzky N.S., Ten O.A., Kramerov D.A. B1 and related SINEs in mammalian genomes. Gene. 2003;319:149–160. doi: 10.1016/s0378-1119(03)00805-9. [DOI] [PubMed] [Google Scholar]
- Verdel A., Moazed D. RNAi-directed assembly of heterochromatin in fission yeast. FEBS Lett. 2005;579:5872–5878. doi: 10.1016/j.febslet.2005.08.083. [DOI] [PubMed] [Google Scholar]
- Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Takeda A., Tsukiyama T., Mise K., Okuno T., Sasaki H., Minami N., Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes & Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Totoki Y., Toyoda A., Kaneda M., Kuramochi-Miyagawa S., Obata Y., Chiba H., Kohara Y., Kono T., Nakano T., et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Weiner A.M. tRNA maturation: RNA polymerization without a nucleic acid template. Curr. Biol. 2004;14:R883–R885. doi: 10.1016/j.cub.2004.09.069. [DOI] [PubMed] [Google Scholar]
- Wianny F., Zernicka-Goetz M. Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- Yang S., Tutton S., Pierce E., Yoon K. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol. Cell. Biol. 2001;21:7807–7816. doi: 10.1128/MCB.21.22.7807-7816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]