Abstract
Segregation of homologous chromosomes during meiosis depends on linkages (chiasmata) created by crossovers and on selective release of a subset of sister chromatid cohesion at anaphase I. During Caenorhabditis elegans meiosis, each chromosome pair forms a single crossover, and the position of this event determines which chromosomal regions will undergo cohesion release at anaphase I. Here we provide insight into the basis of this coupling by uncovering a large-scale regional change in chromosome axis composition that is triggered by crossovers. We show that axial element components HTP-1 and HTP-2 are removed during late pachytene, in a crossover-dependent manner, from the regions that will later be targeted for anaphase I cohesion release. We demonstrate correspondence in position and number between chiasmata and HTP-1/2-depleted regions and provide evidence that HTP-1/2 depletion boundaries mark crossover sites. In htp-1 mutants, diakinesis bivalents lack normal asymmetrical features, and sister chromatid cohesion is prematurely lost during the meiotic divisions. We conclude that HTP-1 is central to the mechanism linking crossovers with late-prophase bivalent differentiation and defines the domains where cohesion will be protected until meiosis II. Further, we discuss parallels between the pattern of HTP-1/2 removal in response to crossovers and the phenomenon of crossover interference.
Keywords: Meiosis, chromosome axes, crossover, sister chromatid cohesion, chromosome remodeling, crossover interference
In sexually reproducing organisms, diploid germ cells produce haploid gametes through the specialized cell division program of meiosis. At the onset of meiosis, DNA is replicated and sister chromatid cohesion (SCC) is established (Nasmyth and Schleiffer 2004). In contrast to mitotic cell cycles, this single round of meiotic DNA replication is followed by two rounds of cell division, the first segregating homologous chromosomes (homologs), and the second segregating sister chromatids (Petronczki et al. 2003). This pattern of segregation requires an extended prophase during which chromosomes must assemble meiosis-specific axial structures, locate, and align with their homologs, stabilize this alignment through assembly of the synaptonemal complex (SC), and undergo crossover recombination events between their DNA molecules (Page and Hawley 2003). Crossovers that form in this context play a crucial role in promoting meiotic chromosome segregation, as they collaborate with SCC (on domains flanking the crossover site) to form the basis of chiasmata, cytologically visible connections between the homologs that are revealed upon SC disassembly and structural remodeling of chromosomes during late prophase (Jones 1987). Chiasmata allow homologs to remain connected while orienting away from each other toward opposite poles of the metaphase I spindle. Subsequently, the SCC that maintains the connections afforded by chiasmata is dismantled in a two-step process to permit homolog separation at meiosis I while maintaining the connections between sisters required to allow congression and bipolar orientation on the meiosis II spindle. Thus crossovers serve as a linchpin in the meiotic program, coupling events that serve to solidify connections between homologs with events that ensure their eventual segregation to yield haploid gametes.
Whereas correct assembly of meiosis-specific chromosome structures during early meiotic prophase is essential for the formation of crossovers, crossovers (or nascent crossovers) in turn have the capacity to alter the properties of chromosomes on which they occur. One well-known manifestation of crossover-associated changes in chromosome properties has been recognized for nearly a century (Muller 1916). Specifically, the presence of a crossover reduces the likelihood of other crossovers nearby on the same chromosome. This phenomenon is known as crossover interference, and it results in the wide spacing between crossovers on the same chromosome pair (Jones and Franklin 2006). Moreover, in some organisms, crossover interference mechanisms limit most or all of their chromosomes to a single crossover per homolog pair in most meioses (Hillers and Villeneuve 2003). Despite the widespread occurrence of crossover interference, however, almost nothing is known about the nature of the physical changes in chromosome state that are associated with this phenomenon.
An additional manifestation of the ability of crossovers (or their precursors) to modulate global properties of chromosomes is observed during meiosis in Caenorhabditis elegans, an organism with holocentric chromosomes. Late prophase/metaphase bivalents (homolog pairs connected by chiasmata) in C. elegans have a cytological appearance that closely resembles the bivalents of mouse telocentric chromosomes at similar stages (Nabeshima et al. 2005), but with one major difference. During C. elegans meiosis, either end of the chromosomes has the capacity to retain cohesion and lead the way poleward at the first meiotic division. Moreover, which end does so in a given meiosis is determined by the position of the crossover (Albertson et al. 1997). Effectively, the single crossover between each chromosome pair subdivides the late prophase bivalent into distinct subdomains with different fates. The domains between the crossover and the nearest chromosome ends will be the last to disassemble the SC during the late pachytene and diplotene stages of meiotic prophase, and during late diakinesis (the last stage of prophase) these regions will concentrate proteins (such as Aurora B kinase AIR-2) that promote localized removal of cohesin at the meiosis I division (Kaitna et al. 2002; Rogers et al. 2002). Conversely, the SC will disassemble first from the chromosomal domains harboring the ends most distant from the crossover, and these regions will retain cohesion until meiosis II.
In the present study, we uncovered features suggestive of a link between these two apparently disparate instances of crossover-triggered changes in chromosome state through analysis of C. elegans HTP-1/2. HTP-1 and HTP-2 are members of the conserved meiosis-enriched HORMA domain protein family (Aravind and Koonin 1998), which includes budding yeast Hop1, Arabidopsis Asy1, rice Pair2, and C. elegans paralogs HIM-3 and HTP-3. Proteins in this family, including all four C. elegans paralogs, have been demonstrated to play related but not identical roles in promoting pairing and synapsis between homologous chromosomes during early meiotic prophase (Hollingsworth et al. 1990; Zetka et al. 1999; Caryl et al. 2000; Couteau and Zetka 2005; Martinez-Perez and Villeneuve 2005; Nonomura et al. 2006). Several have also been implicated in creating or maintaining a barrier to sister chromatid-directed double-strand break repair during meiotic prophase, thereby promoting interhomolog recombination (Couteau et al. 2004; Martinez-Perez and Villeneuve 2005; Niu et al. 2005; Carballo et al. 2008). We show here that HTP-1/2 are components of the axial elements of meiotic chromosomes, as shown previously for HIM-3 and HTP-3. Moreover, we report insights into crossover-triggered changes in the properties and behavior of chromosomes that were revealed through analysis of the dynamic localization of HTP-1/2 during late prophase and the meiotic divisions and through functional analysis of late meiotic roles of HTP-1.
First, we demonstrate that crossovers trigger a major remodeling of chromosome axis composition that begins in late pachytene and culminates in depletion of HTP-1/2 from the chromosomal domains that will later be targeted for cohesion loss at the meiosis I division. We show that HTP-1/2 and SC central region protein SYP-1 are depleted from reciprocal chromosomal domains in response to crossovers, suggesting that the boundaries between these domains can serve as early cytological markers of crossover sites. Further, we show that the ability of a crossover to trigger HTP-1/2 removal diminishes over distance, a feature shared with the phenomenon of crossover interference.
Second, we demonstrate a role for HTP-1 in promoting localized protection of SCC during the meiosis I division. This last finding emphasizes the high degree of interconnectedness between succeeding events in the meiotic program: During early/mid-prophase, HTP-1 functions in developing a chromosome organization that promotes formation of interhomolog crossovers and consequent chiasmata. In turn, the crossover then triggers a relocalization of HTP-1 so that it can be redeployed later to ensure that the connection afforded by the chiasma is dismantled in an orderly fashion that promotes homolog segregation. This interconnectedness makes it possible for the position of the crossover to define the subsequent behavior of bivalent subdomains during the meiotic divisions.
Results
HTP-1 and HTP-2 are components of meiotic chromosome axes
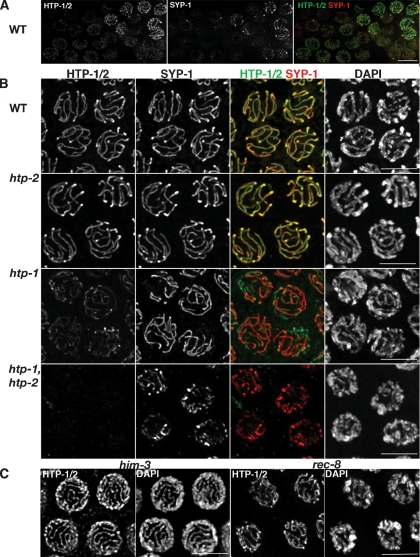
Our previous analysis of htp-1 mutants demonstrated roles for HTP-1 in homolog pairing, in prevention of nonhomologous synapsis and in promoting chiasma formation (Martinez-Perez and Villeneuve 2005). As HTP-1 paralogs HIM-3 and HTP-3 are components of meiotic chromosome axes (Zetka et al. 1999; MacQueen et al. 2005; Goodyer et al. 2008), we raised antibodies against the full-length HTP-1 protein to assess the possible localization of both HTP-1 and HTP-2 to meiotic chromosomes. Since HTP-1 and HTP-2 are 82% identical, the antibodies are predicted to recognize both proteins. α-HTP-1 immunofluorescence (IF) signals are first detected as faint linear stretches in germ cell nuclei that are entering meiosis (Fig. 1A), then become much brighter in regions that are also labeled by antibodies against SC central region component SYP-1 (MacQueen et al. 2002), indicating that the initial loading of HTP-1/2 precedes synapsis. By pachytene, α-HTP-1 and α-SYP-1 IF signals coincide along the full lengths of the chromosomes (Figs. 1B, 2A). This staining pattern is consistent with HTP-1 and/or HTP-2 being components of the axial element of meiotic chromosomes.
Figure 1.
Loading of HTP-1/2 onto meiotic chromosomes. Projections of nuclei from whole-mount gonads labeled with α-HTP-1/2 and α-SYP-1 antibodies. (A) Nuclei entering meiosis, HTP-1/2 are localized to chromosomes prior to SYP-1. (B) Projections of mid-pachytene nuclei. The intensity of the α-HTP-1/2 signals is very similar in wild-type and htp-2 mutant nuclei, but is severely reduced in htp-1 mutants and not detectable in htp-1 htp-2 double mutants. α-SYP-1 antibodies used as an internal control show similar intensity in all genotypes, although some unsynapsed regions (green lines) are present in htp-1 mutants and the SYP-1 tracks present in htp-1 htp-2 double mutants appear more irregular than those observed in wild type and htp-2 and htp-1 single mutants. (C) Mid-pachytene nuclei from him-3(gk179) and rec-8(ok978) mutants showing that HTP-1/2 are associated with pachytene chromosomes of both mutants. Bars, 5 μm.
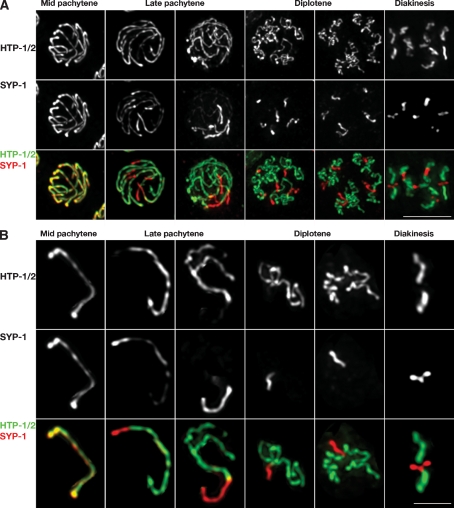
Figure 2.
Late prophase reorganization of SC components in wild-type gonads. (A) Projections of nuclei at the indicated stages labeled with α-HTP-1/2 and α-SYP-1 antibodies. (B) Individual chromosomes cropped from the corresponding nucleus shown in A. (Mid-pachytene) HTP-1/2 and SYP-1 fully colocalize along the SC. (Late pachytene) Each SC track shows a concentration of SYP-1 at one of its ends, which also displays a decrease in the intensity of the α-HTP-1/2 signal. The remaining SC regions are brightly stained by α-HTP-1/2 antibodies but poorly stained by α-SYP-1 antibodies. The axial elements of homologous chromosomes start to come apart in regions in which SYP-1 is not detectable. (Diplotene) Each bivalent is divided into an HTP-1/2-depleted/SYP-1-enriched domain (in which the axial elements of homologous chromosomes remain together) and an HTP-1/2-enriched/SYP-1-depleted domain in which homologous axes are separated and have started to shorten and coil. (Diakinesis) Chromosomes have condensed substantially and can be divided into a pair of short arms, which are strongly stained only by SYP-1, and a pair of long arms, which are only stained by α-HTP-1/2 antibodies. Bars: A, 5 μm; B, 2 μm.
Immunostaining of mutant gonads indicates that these antibodies detect both HTP-1 and HTP-2 (Fig. 1B). First, pachytene nuclei in the htp-1(gk174)-null mutant exhibited a staining pattern that was spatially similar to wild-type, but with a greatly reduced signal intensity; α-SYP-1 IF signals served as an internal control, as SYP-1 was previously shown to localize to chromosomes at similar levels in wild-type and the htp-1 mutant. This indicates that the antibodies recognize HTP-1, and suggests that most of the signal in wild-type pachytene nuclei corresponds to HTP-1. Second, we used a new htp-2 deletion allele (Supplemental Material) to assess whether the residual IF signal in the htp-1 mutant corresponds to HTP-2. No α-HTP-1 IF signal was detected in htp-1 htp-2 double-mutant gonads, confirming that the antibodies detect both proteins. α-SYP-1 staining of this double mutant revealed strong linear SYP-1 stretches that appear more irregular than those observed in wild-type animals, similar to the pattern previously described for htp-1 htp-2RNAi worms (Couteau and Zetka 2005). Finally, we examined α-HTP-1/2 staining in htp-2 single mutant germlines. The htp-2 single mutant does not exhibit overt defects in synapsis or chiasma formation (Fig. 1B; Supplemental Material; Supplemental Fig. S1), indicating that HTP-2 plays a less central role in meiotic prophase than HTP-1 despite the high identity between these two proteins. Both the intensity and spatial pattern of α-HTP-1 IF in the htp-2 single mutant were indistinguishable from wild type, supporting the conclusion that most of the signal in wild-type meiotic nuclei comes from HTP-1. We will operationally use the term “HTP-1/2” to refer to IF signals detected by the α-HTP-1 antibodies to acknowledge the fact that both proteins may be contributing to the observed localization.
Since HTP-1/2 exhibit axial element localization, we assessed whether HTP-1/2 loading requires other known components of meiotic chromosome axes. HTP-1/2 were localized on chromosome axes in a him-3-null mutant (Fig. 1C), indicating that HTP-1/2 loading is not dependent on loading of its paralog HIM-3. We also observed apparently normal staining in rec-8(ok978) mutant gonads, indicating that this mutation does not disrupt HTP-1/2 loading despite causing substantial impairment of chromosome organization during most of prophase.
Localization of HTP-1/2 along chromosome axes is dramatically reorganized beginning in late pachytene
The localization of HTP-1/2 during early meiotic prophase is very similar to that previously described for HIM-3 and HTP-3 (Zetka et al. 1999; Goodyer et al. 2008), with HIM-3, HTP-3, and HTP-1/2 all present along the whole lengths of fully synapsed chromosomes at mid-pachytene (Figs. 1B, 2A,B). However, the staining pattern of HTP-1/2 became strikingly different from those of HIM-3 and HTP-3 at late pachytene. While HIM-3 and HTP-3 remain associated along the whole length of the axes at this stage (Nabeshima et al. 2005; Goodyer et al. 2008), HTP-1/2 levels become severely depleted near one end of each chromosome pair (Fig. 2A,B). Moreover, the domain where HTP-1/2 become depleted is the same domain where SC central region components SYP-1 and SYP-2 start to become concentrated at this same stage (Fig. 2; Nabeshima et al. 2005). By late pachytene, a distinct boundary has developed, dividing each bivalent into a SYP-1-enriched domain and an HTP-1/2-enriched domain (Fig. 2B). Further, in every case, the SYP-1-enriched domain appears substantially shorter than the HTP-1/2-enriched domain. Reduction and loss of IF signals clearly indicates that removal of HTP-1/2 and SYP-1 from chromosomes occurs during this relocalization process; it is also possible that these proteins are being recruited to the domains where they become enriched.
Following redistribution of HTP-1/2 and SYP-1 during late pachytene, desynapsis of the homolog becomes apparent beginning in SYP-1-depleted regions (Fig. 2B). During early stages of desynapsis, axial elements have an extended appearance similar to that observed during mid- and late pachytene (Fig. 2B), while further desynapsis during the diplotene stage is associated with axis coiling. By this stage HTP-1/2 and SYP-1 have acquired an almost mutually exclusive, reciprocal localization, with SYP-1 concentrated in the short regions in which homologs remain associated. By early diakinesis the position of the single chiasma on each bivalent becomes evident, and the bivalents display an asymmetric structure: Each bivalent has a pair of “short arms” (regions now referred to as distal to the chiasma) that retain SYP-1 and are depleted of HTP-1/2 and a pair of “long arms” that retain high levels of HTP-1/2 and are depleted of SYP-1 (Figs. 2A,B, 3A). This localization of HTP-1/2 on diakinesis bivalents stands in striking contrast with the diakinesis localization of HIM-3, REC-8, and HTP-3, which are all retained on both the short and long arms of the bivalents (Fig. 3; Zetka et al. 1999; Pasierbek et al. 2001; Goodyer et al. 2008). This remarkable redistribution of HTP-1/2 during late stages of meiotic prophase, resulting in chromosome axis segments with distinct molecular compositions, represents a novel feature of bivalent differentiation.
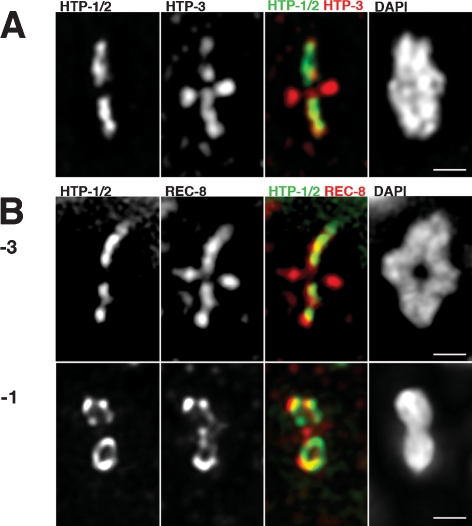
Figure 3.
Localization of axial element components to diakinesis bivalents. Projections of individual bivalents stained with α-HTP-1/2 and α-HTP-3 or α-REC-8 antibodies and DAPI. (A) HTP-3 decorates all arms of the bivalent while HTP-1/2 are only detected on the long arms. (B, top) Bivalent from a −3 oocyte showing that REC-8 is present on both short and long arms, while HTP-1/2 staining is limited to the long arms. (Bottom) In the −1 oocyte, HTP-1/2 and REC-8 colocalize on the long arms of the bivalent in a loop-like organization that extends between the mid-bivalent region and the ends of the long arms. REC-8, but not HTP-1/2, is still present in the short arms of the bivalent. Bars, 1 μm.
Diakinesis organization of meiotic chromosome axes
Our images of bivalents at late diakinesis and during the meiosis I division (see below) highlight an additional feature of meiotic axis organization that had not been appreciated in prior studies. Although many images of diakinesis or meiosis I bivalents give the impression that HTP-1/2 and REC-8 proteins are organized in a roughly linear arrangement at the interfaces between sister chromatids, other images in which orientation of the bivalent is rotated around the long axis reveal that these proteins are actually organized in a loop-like structure on each long arm (Fig. 3; Supplemental Movies; Supplemental Fig. S1D). This loop-like organization becomes apparent in late diakinesis, after the SC central region proteins have fully departed from the bivalents (i.e., −1 and −2 oocytes); it is especially pronounced after kinetochore proteins have assembled onto the highly condensed bivalents (Supplemental Fig. S2) and persists through metaphase I. The loops often exhibit discontinuities, but are continuous near the tips of the bivalent that are most distant from the chiasma; i.e., the ends that will be closest to the spindle poles at the meiosis I division. We interpret this organization to reflect localization of REC-8 and HTP-1/2 to two largely separated chromatid cores (each corresponding to one of the two sister chromatids, which are already substantially resolved) that remain most tightly associated primarily at the bivalent ends.
Patterned reorganization of HTP-1/2 is dependent on meiotic recombination
As asymmetrical disassembly of the SC central region during C. elegans meiosis is dependent on the formation of crossovers (Nabeshima et al. 2005), we investigated whether the late prophase reorganization of HTP-1/2 is also triggered by crossovers. HTP-1/2 remained localized along the full length of chromosomes in late pachytene nuclei and in most early diplotene nuclei in spo-11 mutants (in which meiotic recombination is not initiated) (Fig. 4A,B), and in zhp-3 and msh-5 mutants (which initiate recombination but fail to produce crossovers (Supplemental Fig. S3; Kelly et al. 2000; Jantsch et al. 2004). These results suggest that normal late prophase HTP-1/2 reorganization is dependent on crossovers.
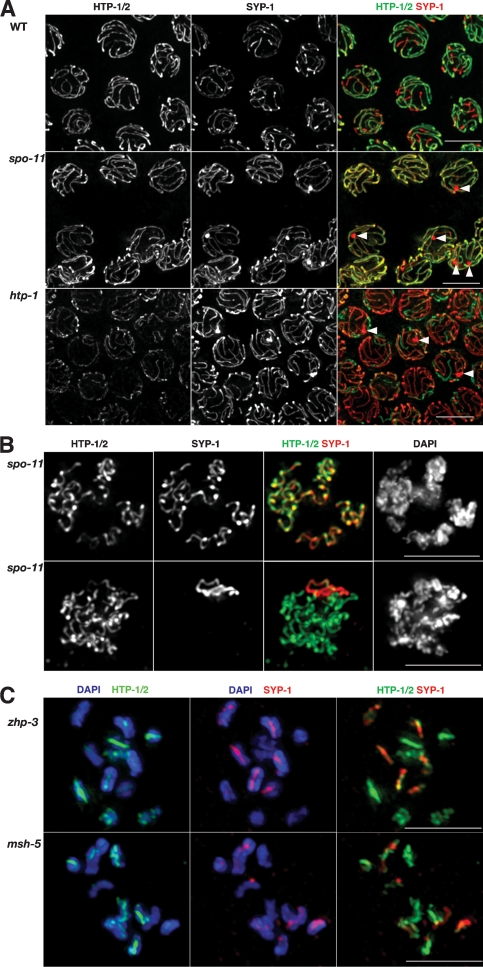
Figure 4.
Patterned reciprocal localization of HTP-1/2 and SYP-1 is dependent on meiotic recombination. (A) Projections of late pachytene nuclei from wild type and spo-11 and htp-1 mutants stained with α-HTP-1/2 and α-SYP-1 antibodies. Wild-type nuclei display the presence of distinctive, reciprocal, bright red (SYP-1) and bright green (HTP-1/2) domains for each SC track. spo-11 mutants lack this patterned reorganization and both HTP-1/2 and SYP-1 remain colocalized along the whole length of the SC. htp-1 mutant nuclei appear mostly red since the residual green signal corresponds solely to HTP-2. Some nuclei in spo-11 and htp-1 mutants display a nonchromosomal aggregation of SYP-1 (arrowheads), which is not detected in wild-type nuclei. (B) Projections of diplotene nuclei from spo-11 mutants stained with α-HTP-1/2 and α-SYP-1 antibodies. The nuclei shown exemplify the two types of staining that we observed: In the top nucleus, HTP-1/2 and SYP-1 colocalize along all chromosomal regions, while in the bottom nucleus HTP-1/2 are present on most chromosomal regions and SYP-1 has been concentrated in the form of a single stretch. (C) Projections of diakinesis nuclei from zhp-3 and msh-5 mutants stained with α-HTP-1/2 and α-SYP-1 antibodies and DAPI. Both nuclei shown contain 12 univalents, although the projection has resulted in some univalents overlapping. On most univalents, regions brightly stained by α-HTP-1/2 antibodies coincide with regions poorly stained by α-SYP-1 antibodies and vice versa. Some univalents are exclusively stained by either α-HTP-1/2 or α-SYP-1 antibodies. Bars, 5 μm.
Despite lack of an orderly patterned reorganization of HTP-1/2 and SYP-1 at late pachytene in recombination mutants, however, several observations suggest that these proteins are mobilized during late prophase even in the absence of crossovers. First, many late pachytene nuclei in spo-11 mutants display a nonchromosomal aggregation of SYP-1 not detected in wild-type germlines (Fig. 4A), suggesting a failure to localize a “mobile” pool of SYP-1 to chromosomes. Second, while HTP-1/2 and SYP-1 remain colocalized along the full length of chromosomes at late pachytene in recombination mutants, during diplotene and diakinesis these proteins acquire an increasingly reciprocal distribution (Fig. 4B,C; Supplemental Fig. S3) so that by diakinesis, the univalent chromosomes display staining patterns in which most regions are predominantly stained either by α-HTP-1/2 antibodies or by α-SYP- 1 antibodies (Fig. 4C).
In summary, patterned reciprocal localization of HTP-1/2 and SYP-1 to distinct chromosomal domains, beginning at late pachytene, is strictly dependent on the presence of crossovers (or crossover precursors). In the absence of crossovers, HTP-1/2 and SYP-1 do eventually achieve a largely reciprocal localization, but this occurs in an apparently stochastic manner, presumably reflecting an inherent incompatibility of these proteins during late prophase.
Evidence of correspondence between crossovers, chiasmata, and zones of HTP-1/2 depletion
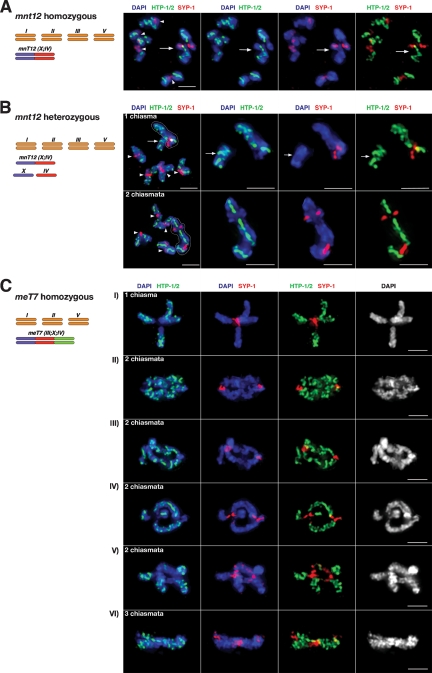
To further investigate the association between crossovers, chiasmata, and patterned reciprocal localization of HTP-1/2 and SYP-1, we analyzed the distributions of these proteins at diplotene/diakinesis in worms carrying two- and three-chromosome fusions. Several additional conclusions regarding late prophase reorganization of HTP-1/2 and SYP-1 emerged from this analysis. First, we analyzed worms homozygous for mnT12, a fusion of chromosomes X and IV. Previously, measurements of recombination frequencies were used to derive a genetic map length of 54 cM for mnT12, indicative of one crossover/homolog pair in most meioses (Hillers and Villeneuve 2003). Further, cytological analysis of DAPI-stained diakinesis nuclei had suggested that bivalents with two chiasmata occurred at a very low frequency (∼3%) (Nabeshima et al. 2004). Consistent with these previous findings, we found that 24 out of 24 diakinesis nuclei in mnT12 homozygotes contained five DAPI-stained bodies, each displaying a single chiasma that coincided with an HTP-1/2-depleted domain. In some mnT12 bivalents (identified based on their larger size), the SYP-1-enriched/ HTP-1/2-depleted domain extended to the ends of the bivalent short arms, as observed for normal-sized chromosomes (Fig. 5A). However, in most cases the chiasma was more centrally located and resulted in a more symmetrical cross-shaped bivalent in which the HTP-1/2-depleted domain did not extend all the way to the chromosome ends (Supplemental Fig. S4). This indicates that each chiasma/crossover is associated with a zone of HTP-1/2 depletion, but further suggests that there are limits to the distance over which a crossover-triggered change in chromosome organization can be propagated.
Figure 5.
Chiasmata coincide with HTP-1/2 depletion domains on bivalents and trivalents containing fused chromosomes. The diagrams to the left of each panel indicate the karyotypes of the worms from which the images on the right were collected. The mnT12 chromosome is a fusion between chromosomes IV and X, while the meT7 chromosome is a fusion of chromosomes III, X, and IV. All nuclei shown were stained with α-HTP-1/2 and α-SYP-1 antibodies and DAPI. (A) Projection of an early diakinesis nucleus from an mnT12 homozygote. Five DAPI-stained bodies are present, and all show a single HTP-1/2-depleted /SYP-1-enriched domain that corresponds with the position of the chiasma. Arrowheads indicate four regular bivalents and an arrow marks the larger mnT12 bivalent. (B) Projections of two early diakinesis nuclei heterozygous for mnT12. Top panels show an example in which mnT12 has formed one chiasma and the bottom panels show an example in which mnT12 has formed two chiasmata. In each case, the first panel shows the whole nucleus and the panels to the right show a zoomed-in image of the mnT12 bivalent or trivalent. (One chiasma panel) Six DAPI-stained bodies are detected: four regular bivalents (arrowheads), a large asymmetric bivalent with a single HTP-1/2 depletion domain (outlined), and a univalent uniformly labeled by HTP-1/2 (arrow). In the later three panels, the univalent has been moved to the left to avoid overlapping with the mnT12 bivalent in the projection. (Two chiasmata panel) Five DAPI-stained bodies presentL four regular bivalents (arrowheads) and a large trivalent (outlined) with two HTP-1/2 depletion domains that correspond with the position of the two chiasmata. (C) Projections of early diakinesis meT7 bivalents. The number of chiasmata observed in each case is indicated on the first panel. All chiasmata correspond to HTP-1/2-depleted/SYP-1-enriched domains. Note that in most cases (all but panel V) when more than one chiasma is present, at least one of them is terminally located, and all terminally located chiasmata coincide with HTP-1/2 depletion domains that extend between the chiasma and the closest telomere. In panel V both internally located chiasmata are accompanied by HTP-1/2 depletion domains that are located between the chiasma and the middle of the bivalent.
Second, we examined diplotene/diakinesis nuclei in mnT12 heterozygotes. Ten out of 22 nuclei contained an asymmetric bivalent (comprised of mnT12 connected by a chiasma to one of its unfused counterparts) plus a univalent lacking a chiasma, again in close agreement with Hillers and Villeneuve (2003). In each of these nuclei, the bivalent had a single HTP-1/2-depleted domain (labeled by SYP-1) corresponding to the chiasma, while the univalent was uniformly labeled by α-HTP-1/2 but lacked SYP-1 (Fig. 5B). This indicates that achiasmate chromosomes occurring in an otherwise wild-type genetic background (i.e., containing a full complement of meiotic recombination proteins) do not undergo redistribution of HTP-1/2, further strengthening the association between crossovers and zones of HTP-1/2 depletion. The other 12 nuclei contained a trivalent comprised of mnT12 connected by chiasmata to both the unfused chromosomes IV and X. In these trivalents, each chiasma corresponded to an HTP-1/2-depleted/SYP-1-enriched zone (Fig. 5B). Moreover, the two chiasmata/HTP-1/2-depleted zones were always widely spaced, located near opposite ends of the fusion chromosomes; wide spacing between HTP-1/2-depleted zones parallels the phenomenon of crossover interference, which is highly robust in this organism (Hillers and Villeneuve 2003).
Finally, we examined worms homozygous for the three-chromosome fusion meT7, which harbors roughly half the genome on a single chromosome pair. Among the 19 nuclei analyzed, we observed three meT7 bivalents with one chiasma, 15 with two chiasmata and one with three (Fig. 5C); again, each chiasma corresponded to a zone of HTP-1/2 depletion. The incidence of two-chiasma meT7 bivalents was higher than initially predicted based on an estimated genetic map of 64 cM (Hillers and Villeneuve 2003). This apparent discrepancy was readily reconciled by examination of the locations of the chiasmata in the two-chiasma and three-chiasma bivalents: The estimated genetic map length was based on the explicit assumption that the recombination frequencies in the unassayed chromosome regions near the chromosome ends (representing 12% of the physical length) were similar to recombination frequencies measured for the assayed 88% of the chromosome, but this expectation was not borne out by the cytological data. Instead, we found that for 11 out of 15 bivalents that had two chiasmata, one or both were located very near to a chromosome end, and in the case of the bivalent with three chiasmata, two were terminally located (Fig. 5C). meT7 bivalents that had a single chiasma were cross-shaped, with the chiasma in the middle of the cross, and the HTP-1/2-depleted domain did not extend all the way to a chromosome end (Fig. 5C, panel I). In meT7 bivalents with two chiasmata, all the terminally located chiasmata were associated with HTP-1/2-depleted domains that extended between the chiasma and the nearest chromosome end (Fig. 5C, panels II,III,IV), whereas HTP-1/2-depleted domains associated with internally located chiasmata failed to extend to a chromosome end (Fig. 5C, panesl III,IV,V). These data reinforce the view that a crossover-triggered zone of HTP-1/2 depletion can extend only over a limited distance from the crossover site. Interestingly, for some of the internally located chiasmata, the HTP-1/2 depletion domain extended toward the middle of the chromosome rather than toward the nearest end (Fig. 5C, panel V). Taken together, our data suggest that crossovers are located near the boundary of the HTP-1/2 depletion domain and that removal of HTP-1/2 usually occurs between a crossover site and the closest chromosome end, via a process whose efficiency decreases with distance in a manner that parallels the properties of crossover interference.
Asymmetric features of bivalent differentiation are lost in htp-1 mutants
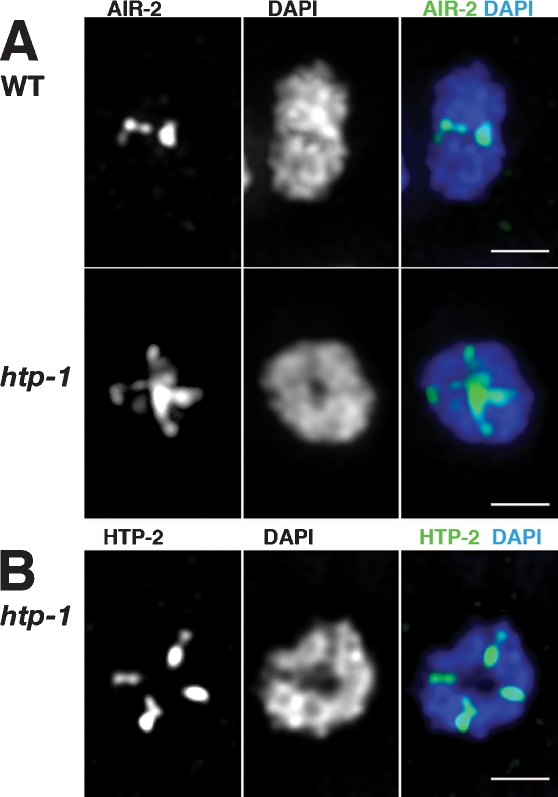
The novel late prophase localization of HTP-1/2 adds a new dimension to a growing body of evidence that, during C. elegans meiosis, crossovers subdivide diakinesis bivalents into highly asymmetric differentiated domains that harbor distinct complements of proteins. We investigated the possibility that HTP-1 might play a role in promoting this differentiation, testing whether asymmetric features of diakinesis bivalents were lost in an htp-1 mutant. Although defective pairing eliminates most autosomal crossovers in the htp-1 mutant, X chromosomes are largely proficient for pairing and form crossovers and chiasmata in ∼20%–35% of meioses (Martinez-Perez and Villeneuve 2005), thereby enabling this analysis. First, we examined localization of Aurora B kinase AIR-2, which can phosphorylate REC-8 in vitro and is required for REC-8 removal and loss of cohesion distal to the chiasma at meiosis I (Kaitna et al. 2002; Rogers et al. 2002). As previously reported, AIR-2 localized specifically to the short arms of wild-type late diakinesis bivalents (i.e., at the midbivalent, in the regions distal to the chiasma) and was excluded from the sister chromatid interfaces of the bivalent long arms (Fig. 6A; Rogers et al. 2002). In the htp-1 mutant, in contrast, AIR-2 was localized at the sister chromatid interfaces of all four arms of the bivalent (Fig. 6A). Further, whereas wild-type late diakinesis bivalents appeared mostly as elongated structures, the bivalents in htp-1 mutants had a more symmetrical appearance, often without clearly distinguishable long and short arms (Fig. 6A,B).
Figure 6.
Loss of asymmetrical features in htp-1 bivalents. (A) Projections of individual late diakinesis bivalents (−1 oocyte) stained with α-AIR-2 antibodies and DAPI. On the wild-type bivalent, α-AIR-2 signals are detected only at the mid-bivalent region (corresponding to the short arms), while in the htp-1 mutant AIR-2 is detected on all four arms of the bivalent. Note that while the DAPI staining of the wild-type bivalent reveals an elongated structure, the htp-1 mutant bivalents display a more rounded structure. (B) Late diakinesis bivalent from an htp-1 mutant, stained with α-HTP-1/2 antibodies and DAPI. HTP-2 protein is detected on all four arms of the bivalent. Bars, 1 μm.
Second, we examined localization of HTP-2 on diakinesis bivalents in htp-1 mutants. Similarly to AIR-2, the HTP-2 protein decorated all arms of the late diakinesis bivalent (Fig. 6B). As these HTP-2 IF signals were readily detected in the htp-1 mutant under conditions where HTP-1/2 IF signals were usually undetectable on the short arms of wild-type bivalents, we interpret this finding as additional evidence of loss of bivalent asymmetry.
The loss of bivalent asymmetry in the htp-1 mutant is not a consequence of crossovers occurring disproportionately near the center of the chromosome, as the distribution of crossovers along the length of the X chromosomes is not altered in the htp-1 mutant (Supplemental Fig. S5; Supplemental Material). Thus, these results support the view that HTP-1 plays a role (beyond its role in promoting crossovers) in setting up the asymmetrical features of late diakinesis bivalents.
HTP-1 is required to prevent premature loss of SCC
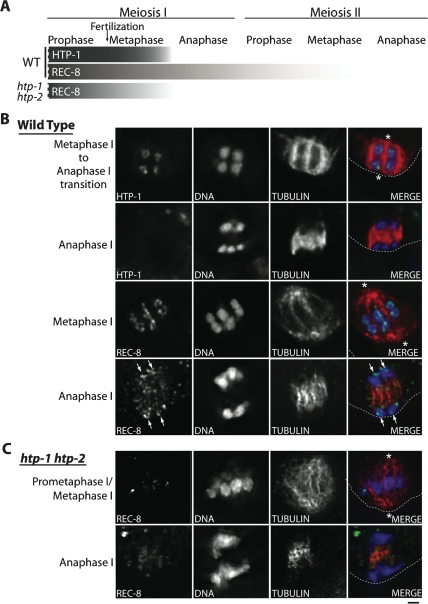
The localization of HTP-1/2 to the domains where cohesion is protected during the first meiotic division, coupled with the mislocalization of AIR-2 to all four arms of the bivalents in htp-1 mutants, prompted us to investigate whether HTP-1 might play a role in regulating loss of cohesion during meiosis. First, we examined the relative timing of departure of REC-8 and HTP-1/2 from the chromosomes during the meiotic divisions (Fig. 7A,B). Chromosome-associated REC-8 diminishes gradually during meiosis I, beginning in late diakinesis. A substantial fraction of REC-8 is removed at anaphase I, but a subset is retained on chromosomes (Rogers et al. 2002). REC-8 continues to be detected on chromosomes through metaphase II, and becomes undetectable at the onset of anaphase II. In contrast, HTP-1/2 are maintained at similar levels from diakinesis through metaphase I, then abruptly disappear at the onset of anaphase I. Simultaneous IF of HTP-1/2 and tubulin (Fig. 7B) showed that HTP-1/2 were readily detected throughout prometaphase I (five out of five embryos) and metaphase I (six out of six embryos), right up until the metaphase/anaphase transition, but were absent at anaphase I (eight out of eight embryos). Moreover, REC-8 was prematurely lost in htp-1 and htp-1 htp-2 mutants (Fig. 7A,C). As in wild-type controls, REC-8 was detected on chromosomes during prometaphase/metaphase I, but in contrast to the wild-type, REC-8 was undetectable on chromosomes between anaphase I and prometaphase II/metaphase II in htp-1 and htp-1 htp-2 mutants (nine out of nine embryos). Together these data suggest that HTP-1/2 play a role in protecting a subset of REC-8 from removal during meiosis I, then are themselves removed at anaphase I to permit removal of the remaining REC-8 during meiosis II.
Figure 7.
Localization of HTP-1/2 and REC-8 during the meiotic divisions. (A) Schematic representation indicating the relative abundance of HTP-1/2 and REC-8 along the long axis of the bivalent (or on univalents) during progression through the meiotic divisions. During wild-type meiosis, the abundance of HTP-1/2 on chromosomes appears constant until the onset of anaphase I, when HTP-1/2 abruptly become undetectable. REC-8 is removed from the chromosomes gradually until it becomes undetectable at the end of metaphase II. In htp-1 htp-2 mutants, REC-8 diminishes during meiosis I and becomes undetectable on chromosomes by anaphase I. (B) Images of partial projections of oocytes undergoing the first meiotic division, stained with Hoechst and α-tubulin and HTP-1/2 antibodies (top two rows) or Hoechst and α-tubulin and REC-8 antibodies (bottom two rows). The top row shows the strong loop-like localization of HTP-1/2 on chromosomes at the metaphase I to anaphase I transition. (Second row) Immediately after this stage, in early anaphase I, HTP-1/2 are absent from chromosomes. Images in the third row show REC-8 localizing in loops on the chromosomes at mid metaphase I; REC-8 is also detected on the chromosomes (arrows), and weakly on the spindle, of an anaphase I oocyte (shown in the bottom row). (C) Images of the first oocyte meiotic division in an htp-1 htp-2 mutant. The top row is labeled “prometaphase I/metaphase I” since congression is impaired by the lack of chiasmata; this combined terminology is used to indicate that chromosomes are mostly, but not fully aligned on the metaphase plate, but the spindle has rotated and is oriented perpendicular to the cortex (characteristic of late metaphase). REC-8 is detected on the univalent chromosomes at prometaphase/metaphase I, but is undetectable on the chromosomes at anaphase I. For B and C, a dotted line marks the edge of the oocyte, and asterisks indicate the spindle poles. Bar, 1 μm.
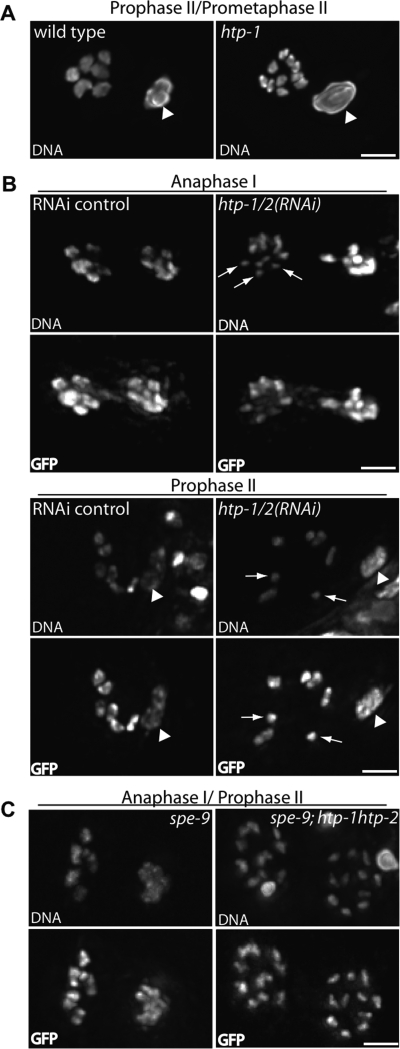
Imaging of chromosome morphology during the meiotic divisions provided direct evidence for premature dissociation of sister chromatids in htp-1 and htp-1 htp-2 mutants (Fig. 8). First, whereas in wild-type embryos at prophase II/prometaphase II, we consistently observed six Hoeschst-stained chromosomes in which sister chromatids remained closely attached to each other (six out of six embryos), normal-appearing chromosomes were not observed in eight out of eight htp-1 and htp-1 htp-2 embryos with a prophase II/prometaphase II chromosome configuration (Fig. 8A). Instead, these embryos either contained well-resolved Hoechst-stained bodies that corresponded in size to single unattached chromatids (four out of eight), or contained a mixture of unattached chromatids and pairs of sister chromatids that were largely separated but remained connected by thin threads (four out of eight).
Figure 8.
Precocious separation of sister chromatids in htp-1 and htp-1 htp-2 mutants. (A) Partial projections of high-resolution images of Hoechst-stained chromosomes from wild-type and htp-1 mutant embryos, showing the first polar body (marked by arrowhead) and the chromosomes in a prophase II/prometaphase II configuration. In the wild-type image, six chromosomes with unseparated sister chromatids can be clearly distinguished. In the htp-1 image, 10 smaller separated Hoechst-stained bodies (of 11 total), corresponding to single chromatids, can be distinguished. (B) Meiotic figures from control embryos and embryos in which HTP-1/2 had been partially depleted by RNAi. Experiments were conducted using a strain expressing GFP∷tubulin (to serve as a staging marker) and GFP∷histone H2B; paired panels show Hoechst (DNA) and GFP fluorescence; polar bodies in the prophase II figures are marked by arrowheads. Examples of isolated single chromatids at anaphase I and prophase II in htp-1/2(RNAi) embryos are indicated by arrows. In control embryos, sister chromatids remain closely associated. (C) Meiotic figures from spe-9 and spe-9; htp-1 htp-2 oocytes, which undergo a prolonged anaphase I and never progress beyond prophase II. Experiments were conducted using worms expressing GFP∷tubulin and GFP∷histone H2B; paired panels show Hoechst (DNA) and GFP fluorescence. In the spe-9 control, six chromosomes with unseparated sister chromatids were easily distinguished in the left cohort of chromosomes. In the spe-9; htp-1 htp-2 oocyte, well-resolved individual chromatids are clearly visible in both cohorts. Bars, 2 μm.
Second, we examined morphology of Hoechst-stained meiotic chromosomes in control and htp-1/2 RNAi embryos from worms expressing GFP∷tubulin (to serve as a staging marker) and GFP∷histone H2B (Fig. 8B). Whereas sister chromatids remained closely associated in 15 out of 15 prophase II control embryos, we observed two or more isolated single chromatids and/or several pairs of sister chromatids connected by thin threads in six out of six prophase II htp-1/2 RNAi embryos. We also scored several anaphase I embryos in which orientation and spacing of chromosomes permitted individual chromosomes to be distinguished; sister chromatids remained closely associated in six out of six controls, while separate single chromatids were detected in two out of two htp-1/2 RNAi embryos.
Finally, we examined meiotic chromosome morphology in oocytes from control and htp-1 htp-2 worms carrying a spe-9 mutation, which results in fertilization-incompetent sperm (Fig. 8C). In spe-9 worms, oocyte meiosis undergoes a prolonged anaphase I and does not progress beyond prophase II; meiosis II spindles do not form, and sister chromatids never separate (McNally and McNally 2005). As expected, meiosis II spindles never formed in either spe-9 or spe-9; htp-1 htp-2 oocytes. However, well-resolved individual chromatids were clearly observed at anaphase I/prophase II in eight out of eight spe-9; htp-1 htp-2 oocytes, whereas sister chromatids remained attached in spe-9 controls (13 out of 13). The presence of separated sister chromatids in htp-1 htp-2 mutant oocytes under conditions where meiosis does not progress beyond prophase II clearly demonstrates a role for HTP-1/2 in preventing premature separation of sister chromatids.
Discussion
Global changes in axis composition triggered by crossovers
We showed that meiotic chromosome axes in C. elegans undergo a striking change in molecular composition that is first visible at late pachytene. Triggering of this remodeling by crossovers (or crossover precursors) results in subdivision of the meiotic bivalent into two distinct domains with different axial composition: a short domain in which HTP-1/2 are depleted and a longer domain in which HTP-1/2 are enriched. The timing of onset of this patterned reorganization of HTP-1/2 coincides with a set of major developmental changes in germline nuclei that includes: (1) activation of the MAP kinase signaling cascade, which induces transit from mid- to late pachytene and progression to later stages of prophase (Church et al. 1995); (2) loss of competence to repair DSBs as interhomolog crossovers (Hayashi et al. 2007), (3) switching off of a specialized meiotic DSBR mode (Hayashi et al. 2007; Smolikov et al. 2007); (4) release from checkpoint-like and structural constraints that favor interhomolog interactions (Martinez-Perez and Villeneuve 2005); and (5) the onset of SC disassembly, beginning with depletion of SC central region proteins from regions where HTP-1/2 become concentrated (Nabeshima et al. 2005). Thus, crossover-triggered reorganization of axial element composition initiated at late pachytene is part of a highly coordinated process by which meiotic chromosomes are transformed from the elongated linear structures visible at mid-pachytene into the condensed, highly asymmetrical cross-shaped bivalent structures present at diakinesis.
Since C. elegans chromosomes range in size from 13 to 21 Mb and the HTP-1/2 depletion domains at late pachytene normally extend for up to one-third of the total SC length, we infer that the patterned changes in axis composition reported here extend over a scale of several Mb. The scale of these changes is substantially larger than crossover-triggered local changes/destabilization of chromosomal axes described in previous reports, as such changes were detected only in close proximity to crossover (or crossover-designated) events. For example, in rat diplotene spermatocytes, bridges between homologs observed at chiasma sites are not stained by REC8 antibodies, but REC8 is present on the axes immediately flanking the chiasma (Eijpe et al. 2003). A similar situation occurs in diplotene bivalents of C. elegans, where the chiasma site coincides with a thinning in REC-8 IF signals (Nabeshima et al. 2005). Finally, EM images of Sordaria pachytene nuclei demonstrate that sister axes are found locally separated at crossover sites (Storlazzi et al. 2008). Thus, our observations of the extensive redistribution of HTP-1/2 during late prophase represent the most striking cytological example of a modification in axis composition associated with crossovers.
Our data contribute to a growing body of evidence supporting a model in which biochemical progression of meiotic recombination is linked to changes in chromosome axis structure (Blat et al. 2002; Kleckner 2006). A recent report showing that C. elegans axial element component HTP-3 forms a complex with DSBs repair proteins RAD-50 and MRE-11 and is proposed to facilitate formation of DSBs by SPO-11 (Goodyer et al. 2008) suggests that meiotic recombination complexes are associated with chromosome axis components from the earliest stages of recombination in C. elegans. Further, both HIM-3 and HTP-1 were previously implicated both in promoting crossovers by inhibiting use of the sister chromatid as a template for DSB repair (Couteau et al. 2004; Martinez-Perez and Villeneuve 2005) and in imposing meiosis-specific requirements for loading of DNA strand exchange proteins (Hayashi et al. 2007). In the present study, we show that patterned reorganization of HTP-1/2 is first detected at the time when recombination events are proposed to mature into crossovers (late pachytene), and this reorganization is coupled to crossover formation. Together, these studies indicate that during C. elegans meiosis, chromosome axis components of the meosis-specific HORMA domain family are intimately associated with the progression of recombination throughout meiotic prophase, and suggest that the maturation of a crossover event is directly linked to chromosome-wide changes in axis composition.
Relationship between HTP-1/2 depletion domains, chiasmata, and crossover sites
Our imaging of diplotene and diakinesis chromosomes has demonstrated a clear correspondence both in position and number between chiasmata and zones of HTP-1/2 depletion. As chiasmata have been demonstrated to correspond to crossover sites in some systems (Tease and Jones 1978) (and are assumed to do so generally), we infer that HTP-1/2 depletion boundaries at diplotene/diakinesis also correspond to crossover sites. Do the boundaries between HTP-1/2-enriched and HTP-1/2-depleted (SYP-1-enriched) domains also correspond to the crossover sites at late pachytene? As soon as they can be distinguished in late pachytene, the HTP-1/2-depleted domain is always much shorter than the HTP-1/2-enriched domain; since C. elegans crossovers occur disproportionately within 30% of the chromosome length from an end (Albertson et al. 1997), this distribution is consistent with crossovers being located near the HTP-1/2 depletion boundary at this stage. Further, a concurrent study has identified C. elegans ZHP-3 as a likely marker of crossover sites, as ZHP-3 localizes to a single bright focus per bivalent during late prophase, located at the position of the chiasma in diplotene bivalents and at the boundary of the SYP-1-enriched (HTP-1/2-depleted) domain on late pachytene chromosomes (Bhalla et al. 2008). Thus, although we cannot independently pinpoint the exact position of crossover events, based on the above observations we suggest that at late pachytene, crossovers are located in close proximity to the boundary of the HTP-1/2 depletion domain.
How is a chromosomal end selected for HTP-1/2 depletion?
In worms with normal karyotypes, HTP-1/2 are consistently retained at the ends of the chromosomes that will lead the way toward the spindle pole at anaphase I and depleted from the ends that localize at the metaphase plate during meiosis I. However, unlike organisms with localized centromeres, during C. elegans meiosis either end of each chromosome has the capacity to lead the way poleward on the meiosis I spindle, and which end does so in a given meiosis is dependent on the position of the crossover (Albertson et al. 1997). This property rules out the possibility that HTP-1/2 are removed from the same chromosomal end in every meiosis, and begs the question of how the domains of HTP-1/2 depletion and retention are determined. It is possible that an inherent asymmetry in the recombination event itself could provide the basis for this decision, as studies in yeast have demonstrated differential processing of sequences flanking opposite sides of Spo11-induced DSBs, suggesting that the ends of DSBs may be biochemically different from the very early stages of recombination (Neale et al. 2005). Although this model could explain why either chromosomal end can be depleted of HTP-1/2, it does not, however, readily explain why the HTP-1/2-depleted domain is normally much shorter than the HTP-1/2-enriched domain. Instead, this property suggests that the HTP-1/2 depletion pattern at late pachytene is defined by the position of the crossover relative to some other feature of global chromosome organization, most likely the nearest chromosome end. Implicit in this model is the existence of a mechanism by which the crossover is able to “communicate” with the rest of the chromosome to identify the closest telomere.
Our observations of two-chromosome (mnT12) and three-chromosome (meT7) fusion bivalents revealed an additional property of the HTP-1/2 depletion process. Whereas HTP-1/2 depletion domains on wild-type diakinesis bivalents invariably extended between the chiasma and a chromosome end, on mnT12 and meT7 bivalents with internally located chiasmata, HTP-1/2 depletion domains often failed to reach a chromosome end. This suggests that although the crossover had induced HTP-1/2 depletion (usually but not always in the direction of the closest telomere), this aspect of axial element remodeling had stopped before reaching a telomere. This feature suggests that crossover-triggered HTP-1/2 depletion can be propagated along a chromosome, but only for a limited distance. For wild-type bivalents, this distance is always sufficient to deplete HTP-1/2 between the crossover site (or its vicinity) and the closest telomere, whereas for fusion chromosome bivalents, the crossover-dependent signal that induces axis remodeling may decay before reaching a telomere. Alternatively, there may be features of chromosome organization present in the fused chromosomes (e.g., internally located subtelomeric regions) that may prevent efficient transmission of the crossover-dependent signal.
HTP-1/2 removal pattern and crossover interference
Several properties of the HTP-1/2 / SYP-1 relocalization process evoke parallels with the phenomenon of crossover interference. In both cases, crossovers (or nascent crossovers) trigger a change in the global properties of chromosomes; i.e., redistribution of HTP-1/2 and SYP-1 proteins, or inhibition of additional nearby crossovers. Likewise, both phenomena appear to involve effects that propagate along the length of a chromosome but that diminish in intensity with distance from the crossover site. Further, a consequence of crossover-triggered axial element remodeling is that it renders chromosomes divided into two domains that are likely unable to support crossover formation, as each domain is depleted for a crossover promoting activity (MacQueen et al. 2002; Martinez-Perez and Villeneuve 2005).
Despite these intriguing parallels, however, there are important distinctions between the two phenomena. In particular, there is a growing body of evidence suggesting that crossover/noncrossover decisions may be made very early in prophase, during the zygotene stage (Bishop and Zickler 2004), whereas the visible patterned redistribution of HTP-1/2 and SYP-1 occurs much later, at late pachytene. This temporal disparity suggests that the creation of two crossover-incompetent domains at late pachytene is unlikely to represent the initial change that establishes interference. However, it may be the case that crossover interference and HTP-1/2/SYP-1 reciprocal localization represent two different downstream manifestations of the same underlying change in chromosome axis organization. Further, reciprocal depletion of HTP-1/2 and SYP-1 at the late pachytene stage may play a role in enforcing pathway decisions made at an earlier stage, by creating a crossover-incompetent environment that functions (in conjunction with removal of the barrier to sister chromatid-directed repair that is proposed to occur at this stage) to ensure that interhomolog crossovers are disfavored.
Parallels with crossover interference also provide a framework for thinking about mechanisms that could lead to the observed pattern of HTP-1/2 and SYP-1 relocalization. In particular, one current class of model for interference may help explain the observed asymmetry in the behavior of chromosomal domains flanking the crossover site with respect to retention or depletion of HTP-1/2. According to the “stress relief” model of interference, meiotic chromosomes are under mechanical stress, which induces crossover designation; crossover designation at a given site in turn relieves stress in flanking regions, thereby inhibiting the formation of further crossovers (Borner et al. 2004; Kleckner et al. 2004). Such relief of stress emanating in both directions from an off-center crossover site could propagate a change in chromosome state that either reaches one chromosome end before the other or that results in a more complete relief of stress on the shorter chromosome segment. This difference, in turn, could form the basis of the differential removal and retention of distinct complements of proteins at the mid–late pachytene transition.
Coupling crossover-triggered differentiation of bivalent subdomains with two-step loss of SCC
A clear consequence of crossover-triggered remodeling of chromosome axis composition is differentiation of the meiotic bivalent into distinct subregions, each harboring distinct complements of proteins that enable two-step loss of cohesion. The short arms of the bivalent, which lack HTP-1/2, define the “midbivalent,” where Aurora B kinase AIR-2 and other components of the chromosome passenger complex will become concentrated just prior to meiosis I. This domain will localize to the metaphase plate at meiosis I, and is fated for REC-8 removal and full SCC loss at the metaphase I/anaphase I transition to allow segregation of homologs to opposite spindle poles. Conversely, the bivalent longs arms, where HTP-1/2 are retained (and colocalize with REC-8) until anaphase I include the domains where sister chromatids will remain conjoined at anaphase I.
The localization of HTP-1/2 on the diakinesis/metaphase I bivalents, the timing of its removal, the loss of bivalent asymmetry in the htp-1 mutant, and the precocious loss of both REC-8 protein and SCC in htp-1 mutants, together indicate that HTP-1 plays a role in promoting the orderly two-step loss of cohesion during the meiotic divisions, by preventing full loss of cohesion at meiosis I. Localized protection of cohesion at meiosis I could be an indirect consequence of the prophase localization of HTP-1, which could function by promoting concentration at the midbivalent of proteins that promote cohesion removal (e.g., AIR-2, which is required for homolog separation and localized REC-8 removal at meiosis I) (Kaitna et al. 2002; Rogers et al. 2002). Alternatively, HTP-1 may also play a more direct, post-prophase role in inhibiting cohesion loss during the meiosis I division. We favor the latter, based on the abrupt disappearance of HTP-1/2 at anaphase I onset. Interestingly, de Carvalho et al. (2008) have independently identified a novel C. elegans protein, LAB-1, that localizes similarly to HTP-1/2 on bivalent long arms and has also been implicated in regulating SCC during meiosis.
A role for HTP-1 in localized protection of sister cohesion at meiosis I is analogous to the role of MEI-S332/Shugoshin proteins, which have been demonstrated to protect cohesion in the vicinity of centromeres during meiosis in fungi, Drosophila, and mammals (Tang et al. 1998; Kitajima et al. 2004; Lee et al. 2008). Although a C. elegans gene encoding a potential shugoshin homolog has been annotated as sgo-1, analysis of sgo-1 mutants has failed to yield evidence of a role for SGO-1 in protecting SCC during meiosis I (de Carvalho et al. 2008; A.F. Dernburg, unpubl.). This suggests either that SGO-1 does not function in this capacity or that another mechanism for localized cohesion protection (e.g., involving HTP-1 and LAB-1) predominates and masks its contribution. Conversely, since HTP-1 belongs to a conserved group of meiosis-enriched HORMA domain proteins, it is possible that other members of this family may contribute to regulated cohesion protection/removal in other organisms. Although such roles have not yet been discovered in other systems, they might be masked by earlier defects in meiosis in the relevant mutants, by the presence of other members of this rapidly evolving gene family (which has undergone gene duplication and functional diversification of paralogs in plants and in multiple animal lineages), or by the predominance of Shugoshin-based cohesion-protection mechanisms.
Concluding remarks
We demonstrated here that formation of crossovers during C. elegans meiosis triggers global changes in chromosome axis organization. These changes both (1) exhibit striking parallels with the phenomenon of crossover interference, and (2) determine later behavior of bivalents subdomains. Coupling of crossover events to patterned redistribution of chromosome axis-associated proteins that subsequently define patterned removal of cohesion provides a mechanism by which organisms with chromosomes that lack localized centromeres can generate “unambivalent bivalents” (Moore and Orr-Weaver 1998) that exhibit predictable and reliable segregation behavior during the meiotic divisions. Conversely, the use of the crossover and emerging chiasma as a trigger for subdividing the bivalent into distinct functional domains important for segregation may have provided the selective pressure for the robust chromosome-wide crossover control mechanism(s) that limit C. elegans chromosomes to a single crossover per homolog pair.
Materials and methods
Genetics
C. elegans strains were cultured at 20°C under standard conditions (Brenner 1974). The following mutations and chromosome rearrangements were used: LG I: zhp-3 (jf61), spe-9(hc88); LG IV: htp-1(gk174), htp-2(tm2543), rec-8 (ok978), spo-11(ok79), him-3(gk149), msh-5(me23), nT1[unc-?(n754) let-?(m435)](IV;V), nT1[unc-?(n754) let-? qIs50](IV;V); LG X: dpy-3(e27), unc-3(e151). Chromosome fusions used were mnT12 (X; IV) and meT7 (III; X; IV).
Cytological analyses
Immunostaining of dissected gonads from 20-h post-L4 adults was performed as in Martinez-Perez and Villeneuve (2005) with modifications described in the Supplemental Material. The following primary antibodies were used at the indicated dilutions: α-HTP-1/2 (1:200) (antibodies were raised in rabbits against the whole HTP-1 protein), α-SYP-1 (1:50) (MacQueen et al. 2002), α-AIR-2 (1:200) (Schumacher et al. 1998), α-HTP-3 (MacQueen et al. 2005). Images were acquired as stacks of optical sections at 0.2-μm intervals using a DeltaVision deconvolution microscopy system
For images in Figures 3B, 7, and 8A, and Supplemental Figure S2, we used fixation procedures optimized for preservation of microtubules (Gonczy et al. 1999; Oegema et al. 2001), with modifications described in the Supplemental Material. Mouse α-REC-8 (CIM, Arizona State University), rabbit α-HTP-1/2, rabbit α-AIR-2, and rat α-HIM-10 (Howe et al. 2001) antibodies were used at 1:200, 1:500, 1:250, and 1:500, respectively. Microtubules were stained with FITC-conjugated anti-α-tubulin monoclonal antibody DM 1A (Sigma).
For experiments in Figure 8B, feeding RNAi was performed at 15°C using worms carrying transgenes expressing GFP∷H2B and GFP∷tubulin (Praitis et al. 2001). Clone JA:F41H10.10 (Kamath and Ahringer 2003) was used to deplete htp-1 and htp-2 RNA; empty vector L4440 was used as the control. For Figure 8C, spe-9(hc88ts) and spe-9(hc88ts); htp-1 htp-2 L4 worms expressing GFP∷H2B and GFP∷tubulin were shifted from 20°C to 25°C for 17–20 h to inactivate sperm, resulting in a block to progression of oocyte meiosis (McNally and McNally 2005). In both cases, whole worms were fixed in ethanol (Bessler et al. 2007) and stained with Hoechst 33258.
Acknowledgments
We thank the Caenorhabditis Genetics Center, the Gene Knockout Consortium and the National Bioresource Project for strains; A. Straight, B. Meyer, and J. Schumacher for antibodies; A. Goldman for critical reading of the manuscript; and S. Wignall for timely experimental assistance in the final stages of this work. This work was supported by a BBSRC David Phillips Fellowship to E.M.-P., a BBSRC post-doctoral contract to C.B., a BBSRC Studentship to J.L., NIH grants R01GM53804 and R01GM67268 to A.M.V., and a CIHR fellowship to M.S.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1694108.
References
- Albertson D.G., Rose A.M., Villeneuve A.M. Chromosome organization, mitosis, and meiosis. In: Riddle D.L., et al., editors. C. elegans II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 47–48. [PubMed] [Google Scholar]
- Aravind L., Koonin E.V. The HORMA domain: A common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem. Sci. 1998;23:284–286. doi: 10.1016/s0968-0004(98)01257-2. [DOI] [PubMed] [Google Scholar]
- Bessler J.B., Reddy K.C., Hayashi M., Hodgkin J., Villeneuve A.M. A role for Caenorhabditis elegans chromatin-associated protein HIM-17 in the proliferation vs. meiotic entry decision. Genetics. 2007;175:2029–2037. doi: 10.1534/genetics.107.070987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N., Wynne D.J., Jantsch V., Dernburg A.F. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 2008. doi: 10.1371/journal.pgen.1000235. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D.K., Zickler D. Early decision; Meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- Blat Y., Protacio R.U., Hunter N., Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Borner G.V., Kleckner N., Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo J.A., Johnson A.L., Sedgwick S.G., Cha R.S. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008;132:758–770. doi: 10.1016/j.cell.2008.01.035. [DOI] [PubMed] [Google Scholar]
- Caryl A.P., Armstrong S.J., Jones G.H., Franklin F.C. A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma. 2000;109:62–71. doi: 10.1007/s004120050413. [DOI] [PubMed] [Google Scholar]
- Church D.L., Guan K.L., Lambie E.J. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development. 1995;121:2525–2535. doi: 10.1242/dev.121.8.2525. [DOI] [PubMed] [Google Scholar]
- Couteau F., Zetka M. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes & Dev. 2005;19:2744–2756. doi: 10.1101/gad.1348205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., Nabeshima K., Villeneuve A., Zetka M. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 2004;14:585–592. doi: 10.1016/j.cub.2004.03.033. [DOI] [PubMed] [Google Scholar]
- de Carvalho C.E., Zaaijer S., Smolikov S., Gu Y., Schumacher J.M., Colaiácovo M.P. LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes & Dev. 2008 doi: 10.1101/gad.1691208. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijpe M., Offenberg H., Jessberger R., Revenkova E., Heyting C. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1β and SMC3. J. Cell Biol. 2003;160:657–670. doi: 10.1083/jcb.200212080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P., Pichler S., Kirkham M., Hyman A.A. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyer W., Kaitna S., Couteau F., Ward J.D., Boulton S.J., Zetka M. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev. Cell. 2008;14:263–274. doi: 10.1016/j.devcel.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Hayashi M., Chin G.M., Villeneuve A.M. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 2007;3:e191. doi: 10.137/journal.pgen.003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers K.J., Villeneuve A.M. Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol. 2003;13:1641–1647. doi: 10.1016/j.cub.2003.08.026. [DOI] [PubMed] [Google Scholar]
- Hollingsworth N.M., Goetsch L., Byers B. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell. 1990;61:73–84. doi: 10.1016/0092-8674(90)90216-2. [DOI] [PubMed] [Google Scholar]
- Howe M., McDonald K.L., Albertson D.G., Meyer B.J. HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J. Cell Biol. 2001;153:1227–1238. doi: 10.1083/jcb.153.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch V., Pasierbek P., Mueller M.M., Schweizer D., Jantsch M., Loidl J. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol. Cell. Biol. 2004;24:7998–8006. doi: 10.1128/MCB.24.18.7998-8006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G.H. Chiasmata. In: Moens P.B., editor. Meiosis. Academic Press; Orlando, FL: 1987. pp. 213–244. [Google Scholar]
- Jones G.H., Franklin F.C. Meiotic crossing-over: Obligation and interference. Cell. 2006;126:246–248. doi: 10.1016/j.cell.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Kaitna S., Pasierbek P., Jantsch M., Loidl J., Glotzer M. The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr. Biol. 2002;12:798–812. doi: 10.1016/s0960-9822(02)00820-5. [DOI] [PubMed] [Google Scholar]
- Kamath R.S., Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kelly K.O., Dernburg A.F., Stanfield G.M., Villeneuve A.M. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics. 2000;156:617–630. doi: 10.1093/genetics/156.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima T.S., Kawashima S.A., Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Chiasma formation: Chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–194. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Zickler D., Jones G.H., Dekker J., Padmore R., Henle J., Hutchinson J. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kitajima T.S., Tanno Y., Yoshida K., Morita T., Miyano T., Miyake M., Watanabe Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat. Cell Biol. 2008;10:42–52. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
- MacQueen A.J., Colaiacovo M.P., McDonald K., Villeneuve A.M. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes & Dev. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Phillips C.M., Bhalla N., Weiser P., Villeneuve A.M., Dernburg A.F. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123:1037–1050. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., Villeneuve A.M. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes & Dev. 2005;19:2727–2743. doi: 10.1101/gad.1338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K.L., McNally F.J. Fertilization initiates the transition from anaphase I to metaphase II during female meiosis in C. elegans. Dev. Biol. 2005;282:218–230. doi: 10.1016/j.ydbio.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Moore D.P., Orr-Weaver T.L. Chromosome segregation during meiosis: Building an unambivalent bivalent. Curr. Top. Dev. Biol. 1998;37:263–299. doi: 10.1016/s0070-2153(08)60177-5. [DOI] [PubMed] [Google Scholar]
- Muller H.J. The mechanisms of crossing over. Am. Nat. 1916;50:193–221. [Google Scholar]
- Nabeshima K., Villeneuve A.M., Hillers K.J. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics. 2004;168:1275–1292. doi: 10.1534/genetics.104.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve A.M., Colaiacovo M.P. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J. Cell Biol. 2005;168:683–689. doi: 10.1083/jcb.200410144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Schleiffer A. From a single double helix to paired double helices and back. Philos. Trans. R. Soc. Lon. B Biol. Sci. 2004;359:99–108. doi: 10.1098/rstb.2003.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M.J., Pan J., Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Wan L., Baumgartner B., Schaefer D., Loidl J., Hollingsworth N.M. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K., Nakano M., Eiguchi M., Suzuki T., Kurata N. PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J. Cell Sci. 2006;119:217–225. doi: 10.1242/jcs.02736. [DOI] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., Hyman A.A. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S.L., Hawley R.S. Chromosome choreography: The meiotic ballet. Science. 2003;301:785–789. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D., Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes & Dev. 2001;15:1349–1360. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Siomos M.F., Nasmyth K. Un menage a quatre: The molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers E., Bishop J.D., Waddle J.A., Schumacher J.M., Lin R. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J. Cell Biol. 2002;157:219–229. doi: 10.1083/jcb.200110045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J.M., Golden A., Donovan P.J. AIR-2: An Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegansembryos. J. Cell Biol. 1998;143:1635–1646. doi: 10.1083/jcb.143.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Eizinger A., Hurlburt A., Rogers E., Villeneuve A.M., Colaiacovo M.P. Synapsis-defective mutants reveal a correlation between chromosome conformation and the mode of double-strand break repair during Caenorhabditis elegans meiosis. Genetics. 2007;176:2027–2033. doi: 10.1534/genetics.107.076968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A., Tesse S., Ruprich-Robert G., Gargano S., Poggeler S., Kleckner N., Zickler D. Coupling meiotic chromosome axis integrity to recombination. Genes & Dev. 2008;22:796–809. doi: 10.1101/gad.459308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T.T., Bickel S.E., Young L.M., Orr-Weaver T.L. Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes & Dev. 1998;12:3843–3856. doi: 10.1101/gad.12.24.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tease C., Jones G.H. Analysis of exchanges in differentially stained meiotic chromosomes of Locusta migratoria after BrdU-substitution and FPG staining. Chromosoma. 1978;69:163–178. [Google Scholar]
- Zetka M.C., Kawasaki I., Strome S., Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes & Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]