Abstract
Na+/K+ ATPase is a plasma membrane-localized sodium pump that maintains the ion gradients between the extracellular and intracellular environments, which in turn controls the cellular resting membrane potential. Recent evidence suggests that the pump is also localized at synapses and regulates synaptic efficacy. However, its precise function at the synapse is unknown. Here we show that two mutations in the α subunit of the eat-6 Na+/K+ ATPase in Caenorhabditis elegans dramatically increase the sensitivity to acetylcholine (Ach) agonists and alter the localization of nicotinic Ach receptors at the neuromuscular junction (NMJ). These defects can be rescued by mutated EAT-6 proteins which lack its pump activity, suggesting the presence of a novel function for Ach signaling. The Na+/K+ ATPase accumulates at postsynaptic sites and appears to surround Ach receptors to maintain rigid clusters at the NMJ. Our findings suggest a critical pump activity-independent, allele –specific role for Na+/K+ ATPase on postsynaptic organization and synaptic efficacy.
Introduction
Clustering of neurotransmitter receptors at appropriate postsynaptic sites is a crucial step in synaptic maturation. Rigid and concentrated localization of receptors enables the effective synaptic transmission and controls neural network activity. Regulated synaptic activity also defines cellular excitability, which affects the survival of neurons as well as maintenance of brain functions. The clustering of nicotinic acetylcholine receptors (nAchRs) at neuromuscular junctions (NMJs) in vertebrates requires the expression of agrin protein in motor neuron terminals, which then activates the postsynaptic protein muscle-specific receptor kinase (MuSK) followed by the accumulation of rapsyn at postsynaptic sites and nAchR concentration (Sanes and Lichtman, 2001). However, it is not fully understood how each receptor is expressed and clustered at specific synapses, which molecules define the regions of clustering, and which mechanisms maintain the overall synaptic efficacy through the excitability of neuronal cells.
Na+/K+ ATPase, a plasmamembrane-localized sodium pump, acts in the maintenance of ion gradients in all animal cells (Kaplan, 2002). Following membrane excitation, the pump restores the ion gradients across the membrane by continually extruding three Na+ ions from the cell and importing two K+ ions into the cell. The sodium pump consists of two essential subunits, α and β; however, the vertebrate Na+/K+ ATPase is predicted to have an additional γ subunit. The catalytic α subunit has ten transmembrane domains and an ATP-binding domain, while the β subunit is thought to be a regulatory subunit for pump maturation, membrane transport and membrane insertion. In the vertebrate brain, several isoforms of the Na+/K+ ATPase are ubiquitously expressed (Mobasheri et al., 2000), but only the α3 isoform is known to be strongly localized at synapses, where it serves as a receptor for neuronal agrin (Hilgenberg et al., 2006; Hoover et al., 2003). Considering the fact that the highest levels of agrin expression in the developing brain coincide with periods of synapse formation (Cohen et al., 1997; Li et al., 1997), these results raise the possibility that the Na+/K+ ATPase has a function for regulation of synapse structure in addition to regulation of the resting-membrane potential.
To understand the roles of the Na+/K+ ATPase in synaptic function, we examined sensitivities to several agonists for synaptic receptors and the localization patterns of those receptors in postsynaptic muscle cells in Caenorhabditis elegans eat-6 mutants. eat-6 encodes an α subunit of Na+/K+ ATPase in C. elegans, and mutations in eat-6 affect the membrane potential in pharyngeal muscle cells (Davis et al., 1995). The C. elegans body-wall muscle cells contain two nAchRs and one GABA receptor (Richmond and Jorgensen, 1999). These nAchRs are discriminated from each other by their sensitivities to different Ach agonists: the levamisole-sensitive receptor and nicotine-sensitive receptor. The levamisole receptor is thought to contain UNC-38/UNC-63 α subunits and UNC-29/LEV-1 non-α subunits (Culetto et al., 2004; Fleming et al., 1997; Jones and Sattelle, 2004; Richmond and Jorgensen, 1999), whereas the nicotine receptor contains ACR-16 α subunit (Francis et al., 2005). Recently, the transmembrane CUB/LDL protein LEV-10 was found to regulate the clustering of levamisole-sensitive receptors at NMJs (Gally et al., 2004), and the receptor tyrosine kinase CAM-1 was required for the localization of the nicotine-sensitive ACR-16 receptor at postsynaptic sites (Francis et al., 2005). Thus, different transmembrane proteins seem to have function to cluster different nAchRs at the C. elegans NMJs, which may have the function equivalent to that of MuSK in the vertebrate NMJ. However, no homologs of vertebrate agrin have been found in C. elegans based on primary sequence analyses, and no rapsyn-like protein has been identified in C. elegans. We hypothesize that there should be another molecular mechanisms exist to build a well-defined configuration of the synapse structure in C. elegans NMJ. Here we provide a line of the evidence that mutations in eat-6 affect synaptic efficacy by altering the expression and localization of nAchRs at the NMJ. Moreover, we show that these defects are allele-specific, and seem to be independent of its pump activity.
RESULTS
Abnormal Ach synaptic transmission in Na+/K+ ATPase mutants
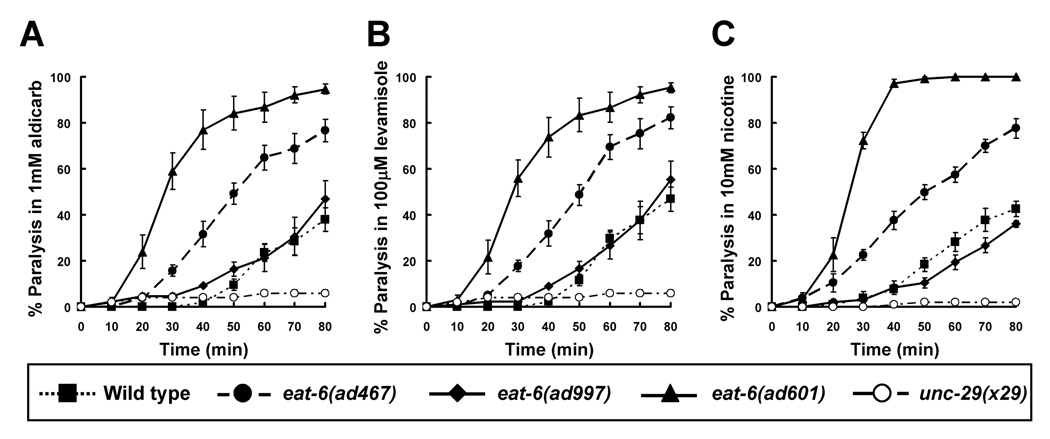
To elucidate the synaptic functions of the Na+/K+ ATPase, we first examined the sensitivity of the C. elegans Na+/K+ ATPase mutant eat-6 to several drugs that target cholinergic synaptic transmission (Fig. 1). In C. elegans, Ach signaling at the NMJ can be indirectly investigated by examining sensitivity to the Ach esterase inhibitor aldicarb. Aldicarb treatment causes acute paralysis in worms by inducing the accumulation of Ach at synaptic clefts. However, mutations that confer reduced Ach signaling causes resistance to aldicarb (Miller et al., 1996; Sieburth et al., 2005). Of the three eat-6 mutant alleles tested (ad467, ad601 and ad997, see Fig. 7 for amino acid substitutions), the eat-6(ad601) mutant was paralyzed by aldicarb significantly faster than wild-type animals (Fig. 1A). The eat-6(ad467) mutants also showed a hypersensitive phenotype, but it was milder than that of the ad601 animals. In contrast, the response of the eat-6(ad997) mutants was similar to that of the wild-type animals. The increased sensitivity of the ad601 and ad467 mutants to aldicarb suggests an elevated Ach signaling at the NMJ.
Figure 1. Drug responses of the eat-6 Na+/K+ ATPase mutants.
(A) Responses to 1 mM aldicarb, an inhibitor of ach esterase. (B) 100 µM levamisole. (C) 10 mM nicotine. Levamisole and nicotine are potent nAchR agonists. The ad601 mutants were significantly sensitive to all drugs, and the ad467 animals were significantly sensitive though (P < 0.05 from 40 to 80 min observation, compared to the wild type, one-way ANOVA with Turkey’s multiple comparisons). The unc-29 mutant was assayed simultaneously as a negative control. Error bars indicate the S.E.M. (n > 4 replicates).
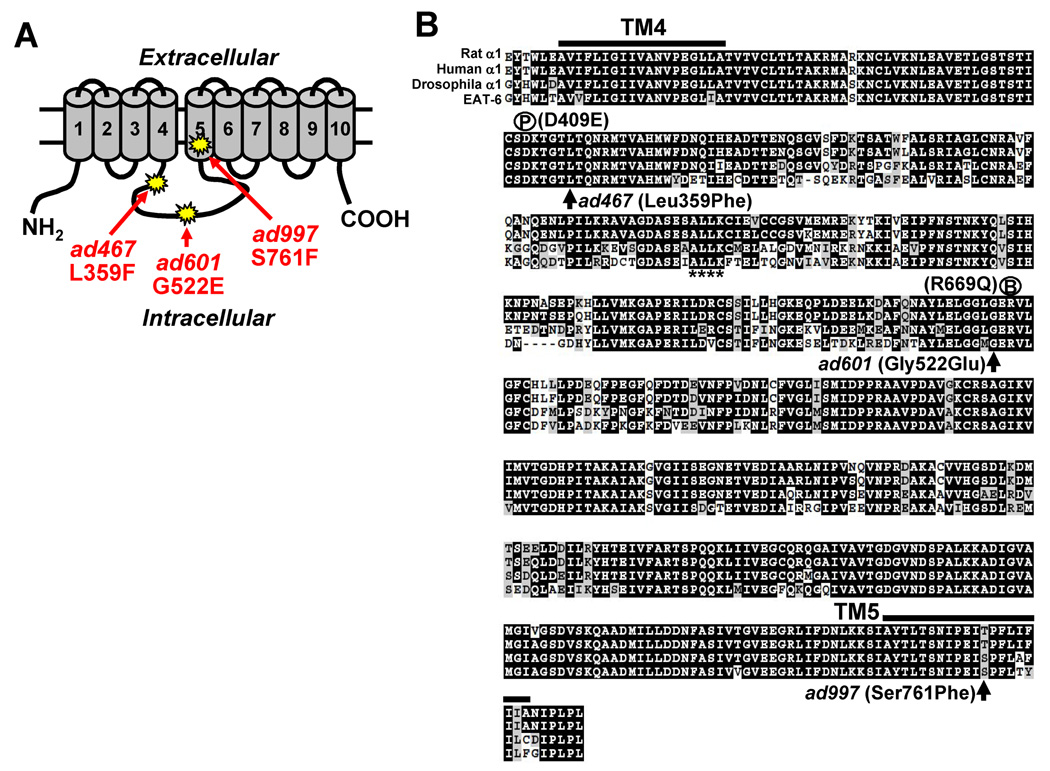
Figure 7. Amino acid alignment of Na+/K+ ATPase α1 subunits.
(A) Schematic representation of the Na+/K+ ATPase α subunit. Columns indicate the transmembrane regions. The sites of three eat-6 mutant alleles are shown. (B) The amino acid sequences of rat α1, human α1, Drosophila α1 and EAT-6 were aligned using ClustalW. The region from the 4th transmembrane domain to the 5th transmembrane domain is shown. Each mutation site is indicated by arrows. P, phosphorylation site; B, ATP-binding site. The ankyrin-binding motif is shown by asterisks under the amino acids.
To determine whether the increases in aldicarb sensitivity were due to altered transmitter release from presynaptic motor neurons or to an increase in postsynaptic muscle sensitivity, each of the mutants was treated with levamisole, a potent Ach agonist in C. elegans (Fleming et al., 1997). Similar to the results seen with aldicarb, the ad601 mutants were significantly hypersensitive to levamisole (Fig. 1B), suggesting that the postsynaptic response to the agonist was increased. We found that the ad601 mutation also caused severe hypersensitivity to nicotine, another agonist of Ach (Fig. 1C). However, the ad997 mutants did not differ from the wild type in their response to levamisole (Fig. 1B), and they looked to be weakly resistant to nicotine (Fig. 1C). The ad467 mutant animals showed an intermediate response to both agonists. These pharmacological data indicate that the ad601 and ad467 mutations in the EAT-6 Na+/K+ ATPase increase the postsynaptic cellular response to Ach agonists at the NMJ. On the other hand, the ad997 mutation does not affect this response and may even lower it, suggesting that these phenotypes are caused in an allele-specific manner.
The responses of the eat-6 mutants to Ach agonists are independent of Na+/K+ ATPase pump activity
One obvious function of the Na+/K+ ATPase is the Na+ pump activity and it is the reasonable hypothesis to explain our observations is that the Na+ pump activity is affected differently by these mutations. To test this hypothesis, we introduced mutated Na+/K+ ATPase, which contained an abolished activity of the sodium pump, and examined their rescue ability to each defect in different eat-6 mutants. We expected that, if the pump activity is necessary to restore the defects we observed in the eat-6 mutants, expression of pump-dead EAT-6 proteins should not restore these defects or even worsen them.
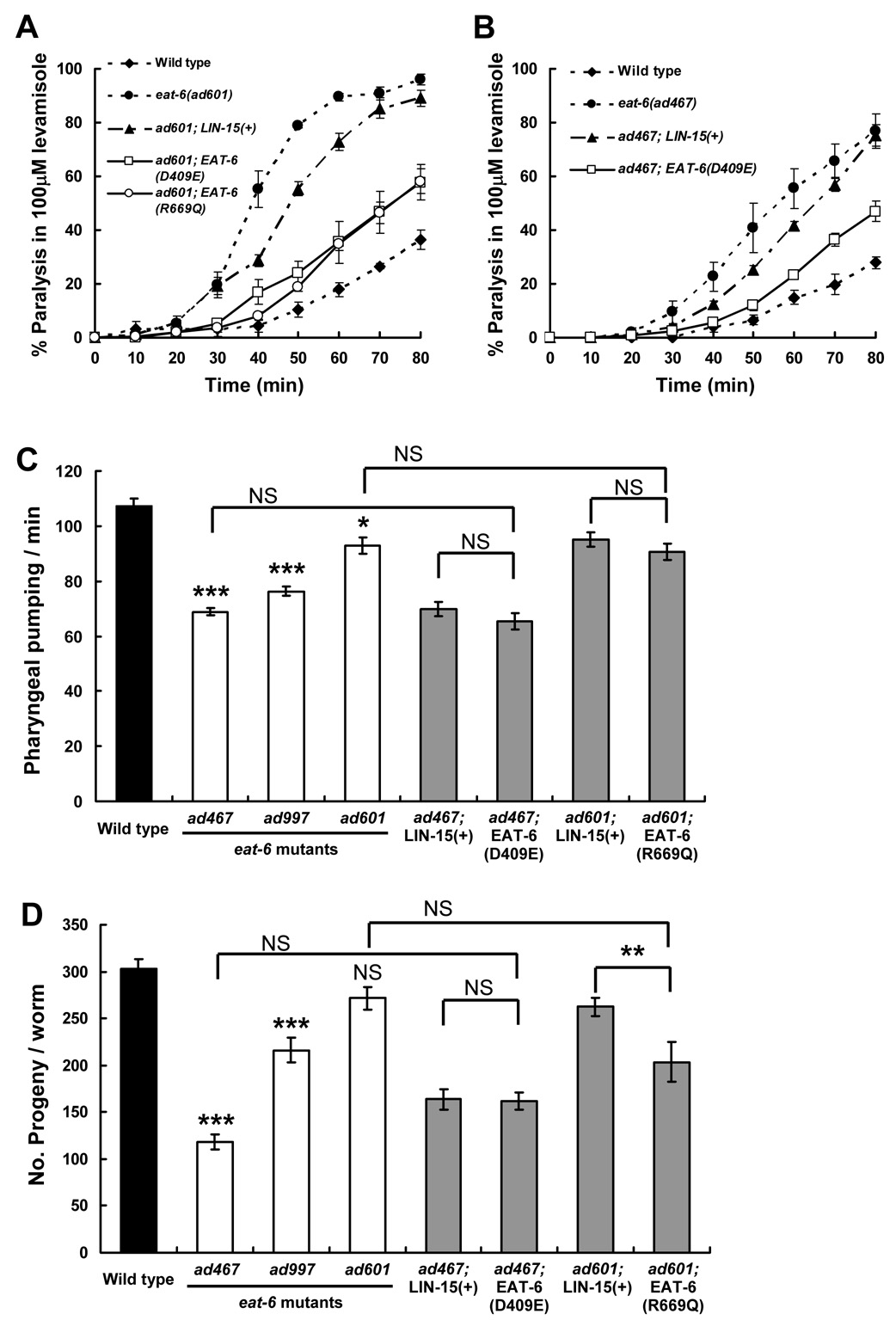
The 354th Asp of EAT-6 protein, which corresponds to the 409th Asp in human α1 subunit of Na+/K+ ATPase, was changed to Glu to substitute the phosphorylation site for the catalytic cycle (hereafter this mutation is referred to as D409E; Liang et al., 2006; Ohtsubo et al., 1990). Similarly, the 524th Arg, which corresponds to the 699th Arg in human α1, was changed to Gln to disrupt ATP binding (R669Q; Jacobsen et al., 2002). Both residues are well conserved from worm to human (see Fig. 7B), and eliminate its pump activity. Unexpectedly, these pump-dead Na+/K+ ATPase restored the increased sensitivity to levamisole and nicotine in both the ad601 and ad467 mutants (Fig. 2A, B, and data not shown). These results strongly indicate that the EAT-6 Na+/K+ ATPase regulates cholinergic synaptic transmission through independent from its pump activity.
Figure 2. Pump activity-dead EAT-6 mutations recover levamisole sensitivity in eat-6 mutants.
(A, B) Responses to 100µM levamisole in the transgenic animals bearing pump-dead EAT-6 transgenes. LIN-15(+) indicates the control transgene containing only the injection marker DNA. Both D409E and R669Q EAT-6 mutations significantly rescued the increased sensitivity of the ad601 (A) and ad467 alleles (B) (P < 0.05 from 50 to 80 min observation, compared to the control transgenic lines, Student’s two-tailed t-test). Error bars indicate SEM. (C) Number of pharyngeal pumping in 30 s. (n = 12). (D) Average number of progeny per worm. (n = 15). Three eat-6 mutant alleles (white bars) were compared to the wild type (black bar); the results of the statistical analyses are indicated above each bar. Transgenic animals (grey bars) bearing pump-dead EAT-6 were compared to both the corresponding mutant and control transgenic animals. *** P < 0.001, ** P < 0.01, * P < 0.05, Student’s two-tailed t-test. NS, not significant. Error bars indicate SEM.
We also investigated whether other defects observed in each eat-6 mutants have similar phenotypic variation with agonist sensitivities and whether these defects can be rescued by pump-dead EAT-6 or not. The eat-6 mutants show an impaired pharyngeal pumping, decreased number of progeny and a defective pharyngeal muscle membrane potential ((Davis et al., 1995) and Fig. 2C, D). It was previously shown that the strength of these phenotypes well correlates with the electrogenic pump activity presumed from the recording of electropharyngeogram (EPG) (Davis et al., 1995). Interestingly, the phenotypic strength of these defects does not correspond to that of agonist sensitivities: the ad467 mutation causes the severest defects and the mildest in the ad601 allele. Furthermore, pump-dead EAT-6 proteins did not rescue both the pumping defects and decreased number of progeny in the ad467 and ad601 mutants at all (Fig. 2C, D). No significant recoveries in the pump-dead EAT-6 transgenic animals were observed compared with the control transgenic animals that bear only injection-marker genes (compare ad467; EAT-6(D409E) or ad601; EAT-6(R669Q) vs. ad467; Lin-15(+) or ad601; Lin-15(+) in Fig. 2C, D). This suggests that introduced mutations do not recover all the defects observed in eat-6 mutants, and that these defects are presumably dependent on the pump activity. All of these results using the pump-dead EAT-6 indicate that the altered Ach signaling in eat-6 mutants is specifically independent on its electrogenic pump activity and that a novel function for Na+/K+ ATPase may be involved in Ach signaling at the NMJ.
Mutations in the Na+/K+ ATPase alter the expression and localization patterns of nAch receptors
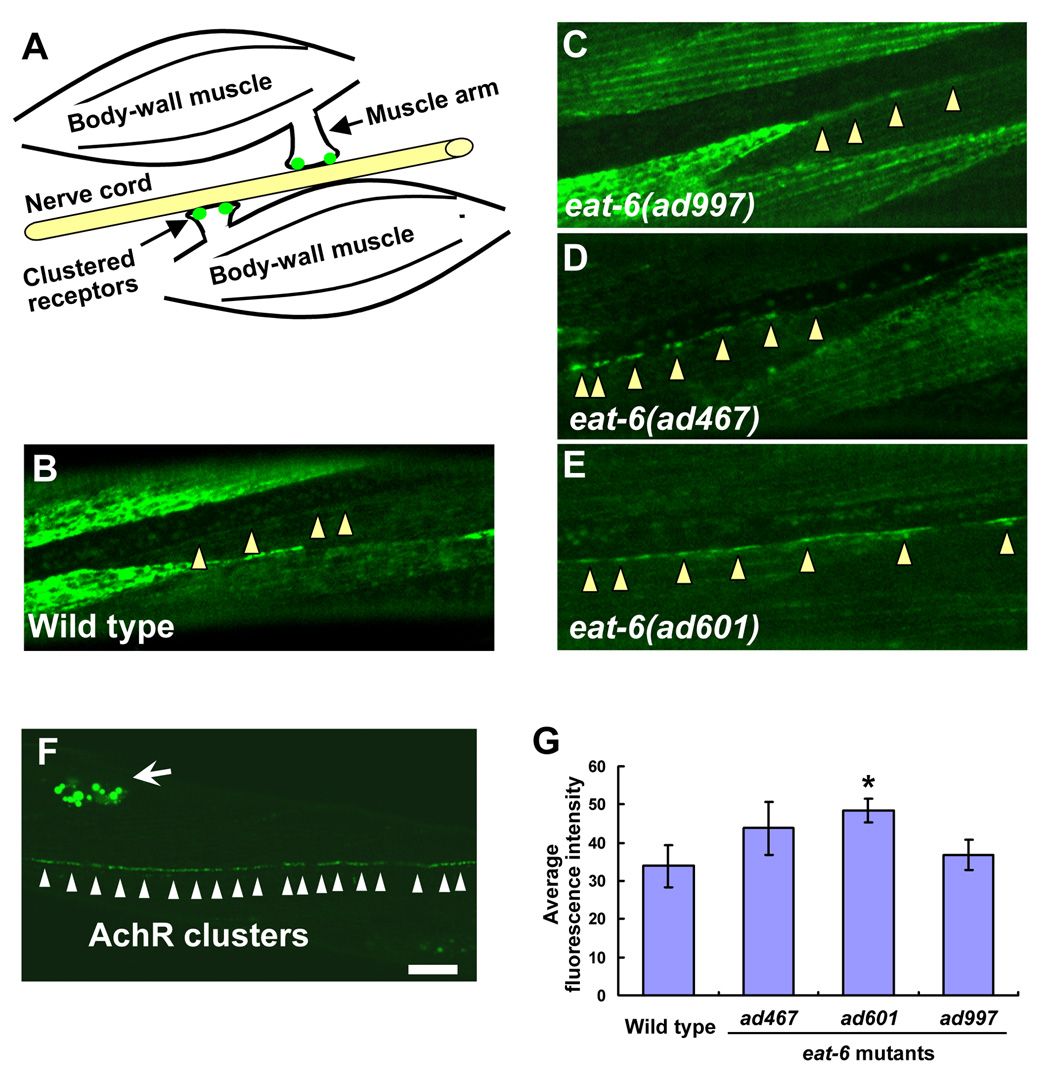
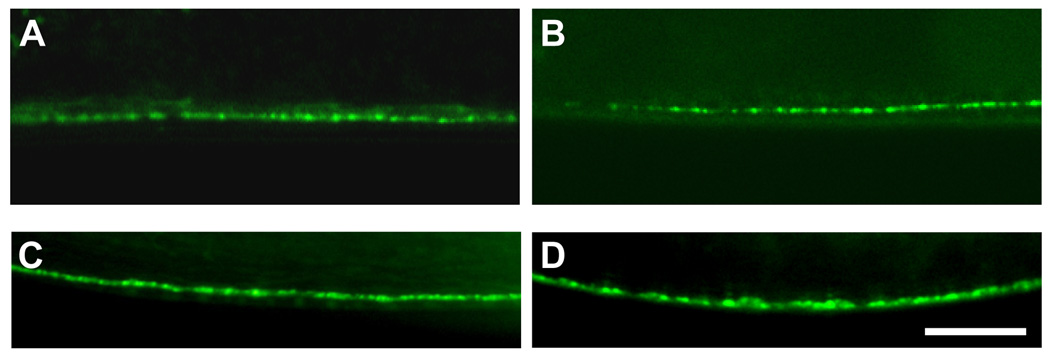
To further clarify the effects of the ad467 and ad601 mutations on Ach signaling, we examined whether nAchR expression was altered in the postsynaptic body-wall muscles. The nAchRs are discriminated by their different sensitivities to Ach agonists: UNC-29 is the levamisole-sensitive receptor and ACR-16 is the nicotine-sensitive receptor. We expressed UNC-29::GFP and ACR-16::GFP fusion proteins under the control of the muscle-specific promoter myo-3 (Okkema et al., 1993), and their expression and localization patterns at the NMJ were examined. At the C. elegans NMJ, each body-wall muscle cell innervates muscle arms toward the axon of presynaptic motor neurons, which lie in the ventral or dorsal nerve cord. Ach receptors accumulate at the tips of muscle arms to form mature synapses aligned with presynaptic sites (Fig. 3A). Slight accumulation of the UNC-29 fusion protein was observed at the NMJ in the wild-type animals (Fig. 3B), whereas strong intracellular localization of the fusion protein was detected in the muscle cells, consistent with the data previously shown by the serial confocal microscopic sections (Fleming et al., 1997). The ad997 mutants also showed strong intracellular accumulation of the fusion protein (Fig. 3C). In the ad467 and ad601 mutants, however, a strong GFP signal was observed along the ventral nerve cord, while the intracellular localization of the GFP fusion protein was substantially decreased in the muscle cells compared to that in the wild-type and in ad997 animals (Fig. 3D, E). This suggests that these mutations may affect the intracellular transport of the receptor to or from the synaptic membrane. However, the fusion of GFP to UNC-29 might influence receptor transport or its insertion into the membrane because GFP itself is relatively large. To address this concern, we took another approach. We generated strains expressing the non-α subunit LEV-1 tagged with 3xHA (hemagglutinin) at the C-terminal end which is predicted to be exposed to the extracellular environment. We detected the membrane-inserted levamisole-sensitive receptor at the NMJ by injecting fluorescently-labeled anti-HA antibodies into the body cavity (Gottschalk et al., 2005; Gottschalk and Schafer, 2006). Six hours after antibody injection, small fluorescent puncta were observed along the ventral nerve cord of the wild-type animals, indicating the localization of the membrane-inserted LEV-1 receptors (Fig. 3F). The excess antibodies were taken up via endocytsis into the coelomocytes. We quantified the amount of fluorescence along the ventral nerve cord in each eat-6 mutant, and found that postsynaptic expression of the levamisole-sensitive receptors was significantly increased in the ad601 mutants (Fig. 3G). Although a significant difference was not seen in the ad467 mutants, both mutations increased the membrane-transport and/or –insertion of nAchRs. The level of expression in the ad997 mutants was similar to that in the wild-type animals.
Figure 3. Increased expression and localization of levamisole-sensitive nAchRs in the eat-6 mutants.
(A) Schematic drawing of a C. elegans NMJ. Each muscle cell in the left or right ventral muscle quadrant innervates muscle arms and forms synapses on the axon of motor neurons. Clustered receptors are localized on the distal part of the muscle arms. (B – E) Localization patterns of UNC-29:: GFP fusion protein at NMJs along the ventral nerve cord. (B) Wild type. (C) eat-6(ad997). (D) eat-6(ad467). (E) eat-6(ad601). Arrowheads indicate NMJs along the ventral nerve cord. Note the strong GFP signal in the ad467 and ad601 animals compared to undetectable signal in the wild-type and ad997 animals. (F) A staining pattern for LEV-1:: 3xHA fusion protein using fluorescently-labeled anti-HA antibodies in the wild-type animal. Six hours after the injection of the antibody, puncta were observed along the ventral nerve cord. Excess antibodies were removed by coelomocytic endocytosis (arrow). (G) Average fluorescence intensity in the wild-type and mutant animals (n = 8–12). * P < 0.05, compared to the wild type, two-tailed Mann-Whitney U-test. Error bars indicate SEM. Scale bar, 10 µm.
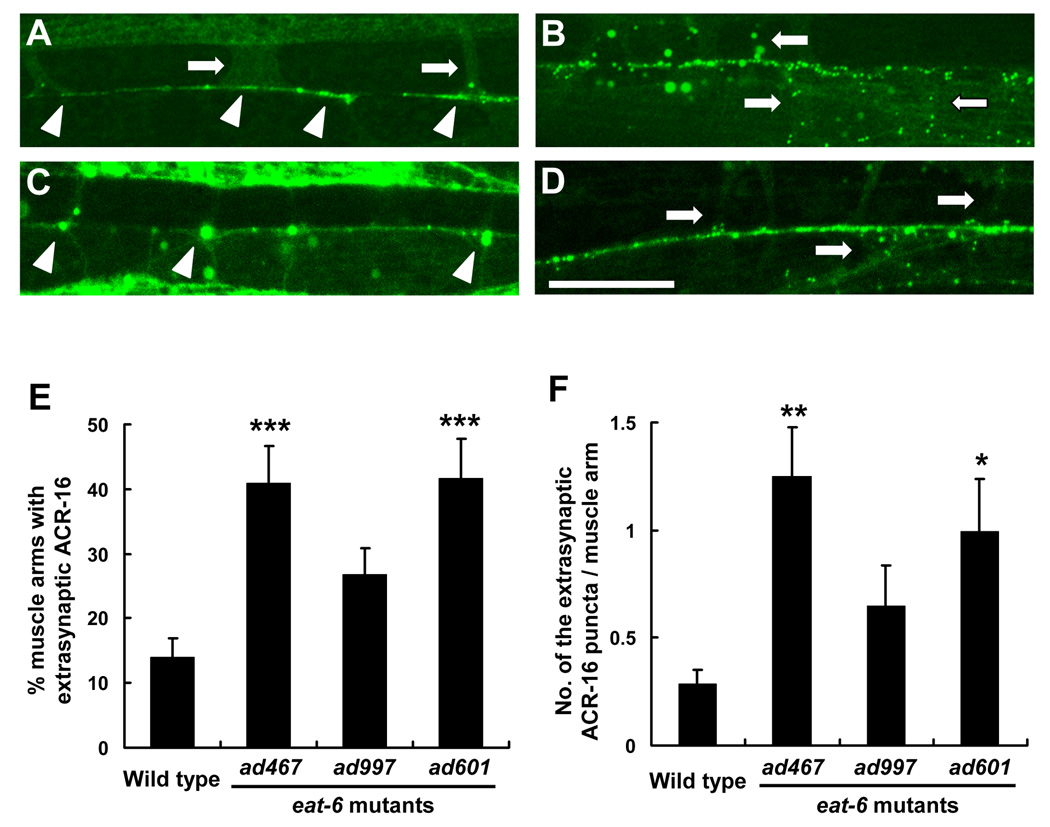
In these mutants, the localization of ACR-16 nicotine-sensitive receptor was also altered, but its localization pattern was different from that of UNC-29 (Fig. 4). In wild-type animals, the ACR-16:: GFP fusion protein was strongly accumulated at the tips of the muscle arms and in punctated clusters along the nerve cords (Francis et al., 2005). However, in the ad467 and ad601 mutants, more GFP puncta were mislocalized to the trunks of the muscle arms compared to the wild type (Fig. 4B, D). We quantitatively analyzed both the percentage of muscle arms containing extrasynaptic GFP (Fig. 4E) and the number of extrasynaptic GFP puncta in one muscle arm (Fig. 4F), and found that both were significantly increased in the ad467 and ad601 mutants (Fig. 4E, F). We noticed that, in the ad997 mutants, large aggregates of the ACR-16:: GFP fusion protein were observed in muscle cells (Fig. 4C). These observations suggest that the ad467 and ad601 alter the expression of levamisole-sensitive receptors at postsynaptic NMJs and increase the formation of the mislocalized, irregular clusters of nicotine-sensitive receptors in the extrasynaptic regions of the NMJ.
Figure 4. Extrasynaptic localization of nicotine-sensitive AchRs in the eat-6 mutants.
(A – D) Localization patterns of ACR-16 (nicotine-sensitive AchR α subunit) and the GFP fusion protein. (A) Wild type. (B) ad467. (C) ad997. (D) ad601. Arrowheads indicate the NMJ at which GFP fused receptors are concentrated. Arrows indicate the muscle arms innervated from the body-wall muscles. Scale bar, 20 µm. Note that small GFP puncta can be observed not only at the NMJ but also on the extrasynaptic muscle arm trunk in the ad467 and ad601 mutants. (E and F) Quantitative analysis of extrasynaptic ACR-16 localization. Extrasynaptic GFP puncta on muscle arms were counted in wild-type animals and each eat-6 mutant animals. GFP puncta not localized at the NMJ were defined as extrasynaptic. (E) The ratio of the muscle arms containing extrasynaptic GFP to the total number of muscle arms is shown (n = 20–30 animals). ** P < 0.01, compared to the wild type, two-tailed Mann-Whitney U-test. Error bars indicate SEM. (F) Average number of extrasynaptic GFP puncta in one muscle arm. ** P < 0.01, * P < 0.05, compared to the wild type, two-tailed Mann-Whitney U-test. Error bars indicate SEM.
eat-6 mutations do not affect GABAergic synapse function and the formation of presynaptic structure
We next asked whether Na+/K+ ATPase specifically regulates the expression and localization of the nAchR or more broadly affects localizations of additional synaptic proteins. The UNC-49 GABA receptor is expressed in the body-wall muscles, and a UNC49B:: GFP fusion protein is strongly localized along the ventral nerve cord in wild-type NMJs (Bamber et al., 1999). We examined the localization patterns of UNC-49B:: GFP fusion protein in the eat-6 mutants and found that the expression and localization of the GABA receptor at the NMJ was unaffected (Figs. 5A, B). It, therefore, appears that mutations in eat-6 specifically affect both the expression and localization of nAchRs. Consistent with this observation, all eat-6 mutants responded normally to the GABA agonist muscimol (data not shown). These observations also suggest that these eat-6 mutations do not affect GABA receptor function or GABA synaptic transmission. Similarly, the eat-6 mutants responded normally to serotonin (5-HT) (data not shown).
Figure 5. Normal localization of GABA receptor and presynaptic synaptobrevin in the eat-6 mutants.
(A and B) Localization pattern of the UNC-49B:: GFP fusion protein. unc-49B encodes the C. elegans GABAA receptor. (A) Wild type. (B) eat-6(ad467). A similar localization pattern was observed along the ventral nerve cord in both the wild-type and eat-6 animals, suggesting normal GABA receptor localization in the eat-6 inhibitory synapses. (C and D) Localization pattern of the SNB-1:: GFP fusion protein. SNB-1 is the C. elegans homolog of the synaptic vesicle protein synaptobrevin (VAMP). The puncta represent presynaptic terminals on the axons of the ventral nerve cord motor neurons. Similar localization patterns indicate normal presynaptic organization in the eat-6 mutants. Scale bar, 10 µm.
We also examined the presynaptic architecture of the ventral nerve cord using the synaptic vesicle protein SNB-1 fused with GFP under the control of the snb-1 (pan-neuronal) or unc-25 promoter (GABAergic). No gross differences were observed between the wild type and eat-6 mutants (Figs. 5C, D and data not shown), suggesting that normal cholinergic and GABAergic presynapses were formed in eat-6 mutants. This is consistent with the notion that the hypersensitivities of the mutants to Ach agonists are not due to presynaptic abnormalities, such as an increased number of release sites, or an altered number of clustered vesicles at the presynaptic release sites. These results strongly indicate that mutations in eat-6 specifically affect cholinergic synaptic transmission by disrupting postsynaptic nAchR dynamics.
Postsynaptic Na+/K+ ATPase confers hypersensitivity to Ach agonists
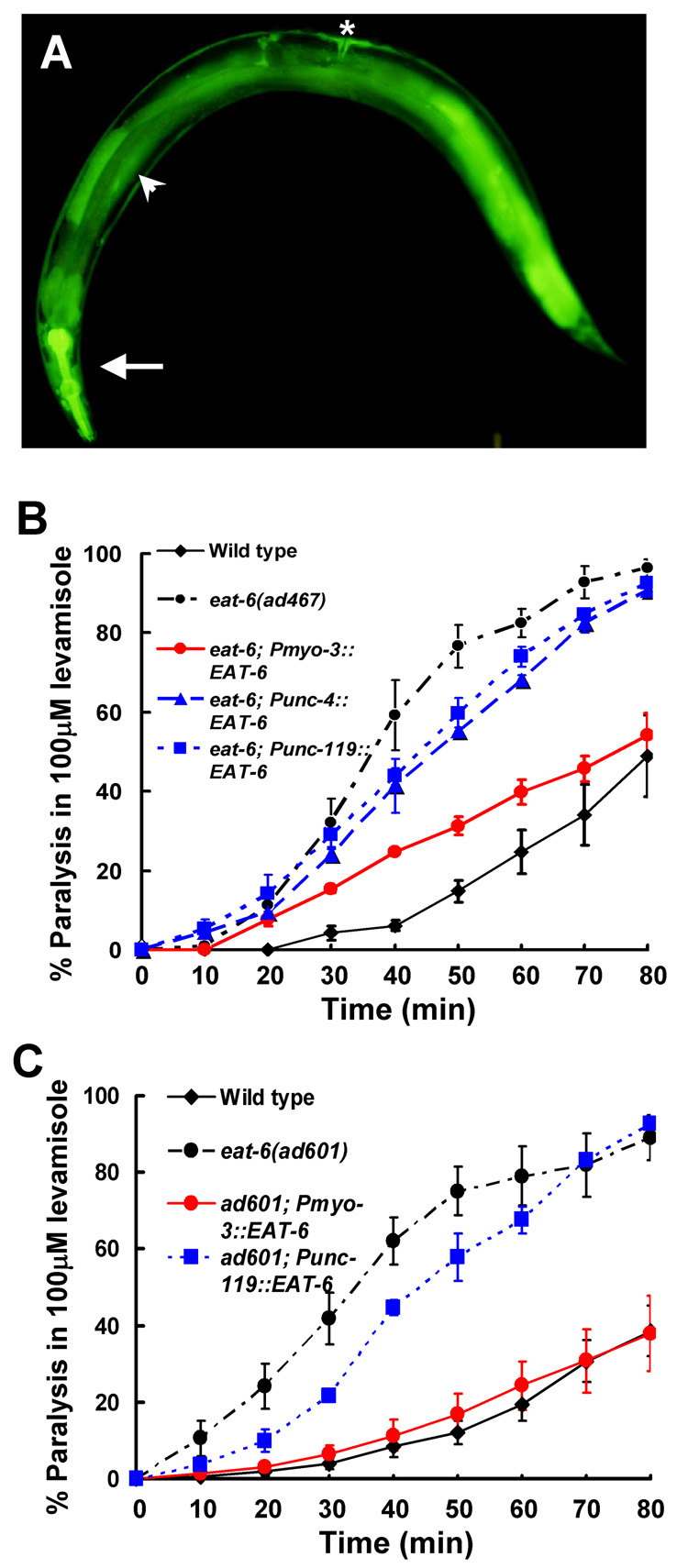
To determine whether increased Ach transmission in the eat-6 mutants corresponds to defects in the EAT-6 Na+/K+ ATPase in the postsynaptic body-wall muscles, we examined the site of action for EAT-6 protein. Endogenous eat-6 promoter activity was observed in most muscular tissues, including body-wall muscles while neuronal expression was barely detectable (Fig. 6A). To clarify the tissue-specific expression profile of eat-6, we also examined the expression patterns of other Na+/K+ ATPase α subunits in the C. elegans genome. The C. elegans genome encodes five Na+/K+ ATPase α subunits, EAT-6, C01G12.8, C09H5.2, C02E7.1 and CATP-1 (Okamura et al., 2003; Ruaud and Bessereau, 2007). catp-1 is expressed in the epidermal cells and the excretory duct cell (Ruaud and Bessereau, 2007). Using GFP fusion constructs, we found that C01G12.8 was expressed in the germline, C09H5.2 in hypodermal cells, and C02E7.1 undetected in any tissues. Thus, any α subunits expression was not clearly detected in the nervous system (MD and KI, unpublished data). Furthermore, the α subunit mutant animals, except for the eat-6 mutants, did not show obvious behavioral defects; thus, we conclude that eat-6 is broadly expressed not only in muscle but also in neuronal cells. To address which cells really require the EAT-6 activity for proper cholinergic transmission, we expressed EAT-6 in a tissue-specific manner in the eat-6 mutants and examined levamisole sensitivity in the resulting transgenic animals (Figs. 6B, C). Pan-neuronal (unc-119 promoter) or cholinergic motor-neuronal expression (unc-4 promoter) minimally affected levamisole sensitivity in both the ad467 and ad601 mutants compared to the wild type. On the other hand, muscle-specific expression (myo-3 promoter) of EAT-6 conferred a normal sensitivity to levamisole in both ad601 and ad467 mutants (Figs. 6B, C). These results suggest that muscle cells are the sites of action for EAT-6 Na+/K+ ATPase in the regulation of cholinergic synaptic transmission.
Figure 6. EAT-6 functions in postsynaptic muscles for cholinergic synaptic transmission.
(A) Expression pattern of the eat-6 promoter and GFP fusion construct. The arrow indicates the pharynx, the arrowhead indicates the body-wall muscle, and a star indicates the vulval muscle. GFP fluorescence was also observed in the intestine, hypodermis, and coelomocytes. (B, C) Tissue-specific rescue of levamisole sensitivity in the eat-6 mutants. Muscle-specific EAT-6 expression confers levamisole sensitivity on the ad467 (B) and ad601 mutants (C). For both alleles, weak rescue by neuronal EAT-6 expression was observed. Error bars indicate the S.E.M. (n > 4).
Our results show that the ad601 and ad467 mutant alleles alter nAchR localization and cholinergic transmission, whereas the ad997 mutation does not affect either no abnormalities in both. What kinds of amino acid changes in the EAT-6 protein do produce such phenotypic variation among three alleles? The ad467 mutation has been known as a single amino acid substitution of Phe for Leu at the residue 359 (Shima et al., 1998), which is close to the phosphorylation site (D409E) in the large intracellular loop. However, no information of other mutational sites was reported before. Sequencing of the other mutants revealed that the ad601 mutation is also a single amino acid substitution (Gly522 to Glu) close to the ATP binding site (R669Q), while the ad997 mutation is located in the 5th transmembrane region (Ser761 to Phe; Fig. 7). The substituted amino acids in ad467 and ad601 are well conserved from nematodes to vertebrates, and Ser761 is functionally related to Thr in vertebrates (Fig. 7B). The functions of these amino acids in the Na+/K+ ATPase are unknown; however, the possible role of this region is discussed below (see Discussion).
Distinct localization of Na+/K+ ATPase and nAchR at postsynaptic sites
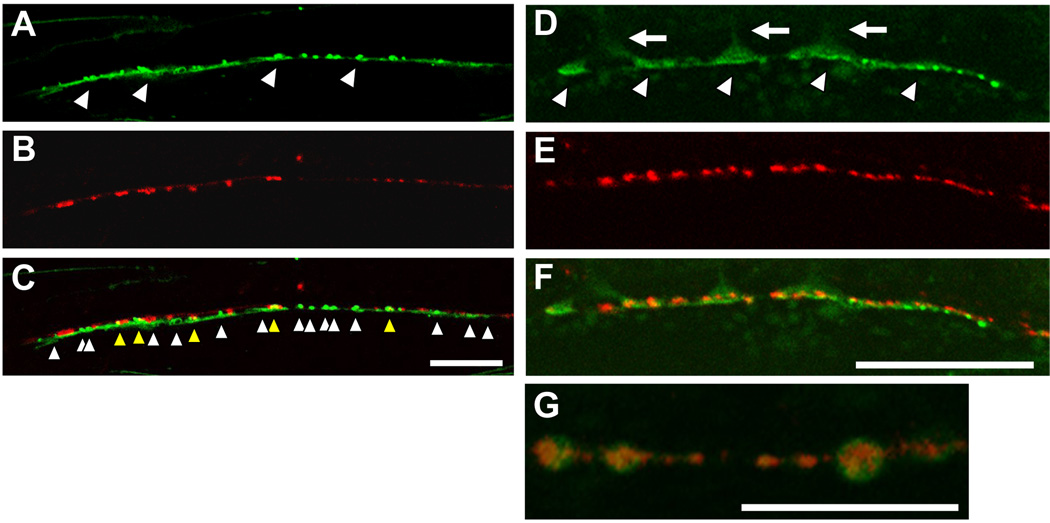
Our analyses strongly suggest a novel role for Na+/K+ ATPase in regulating nAchR localization at postsynaptic sites in the NMJ, independent from its pump activity. To further understand the role of Na+/K+ ATPase at the synapse, we examined the localization patterns of Na+/K+ ATPase in the postsynaptic body-wall muscles. Both Na+/K+ ATPase α (EAT-6) and β (NKB-1) subunits were fused to GFP protein, and the fusion constructs were specifically expressed in the body-wall muscles. NKB-1 β subunit is the most widely expressed β subunit in three C. elegans β subunits. NKB-1 protein physically binds EAT-6 directly, and nkb-1 mutant animals show defects that are similar to those in eat-6 mutants, including defects in pumping, reduced numbers of progeny, and small body size (MD and KI, in preparation). This suggests that EAT-6 and NKB-1 form a functional Na+/K+ ATPase in vivo; thus, we examined the localization patterns of both fusion proteins. In transgenic animals expressing either fusion protein, a fluorescent signal was observed at the muscle cell surface, and a particularly bright signal was detected at the NMJ along the ventral nerve cord (Figs. 8A, Fig. S1A). This means that the fusion proteins were highly concentrated at the postsynaptic regions of the synapse. For unknown reasons, NKB-1 exhibited a more punctated localization than EAT-6 (compare Fig. 8A and Fig. S1A). To know whether these Na+/K+ ATPase subunits are localized at either cholinergic synapses or GABAergic synapses, or both, we co-expressed the NKB-1:: GFP fusion protein with mRFP-labeled synaptogyrin, a presynaptic marker that is specifically expressed in inhibitory GABAergic neurons. About 20 % GABAergic presynaptic terminals (RFP puncta) co-localized with postsynaptic NKB-1 (GFP) localization (103 co-localization with GFP puncta in totally observed 470 RFP puncta (n = 20 animals)). Thus, the majority of the postsynaptic Na+/K+ ATPase localization did not overlap with that of GABAergic synapses, suggesting a predominant localization of the Na+/K+ ATPase at cholinergic postsynaptic sites (Fig. 8C).
Figure 8. Synaptic localization of Na+/K+ ATPase and the distinct localization of the nAchR.
(A) Localization pattern of the NKB-1::GFP fusion protein around the anterior ventral nerve cord. The fusion protein formed a punctate pattern along the ventral nerve cord. (B) Localization pattern of the presynaptic SNG-1::mRFP fusion protein expressed under the control of the unc-25 promoter. The puncta indicate GABAergic presynaptic sites. (C) Merged image. Although several of the mRFP and GFP puncta were colocalized (yellow arrowhead), a lot of GFP puncta did not colocalize with mRFP (white arrowhead), suggesting a predominant localization of the Na+/K+ ATPase at cholinergic postsynaptic sites. Scale bars, 20 µm. (D) Localization pattern of the NKB-1::GFP fusion protein. Arrowheads indicate the NMJs. Arrows indicate the muscle arms innervated from the muscle cells. (E) Localization pattern of the LEV-1:: 3xHA fusion protein. LEV-1 is a nAchR non-α subunit expressed in body-wall muscles, and membrane-inserted LEV-1 protein was visualized by staining with Alexa-594-labelled anti-HA antibodies. (F) Merge image. Scale bars, 20 µm. (G) In some cases, nAchR clusters (UNC-38:: myc, red) were surrounded by Na+/K+ ATPase (EAT-6:: GFP, green). Scale bars, 10 µm. All pictures show the ventral view.
At the postsynaptic sites, however, the Na+/K+ ATPase fusion protein was concentrated in distinct regions from the nAchR clusters (Fig. 8D–8G and Fig. S1). We co-expressed GFP-fused EAT-6 or NKB-1 and nAchR (HA-tagged LEV-1 or myc-tagged UNC-38; see Fig. 3), and receptor clustering was visualized by injecting fluorescently-labeled HA or myc antibodies. The membrane-inserted nAch receptors were tightly clustered at the distal regions of the muscle arms, along the ventral nerve cord (Fig. 8E). GFP-tagged Na+/K+ ATPase, however, was localized more diffusely and at proximal regions of the postsynaptic muscle arms, compared with fluorescence-tagged nAchR. At a higher magnification, the Na+/K+ ATPase fusion proteins were occasionally observed surrounding the clustered nAchRs (Fig. 8G). The distinct and sub-lateral localization of the Na+/K+ ATPase around the nAchRs suggest that the Na+/K+ ATPase may function to compartmentalize the region in which nAchRs are rigidly clustered for efficient synapses at the NMJ.
DISCUSSION
In this report, we showed that mutations in the EAT-6 Na+/K+ ATPase α subunit in C. elegans alter the response to Ach agonists. These defects were allele-specific; mutations in the large intracellular loop caused agonist hypersensitivity, but the mutation in the 5th transmembrane region did not. Alternatively, it decreased sensitivity to nicotine. We also showed that expression and localization of the nAchRs at the NMJ were altered. The Na+/K+ ATPase subunits are concentrated in the postsynaptic regions of the body-wall muscles, however, the distinct postsynaptic localization of the Na+/K+ ATPase and nAchRs suggests that the sodium pump may have a novel function like a scaffolding protein to help establish a rigid receptor cluster just beneath of presynaptic release site.
Pump activity-independent regulation of Ach signaling by Na+/K+ ATPase
Our results strongly indicate that Na+/K+ ATPase regulates Ach signaling at the C. elegans NMJ by affecting the expression and localization of postsynaptic nAchRs. This function is probably independent of the activity of the pump and the resulting of resting membrane potential of the postsynaptic muscle cells. Although we were unable to record resting membrane potentials directly from the intact body-wall muscles of the mutants due to the quite thin structure of the cells, several lines of evidence support our conclusions. First, the phenotypic variation in terms of Ach signaling was not coincident with that of several other phenotypes including pharyngeal pumping, the number of progeny, and EPG data. For example, the ad601 mutation, which caused dramatic changes in both agonist sensitivity and receptor expression, does not seem to greatly affect depolarization of the membrane potential because several phenotypes were quite similar to those of the wild-type animals compared to the other eat-6 mutants. On the other hand, the ad997 mutation, which did not have any effect on agonist sensitivity, had relatively large effects on other phenotypes and the EPG data (Davis et al., 1995). This observation is quite important because the ad997 mutation probably affects resting membrane potential relatively large. However this alteration does not seems to simply correspond to agonist sensitivities. Thus, it is highly possible that membrane-potential changes by altered pump activity are not the primary cause of altered Ach signaling at the NMJ.
Second, pump activity-dead mutations in EAT-6 significantly recovered the agonist sensitivities in the ad467 and ad601 mutants. In mammalian cells, these mutations clearly disrupt pump activity (Jacobsen et al., 2002; Liang et al., 2006; Ohtsubo et al., 1990), and the mutated amino acids in the Na+/K+ ATPase α subunits are well conserved between C. elegans and vertebrates (Fig. 7B). Interestingly, the mutant EAT-6 proteins did not rescue other defects, such as impaired pharyngeal pumping and small numbers of progeny, suggesting that the recovery of these phenotypes requires sufficient ATPase activity. Rescue by pump-dead EAT-6 means that EAT-6 or Na+/K+ ATPase itself (the EAT-6 α subunit complexed with the NKB-1 β subunit), not the function of the sodium pump, is required for proper Ach signaling at the C. elegans NMJ.
Third, recent emerging evidence also supports the possibility of a pump-independent function for Na+/K+ ATPase function in vivo. In C. elegans, another P-type ATPase CATP-1, which is distantly related to EAT-6 and vertebrate Na+/K+ ATPase, regulates larval developmental timing through the insulin/IGF and Ras-MAPK pathways (Ruaud and Bessereau, 2007). Pump activity is not required for this regulation because pump-dead CATP-1 rescues the developmental delay in catp-1 mutants. The Drosophila Na+/K+ ATPase also has a pump-independent function in epithelial and septate junction formation (Paul et al., 2007; Paul et al., 2003). This function in cell junction appears to be conserved in vertebrates because the rat α1 isoform can rescue all junctional defects in Drosophila Atpα-null mutants. The effects of Na+/K+ ATPase mutations are variable, but they strongly suggest the possibility that the Na+/K+ ATPase functions as a scaffold protein for other membrane proteins, such as Ach receptor in the C. elegans NMJ.
Both the partial rescue by pump-dead EAT-6 and the weak recoveries by neuronal expressions of EAT-6 in levamisole sensitivity may suggest a requirement of wild-type EAT-6 in presynaptic neurons for proper Ach signaling. It is possible that the presynaptic EAT-6 functions as a sodium pump for the maintenance of normal resting potential and that this regulates normal transmitter release from presynaptic neurons. Thus, we conclude that the pump activity of EAT-6 is required for Ach signaling either presynaptically or postsynaptically, but that its contribution in presynaptic neurons is relatively weak. Moreover, we conclude that Na+/K+ ATPase has a novel function in nAch receptor expression and clustering in a pump activity-independent, allele-specific.
Possible mechanisms for nAchR clustering by Na+/K+ ATPase
Our results suggest a novel function for the EAT-6 Na+/K+ ATPase in nAchR clustering. How does Na+/K+ ATPase regulate nAchR dynamics? There is no clear evidence to show that these two molecules directly interact physically or genetically in C. elegans and other species; in addition, to our knowledge, our study is the first to describe the functional interaction of these two membrane proteins. However, we failed to detect a direct protein-protein interaction between EAT-6 and the UNC-38, UNC-29 and LEV-1 levamisole-sensitive receptor subunits by yeast two-hybrid assay (data not shown). These preliminary results and the distinct localization of these molecules imply indirect regulation of receptor expression and localization. For example, Na+/K+ ATPase may be able to modulate nAchR-membrane trafficking through the activation/inactivation of Src tyrosine kinase. Recently, Tian et. al. (2006) showed that Na+/K+ ATPase and Src were colocalized at the plasma membrane in several different cell lines, and that binding of Na+/K+ ATPase to Src kept the kinase in an inactivated state. They also showed that the kinase domain of Src specifically binds the large third intracellular loop of ATPase, in which the ad601 and ad467 mutations were located. The Na+/K+ ATPase inhibitor ouabain induces the release of the kinase from the pump, and released active Src facilitates the subsequent phosphorylation of several target molecules. In our case, it is possible that the ad601 and/or ad467 mutations affect EAT-6 binding to the worm Src. nAchRs are also one of the target molecules of Src in the peripheral and central synapses of vertebrates (Wiesner and Fuhrer, 2006). Further biochemical analyses of the phosphorylation levels of the nAchRs in the eat-6 mutants will clarify the relationship between the distinct localization patterns of Na+/K+ ATPase and the phosphorylation state of the nAchRs.
Another possible mechanism for the regulation of nAchR expression and localization, which is distinct from Na+/K+ ATPase localization, can be deduced form the molecular interaction between Na+/K+ ATPase and ankyrin adapter protein, which mediates the linkage of membrane proteins with the spectrin-based cytoskeleton. The α subunit of Na+/K+ ATPase interacts strongly with ankyrin at its second intracellular loop and weakly at the third intracellular loop (Devarajan et al., 1994; Jordan et al., 1995; Zhang et al., 1998). This interaction maintains the polarized distribution of Na+/K+ ATPase in epithelial cells (Nelson and Veshnock, 1987). At developing rat NMJs, voltage-gated sodium channels (Nav1) and ankyrinG occupy a distinct postsynaptic domain from the AchR clusters (Bailey et al., 2003). At the C. elegans NMJ, the sodium channel could be replaced by Na+/K+ ATPase, because there is no candidate gene corresponding to a voltage-gated sodium channel in the C. elegans genome. Because neither the ad601 nor ad467 mutation is located close to the identified ankyrin-binding motif in the third intracellular loop (Fig. 7B), these two amino acid substitutions in the mutants likely have a novel function in either the interaction with ankyrin or nAchR localization through the Na+/K+ ATPase/ankyrin complex.
A surprising finding in the eat-6 mutants was that the levamisole and nicotine receptors were differently affected in their expression and localization at the NMJ. For the levamisole receptors, the localizations of both GFP-fused UNC-29 and HA-tagged LEV-1 were strongly increased in the ad467 and ad601 mutants (Fig. 3). This suggests that Na+/K+ ATPase may control the amount of membrane-inserted levamisole-sensitive receptors by negatively regulating receptor transport from intracellular compartments or by positively regulating receptor internalization from the synaptic membrane. Interestingly, few extrasynaptic receptor clusters, like those seen with ACR-16 in the eat-6 mutants, were observed in either the UNC-29:: GFP or HA-tagged LEV-1 localization analyses, suggesting that Na+/K+ ATPase may not regulate the clustering of levamisole-sensitive receptors. On the other hand, in the case of the nicotine-sensitive receptor ACR-16, several extrasynaptic clusters were observed not only in the postsynaptic areas but also on the trunks of the muscle arms in the ad467 and ad601 mutants (Fig. 4). However, in the ad997 mutant, the number of receptors clustered at the NMJ looks to be decreased and the fusion protein tends to form large intracellular aggregates (Fig. 4C). Na+/K+ ATPase may regulate the site of nicotine receptor clustering by functioning such as a scaffold protein. These results are consistent with the responses of the receptors to various agonists (Fig. 1), suggesting that the fluorescently-labeled, localized receptors were probably functional, and that the dynamics of these fusion receptors likely reflect the in vivo functions of the native receptors. Thus, the EAT-6 Na+/K+ ATPase may differentially regulate the cellular dynamics of each receptor, such as the transport of levamisole receptors to or from the synapse, and the localization of nicotine receptors to appropriate synaptic sites. For example, these receptors may use distinct motor proteins or vesicles as part of their transport machinery. Alternatively, it is possible that they use different scaffold proteins for their localization, given that the mutation in cam-1 receptor tyrosine kinase specifically affects the localization of ACR-16 (Francis et al., 2005). Currently we have no evidence to show how these eat-6 mutant alleles lead to different phenotypes in the two receptors; additional molecular analyses are thus necessary to understand the regulatory machinery involved in the expression and/or localization of each nAchR.
Recent analyses have shown that disrupting Na+/K+ ATPase function causes neuronal degeneration in the Drosophila brain (Palladino et al., 2003) and neuronal cell death in cultured vertebrate neurons (de Carvalho Aguiar et al., 2004). Furthermore, mutations in the human Na+/K+ ATPase subunits are associated with several neuronal diseases such as familial migrate hemigrate (De Fusco et al., 2003) and rapid-onset of dystonia parkinsonism (de Carvalho Aguiar et al., 2004). The amino acid changes associated with human neuronal disorders are strongly believed to inhibit Na+/K+ ATPase pump activity, resulting in a depolarized membrane potential and increased intracellular Na+ and Ca2+ ion concentrations, based on experiments in cultured HEK293 cells (de Carvalho Aguiar et al., 2004). We speculate that a subtle balance between synaptic activity and the resting-membrane potential may control the intrinsic intracellular ionic concentrations and the subsequent cell survival rate in vivo. It is also possible that an imbalance in the resting membrane potential and altered synaptic efficacy caused by defective nAchR localization like eat-6 mutants may accelerate neuronal cell death in vivo nervous system. Our findings imply a novel role for Na+/K+ ATPase in cholinergic synapses, and by applying a genetic approach to these eat-6 mutants, we may further elucidate the synaptic functions of Na+/K+ ATPase and its subsequent effects on neuronal dysfunction or neuronal cell survival.
Experimental Methods
General methods and C. elegans strains
Worms were kept on the standard NGM plates seeded with bacterial OP50. Strains used in this study are follows: eat-6(ad467, ad601, ad997), nkb-1(tg91), unc-29(x29), ZZ2171 (unc-29(zz29); Is[Pmyo-3:: UNC-29:: GFP]), oxIs22 (Punc-49:: UNC-49B:: GFP), jsIs1 (Psnb-1:: SNB-1:: GFP), juIs1 (Punc-25:: SNB-1:: GFP). Some strains were obtained from the CGC.
Labeling of the postsynaptic nAchR subunits and Na+/K+ ATPase
To observe the localization of UNC-29 protein, strain ZZ2171 (unc-29(zz29); Is[Pmyo-3:: UNC-29:: GFP]) was backcrossed to wild-type N2 to remove the unc-29 mutation, and the resulting strain was crossed to each eat-6 mutant. The homozygous mutants were confirmed by sequencing. For ACR-16 receptor localization, the pMD906 (Pmyo-3:: ACR-16:: GFP) plasmid was injected into wild-type or eat-6 mutant animals, and at least three independent, stable transgenic lines for each genotype were used for the analyses. For double-labeling of both Na+/K+ ATPase and nAchR, transgenic animals bearing either EAT-6:: GFP/UNC-38:: 3xmyc or NKB-1:: GFP/LEV-1:: 3xHA were generated by injecting the corresponding plasmids (pDK43 and pAG9, pDK91 and pAG8, respectively) into wild-type animals. Staining for the HA-labeled LEV-1 and myc-tagged UNC-38 was done as descried (Gottschalk et al., 2005; Gottschalk and Schafer, 2006). Strains jsIs1 (Psnb-1:: SNB-1:: GFP), juIs1 (Punc-25:: SNB-1:: GFP) and oxIs22 (Punc-49:: UNC-49B:: GFP) were also crossed to the eat-6 mutants to determine the localization of the other synaptic proteins.
Microscopic observation
The localization patterns of the fluorescently labeled synaptic proteins at the NMJ were observed using confocal microscopy (Zeiss LSM Pascal 5), using a 63 or 100× oil PlanApo objective lens (Zeiss). Images were collected with the same laser power and detector settings using Pascal software version 3.2. Most of the transgenic animals were generated using a wild-type rol-6 plasmid (pRF4) as an injection marker; consequently, the roller (i.e., transgenic) worms mounted on the slides occasionally faced their ventral sides to the objective lens, allowing a clear view of the NMJ around the ventral nerve cord. Each observation was performed at the anterior region of the vulva in young-adult animals.
Quantification of the membrane-inserted nAchRs at the NMJ
Quantification of the amount of membrane-inserted, synaptic nAchR was determined as described previously (Gottschalk et al., 2005; Gottschalk and Schafer, 2006). Briefly, an integrated line of LEV-1:: 3xHA and Rol-6d (zxIs1) was crossed to each eat-6 mutant. The worms from each strain were then mounted on 2 % agarose-pad cover glasses, and Alexa594-labeled anti-HA antibodies (1:200 dilution in injection buffer; Molecular Probes) were injected into the body cavity. The injected worms were then transferred onto NGM plates and kept undisturbed at room temperature. Six hours later, the worms were mounted on 5 % agarose-pad glass slides, and the level of fluorescence along the ventral nerve cord was observed as descried above. To quantify the fluorescence intensity of the labeled receptors, the images were projected into a single plane. The background signal intensity was set as the minimum threshold, and the pixel intensity/count table was generated in 1-pixel increments using Pascal software. The data were then exported to Microsoft Excel, and the total and average pixel intensities were calculated. Statistical analysis was performed using Student’s two-tailed t-test.
Molecular biology
To analyze eat-6 expression, the 4-kb upstream from the EAT-6 start codon (−4121 to +3 from the ATG) was amplified using primers with restriction enzyme sites and the resulting fragment was inserted between the SalI and BamHI sites of the pPD95.77 to generate pMD136.
For the tissue-specific expression and localization of EAT-6, full-length eat-6 cDNA was amplified by RT-PCR using primers with restriction enzyme sites and then fully sequenced to check for PCR errors. The fragment was then inserted between the XbaI and KpnI sites of the plasmid pPD95.79, in which GFP was exchanged with Venus fragment, to generate pDK40. The myo-3 promoter fragment was excised from pPD96.52 and subcloned between the HindIII and XbaI sites in pDK40 to generate pDK43. For pan-neuronal and motor-neuronal expression, the unc-119 and unc-4 promoter regions were amplified uisng restriction site-attached primers and subcloned between the PstI and XbaI sites of pDK40 to generate pDK70 and pDK69, respectively. The following promoter regions were used: unc-119 (−2194 to −1) and unc-4 (−2947 to −1).
For the rescue experiments using pump activity-dead EAT-6, two mutant eat-6 cDNAs were generated using mutational primers corresponding to D409E (D365 in EAT-6) and R669Q (R556 in EAT-6), respectively. The amplified fragments were then replaced with the wild-type eat-6 cDNA fragment from pDK43, and the myo-3 promoter region was replaced with the eat-6 promoter region from pMD136 to generate the plasmid pDK347 (D409E) and pDK349 (R669Q), respectively.
For GABAergic synapse-synaptic labeling by RFP, full-length sng-1 cDNA was amplified by RT-PCR and then subcloned between the BamHI and KpnI sites in pPD95.75/mRFP plasmid, in which the GFP sequence was replaced with the mRFP sequence. The promoter region of unc-25 (−1935 to −1) was then amplified and subcloned between the SalI and BamHI sites in the plasmid to generate pDK96. The full-length NKB-1:: GFP fusion construct (pDK91) was generated as follows. The myo-3 promoter region of pPD96.52 and full-length nkb-1 cDNA were subcloned between the HindIII and KpnI sites of the plasmid. The primers used in this study are divided upon request.
Behavioral analysis
The drug responses of the worms were determined as previously described (Doi and Iwasaki, 2002). Briefly, 25 young-adult animals were placed on an agar plate containing each drug. Paralysis was examined by probing the animals with a thin glass picker every 10 min. The animals were defined as ‘paralyzed’ if they showed no body-bending across the midline. All experiments were performed blindly with at least four replicates per strain.
Supplementary Material
(A) Expression pattern of the EAT-6:: GFP fusion proteins at the ventral nerve cord. A strong GFP signal was observed along the nerve cord, but which was more diffuse than the localization of NKB-1 (Fig. 8). Arrowheads indicate the NMJs. Arrows indicate the muscle arms. (B) Staining of membrane-inserted myc-tagged UNC-38, which was visualized with Cy3-labeled anti-myc antibodies. (C) Merged image. UNC-38 was localized to the more distal part of the muscle arm (arrowheads). Scale bars, 20 µm.
Acknowledgements
We are grateful to the followings for providing strains and plasmids: W. Schafer for the ZZ2171 (Is[Pmyo-3::UNC-29::GFP]) strain, M Fransis and V. Maricq for the pDM906 plasmid, Y. Jin for the juIs1 strain, M. Nonet for the jsIs1 strain, B. Bamber for the oxIs22 strain, J-L. Bessereau for the anti-UNC-29 antibodies, A. Gottschalk for the zxIs1 strain and the pAG8 and pAG9 plasmids, A. Fire for the pPD plasmids, and Y. Kohara for the cDNA clones. We also thank J-L. Bessereau, H. Okamoto, K. Kameyama, H. Nishimaru, K. Kiyosue, and T. Teramoto for helpful suggestions and discussions. Some strains were obtained from the Caenorhabditis Genetic Center (CGC). This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to M.D, and NIH R01 GM082133-02 to K.I..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey SJ, Stocksley MA, Buckel A, Young C, Slater CR. Voltage-gated sodium channels and ankyrinG occupy a different postsynaptic domain from acetylcholine receptors from an early stage of neuromuscular junction maturation in rats. J Neurosci. 2003;23:2102–2111. doi: 10.1523/JNEUROSCI.23-06-02102.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamber BA, Beg AA, Twyman RE, Jorgensen EM. The Caenorhabditis elegans unc-49 locus encodes multiple subunits of a heteromultimeric GABA receptor. J Neurosci. 1999;19:5348–5359. doi: 10.1523/JNEUROSCI.19-13-05348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NA, Kaufmann WE, Worley PF, Rupp F. Expression of agrin in the developing and adult rat brain. Neuroscience. 1997;76:581–596. doi: 10.1016/s0306-4522(96)00345-4. [DOI] [PubMed] [Google Scholar]
- Culetto E, Baylis HA, Richmond JE, Jones AK, Fleming JT, Squire MD, Lewis JA, Sattelle DB. The Caenorhabditis elegans unc-63 gene encodes a levamisole-sensitive nicotinic acetylcholine receptor alpha subunit. J Biol Chem. 2004;279:42476–42483. doi: 10.1074/jbc.M404370200. [DOI] [PubMed] [Google Scholar]
- Davis MW, Somerville D, Lee RY, Lockery S, Avery L, Fambrough DM. Mutations in the Caenorhabditis elegans Na,K-ATPase alpha-subunit gene, eat-6, disrupt excitable cell function. J Neurosci. 1995;15:8408–8418. doi: 10.1523/JNEUROSCI.15-12-08408.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho Aguiar P, Sweadner KJ, Penniston JT, Zaremba J, Liu L, Caton M, Linazasoro G, Borg M, Tijssen MA, Bressman SB, Dobyns WB, Brashear A, Ozelius LJ. Mutations in the Na+/K+ -ATPase alpha3 gene ATP1A3 are associated with rapid-onset dystonia parkinsonism. Neuron. 2004;43:169–175. doi: 10.1016/j.neuron.2004.06.028. [DOI] [PubMed] [Google Scholar]
- De Fusco M, Marconi R, Silvestri L, Atorino L, Rampoldi L, Morgante L, Ballabio A, Aridon P, Casari G. Haploinsufficiency of ATP1A2 encoding the Na+/K+ pump alpha2 subunit associated with familial hemiplegic migraine type 2. Nat Genet. 2003;33:192–196. doi: 10.1038/ng1081. [DOI] [PubMed] [Google Scholar]
- Devarajan P, Scaramuzzino DA, Morrow JS. Ankyrin binds to two distinct cytoplasmic domains of Na,K-ATPase alpha subunit. Proc Natl Acad Sci U S A. 1994;91:2965–2969. doi: 10.1073/pnas.91.8.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Iwasaki K. Regulation of retrograde signaling at neuromuscular junctions by the novel C2 domain protein AEX-1. Neuron. 2002;33:249–259. doi: 10.1016/s0896-6273(01)00587-6. [DOI] [PubMed] [Google Scholar]
- Fleming JT, Squire MD, Barnes TM, Tornoe C, Matsuda K, Ahnn J, Fire A, Sulston JE, Barnard EA, Sattelle DB, Lewis JA. Caenorhabditis elegans levamisole resistance genes lev-1, unc-29, and unc-38 encode functional nicotinic acetylcholine receptor subunits. J Neurosci. 1997;17:5843–5857. doi: 10.1523/JNEUROSCI.17-15-05843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis MM, Evans SP, Jensen M, Madsen DM, Mancuso J, Norman KR, Maricq AV. The Ror receptor tyrosine kinase CAM-1 is required for ACR-16-mediated synaptic transmission at the C. elegans neuromuscular junction. Neuron. 2005;46:581–594. doi: 10.1016/j.neuron.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Gally C, Eimer S, Richmond JE, Bessereau JL. A transmembrane protein required for acetylcholine receptor clustering in Caenorhabditis elegans. Nature. 2004;431:578–582. doi: 10.1038/nature02893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A, Almedom RB, Schedletzky T, Anderson SD, Yates JR, 3rd, Schafer WR. Identification and characterization of novel nicotinic receptor-associated proteins in Caenorhabditis elegans. Embo J. 2005;24:2566–2578. doi: 10.1038/sj.emboj.7600741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A, Schafer WR. Visualization of integral and peripheral cell surface proteins in live Caenorhabditis elegans. J Neurosci Methods. 2006;154:68–79. doi: 10.1016/j.jneumeth.2005.11.016. [DOI] [PubMed] [Google Scholar]
- Hilgenberg LG, Su H, Gu H, O'Dowd DK, Smith MA. Alpha3Na+/K+-ATPase is a neuronal receptor for agrin. Cell. 2006;125:359–369. doi: 10.1016/j.cell.2006.01.052. [DOI] [PubMed] [Google Scholar]
- Hoover CL, Hilgenberg LG, Smith MA. The COOH-terminal domain of agrin signals via a synaptic receptor in central nervous system neurons. J Cell Biol. 2003;161:923–932. doi: 10.1083/jcb.200301013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen MD, Pedersen PA, Jorgensen PL. Importance of Na,K-ATPase residue alpha 1-Arg544 in the segment Arg544-Asp567 for high-affinity binding of ATP, ADP, or MgATP. Biochemistry. 2002;41:1451–1456. doi: 10.1021/bi015891h. [DOI] [PubMed] [Google Scholar]
- Jones AK, Sattelle DB. Functional genomics of the nicotinic acetylcholine receptor gene family of the nematode, Caenorhabditis elegans. Bioessays. 2004;26:39–49. doi: 10.1002/bies.10377. [DOI] [PubMed] [Google Scholar]
- Jordan C, Puschel B, Koob R, Drenckhahn D. Identification of a binding motif for ankyrin on the alpha-subunit of Na+,K(+)-ATPase. J Biol Chem. 1995;270:29971–29975. doi: 10.1074/jbc.270.50.29971. [DOI] [PubMed] [Google Scholar]
- Kaplan JH. Biochemistry of Na,K-ATPase. Annu Rev Biochem. 2002;71:511–535. doi: 10.1146/annurev.biochem.71.102201.141218. [DOI] [PubMed] [Google Scholar]
- Li Z, Massengill JL, O'Dowd DK, Smith MA. Agrin gene expression in mouse somatosensory cortical neurons during development in vivo and in cell culture. Neuroscience. 1997;79:191–201. doi: 10.1016/s0306-4522(96)00654-9. [DOI] [PubMed] [Google Scholar]
- Liang M, Cai T, Tian J, Qu W, Xie ZJ. Functional characterization of Src-interacting Na/K-ATPase using RNA interference assay. J Biol Chem. 2006;281:19709–19719. doi: 10.1074/jbc.M512240200. [DOI] [PubMed] [Google Scholar]
- Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, Rand JB. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc Natl Acad Sci U S A. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobasheri A, Avila J, Cozar-Castellano I, Brownleader MD, Trevan M, Francis MJ, Lamb JF, Martin-Vasallo P. Na+, K+-ATPase isozyme diversity; comparative biochemistry and physiological implications of novel functional interactions. Biosci Rep. 2000;20:51–91. doi: 10.1023/a:1005580332144. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Veshnock PJ. Ankyrin binding to (Na+ + K+)ATPase and implications for the organization of membrane domains in polarized cells. Nature. 1987;328:533–536. doi: 10.1038/328533a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Noguchi S, Takeda K, Morohashi M, Kawamura M. Site-directed mutagenesis of Asp-376, the catalytic phosphorylation site, and Lys-507, the putative ATP-binding site, of the alpha-subunit of Torpedo californica Na+/K(+)-ATPase. Biochim Biophys Acta. 1990;1021:157–160. doi: 10.1016/0005-2736(90)90028-m. [DOI] [PubMed] [Google Scholar]
- Okamura H, Yasuhara JC, Fambrough DM, Takeyasu K. P-type ATPases in Caenorhabditis and Drosophila: implications for evolution of the P-type ATPase subunit families with special reference to the Na,K-ATPase and H,K-ATPase subgroup. J Membr Biol. 2003;191:13–24. doi: 10.1007/s00232-002-1041-5. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics. 1993;135:385–404. doi: 10.1093/genetics/135.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MJ, Bower JE, Kreber R, Ganetzky B. Neural dysfunction and neurodegeneration in Drosophila Na+/K+ ATPase alpha subunit mutants. J Neurosci. 2003;23:1276–1286. doi: 10.1523/JNEUROSCI.23-04-01276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul SM, Palladino MJ, Beitel GJ. A pump-independent function of the Na,K-ATPase is required for epithelial junction function and tracheal tube-size control. Development. 2007;134:147–155. doi: 10.1242/dev.02710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul SM, Ternet M, Salvaterra PM, Beitel GJ. The Na+/K+ ATPase is required for septate junction function and epithelial tube-size control in the Drosophila tracheal system. Development. 2003;130:4963–4974. doi: 10.1242/dev.00691. [DOI] [PubMed] [Google Scholar]
- Richmond JE, Jorgensen EM. One GABA and two acetylcholine receptors function at the C. elegans neuromuscular junction. Nat Neurosci. 1999;2:791–797. doi: 10.1038/12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruaud AF, Bessereau JL. The P-type ATPase CATP-1 is a novel regulator of C. elegans developmental timing that acts independently of its predicted pump function. Development. 2007;134:867–879. doi: 10.1242/dev.02790. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Lichtman JW. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat Rev Neurosci. 2001;2:791–805. doi: 10.1038/35097557. [DOI] [PubMed] [Google Scholar]
- Shima Y, Tada Y, Furuki M, Hara Y, Ohta H. A missense mutation of the gene for Na+,K(+)-ATPase alpha-subunit causes abnormal feeding behavior in Caenorhabditis elegans. Biochem Biophys Res Commun. 1998;248:778–782. doi: 10.1006/bbrc.1998.8981. [DOI] [PubMed] [Google Scholar]
- Sieburth D, Ch'ng Q, Dybbs M, Tavazoie M, Kennedy S, Wang D, Dupuy D, Rual JF, Hill DE, Vidal M, Ruvkun G, Kaplan JM. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- Tian J, Cai T, Yuan Z, Wang H, Liu L, Haas M, Maksimova E, Huang XY, Xie ZJ. Binding of Src to Na+/K+-ATPase forms a functional signaling complex. Mol Biol Cell. 2006;17:317–326. doi: 10.1091/mbc.E05-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner A, Fuhrer C. Regulation of nicotinic acetylcholine receptors by tyrosine kinases in the peripheral and central nervous system: same players, different roles. Cell Mol Life Sci. 2006;63:2818–2828. doi: 10.1007/s00018-006-6081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Devarajan P, Dorfman AL, Morrow JS. Structure of the ankyrin-binding domain of alpha-Na,K-ATPase. J Biol Chem. 1998;273:18681–18684. doi: 10.1074/jbc.273.30.18681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Expression pattern of the EAT-6:: GFP fusion proteins at the ventral nerve cord. A strong GFP signal was observed along the nerve cord, but which was more diffuse than the localization of NKB-1 (Fig. 8). Arrowheads indicate the NMJs. Arrows indicate the muscle arms. (B) Staining of membrane-inserted myc-tagged UNC-38, which was visualized with Cy3-labeled anti-myc antibodies. (C) Merged image. UNC-38 was localized to the more distal part of the muscle arm (arrowheads). Scale bars, 20 µm.