Abstract
The turnover of collagen triple-helical structure (collagenolysis) is a tightly regulated process in normal physiology, and has been ascribed to small number of proteases. Several members of the matrix metalloproteinase (MMPs) family possess collagenolytic activity, and the mechanisms by which these enzymes process triple-helices are beginning to be unraveled. The present study has utilized 2 triple-helical sequences to compare the cleavage site specificities of 10 MMPs. One substrate featured a continuous Gly-Xxx-Yyy sequence (Pro-Leu-Gly~Met-Arg-Gly) while the other incorporated an interruption in the Gly-Xxx-Yyy repeat (Pro-Val-Asn~Phe-Arg-Gly). Both sequences were selectively cleaved by MMP-13 while in linear form, but neither proved to be selective within a triple-helix. This suggests that the conformational presentation of substrate sequences to an MMP active site is critical for enzyme specificity, in that activities differ when sequences are presented from an unwound triple-helix versus an independent single strand. Differences in specificity between secreted and membranetype (MT) MMPs were also observed for both sequences, where MMP-2 and MT-MMPs showed an ability to hydrolyze a triple-helix at an additional site (Gly-Gln bond). Interruption of the triple-helix had different effects on secreted MMPs and MT-MMPs, as MT-MMPs could not hydrolyze the Asn-Phe bond, but instead cleaved the triple-helix nearer the C-terminus at a Gly-Gln bond. It is possible that MT-MMPs have a requirement for Gly in the P1 subsite in order to be able to efficiently process a triple-helical molecule. Analysis of individual kinetic parameters and activation energies indicated different substrate preferences within secreted MMPs, as MMP-13 preferred the interrupted sequence while MMP-8 showed little discrimination between non-interrupted and interrupted triple-helices. Based on the present and prior studies, we can assign unique triple-helical peptidase behaviors to the collagenolytic MMPs. Such differences may be significant for understanding MMP mechanisms of action and aid in the development of selective MMP inhibitors.
Keywords: Triple-helix, matrix metalloproteinase, fluorogenic substrate, collagen
Collagen serves as a structural scaffold and a barrier between tissues, and thus collagen catabolism (collagenolysis) is required to be a tightly regulated process in normal physiology. In turn, the destruction or damage of collagen during pathological states plays a role in tumor cell invasion or atherosclerotic plaque formation and rupture. Only a small number of proteases have been identified capable of efficient processing of triple-helical regions of collagens. A mechanistic understanding of the cleavage of intact collagens has been pursued for many years. Several members of the zinc metalloenzyme family, specifically matrix metalloproteinases (MMPs),1 possess collagenolytic activity. For example, one or more of the interstitial collagens (types I–III) is hydrolyzed within their triple-helical domain by MMP-1, MMP-2, MMP-8, MMP-13, MMP-18, MT1-MMP (MMP-14), and MT2-MMP (MMP-15) (1–3). The mechanism of MMP-catalyzed collagenolysis is beginning to be unraveled, with the interactive roles of the enzyme and the substrate becoming better understood (4–6). For example, to access the individual strands of collagen, the MMP unwinds the substrate, and unwinding activity is present in multiple MMP domains (4, 6). However, there are subtle differences in MMP collagenolytic activities. MMP-1 hydrolyzes type III collagen more rapidly than type I, while MMP-8 and MT1-MMP show a slight preference for type I collagen compared to type III (7, 8). Neither MMP-1 nor MMP-8 hydrolyze type II collagen efficiently (8). Conversely, MMP-13 prefers type II collagen and hydrolyzes this collagen much more rapidly than MMP-1 or MMP-8 (8). Why MMPs exhibit collagen preferences is far from completely understood, although recent studies have suggested that the combination of substrate sequence and stability can play a role (5, 9).
The sequence specificities of collagenolytic MMPs have been extensively examined utilizing single-stranded peptides, peptide libraries, and phage display libraries. These include studies of MMP-1, MMP-2, and MMP-8 (10, 11), MMP-13 (12, 13), and MT1-MMP (11, 14–17). However, there are only a few studies that described the effects of amino acid substitutions within a triple-helical context on MMP activity (5, 9, 18, 19). The influence of triple-helical structure on the interaction between MMP subsites and individual substrate residues has not been explored, but may provide additional information for the mechanism of collagenolysis, the understanding of collagen specificity, and the design of selective inhibitors.
Collagenolytic activity may also be considered in light of an array of diseases caused by mutations occurring in collagen, such as osteogenesis imperfecta (20) or Ehlers-Danlos syndrome (21).2 The majority of these mutations result in the replacment of Gly by another amino acid, thus causing an interruption of the Gly-Xxx-Yyy register found within the triple-helix. Of direct relevance to collagenolysis is the mutation of Gly775 to Glu in the α1 chain of type III collagen, leading to Ehlers-Danlos syndrome IV and aortic aneurism (22). The α1(III) Gly775-Leu776 bond is normally cleaved by collagenolytic MMPs (23). Collagenase (MMP-1, MMP-8, MMP-13, and MT1-MMP) cleavage of non-Gly-Xxx-Yyy repeating sequences has been examined, but only with linear substrates (10–17). Within the triple-helix, the enhancement or inhibition of collagenolysis due to Gly-Xxx-Yyy interruptions has not been explored.
The present study has considered MMP sequence specificities in a triple-helical context in a further attempt to evaluate the role of substrate characteristics on collagenolysis. More precisely, we have utilized prior studies on MMP-13 selectivity for single-stranded sequences (12, 13) to evaluate the specificity of such sequences in a triple-helix. We have examined changes in MMP activity for both continuous Gly-Xxx-Yyy sequences and as a result of interruption in the Gly-Xxx-Yyy motif. MMP triple-helical peptidase activities were compared using fluorescent resonance energy transfer (FRET) triple-helical substrates. The general considerations for the design and synthesis of such substrates, including (7-methoxycoumarin-4-yl)acetyl (Mca) as a fluorophore and 2,4-dinitrophenyl (Dnp) as a quencher, were reported previously (24–26). The effects of enhanced substrate thermal stability on MMP activity has also been evaluated, based on prior work showing that collagenolytic MMPs have different tolerances for more thermally stable substrates (5, 9).
MATERIALS AND METHODS
All standard chemicals were peptide synthesis or molecular biology grade and purchased from Fisher Scientific. 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate, 1-hydroxybenzotriazole, and 9-fluorenylmethoxycarbonyl (Fmoc)-amino acid derivatives [including Fmoc-Lys(Mca) and Fmoc-Lys(Dnp)] were obtained from Novabiochem (San-Diego, CA). Amino acids are of L-configuration (except for Gly). Mca-Lys-Pro-Leu-Gly-Leu-Lys(Dnp)-Ala-Arg-NH2 and NFF-3 were synthesized by methods described previously (27, 28). The synthesis and characterization of fTHP-4 was also described previously (5).
Peptide Synthesis
Peptide-resin assembly of fluorogenic triple-helical substrates (fTHPs) was performed by Fmoc solid-phase methodology on an ABI 433A Peptide Synthesizer (24). All peptides were synthesized as C-terminal amides to prevent diketopiperazine formation (29). Peptide-resins were lipidated with hexanoic acid [CH3(CH2)4CO2H, designated C6] as described (30, 31). Cleavage and side-chain deprotection of peptide-resins proceeded for at least 3 h using thioanisole-water-TFA (5:5:90) (32). Cleavage solutions were extracted with methyl tBu ether prior to purification.
Peptide Purification
RP-HPLC purification was performed on a Rainin AutoPrep System with a Vydac 218TP152022 C18 column (15–20 µm particle size, 300 Å pore size, 250 × 22 mm) at a flow rate of 10.0 mL/min. Eluants were 0.1% TFA in water (A) and 0.1% TFA in acetonitrile (B). The elution gradient was adjusted as required. Detection was at λ = 220 nm. Analytical RP-HPLC and MALDI-TOF MS (see below) were used to identify fractions of homogenous product.
Peptide Analyses
Analytical RP-HPLC was performed on a Hewlett-Packard 1100 Liquid Chromatograph equipped with a Vydac 218TP5415 C18 RP column (5 µm particle size, 300 Å pore size, 150 × 4.6 mm). Eluants were 0.1% TFA in water (A) and 0.1% TFA in acetonitrile (B). The elution gradient was 0–100% B in 20 min with a flow rate of 1.0 mL/min. Detection was at λ = 220, 324, and 363 nm. MALDI-TOF-MS was performed on a Applied Biosystems Voyager MALDI-TOF mass spectrometer using α-cyano-4-hydroxycinnamic acid matrix (33). fTHP mass values were as follows: fTHP-12, [M+H]+ 4594 Da (theoretical 4592.0 Da); C6-fTHP-12, [M+H]+ 4697 Da (theoretical 4690.2 Da); fTHP-13, [M+H]+ 4651 Da (theoretical 4648.0 Da); C6-fTHP-13, [M+H]+ 4748 Da (theoretical 4749.2 Da); and C6-fTHP-14, [M+H]+ 4722.4 Da (theoretical 4718.2 Da). All mass values were within 0.14% of theoretical.
Circular Dichroism (CD) Spectroscopy
CD spectra were recorded over the range λ = 190–250 nm on a JASCO J-810 spectropolarimeter using a 1.0 cm path-length quartz cell. Thermal transition curves were obtained by recording the molar ellipticity ([Θ]) at λ = 222 nm while the temperature was continuously increased in the range of 5–95 °C at a rate of 0.2 °C/min. Temperature was controlled using a JASCO PFD-425S temperature control unit. For samples exhibiting sigmoidal melting curves, the inflection point in the transition region (first derivative) is defined as the melting temperature (Tm).
Matrix Metalloproteinases
ProMMP-1 and proMMP-3 were expressed in E. coli and folded from the inclusion bodies as described previously (34, 35). ProMMP-1 was activated by reacting with 1 mM 4-aminophenyl mercuric acetate (APMA) and 0.1 equiv of MMP-3(Δ248–460) at 37 °C for 6 h. After activation, MMP-3(Δ248–460) was completely removed from MMP-1 by affinity chromatography using an anti-MMP-3 IgG Affi-Gel 10 column. ProMMP-3 was activated by reacting with 5 µg/mL chymotrypsin at 37 °C for 2 h. Chymotrypsin was inactivated with 2 mM diisopropylfluorophosphate. ProMMP-2 was purified from the culture medium of human uterine cervical fibroblasts (36) and activated by incubating with 1 mM APMA for 2 h at 37 °C. ProMMP-8 was expressed in CHO-K1 cells as described previously (37). ProMMP-8 was activated by incubating with 1 mM APMA for 2 h at 37 °C. Recombinant MMP-9 with the linker and the C-terminal hemopexin-like domain deleted [residues 444–707; designated MMP-9(Δ444–707)] was expressed in E. coli in the active form with Phe107 the N-terminus as described elsewhere.3 The resulting MMP-9(Δ444–707) was similar to that previously documented (38). ProMMP-13 was purchased from R&D Systems (Minneapolis, MN) and activated by incubating with 1 mM APMA for 2 h at 37 °C. The concentrations of active MMP-1, MMP-2, MMP-3, MMP-8, MMP-9(Δ444–707), and MMP-13 were determined by titration with recombinant tissue inhibitor of metalloproteinases 1 (TIMP-1) or N-TIMP-1 over a concentration range of 0.1–3 µg/mL (39). Recombinant MT1-MMP with the linker and C-terminal hemopexin-like domains deleted [residues 279–523; designated MT1-MMP(Δ279–523)] was purchased from Chemicon International (Temecula, CA). MT1-MMP(Δ279–523) was expressed and activated, resulting in Tyr112 at the N-terminus. MT1-MMP(Δ279–523), which, in contrast to MT1-MMP, does not undergo rapid autoproteolysis, was used in the present studies due to the relatively small differences in MT1-MMP(Δ279–523) and MT1-MMP triple-helical peptidase activities noted previously (40). Recombinant MT2-MMP with the linker and C-terminal hemopexin-like domains deleted [residues 268–628; designated MT2-MMP(Δ268–628)] was also purchased from Chemicon International. MT2-MMP(Δ268–628) was expressed and activated, resulting in Tyr91 at the N-terminus. Recombinant MT5-MMP and MT6-MMP were expressed in MDCK cells and E. coli, respectively, as described previously (41, 42). The concentrations of active MT1-MMP(Δ279–523), MT2-MMP(Δ268–628), MT5-MMP, and MT6-MMP were determined by titration with recombinant TIMP-2, N-TIMP-2, or N-TIMP-3 (40, 42–45). ProMMP-3(Δ248–460) was expressed in E. coli using the expression vector pET3a (Novagen), folded from inclusion bodies and purified as described previously (46). ProMMP-3(Δ248–460) was activated by reacting with 5 µg/mL chymotrypsin at 37 °C for 2 h. Chymotrypsin was inactivated with 2 mM diisopropylfluorophosphate. Active site titrations utilized either Mca-Lys-Pro-Leu-Gly-Leu-Lys(Dnp)-Ala-Arg-NH2 or NFF-3 as substrate (27, 28, 47).
Assays
Substrate stock solutions were prepared at various concentrations with 0.25–1.0% DMSO in EAB buffer (50 mM Tricine, 50 mM NaCl, 10 mM CaCl2, 0.005% Brij-35, pH 7.5). MMP assays were conducted in EAB buffer by incubating a range of substrate concentrations with 10 nM enzyme at 30 °C. Fluorescence was measured on a Molecular Devices SPECTRAmax Gemini EM Dual-Scanning Microplate Spectrofluoremeter using λexcitation = 324 nm and λemission = 393 nm. Initial velocities were obtained from plots of fluorescence versus time, using data points from only the linear portion of the hydrolysis curve. The slope from these plots was divided by the fluorescence change corresponding to complete hydrolysis and then multiplied by the substrate concentration to obtain initial velocity in units of micromolar per second. Kinetic parameters were evaluated by Lineweaver-Burk, Eadie-Hofstee, and Hanes-Woolf analyses. To determine activation energies (Ea), kinetic parameters were determined over the range of 24–37 °C (based on the Tm of the substrate), and an Arrhenius plot of log kcat versus 1/temperature (K) was constructed where the slope = -Ea/2.3R and R is the molar gas constant. MMP substrate cleavage sites were established by MALDI-TOF MS. Cleavage sites were not altered by peptide lipidation with hexanoic acid, as previously observed (24, 33).
RESULTS AND DISCUSSION
Design and Structural Characterization of Fluorogenic Triple-Helical (fTHP) Substrates
fTHP-4 (Table 1) is a model of the MMP consensus cleavage site in human types I–III collagens (5). Important features of fTHP-4 include the Gly~Leu cleavage site and Gly-Pro-Hyp repeats to enhance triple-helical stability. To serve as the FRET fluorophore-quencher pair, Mca and Dnp are linked to Lys side-chains in the P5 and P5’ subsites, respectively. The fTHP-4 sequence was modified subsequently in the P2, P1, and/or P1’ subsites to create fTHP-12 and fTHP-13. For fTHP-12, the P2 subsite Gln and the P1’ subsite Leu from fTHP-4 were replaced with Leu and Met, respectively (Table 1). Deng et al. utilized phage display to identify selective MMP-13 substrates, and found that the sequence Pro-Leu-Gly~Met-Arg-Gly was hydrolyzed by MMP-13 820 times more rapidly than by MMP-1, 1300 times more rapidly than by MMP-3, and 11 times more rapidly than by MMP-9 (12). Thus, fTHP-12 was constructed to determine if selectively observed by MMP subsite interactions with single-stranded substrates was retained in triple-helical substrates. A similar prior study showed that Pro-Val-Asn-Phe-Arg was hydrolyzed by MMP-13 >17000 times more rapidly than by MMP-1, 400 times more rapidly than by MMP-3, 220 times more rapidly than by MMP-9, and 88 times more rapidly than by MMP-2 (13). Thus, replacement of the P2-P1’ subsite Gln-Gly-Leu triplet from fTHP-4 with Val-Asn-Phe resulted in fTHP-13 (Table 1). fTHP-13 was further modified in the P4’ subsite, where Gln was replaced by Pro to remove the MT1-MMP(Δ279–523) and MT2-MMP(Δ268–628) Gly-Gln cleavage site (see below). The modified sequence was designated fTHP-14 (Table 1).
Table 1.
Sequences and Stabilities of (Gly-Pro-Hyp)5-Gly-Pro-Lys(Mca)-Gly-Pro-P2-P1~P1’-P2’-P3’-P4’-Lys(Dnp)-Gly-Val-Arg-(Gly-Pro-Hyp)5-NH5 fTHPs.
| fTHP | P2-P1~P1’-P2’-P3’-P4 | Peptide | C6-Peptide |
|---|---|---|---|
| Sequence | Tm (°C) | Tm (°C) | |
| fTHP-4 | Gln- Gly~Leu-Arg-Gly-Gln | 36.5 | ND |
| fTHP-12 | Leu-Gly~Met-Arg-Gly-Gln | 45.0 | 51.0 |
| fTHP-13 | Val-Asn~Phe-Arg-Gly-Gln | 39.0 | 50.0 |
| fTHP-14 | Val-Asn~Phe-Arg-Gly-Pro | ND | 46.0 |
Substitutions relative to fTHP-4 are indicated in bold. ND = not determined
fTHP-12, fTHP-13, and fTHP-14 were acylated with hexanoic (C6) acid to provide a second series of substrates with differing thermal stability, as addition of hexanoic acid to the N-terminus of triple-helical peptides was found previously to increase the triple-helix thermostability by 6.6–8.5 °C (5, 24, 31). MMP-1 and MT1-MMP(Δ279–523) triple-helical peptidase activities have been shown to be dependant on substrate thermal stability (5), and thus we wished to evaluate the role of substrate thermal stability on activity for a large number of MMPs. fTHP-14 could not be obtained in high purity, but C6-fTHP-14 could, and thus only C6-fTHP-14 was used in the present studies. The purified, lipidated substrates exhibited decreased solubility in assay buffer compared with the non-lipidated substrates, and thus DMSO was utilized for improved solubility.
CD spectra were obtained for all THPs, and were found to be characteristic of triple-helices (data not shown). To quantify the thermal stability of potential substrates, [Θ] at λ = 222 nm was monitored as a function of increasing temperature. All structures exhibited cooperative transitions, indicative of the melting of a triple-helix to a single-stranded structure (data not shown, but comparable to previously published fTHP melting curves (24–26)). Melting temperatures ranged from 36.5 to 51.0 °C (Table 1). The decrease in triple-helical thermal stability of 6 °C going from a continuous triple-helical sequence (fTHP-12) to an interrupted one (fTHP-13) was not substantial. As shown previously, the use of triple-helical stabilizing regions [such as (Gly-Pro-Hyp)n] on both the N- and C-termini can fold and order a central non-Gly-Xxx-Yyy region, minimizing relative thermal destabilization (48, 49). All fTHPs had appropriate thermal stabilities for examination of substrate hydrolysis at 30 °C.
Cleavage Site Specificity and Selectivity
All fTHPs were initially examined for their MMP cleavage site specificities (Table 2) and rates of hydrolysis (Figure 1 and Figure 2). MMP-1, MMP-8, MMP-13, and MT1-MMP were chosen based on their ability to cleave types I–III collagen (7, 50–54). Of the gelatinase family members, MMP-2 is also known to cleave types I and III collagen (55, 56), in contrast to MMP-9, which does not process types I–III collagens (57). It is also of interest to compare the capabilities of gelatinases and collagenases to process triple-helical substrates, and thus both gelatinases were included in these studies. MT2-MMP was included because, while the full range of activity is unknown, it has been reported to cleave type I collagen (3, 58). MMP-3 was chosen to provide a negative control for secreted MMPs, since it was previously shown to have poor or no triple-helical peptidase activities (25, 59). MT5-MMP and MT6-MMP were used as non-collagenolytic controls for transmembrane type and GPI-anchored MMPs, respectively. The 10 MMPs were either of mammalian (MMP-2, MMP-8, MMP-13, MT1-MMP, MT2-MMP, MT5-MMP) or bacterial (MMP-1, MMP-3, MMP-9, MT6-MMP) origin. Prior studies had shown no difference in proteolytic activities based on MMP glycosylation (60, 61), and thus convenient and well-characterized expression systems for each MMP were used here.
Table 2.
Cleavage Site Specificity for MMP Hydrolysis of fTHP-4, fTHP-12, fTHP-13, and fTHP-14 at 30 °C.
| Enzyme | fTHP-4 | fTHP-12 | fTHP-13 | C6-fTHP-14 |
|---|---|---|---|---|
| MMP-1 | Gly-Leu | ND | ND | NC |
| MMP-2 | Gly-Leu, Gly-Gln | Gly-Met | Asn-Phe | NC |
| MMP-3 | NC | ND | ND | ND |
| MMP-8 | Gly-Leu | Gly-Met | Asn-Phe | Asn-Phe |
| MMP-9(Δ444–707) | Gly-Leu | Gly-Met | ND | NC |
| MMP-13 | Gly-Leu | Gly-Met | Asn-Phe | Asn-Phe |
| MT1-MMP(Δ279–523) | Gly-Leu, Gly-Gln | Gly-Met, Gly-Gln | Gly-Gln | Asn-Phe |
| MT2-MMP(Δ268–628) | Gly-Leu | Gly-Met, Gly-Gln | Gly-Gln | ND |
| MT5- MMP | NC | Gly-Met, Gly-Gln | NC | NC |
| MT6- MMP | NC | NC | NC | NC |
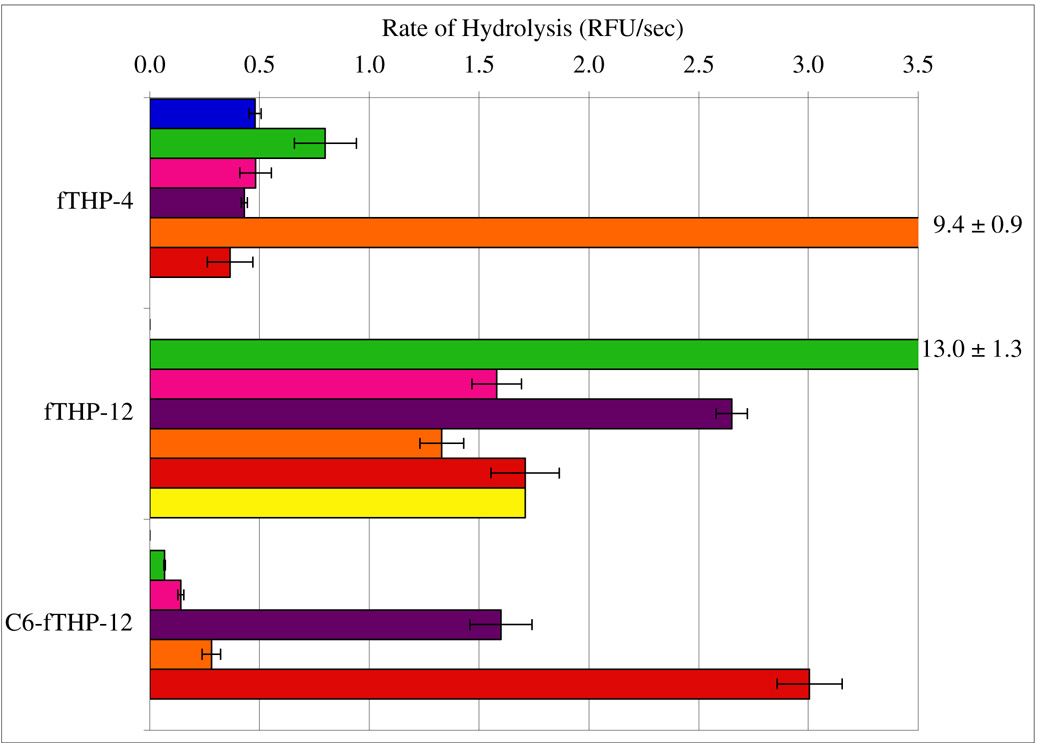
Figure 1. Rates of MMP hydrolysis for fTHP-4 and fTHP-12.
Hydrolysis was examined using 10 nM of each MMP and 4 µM substrate (fTHP-4, fTHP-12, or C6-fTHP-12). Fluorescence was measured using λexcitation = 324 nm and λemission = 393 nm. Rates of hydrolysis were obtained from plots of fluorescence versus time, using data points from only the linear portion of the hydrolysis curve. The MMPs examined were MMP-1 (blue), MMP-2 (green), MMP-8 (pink), MMP-13 (purple), MT1-MMP (orange), MT2-MMP (red), and MT5-MMP (yellow). Numbers at the far right indicate off scale hydrolysis values and standard deviations. Data is not shown in cases where hydrolysis rates were negligible.
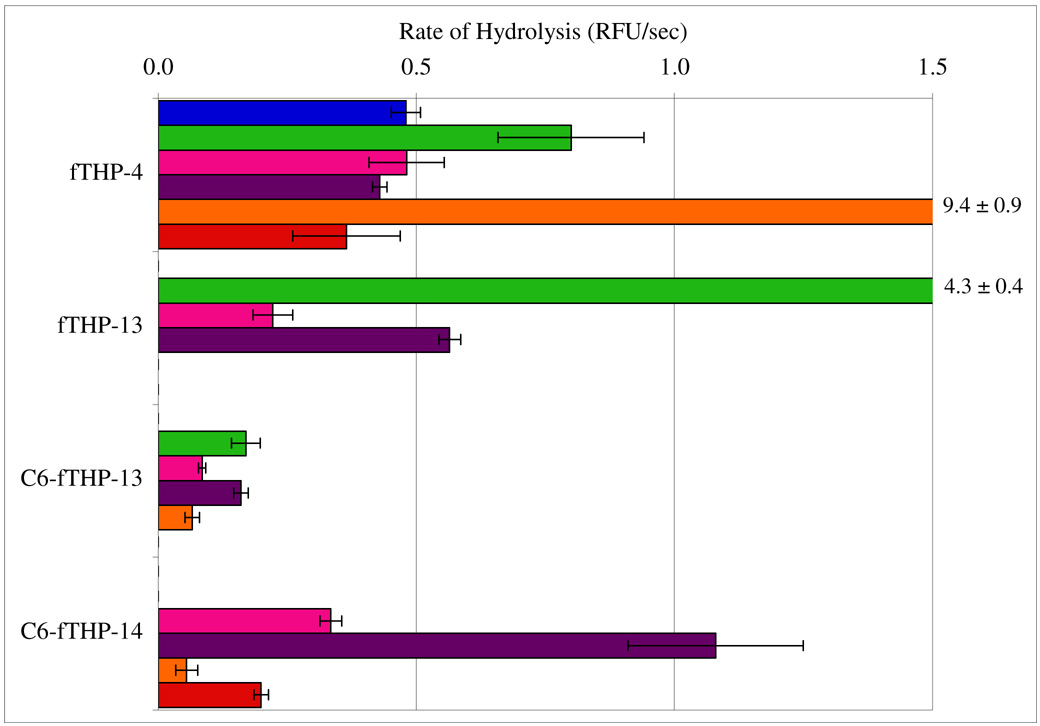
Figure 2. Rates of MMP hydrolysis for fTHP-4, fTHP-13, and fTHP-14.
Hydrolysis was examined using 10 nM of each MMP and 4 µM substrate (fTHP-4, fTHP-13, C6-fTHP-13, or C6-fTHP-14). Fluorescence was measured using λexcitation = 324 nm and λemission = 393 nm. Rates of hydrolysis were obtained from plots of fluorescence versus time, using data points from only the linear portion of the hydrolysis curve. The MMPs examined were MMP-1 (blue), MMP-2 (green), MMP-8 (pink), MMP-13 (purple), MT1-MMP (orange), MT2-MMP (red), and MT5-MMP (yellow). Numbers at the far right indicate off scale hydrolysis values and standard deviations. Data is not shown in cases where hydrolysis rates were negligible.
Three MMPs lacking their hemopexin-like C-terminal domains were studied: MT1-MMP(Δ279–523), MT2-MMP(Δ268–628), and MMP-9(Δ444–707). A prior comparison of the MT1-MMP catalytic domain [MT1-MMP(Δ319–523)] with the MT1-MMP ectodomain (residues 112–523; designated ΔTM-MT1 MMP) showed that MT1-MMP(Δ319–523) was more efficient at single-stranded substrate hydrolysis than ΔTM MT1 MMP, but differences in triple-helical peptidase activity were only slight (40). One may assume that MT2-MMP behaves in a similar fashion as MT1-MMP in terms of the role of the C-terminal domain. The C-terminal domain of MMP-9 does not modulate proteolytic activities of the enzyme (61, 62). The C-terminal domain is not critical for triple-helical peptidase activity, as noted previously (5, 24, 40, 59, 63–65).
Secreted MMPs (MMP-1, MMP-2, MMP-8, and MMP-13) typically cleaved only one bond in fTHP-4, fTHP-12, fTHP-13, and fTHP-14 (Table 2). In some cases, cleavage sites could not be determined due to slow rates of hydrolysis. MMP-3 was not efficient at cleaving triple-helical substrates, consistent with prior results for MMP-3 hydrolysis of fTHP-4 (9). The Pro-Leu-Gly-Met-Arg-Gly-Gln sequence (fTHP-12), which proved to be MMP-13 selective as a phage display derived linear sequence, was neither specific nor selective when contained in a triple-helix. fTHP-12 was cleaved by almost all MMPs at significant rates (Figure 1). The Pro-Leu-Gly~Met-Arg-Gly motif was previously shown to be greatly favored by MMP-13 over MMP-1 and MMP-9 in a linear context (13). We found the same pattern to exist for this motif in a triple-helical context (Figure 1). However, MMP-2 favored Pro-Leu-Gly~Met-Arg-Gly over MMP-13 in the triple-helix, the opposite of the behavior observed for a linear construct (13). Thus, the ability of MMP-2 to process the Gly-Met bond was sensitive to the conformation of the substrate. We previously found that the α1(V) collagen-derived sequence Gly-Pro-Pro-Gly~Val-Val-Gly-Glu sequence was efficiently hydrolyzed by MMP-2 and MMP-9 in a triple-helical context, but not as a single-stranded substrate (25).
There was a clear difference in fTHP-12 specificity between secreted and MT-MMPs. All MT-MMPs that cleaved fTHP-12 did so at two bonds, Gly-Met and Gly-Gln (Table 2). The secreted MMPs cleaved fTHP-12 only at the Gly-Met bond (Table 2). This substrate was also the only triple-helical sequence cleaved by MT5-MMP. Addition of hexanoic acid (C6) to fTHP-12 made this substrate more selective towards MMP-13 and MT2-MMP(Δ268–628) (Figure 1). MT2-MMP(Δ268–628) actually showed greater activity towards the C6 variant of this substrate. MMP-2 was affected the most by the increase of thermal stability of the substrate, as activity was virtually lost in the C6 variant (Figure 1).
It stands to reason that since different MMPs are affected by triple-helical stability to a different degree, then interruption of the triple-helical sequence due to the Gly mutation might have similar effects. Introduction of a Gly interruption (Pro-Val-Asn-Phe-Arg-Gly-Gln; fTHP-13) did not seem to shift the “reading frame” of secreted MMPs, as cleavage still occurred at the same relative P1-P1’ subsite location as in the uninterrupted sequences (Table 2). Selectivity, however, increased for MMP-13 and MMP-2 as compared to the consensus type I–III sequence (fTHP-4; see Figure 2). MT-MMPs were more effected than secreted MMPs by the introduction of a Gly interruption, which apparently resulted in an inability of MT-MMPs to cleave at the putative P1-P1’ bond (Asn-Phe). A secondary cleavage site (Gly-Gln) found in fTHP-4 and fTHP-12 became the primary MT1-MMP(Δ279–523) and MT2-MMP(Δ268–628) cleavage site in fTHP-13 (Table 2). This suggests either the necessity of Gly present in P1 subsite or an intact Gly-Xxx-Yyy register in order for MT-MMPs to bind and process the triple-helical molecule. The addition of hexanoic acid (C6) to the Gly interrupted sequence (C6-fTHP-13) practically abolished the activities of MT1-MMP(Δ279–523), MT2-MMP(Δ268–628), and MMP-8 and significantly lowered MMP-2 and MMP-13 activities (Figure 2). The subsequent substitution of Gln in the P4’ subsite for Pro (fTHP-14) led to the more selective substrate (Figure 2). Interestingly, MT1-MMP(Δ279–523) was able to cleave the Asn-Phe bond in this case, but at a very insignificant rate.
Based on prior studies, it is not readily apparent why MT1-MMP(Δ279–523) does not cleave the Asn-Phe bond in fTHP-13. MT1-MMP has activity towards a great variety of proteins beyond collagen (66, 67). No significant differences are observed in the S1 pocket of MT1-MMP compared to other MMPs (68), and thus Asn should be as well tolerated as in the secreted MMPs. A variety of linear substates containing Asn~Phe-Arg motifs were previously shown to be hydrolyzed efficiently (kcat/KM = 5,900–49,600 sec−1M−1) by MT1-MMP (16). Triple-helical peptide, linear peptide, and phage display peptide library studies indicate that MT1-MMP prefers long chain, hydrophobic residues in the P1’ subsite (5, 14–16). More specifically, Phe in the P1’ subsite should also be well tolerated, since both MT1-MMP and MT2-MMP have deep hydrophobic S1’ pockets as evidenced by their ability to cleave Gly-Cys(Mob) bonds (5, 9). However, the above mentioned linear peptides all contained Glu in P2, Thr in P3’, and Ala in P4’, which are not seen in fTHP-13. Thus, the lack of MT-MMP hydrolysis of the fTHP-13 Asn-Phe bond may be due to the influence of neighboring residues, triple-helical context, or both. It is conceivable that triple-helical peptides incorporating the Gly-Pro-Val-Asn-Phe-Arg-Gly-Gln motif can be useful as an active MMP profiling tool due to the presence of two distinct cleavage sites, one of which is amenable to cleavage by secreted MMPs and another by MT-MMPs.
Kinetic Parameters and Activation Energies
Kinetic parameters and activation energies for substrate hydrolysis were determined for two secreted enzymes, MMP-8 and MMP-13, as these MMPs exhibited reasonable rates of hydrolysis for all potential substrates (Figure 1 and Figure 2). Initially, a comparison was made between the consensus triple-helical sequence (fTHP-4) and the interrupted triple-helical sequence (fTHP-13). MMP-8 showed a marginally higher kcat/KM with fTHP-4 than fTHP-13 (4490 versus 3010 sec−1M−1; Table 3). MMP-13 exhibited the reverse kcat/KM trends for fTHP-4 and fTHP-13, with values of 1735 and 6116 sec−1M−1, respectively. This indicates that among the secreted MMPs differences exist in specificity towards triple-helical sequences with an interrupted Gly register. In the case of MMP-13, the difference in specificity is primarily due to an increase in kcat.
Table 3.
Kinetic Parameters for fTHP-4, fTHP-12, and fTHP-13 Hydrolysis by MMP-8 and MMP-13 at 30 °C.
| Enzyme | Substrate | kcat/KM | kcat | KM |
|---|---|---|---|---|
| (sec−1M−1) | (sec−1) | (µM) | ||
| MMP-8 | fTHP-4 | 4,500a | 0.035 ± 0.016a | 7.60 ± 2.40a |
| MMP-8 | C6-fTHP-12 | 5,090 | 0.070 ± 0.020 | 14.50 ± 4.80 |
| MMP-8 | fTHP-13 | 3,010 | 0.028 ± 0.004 | 9.30 ± 1.84 |
| MMP-8 | C6-fTHP-13 | 3,870 | 0.020 ± 0.010 | 3.90 ± 2.30 |
| MMP-13 | fTHP-4 | 1,600a | 0.015 ± 0.005a | 8.60 ± 1.70a |
| MMP-13 | C6-fTHP-12 | 9,710 | 0.060 ± 0.030 | 6.35 ± 3.0 |
| MMP-13 | fTHP-13 | 6,116 | 0.127 ± 0.046 | 20.50 ± 6.22 |
| MMP-13 | C6-fTHP-13 | 5,800 | 0.060 ± 0.040 | 9.95 ± 0.35 |
(9).
A second comparison was made for the MMP-13 derived triple-helical sequence (fTHP-12) and the interrupted triple-helical sequence (fTHP-13). In this case, the C6 variants of the substrates were used, as their thermal stabilities were very similar (Table 1). Comparison of kinetic parameters for MMP-8 and MMP-13 hydrolysis of C6-fTHP-12 and C6-fTHP-13 (Table 3) indicated similar activities towards both substrates, but MMP-8 had distinctly different individual kcat and KM values for each substrate. For example, C6-fTHP-12 was turned over more rapidly than C6-fTHP-13 by MMP-8. MMP-8 had a much worse KM value for the uninterrupted sequence (C6-fTHP-12), while MMP-13 had a worse KM value for the interrupted sequence (C6-fTHP-13).
The Gly interruption of fTHP-13 had opposite effects on MMP-8 and MMP-13 activation energies. MMP-8 showed a lower Ea for hydrolysis of an uninterrupted sequence (fTHP-4), whereas MMP-13 had a lower Ea with the interrupted fTHP-13 sequence (Table 4). These activation energy trends correlate with the sequence specificities of the enzymes. Overall, MMP-8 showed much closer Ea values and individual kinetic parameters for both sequences than MMP-13. As activation energies reflect the difficulty in accessing water at the site of hydrolysis in native collagen, i.e. reaching the transition state (69, 70), differences between MMP-8 and MMP-13 may well reflect the collagenolytic potential of the two enzymes. Considering the present and prior studies (5), MT1-MMP exhibited the lowest activation energy among all tested collagenolytic MMPs for the hydrolysis of the consensus type I–III collagen sequence (fTHP-4). In light of this result, the inability of MT1-MMP to cleave an interrupted sequence is even the more intriguing.
Table 4.
Activation Energies for Substrate Hydrolysis by MMPs.
| Enzyme | Substrate | Substrate | Ea |
|---|---|---|---|
| Tm (°C) | (kcal/mol) | ||
| MMP-8 | fTHP-4 | 36.3 | 12.8 ± 2.1 |
| MMP-8 | fTHP-13 | 39.0 | 15.5 ± 1.0 |
| MMP-13 | fTHP-4 | 36.3 | 17.4 ± 2.0 |
| MMP-13 | fTHP-13 | 39.0 | 12.7 ± 0.6 |
| MMP-1 | fTHP-4 | 36.3 | 20.0 ± 1.6a |
| MMP-1(Δ243–450) | fTHP-4 | 36.3 | 29.0 ± 0.6a |
| MMP-14(Δ279–523) | fTHP-4 | 36.3 | 8.8 ± 0.1a |
(5).
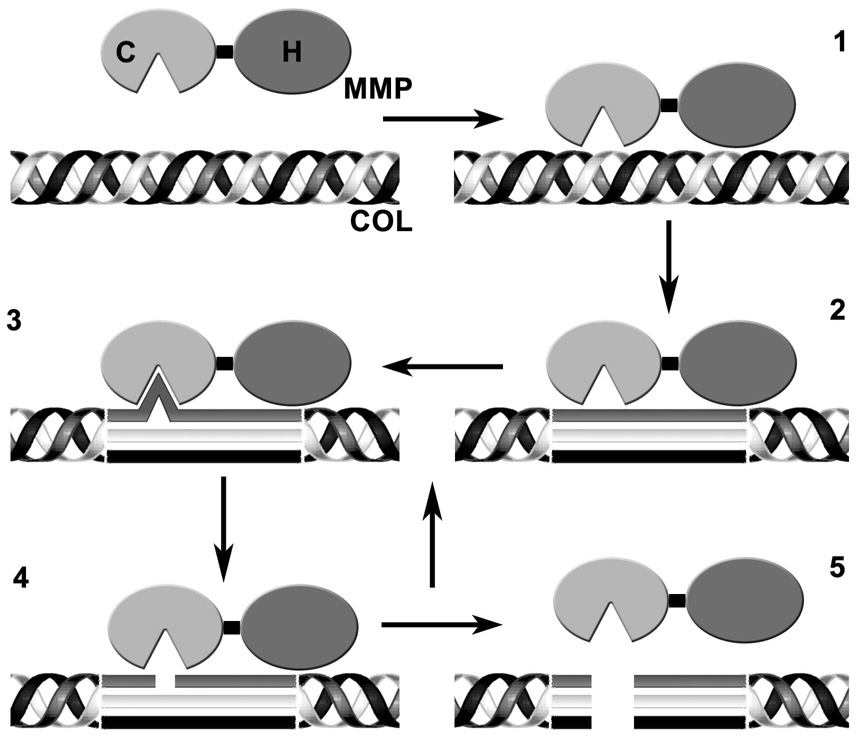
A putative mechanism by which an MMP catalyzes hydrolysis of a triple-helix can be divided into several distinct steps (Figure 3). First is an initial binding of the MMP to a triple-helix. Many MMPs are capable of binding to triple-helices, including some that do not cleave collagen (71, 72). Second, an unwinding of the triple-helix allows access to single strands (4). The unwinding step appears to differentiate collagenolytic from non-collagenolytic MMPs (4). Third, a single strand binds to the active site. The binding of the single strand to the active site appears to be the last step in the triple-helix unwinding process (4). Fourth, a single strand is cleaved and product released. Fifth, in rapid succession, the other two strands are bound and cleaved and the products are released. The results of the present study suggest that the third and fourth step occur at a different rate than if the single strand is introduced independently (i.e., not part of an initial triple-helix). For example, the Pro-Leu-Gly-Met-Arg-Gly-Gln sequence was not selective for MMP-13 when presented in a triple-helix, but was as a single-stranded sequence (12). In similar fashion, the Asn-Phe-Arg motif, when part of a triple-helix, was not hydrolyzed by MT1-MMP, while single-stranded sequences containing this motif were (16). Although collagenases have different collagen specificities, KM values for hydrolysis of types I–III collagen are similar (73, 74). Overall, this suggests that the conformational presentation of substrate sequences to an MMP active site is critical for enzyme specificity, in that activities differ when sequences are presented from an unwound triple-helix versus an independent single strand.
Figure 3. Putative stepwise mechanism for MMP catalyzed triple-helix hydrolysis.
Step 1 is the binding of the triple-helix to the MMP. Step 2 is the unwinding of the triple-helix, allowing access to individual strands. Step 3 is the binding of a single strand to the MMP active site. Step 4 is the hydrolysis of a single strand and release of product. For step 5, in rapid succession, the other two strands are bound and cleaved and the products are released. COL = collagen, C = catalytic domain, and H = hemopexin-like domain.
The present study has delineated secreted MMP from MT-MMP triple-helical peptidase activity, in terms of sequence specificity (cleavage of Gly-Gln bonds) and hydrolysis of interrupted triple-helical sequences. In turn, amongst the secreted collagenolytic MMPs, MMP-8 and MMP-13 show different relative preferences for interrupted versus uninterrupted triple-helices. Based on the present and prior studies (5, 9, 24, 25, 33, 59), we can assign unique triple-helical peptidase behaviors to most of the collagenolytic MMPs (Figure 4). MMP-1 has the weakest triple-helical peptidase activity, and has great difficulty in hydrolyzing more thermally stable substrates. MMP-1 does not tolerate long chain, hydrophobic residues interacting with its S1’ subsite nor positively charged residues interacting with the S2 subsite. MMP-2 possesses many of the characteristics described for MMP-1, but additionally will cleave Gly-Gln bonds. Unlike MMP-1, MMP-2 prefers the combination of Leu in the substrate P2 subsite and Met in the P1’ subsite compared to Gln and Leu in these respective subsites. MMP-8 is a robust triple-helical peptidase that favors long chain, hydrophobic residues interacting with its S1’ subsite. MMP-9 activity is similar to that of MMP-2, except that cleavage of Gly-Gln bonds is not observed. In contrast to MMP-2, MMP-9 does not cleave type I collagen although it does bind to it (72, 75). As MMP-9 is active against other collagen types (8), specific features of type I collagen may prevent the enzyme from hydrolyzing it. Alternatively, the heavily glycosylated linker of MMP-9 may interfere with hydrolysis of type I collagen, although MMP-9 glycosylation has not yet been shown to play a role in proteolytic activities (61). MMP-13 processes thermally stable sequences efficiently, with little effect of sequence. In contrast to MMP-1, MMP-8, and MT1-MMP, MMP-13 prefers a positively charged residue interacting with the S2 subsite, and may favor certain interrupted triple-helical sequences over uninterrupted ones. MT1-MMP is the most robust triple-helical peptidase, and is reasonably sensitive to substrate thermal stability. It disfavors positively charged residues interacting with the S2 subsite, and is also relatively ineffectual at processing interrupted triple-helical sequences that are cleaved by soluble MMPs. MT2-MMP behaves similarly to MT1-MMP, except that it is less active.
Figure 4. Flowchart depicting MMP triple-helical peptidase activities.
The 10 MMPs examined in the present study are initially sub-divided based upon their ability to cleave a triple-helical substrate. A sub-division within the triple-helical peptidase MMPs occurs upon their ability to process more thermally stable substrates. The sequence specificity of each MMP, initially based upon individual substitutions within the P1’ and P2 subsites (9), allows for the next sub-division. The sequence specificity of each MMP, based upon combined substitutions within the P1’ and P2 subsites, followed by preferences for an interrupted sequence, allows for a final sub-division. Ultimately, all “collagenolytic” MMPs (MMP-1, MMP-2, MP-8, MMP-13, MT1 MMP, and MT2-MMP) display distinct behaviors.
Phage display and peptide libraries have demonstrated that many members of the MMP family not only cleave, but may hydrolyze more rapidly, non-Gly-Xxx-Yyy sequences (11). As discussed earlier, the Pro-Val-Asn~Phe-Arg motif was hydrolyzed more rapidly by MMP-13 than MMP-2 in linear form (13). However, this same motif was hydrolyzed more rapidly by MMP-2 than MMP-13 when incorporated into a triple-helical construct. Thus, while phage display can be informative, it is not necessarily predictive of interactions between enzymes and proteins due to conformational influences. Such behaviors may be quite significant for understanding MMP mechanisms of action, and aid in the development of selective MMP inhibitors.
ACKNOWLEDGMENTS
We gratefully acknowledge Dr. Hideaki Nagase and his laboratory for supplying active MMP-1 and proMMP-2 and Ms. Orsi Giricz for constructing Figure 3.
Footnotes
This work was supported by the National Institutes of Health (AR 40994 to K.B., CA 98799 and EB 000289 to G.B.F.) and the FAU Center of Excellence in Biomedical and Marine Biotechnology.
Abbreviations: APMA, 4-aminophenyl mercuric acetate; CD, circular dichroism; DMSO, dimethylsulfoxide; Dnp, 2,4-dinitrophenyl; Fmoc, 9-fluorenylmethoxycarbonyl; FRET, fluorescence resonance energy transfer; fTHP, fluorogenic triple-helical peptide; Hyp, 4-hydroxy-L-proline; MALDI-TOF-MS, matrix-assisted laser desorption ionization time-of-flight mass spectrometry; Mca, (7-methoxycoumarin-4-yl)acetyl; MMP, matrix metalloproteinase; NC, not cleaved; ND, not determined; RP-HPLC, reversed-phase high-performance liquid chromatography; TFA, trifluoroacetic acid; TIMP, tissue inhibitor of metalloproteinases.
A list of known mutations can be found at Database of Human Collagen Mutations, www.le.ac.uk/genetics/collagen.
S. Wei et al., manuscript in preparation.
REFERENCES
- 1.McCawley LJ, Matrisian LM. Matrix metalloproteinases: They're not just for matrix anymore! Curr. Opin. Cell Biol. 2001;13:534–540. doi: 10.1016/s0955-0674(00)00248-9. [DOI] [PubMed] [Google Scholar]
- 2.Overall CM. Molecular determinants of metalloproteinase substrate specificity. Mol. Biotech. 2002;22:51–86. doi: 10.1385/MB:22:1:051. [DOI] [PubMed] [Google Scholar]
- 3.Morrison CJ, Overall CM. TIMP-independence of MMP-2 activation by MT2-MMP is determined by contributions of both the MT2-MMP catalytic and hemopexin domains. J. Biol. Chem. 2006;281:26528–26539. doi: 10.1074/jbc.M603331200. [DOI] [PubMed] [Google Scholar]
- 4.Chung L, Dinakarpandian D, Yoshida N, Lauer-Fields JL, Fields GB, Visse R, Nagase H. Collagenase unwinds triple helical collagen prior to peptide bond hydrolysis. EMBO J. 2004;23:3020–3030. doi: 10.1038/sj.emboj.7600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minond D, Lauer-Fields JL, Nagase H, Fields GB. Matrix metalloproteinase triple-helical peptidase activities are differentially regulated by substrate stability. Biochemistry. 2004;43:11474–11481. doi: 10.1021/bi048938i. [DOI] [PubMed] [Google Scholar]
- 6.Tam EM, Moore TR, Butler GS, Overall CM. Characterization of the distinct collagen binding, helicase and cleavage mechanisms of matrix metalloproteinase 2 and 14 (gelatinase A and MT1-MMP): The differential roles of the MMP hemopexin C domains and the MMP-2 fibronectin type II modules in collagen triple helicase activities. J. Biol. Chem. 2004;279:43336–43344. doi: 10.1074/jbc.M407186200. [DOI] [PubMed] [Google Scholar]
- 7.Ohuchi E, Imai K, Fujii Y, Sato H, Seiki M, Okada Y. Membrane type I matrix metalloproteinase digests interstitial collagens and other extracellular matrix macromolecules. J. Biol. Chem. 1997;272:2446–2451. doi: 10.1074/jbc.272.4.2446. [DOI] [PubMed] [Google Scholar]
- 8.Woessner JF, Nagase H. Matrix Metalloproteinases and TIMPs. Oxford: Oxford University Press; 2000. [Google Scholar]
- 9.Minond D, Lauer-Fields JL, Cudic M, Overall CM, Pei D, Brew K, Visse R, Nagase H, Fields GB. The roles of substrate thermal stability and P2 and P1' subsite identity on matrix metalloproteinase triple-helical peptidase activity and collagen specificity. J. Biol. Chem. 2006;281:38302–38313. doi: 10.1074/jbc.M606004200. [DOI] [PubMed] [Google Scholar]
- 10.Nagase H, Fields GB. Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides. Biopolymers. 1996;40:399–416. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C399::AID-BIP5%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 11.Turk BE, Huang LL, Piro ET, Cantley LC. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat. Biotech. 2001;19:661–667. doi: 10.1038/90273. [DOI] [PubMed] [Google Scholar]
- 12.Deng S-J, Bickett DM, Mitchell JL, Lambert MH, Blackburn RK, Carter HL, III, Neugebauer J, Pahel G, Weiner MP, Moss ML. Substrate specificity of human collagenase 3 assessed using a phage-displayed peptide library. J. Biol. Chem. 2000;275:31422–31427. doi: 10.1074/jbc.M004538200. [DOI] [PubMed] [Google Scholar]
- 13.Rasmussen FH, Yeung N, Kiefer L, Murphy G, Lopez-Otin C, Vitek MP, Moss ML. Use of a multiple-enzyme/multiple reagent assay system to quantify activity levels in samples containing mixtures of matrix metalloproteinases. Biochemistry. 2004;43:2987–2995. doi: 10.1021/bi036063m. [DOI] [PubMed] [Google Scholar]
- 14.Mucha A, Cuniasse P, Kannan R, Beau F, Yiotakis A, Basset P, Dive V. Membrane type-1 matrix metalloproteinase and stromelysin-3 cleave more efficiently synthetic substrates containing unusual amino acids in their P1' positions. J. Biol. Chem. 1998;273:2763–2768. doi: 10.1074/jbc.273.5.2763. [DOI] [PubMed] [Google Scholar]
- 15.Ohkubo S, Miyadera K, Sugimoto Y, Matsuo K-i, Wierzba K, Yamada Y. Identification of substrate sequences for membrane type-1 matrix metalloproteinase using bacteriophage peptide display library. Biochem. Biophys. Res. Commun. 1999;266:308–313. doi: 10.1006/bbrc.1999.1816. [DOI] [PubMed] [Google Scholar]
- 16.Kridel SJ, Sawai H, Ratnikov BI, Chen EI, Li W, Godzik A, Strongin AY, Smith JW. A unique substrate binding mode discriminates membrane type 1-matrix metalloproteinase (MT1-MMP) from other matrix metalloproteinases. J. Biol. Chem. 2002;277:23788–23793. doi: 10.1074/jbc.M111574200. [DOI] [PubMed] [Google Scholar]
- 17.Pan W, Arnone M, Kendall M, Grafstrom RH, Seitz SP, Wasserman ZR, Albright CF. Identification of peptide substrates for human MMP-11 (stromelysin-3) using phage display. J. Biol. Chem. 2003;278:27820–27827. doi: 10.1074/jbc.M304436200. [DOI] [PubMed] [Google Scholar]
- 18.Wu H, Byrne MH, Stacey A, Goldring MB, Birkhead JR, Jaenisch R, Krane SM. Generation of collagenase-resistant collagen by site-directed mutagenesis of murine pro α1(I) collagen gene. Proc. Natl. Acad. Sci. USA. 1990;87:5888–5892. doi: 10.1073/pnas.87.15.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasty KA, Wu H, Byrne M, Goldring MB, Seyer JM, Jaenisch R, Krane SM, Mainardi CL. Susceptibility of type I collagen containing mutated α1(I) chains to cleavage by human neutrophil collagenase. Matrix. 1993;13:181–186. doi: 10.1016/s0934-8832(11)80001-6. [DOI] [PubMed] [Google Scholar]
- 20.Byers PH, Cole WG. Osteogenesis imperfecta. In: Royce PM, Steinmann B, editors. Connective Tissue and Its Hereditable Disorders. Molecular, Genetic, and Medical Aspects. 2nd ed. New York: Wiley-Liss; 2002. pp. 385–430. [Google Scholar]
- 21.Steinmann B. The Ehlers-Danlos Syndrome. In: Royce PM, Steinmann B, editors. Connective Tissue and Its Heritable Disorders. Molecular, Genetic, and Medical Aspects. 2nd ed. New York: Wiley-Liss; 2002. pp. 431–523. [Google Scholar]
- 22.Pepin M, Schwarze U, Superti-Furga A, Byers PH. Clinical and genetic features of Ehlers-Danlos syndrome type IV, the vascular type. N. Engl. J. Med. 2000;342:673–680. doi: 10.1056/NEJM200003093421001. [DOI] [PubMed] [Google Scholar]
- 23.Fields GB. A model for interstitial collagen catabolism by mammalian collagenases. J. Theor. Biol. 1991;153:585–602. doi: 10.1016/s0022-5193(05)80157-2. [DOI] [PubMed] [Google Scholar]
- 24.Lauer-Fields JL, Broder T, Sritharan T, Nagase H, Fields GB. Kinetic analysis of matrix metalloproteinase triple-helicase activity using fluorogenic substrates. Biochemistry. 2001;40:5795–5803. doi: 10.1021/bi0101190. [DOI] [PubMed] [Google Scholar]
- 25.Lauer-Fields JL, Sritharan T, Stack MS, Nagase H, Fields GB. Selective hydrolysis of triple-helical substrates by matrix metalloproteinase-2 and -9. J. Biol. Chem. 2003;278:18140–18145. doi: 10.1074/jbc.M211330200. [DOI] [PubMed] [Google Scholar]
- 26.Lauer-Fields JL, Kele P, Sui G, Nagase H, Leblanc RM, Fields GB. Analysis of matrix metalloproteinase activity using triple-helical substrates incorporating fluorogenic L- or D-amino acids. Anal. Biochem. 2003;321:105–115. doi: 10.1016/s0003-2697(03)00460-3. [DOI] [PubMed] [Google Scholar]
- 27.Nagase H, Fields CG, Fields GB. Design and characterization of a fluorogenic substrate selectively hydrolyzed by stromelysin 1 (matrix metalloproteinase-3) J. Biol. Chem. 1994;269:20952–20957. [PubMed] [Google Scholar]
- 28.Neumann U, Kubota H, Frei K, Ganu V, Leppert D. Characterization of Mca-Lys-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2, a fluorogenic substrate with increased specificity constants for collagenases and tumor necrosis factor converting enzyme. Anal. Biochem. 2004;328:166–173. doi: 10.1016/j.ab.2003.12.035. [DOI] [PubMed] [Google Scholar]
- 29.Fields GB, Lauer-Fields JL, Liu R-q, Barany G. Principles and Practice of Solid-Phase Peptide Synthesis. In: Grant GA, editor. Synthetic Peptides: A User's Guide. 2nd Edition. New York: W.H. Freeman & Co.; 2001. pp. 93–219. [Google Scholar]
- 30.Yu Y-C, Berndt P, Tirrell M, Fields GB. Self-assembling amphiphiles for construction of protein molecular architecture. J. Am. Chem. Soc. 1996;118:12515–12520. [Google Scholar]
- 31.Yu Y-C, Tirrell M, Fields GB. Minimal lipidation stabilizes protein-like molecular architecture. J. Am. Chem. Soc. 1998;120:9979–9987. [Google Scholar]
- 32.Fields CG, Fields GB. Minimization of tryptophan alkylation following 9-fluorenylmethoxycarbonyl solid-phase peptide synthesis. Tetrahedron Lett. 1993;34:6661–6664. [Google Scholar]
- 33.Lauer-Fields JL, Nagase H, Fields GB. Use of Edman degradation sequence analysis and matrix-assisted laser desorption/ionization mass spectrometry in designing substrates for matrix metalloproteinases. J. Chromatogr. A. 2000;890:117–125. doi: 10.1016/s0021-9673(00)00396-4. [DOI] [PubMed] [Google Scholar]
- 34.Chung L, Shimokawa K, Dinakarpandian D, Grams F, Fields GB, Nagase H. Identification of the RWTNNFREY(183–191) region as a critical segment of matrix metalloproteinase 1 for the expression of collagenolytic activity. J. Biol. Chem. 2000;275:29610–29617. doi: 10.1074/jbc.M004039200. [DOI] [PubMed] [Google Scholar]
- 35.Lauer-Fields JL, Nagase H, Fields GB. Development of a Solid-Phase Assay for Analysis of Matrix Metalloproteinase Activity. J. Biomolecular Techniques. 2004;15:305–316. [PMC free article] [PubMed] [Google Scholar]
- 36.Itoh Y, Binner S, Nagase H. Steps involved in activation of the complex of pro-matrix metalloproteinase 2 (progelatinase A) and tissue inhibitor of metalloproteinases (TIMP)-2 by 4-aminophenylmercuric acetate. Biochem. J. 1995;308:645–651. doi: 10.1042/bj3080645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pelman GR, Morrison CJ, Overall CM. Pivotal molecular determinants of peptidic and collagen triple helicase activities reside in the S3' subsite of matrix metalloproteinase 8 (MMP-8) J. Biol. Chem. 2005;280:2370–2377. doi: 10.1074/jbc.M409603200. [DOI] [PubMed] [Google Scholar]
- 38.Elkins PA, Ho SH, Smith WW, Janson CA, D'Alessio KJ, McQueney MS, Cummings MD, Romanic AM. Structure of the C-teminally truncated human proMMP9, a gelatin-binding matrix metalloproteinase. Acta Cryst. 2002;D58:1182–1192. doi: 10.1107/s0907444902007849. [DOI] [PubMed] [Google Scholar]
- 39.Huang W, Suzuki K, Nagase H, Arumugam S, Van Doren S, Brew K. Folding and characterization of the amino-terminal domain of human tissue inhibitor of metalloproteinases-1 (TIMP-1) expressed at high yield in E. coli. FEBS Lett. 1996;384:155–161. doi: 10.1016/0014-5793(96)00304-3. [DOI] [PubMed] [Google Scholar]
- 40.Hurst DR, Schwartz MA, Ghaffari MA, Jin Y, Tschesche H, Fields GB, Sang Q-XA. Catalytic- and ecto-domains of membrane type 1-matrix metalloproteinase have similar inhibition profiles but distinct endopeptidase activities. Biochem. J. 2004;377:775–779. doi: 10.1042/BJ20031067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pei D. Identification and characterization of the fifth membrane-type matrix metalloproteinase MT5-MMP. J. Biol. Chem. 1999;274:8925–8932. doi: 10.1074/jbc.274.13.8925. [DOI] [PubMed] [Google Scholar]
- 42.Nie J, Pei D. Direct activation of pro-matrix metalloproteinase-2 by leukolysin/membrane-type 6 matrix metalloproteinase/matrix metalloproteinase 25 at the Asn(109)-Tyr bond. Cancer Res. 2003;63:6758–6762. [PubMed] [Google Scholar]
- 43.d'Ortho MP, Will H, Atkinson S, Butler G, Messent A, Gavrilovic J, Smith B, Timpl R, Zardi L, Murphy G. Membrane-type matrix metalloproteinases 1 and 2 exhibit broad-spectrum proteolytic capacities comparable to many matrix metalloproteinases. Eur. J. Biochem. 1997;250:751–757. doi: 10.1111/j.1432-1033.1997.00751.x. [DOI] [PubMed] [Google Scholar]
- 44.Llano E, Pendas AM, Freije JP, Nakano A, Knauper V, Murphy G, Lopez-otin C. Identification and characterization of human MT5-MMP, a new membrane-bound activator of progelatinase a overexpressed in brain tumors. Cancer Res. 1999;59:2570–2576. [PubMed] [Google Scholar]
- 45.English WR, Velasco G, Stracke JO, Knauper V, Murphy G. Catalytic activities of membrane-type 6 matrix metalloproteinase (MMP25) FEBS Lett. 2001;491:137–142. doi: 10.1016/s0014-5793(01)02150-0. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki K, Kan C-C, Huang W, Gehring MR, Brew K, Nagase H. Expression of human pro-matrix metalloproteinase 3 that lacks the N-terminal 34 residues in Escherichia coli: Autoactivation and interaction with tissue inhibitor of metalloproteinase 1 (TIMP-1) Biol. Chem. 1998;379:185–191. doi: 10.1515/bchm.1998.379.2.185. [DOI] [PubMed] [Google Scholar]
- 47.Knight CG, Willenbrock F, Murphy G. A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Lett. 1992;296:263–266. doi: 10.1016/0014-5793(92)80300-6. [DOI] [PubMed] [Google Scholar]
- 48.Mohs A, Popiel M, Li Y, Baum J, Brodsky B. Conformational features of a natural break in the type IV collagen Gly-X-Y repeat. J. Biol. Chem. 2006;281:17197–17202. doi: 10.1074/jbc.M601763200. [DOI] [PubMed] [Google Scholar]
- 49.Hyde TJ, Bryan M, Brodsky B, Baum J. Sequence dependence of renucleation after a Gly mutation in model collagen peptides. J. Biol. Chem. 2006;281:36937–36943. doi: 10.1074/jbc.M605135200. [DOI] [PubMed] [Google Scholar]
- 50.Gross J, Harper E, Harris ED, McCroskery PA, Highberger JH, Corbett C, Kang AH. Animal collagenases: Specificity of action, and structures of the substrate cleavage site. Biochem. Biophys. Res. Commun. 1974;61:605–612. doi: 10.1016/0006-291x(74)91000-6. [DOI] [PubMed] [Google Scholar]
- 51.Miller EJ, Harris J, E D, Chung E, Finch J, J E, McCroskery PA, Butler WT. Cleavage of type II and III collagens with mammalian collagenase: Site of cleavage and primary structure at the NH2-terminal portion of the smaller fragment released from both collagens. Biochemistry. 1976;15:787–792. doi: 10.1021/bi00649a009. [DOI] [PubMed] [Google Scholar]
- 52.Horwitz AL, Hance AJ, Crystal RG. Granulocyte collagenase: Selective digestion of type I relative to type III collagen. Proc. Natl. Acad. Sci. USA. 1977;74:897–901. doi: 10.1073/pnas.74.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baici A, Salgam P, Cohen G, Fehr K, Boni A. Action of collagenase and elastase from human polymorphonuclear leukocytes on human articular cartilage. Rheumatol. Int. 1982;2:11–16. doi: 10.1007/BF00541264. [DOI] [PubMed] [Google Scholar]
- 54.Welgus HG, Fliszar CJ, Seltzer JL, Schmid TM, Jeffrey JJ. Differential susceptibility of type X collagen to cleavage by two mammalian interstitial collagenases and 72-kDa type IV collagenase. J. Biol. Chem. 1990;265:13521–13527. [PubMed] [Google Scholar]
- 55.Aimes RT, Quigley JP. Matrix metalloproteinase-2 is an interstitial collagenase. J. Biol. Chem. 1995;270:5872–5876. doi: 10.1074/jbc.270.11.5872. [DOI] [PubMed] [Google Scholar]
- 56.Yasumitsu H, Miyazaki K, Umenishi F, Koshikawa N, Umeda M. Comparison of extracellular matrix-degrading activities between 64-kDa and 90-kDa gelatinases purified in inhibitor-free forms from human schwannoma cells. J. Biochem. 1992;111:74–80. doi: 10.1093/oxfordjournals.jbchem.a123721. [DOI] [PubMed] [Google Scholar]
- 57.Murphy G, Reynolds JJ, Bretz U, Baggiolini M. Partial purification of collagenase and gelatinase from human polymorphonuclear leucocytes: Analysis of their actions on soluble and insoluble collagens. Biochem. J. 1982;203:209–221. doi: 10.1042/bj2030209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hotary K, Allen E, Punturieri A, Yana I, Weiss SJ. Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J. Cell Biol. 2000;149:1309–1323. doi: 10.1083/jcb.149.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lauer-Fields JL, Tuzinski KA, Shimokawa K, Nagase H, Fields GB. Hydrolysis of triple-helical collagen peptide models by matrix metalloproteinases. J. Biol. Chem. 2000;275:13282–13290. doi: 10.1074/jbc.275.18.13282. [DOI] [PubMed] [Google Scholar]
- 60.Wu YI, Munshi HG, Sen R, Snipas SJ, Salvesen GS, Fridman R, Stack MS. Glycosylation broadens the substrate profile of membrane type 1-matrix metalloproteinase. J. Biol. Chem. 2004;279:8278–8289. doi: 10.1074/jbc.M311870200. [DOI] [PubMed] [Google Scholar]
- 61.Van den Steen PE, Van Aeist I, Hvidberg V, Piccard H, Fiten P, Jacobsen C, Moestrup SK, Fry S, Royle L, Wormald MR, Wallis R, Rudd PM, Dwek RA, Opdenakker G. The hemopexin and O-glycosylated domains tune gelatinase B/MMP-9 bioavailability via inhibition and binding to cargo receptors. J. Biol. Chem. 2006;281:18626–18637. doi: 10.1074/jbc.M512308200. [DOI] [PubMed] [Google Scholar]
- 62.Kroger M, Tschesche H. Cloning, expression and activation of a truncated 92-kDa gelatinase minienzyme. Gene. 1997;196:175–180. doi: 10.1016/s0378-1119(97)00223-0. [DOI] [PubMed] [Google Scholar]
- 63.Ottl J, Battistuta R, Pieper M, Tschesche H, Bode W, Kühn K, Moroder L. Design and synthesis of heterotrimeric collagen peptides with a built-in cystine-knot. FEBS Lett. 1996;398:31–36. doi: 10.1016/s0014-5793(96)01212-4. [DOI] [PubMed] [Google Scholar]
- 64.Ottl J, Gabriel D, Murphy G, Knäuper V, Tominaga Y, Nagase H, Kröger M, Tschesche H, Bode W, Moroder L. Recognition and catabolism of synthetic heterotrimeric collagen peptides by matrix metalloproteinases. Chem. Biol. 2000;7:119–132. doi: 10.1016/s1074-5521(00)00077-6. [DOI] [PubMed] [Google Scholar]
- 65.Gioia M, Fasciglione GF, Marini S, D'Alessio S, De Sanctis G, Diekmann O, Pieper M, Politi V, Tschesche H, Coletta M. Modulation of the catalytic activity of neutrophil collagenase MMP-8 on bovine collagen I. J. Biol. Chem. 2002;277:23123–23130. doi: 10.1074/jbc.M110873200. [DOI] [PubMed] [Google Scholar]
- 66.Hwang IK, Park SM, Kim SY, Lee S-T. A proteomic approach to identify substrates of matrix metalloproteinase-14 in human plasma. Biochim. Biophys. Acta. 2004;1702:79–87. doi: 10.1016/j.bbapap.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 67.Tam EM, Morrison CJ, Wu YI, Stack MS, Overall CM. Membrane protease proteomics: Isotope-coded affinity tag MS identification of undescribed MT1-matrix metalloproteinase substrates. Proc. Natl. Acad. Sci. USA. 2004;101:6917–6922. doi: 10.1073/pnas.0305862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Terp GE, Cruciani G, Christensen IT, Jorgensen FS. Structural differences of matrix metalloproteinases with potential implications for inhibitor selectivity examined by the GRID/CPCA approach. J. Med. Chem. 2002;45:2675–2684. doi: 10.1021/jm0109053. [DOI] [PubMed] [Google Scholar]
- 69.Welgus HG, Jeffrey JJ, Eisen AZ. Human skin fibroblast collagenase: Assessment of activation energy and deuterium isotope effect with collagenous substrates. J. Biol. Chem. 1981;256:9516–9521. [PubMed] [Google Scholar]
- 70.Lauer-Fields JL, Fields GB. Triple-helical peptide analysis of collagenolytic protease activity. Biol. Chem. 2002;383:1095–1105. doi: 10.1515/BC.2002.118. [DOI] [PubMed] [Google Scholar]
- 71.Murphy G, Allan JA, Willenbrock F, Cockett MI, O'Connell JP, Docherty AJP. The role of the C-terminal domain in collagenase and stromelysin specificity. J. Biol. Chem. 1992;267:9612–9618. [PubMed] [Google Scholar]
- 72.Allan JA, Docherty AJP, Barker PJ, Huskisson NS, Reynolds JJ, Murphy G. Binding of gelatinases A and B to type-I collagen and other matrix components. Biochem. J. 1995;309:299–306. doi: 10.1042/bj3090299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Welgus HG, Jeffrey JJ, Eisen AZ. The collagen substrate specificity of human skin fibroblast collagenase. J. Biol. Chem. 1981;256:9511–9515. [PubMed] [Google Scholar]
- 74.Mallya SK, Mookhtiar KA, Gao Y, Brew K, Dioszegi M, Birkedal-Hansen H, Van Wart HE. Characterization of 58-kilodalton human neutrophil collagenase: Comparison with human fibroblast collagenase. Biochemistry. 1990;29:10628–10634. doi: 10.1021/bi00499a008. [DOI] [PubMed] [Google Scholar]
- 75.Xu X, Chen Z, Wang Y, Yamada Y, Steffensen B. Functional basis for the overlap in ligand interactions and substrate specificities of matrix metalloproteinases-9 and -2. Biochem. J. 2005;392:127–134. doi: 10.1042/BJ20050650. [DOI] [PMC free article] [PubMed] [Google Scholar]