Abstract
Production of recombinant proteins in mammalian cells is a successful technology that delivers protein pharmaceuticals for therapies and for diagnosis of human disorders. Cost effective production of protein biopharmaceuticals requires extensive optimization through cell and fermentation process engineering at the upstream and chemical engineering of purification processes at the downstream side of the production process. The majority of protein pharmaceuticals are secreted proteins. Accumulating evidence suggests that the folding and processing of these proteins in the endoplasmic reticulum (ER) is a general rate- and yield limiting step for their production. We will summarize our knowledge of protein folding in the ER and of signal transduction pathways activated by accumulation of unfolded proteins in the ER, collectively called the unfolded protein response (UPR). On the basis of this knowledge we will evaluate engineering approaches to increase cell specific productivities through engineering of the ER-resident protein folding machinery and of the UPR.
Keywords: Endoplasmic reticulum associated protein degradation, Heterologous protein production, Molecular chaperone, Protein folding, Unfolded protein response
Introduction
The world market for biopharmaceuticals is currently 48 billion US$ and has been growing annually by 19% over the last five years. Most biopharmaceuticals are therapeutic proteins, such as erythropoietin, blood coagulation factors, and monoclonal antibodies (Werner 2004). Mammalian cells are often the host of choice because of their ability to correctly fold, assemble, glycosylate, and secrete these proteins. Correct folding and a posttranslational modification pattern identical to the human protein are critical determinants of the activity, immunogenicity, and in vivo clearance rates of therapeutic proteins. Monoclonal antibodies are administered at high doses ranging from 25–40 mg every other week to 100 mg per day (Werner 2004). Protein production at multi-kilogram scales to sustain such therapies is required. The advent of these therapies has triggered speculations about capacity shortages and has highlighted that even modest increases in product yield will significantly decrease capital investment and the cost of goods (Werner 2004).
For all these reasons improvement is desirable at the upstream and downstream stages of the production process. Up to ~50% of the total production costs are associated with the upstream side, providing a strong rationale for upstream process engineering. Over the last decade product titers have increased from ~20 mg L−1 to 2 g L−1 (Werner 2004, Wurm 2004). These increases have been largely driven by increases in viable cell densities. Cell specific productivities nowadays usually reach 30–80 pg cell−1 day−1 (Dinnis and James 2005). A plasma cell secretes several thousand IgM molecules s−1 (de StGroth and Scheidegger 1980; Randall et al. 1992), indicating that productivities of 200–400 pg1 cell−1 day−1 and 2.5–12 fold increases in cell specific productivities are possible, which will translate into significant savings in capital investment and the cost of goods. Considering the already high cell specific productivities of many (Dinnis and James 2005; Robinson and Memmert 1991; Wurm 2004), but not all (Ailor and Reff 2005), antibody expression systems, engineering of ER-resident chaperone systems and the UPR to increase cell specific productivities will be especially important for production of non-antibody proteins in animal cells, where cell specific productivities tend to be much lower (Hippenmeyer and Highkin 1993; Jeon et al. 2003; Lee et al. 2001; Park et al. 1999; Zang et al. 1995). For these reasons, we will summarize our understanding of ER-resident protein folding machineries and of the UPR, and discuss animal cell engineering approaches aimed at improving cell specific productivities by engineering of these protein folding machineries and of the UPR to alleviate a potential bottleneck in eukaryotic protein production factories, the folding and processing of the polypeptide chain in the endoplasmic reticulum (ER).
The rate- and yield limiting step for heterologous protein production
Several lines of evidence suggest that folding and posttranslational modification of newly synthesized proteins in the ER becomes rate- and yield-limiting in many eukaryotic expression systems as cell specific productivities approach the productivities of plasma cells. For example, the amount of secreted heterologous protein does not increase proportionally with gene copy number (Parekh et al. 1995), mRNA (Barnes et al. 2004; Fann et al. 1999; Pendse et al. 1992; Schröder et al. 1999), or even the intracellular amount of the heterologous protein (Pendse et al. 1992; Schröder and Friedl 1997). Further, intracellular aggregation (Hasemann and Capra 1990; Hsu and Betenbaugh 1997; Hsu et al. 1996; Hsu et al. 1994; Schröder et al. 2002), association of heterologous proteins with the molecular chaperone BiP/GRP78 (Dorner et al. 1987; Hurtley et al. 1989; Jarvis and Summers 1989; Kaufman et al. 1988; Machamer et al. 1990; Suzuki et al. 1991), and dilation of the ER (Dorner et al. 1989; Gennaro et al. 1991) have been reported. BiP induction, a hallmark of ER stress and UPR activation, has been reported for many expression systems (Alete et al. 2005; Downham et al. 1996; Jones et al. 2005; Miura et al. 2001; Smales et al. 2004). Further, even in secretory cell types such as plasma cells activation of the UPR has been observed (van Anken et al. 2003; Zhang et al. 2005). These data demonstrate that the rate- and yield limiting step for production of recombinant secretory proteins in many eukaryotic expression systems is exit of the correctly folded polypeptide chain from the ER and highlight the critical roles of the UPR in professional secretory cells and in engineering of host cells with increased cell specific productivities.
The ER-resident protein folding machinery
In mammalian cells the ER functions as Ca2+ store, plays important roles in intracellular Ca2+ signaling, and is the site for the synthesis of phospholipids, sterols, secretory and transmembrane proteins (Bernales et al. 2006; Ron and Walter 2007; Schröder 2008a). The ER harbors a protein folding machinery that folds client proteins, quality controls the folding process, and activates the UPR to maintain the balance between the folding capacity of the ER and the folding demand imposed on this organelle (Strudwick and Schröder 2007). This folding demand arises from nascent polypeptide chains entering the ER in an unfolded conformation through the SEC61 translocation channel. Two classes of proteins, molecular chaperones, and protein foldases assist protein folding in the cell. Molecular chaperones interact with hydrophobic surfaces on unfolded proteins, thereby shielding them from engaging in non-productive interactions with other polypeptide chains. Chaperone foldases promote folding of client proteins through ATP consuming cycles in which they repeatedly bind to and dissociate from their substrates. Chaperone holdases bind to their substrates. Structural maturation of the substrate terminates the holdase-substrate interaction. The protein foldase families of protein disulfide isomerases (PDIs) and cis–trans peptidyl prolyl isomerases (PPIs) catalyze the formation and isomerization of disulfide bonds and the cis–trans isomerization of peptidyl prolyl bonds, respectively. Molecular chaperones and protein foldases participate in protein folding quality control, allowing only correctly folded proteins to exit the ER. Proteins recognized as unfolded are retained in the ER, and if judged to be folding incompetent removed from the folding cycles and targeted for proteasomal degradation in a process called ER associated protein degradation (ERAD). Autophagy also contributes to disposal of aggregated ER luminal proteins (Perlmutter 2006).
ER-resident molecular chaperone systems
Three general chaperone systems operate in the ER (Strudwick and Schröder 2007). The HSP70 chaperones BiP/GRP78 and Lhs1p/GRP170/ORP150 and their co-chaperones constitute the first, the HSP90 chaperone GRP94 the second, and the lectin chaperones calnexin, calreticulin, and calmegin the third chaperone system (Fig. 1). BiP assists completely unfolded polypeptide chains to fold, is involved in translocation of nascent polypeptide chains into the ER, gating of the SEC61 translocation channel, quality control of protein folding reactions in the ER, and activation of the UPR. GRP94 and the lectin chaperones preferentially interact with partially folded substrates. The viability of grp94−/− mouse embryonic fibroblasts (MEFs) (Biswas et al. 2007) suggests that GRP94 has a limited substrate spectrum.
Fig. 1.
Chaperone foldase systems in the mammalian ER. The relationship between the GRP94 and calnexin/calreticulin chaperone is not established. Abbreviations: Glc—d-glucose, Pi—inorganic phosphate (HPO42−)
BiP and Lhs1p—HSP70 chaperones of the ER
HSP70 chaperones consist of an N-terminal ATPase and a C-terminal substrate binding domain. In protein folding BiP cycles through rounds of ATP hydrolysis and ADP-ATP exchange reactions (Fig. 1). ATP hydrolysis and ADP-ATP exchange are stimulated by co-chaperones. DnaJ or HSP40 co-chaperones stimulate the ATPase activity of HSP70s. GrpE co-chaperones stimulate the ADP-ATP exchange reaction. At least six different DnaJ co-chaperones, ERdj1/MTJ1, ERdj2/SEC63, ERdj3/HEDJ, ERdj4/MDG1, ERdj5, and p58IPK, and one GrpE protein, BAP/SIL1, exist in the mammalian ER (Strudwick and Schröder 2007). HSP70 chaperones co-ordinate their functions. Lhs1p is a nucleotide exchange factor for BiP. BiP stimulates the ATPase activity of Lhs1p. In the ADP-bound form BiP has high affinity for unfolded substrates. Substrates bound to BiP are conformationally locked. ADP-ATP exchange releases the substrate from BiP, allowing the substrate to continue to fold. Substrate binding to BiP stimulates the ATPase activity of BiP, converting it to the ADP-bound form with high affinity for substrates. This ATP consumption cycle explains longstanding observations that protein folding consumes ATP (Dorner and Kaufman 1994).
GRP94—an HSP90 chaperone of the ER
HSP90 chaperones consist of an N-terminal ATPase domain, followed by a charged region, a dimerization domain and a C-terminal ER retention motif (Strudwick and Schröder 2007). The crystal structure of GRP94 is very similar to crystal structures of cytosolic HSP90s, suggesting a common mechanism of action (Dollins et al. 2007). In cytosolic and bacterial HSP90s ATP binding induces a closed conformation of the HSP90 dimer, in which the N-terminal domains of two protomers assemble into an active ATPase domain. This conformational change triggers substrate release. ATP hydrolysis and dissociation of ADP from HSP90s regenerate an ‘open’ conformation capable of interacting with client proteins (Fig. 1). GRP94, however, is insensitive toward the nature of the adenine nucleotide (Rosser et al. 2004) and adenine nucleotide binding does not trigger the change to the closed, ATP hydrolysis competent conformation, or substrate release (Rosser et al. 2004). Structural maturation of interacting substrates or regulatory interactions with yet to be identified co-chaperones may trigger substrate release and the ensuing conformational change in GRP94. What is then the role of the ATP hydrolysis cycle in GRP94 function? Whether GRP94 has ATPase activity has long been controversial (Dollins et al. 2007). Point mutations in the ATPase domain inactivate GRP94 (Randow and Seed 2001). GRP94 has a low ATPase activity that is comparable to the ATPase activity of yeast and human HSP90s (Dollins et al. 2007; Frey et al. 2007). The overall low ATPase activity of GRP94 may in part reflect the low probability of the chaperone sampling the hydrolytically productive closed conformation. Point mutations in the ATPase domain that diminish adenine nucleotide binding more severely compromise GRP94 function than point mutations predicted to render the ATPase domain inactive (Randow and Seed 2001). GRP94 is a labile protein that looses activity above 30 °C (Frey et al. 2007), which is well below the temperature optimum for mammalian cells, suggesting that adenine nucleotide binding stabilizes GRP94, and that ATP hydrolysis in the closed conformation provides the energy to return GRP94 into its open conformation to initiate a second round of substrate interaction.
The lectin chaperones calnexin, calreticulin, and calmegin
Many proteins entering the ER are posttranslationally modified by transfer of the oligosaccharide Glc3Man9GlcNAc2 (Glc—d-glucose, Man—d-mannose, GlcNAc—N-acetyl-2-d-glucosamine) to asparagine residues in the consensus sequence Asn-X-Ser/Thr (X = any amino acid except proline). Sequential action by two glucosidases, α-glucosidases I and II, removes the two terminal d-glucose residues (Fig. 2). The monoglucosylated species is recognized by the lectin chaperones calnexin, calreticulin, and the testis specific calnexin paralog calmegin/calnexin-t. These lectin chaperone foldases consist of an N-terminal globular lectin domain and a C-terminal extended hairpin loop, the P domain. Calnexin and calmegin are transmembrane proteins, calreticulin a soluble protein. Calnexin and calreticulin are chaperones for ~100 newly synthesized glycoproteins and have an overlapping, but not identical substrate spectrum (Williams 2006). The lectin domain recognizes monoglucosylated N-linked oligosaccharides. The tip of the P domain mediates association with the thiol oxidoreductase ERp57 (Williams 2006). The chaperone function is provided by both domains (Xu et al. 2004). Hydrolysis of the remaining third d-glucose residue from the oligosaccharide by α-glucosidase II terminates the substrate-chaperone interaction. If unfolded, the substrate is reglucosylated by UDP-glucose:glycoprotein glucosyl transferase (UGGT) triggering another round of interaction with calnexin or calreticulin (Fig. 2) (Dejgaard et al. 2004). This reglucosylation reaction consumes UDP-glucose and ultimately ATP. Proteins are extracted from the calnexin cycle by slow demannosylation reactions catalyzed by α-(1,2)-mannosidases I and II (Fig. 2). These demannosylation reactions generate oligosaccharide structures that serve as targeting signals for export from the ER or toward ERAD. Removal of mannoses B and C facilitates export to the Golgi complex via lectin receptors such as ERGIC-53 (Fig. 2). Removal of mannose A inhibits interaction with ER export receptors and targets the protein toward ERAD.
Fig. 2.
Recognition of N-linked oligosaccharides as a marker for protein folding status (Lederkremer and Glickman 2005, Moremen and Molinari 2006). Oligosaccharyltransferase transfers the oligosaccharide Glc3Man9GlcNAc2 (left, ▲—d-glucose, ○—d-mannose, □—2-N-acetyl-d-glucosamine) onto a newly synthesized polypeptide chain. α-glucosidases I and II (α-Glc I, α-Glc II) remove the two terminal d-glucose moieties. The monoglucosylated form is recognized by calnexin, calreticulin, or calmegin and retained in the ER. Upon removal of the terminal d-glucose moiety by α-glucosidase II, the protein is released from the lectin chaperone, but if still unfolded is reglucosylated by UDP-glucose:glycoprotein glucosyl transferase (UGGT). Removal of d-mannoses B and C converts the oligosaccharide into a poorer substrate for UGGT and α-glucosidase II and decreases the affinity of the oligosaccharide for calnexin and calreticulin and indirectly promotes export to the Golgi complex via recognition of oligosaccharides containing α(1,2)-linked d-mannose residues by export lectin receptors such as ERGIC-53. Trimming of d-mannose A removes the d-glucose acceptor from the oligosaccharide, efficiently blocks the reglucosylation reaction, inhibits the interaction with ERGIC-53, and triggers retrotranslocation and proteasomal degradation of slowly folding proteins. The demannosylation reactions are slower than the deglucosylation reactions. α(1,2)-ER mannosidase I preferentially removes d-mannose B, and more slowly the other d-mannose moieties. Golgi resident mannosidases exhibit specificity for d-mannoses A, C, and D and may be involved in trimming of the α(1,2)-linked d-mannose residues. Reprinted from Fig. 3 published in Schröder (2008a), copyright 2007, Birkhäuser Basel, with kind permission of Springer Science and Business
Oxidative protein folding in the ER
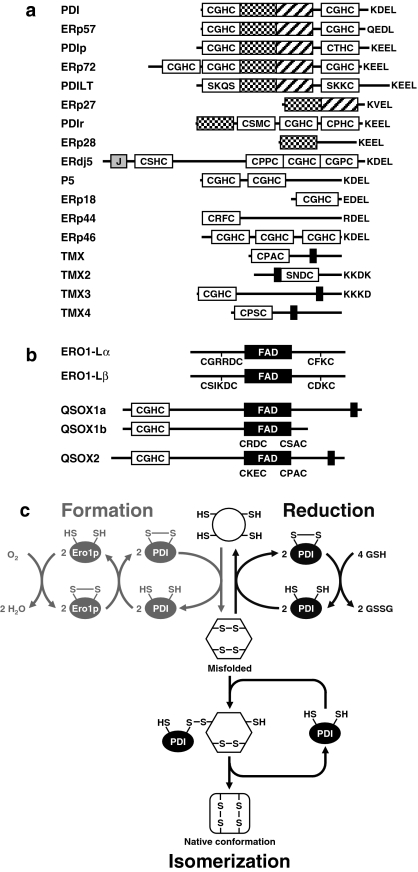
PDIs form a family of at least 17 proteins characterized by one or more CXXC amino acid motifs (Fig. 3a) (Ellgaard and Ruddock 2005). These proteins have thiol oxidase, disulfide isomerase, and chaperone activities. In de novo disulfide bond formation the oxidized form of a PDI is regenerated by disulfide bond shuffling between PDIs and FAD-dependent PDI-like proteins, yeast Ero1p, its mammalian orthologs ERO1-Lα and ERO1-Lβ, and Erv2p (Fig. 3b, c; Tu and Weissman 2002; Tu and Weissman 2004). Pdi1p is a poor substrate for Erv2p in vitro (Thorpe and Coppock 2007), suggesting that additional factors are involved in Pdi1p oxidation by Erv2p in vivo or that the physiological substrates of Erv2p are other redox proteins in the yeast ER. The final electron acceptors for Ero1p are molecular oxygen and to a lesser degree peroxide and superoxide (Tu and Weissman 2002). Two large mammalian transmembrane-anchored proteins, quiescin Q6/QSOX1/QSCN6 and QSOX2/QSCN6L1/SOXN, contain an Erv-like domain in addition to a thioredoxin domain (Fig. 3b; Thorpe and Coppock 2007). These sulfhydryl oxidases can directly transfer electrons from sulfhydryl groups to molecular oxygen via their thioredoxin and Erv-like domains. PDI is a poor substrate for QSOX in vitro. In vivo the bulk of PDI may be involved in disulfide bond isomerization requiring only a small amount of QSOX or ERO1-Lα/ERO-1Lβ for recycling PDI involved in disulfide bond formation (Thorpe and Coppock 2007). The ratio of oxidized to reduced PDI is 1:5 in vivo (Thorpe and Coppock 2007), supporting preferential engagement of PDI in disulfide bond isomerization. Alternatively, QSOX may directly introduce disulfide bonds into substrate proteins which are then reshuffled by PDI (Thorpe and Coppock 2007). Introduction of disulfide bonds into newly synthesized proteins has to intersect with conformational folding of the polypeptide chain. In vitro folding experiments favor a quasi-stochastic coupling mechanism in which formation of a conformationally stable set of disulfide bonds is followed by conformational folding of the polypeptide chain (Welker et al. 2001). This mechanism of coupling of folding of the polypeptide chain to disulfide bond formation requires extensive collaboration between molecular chaperones and PDIs. Therefore, it is not surprising that many PDIs have chaperone activity or form stable complexes with molecular chaperones (Ellgaard and Ruddock 2005). A second important consequence of disulfide bond formation is generation of one molecule H2O2 for every disulfide bond formed. Disulfide bond formation accounts for ~25% of all reactive oxygen species (ROS) in an unstressed cell (Tu and Weissman 2004). During ER stress ROS accumulate in an ERO1 dependent manner (Harding et al. 2003, Haynes et al. 2004). ROS adversely affect cell viability and possibly the quality of the protein product, trigger apoptosis, and function as signaling molecules in the UPR.
Fig. 3.
Disulfide bond formation in the mammalian ER. (a) Human PDIs. Rectangles represent thioredoxin-like domains. Black rectangles represent transmembrane domains, rectangles labeled ‘J’ DnaJ domains, and labeled ‘FAD’ sequences involved in FAD binding. Catalytic site sequences and known ER retention signals are shown in the single letter amino acid code. (b) Human ERO and Erv-like proteins. QSOX1a and QSOX1b are products of alternative splicing. (c) Schematic of disulfide bind formation and isomerization reactions catalyzed by PDIs. Reprinted from (Schröder 2008b)
ERAD and autophagy
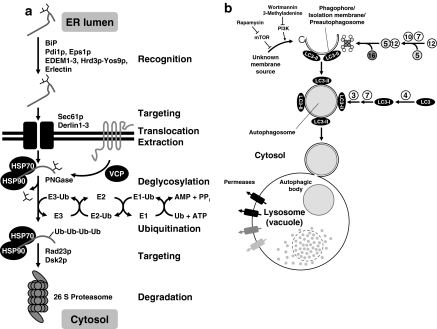
Correctly folded proteins are released from folding cycles and transported to the Golgi complex by packaging into COPII-coated vesicles at ER exit sites (Bonifacino and Glick 2004). For several secretory proteins cargo receptors have been identified. One example is ERGIC-53 which is required for ER to Golgi transport of blood coagulation factors V and VIII (Fig. 2; Nichols et al. 1998). Proteins that remain un- or misfolded after numerous folding attempts are extracted from the folding machinery to prevent its poisoning. These proteins are directed toward ERAD (Fig. 4a; Meusser et al. 2005). ERAD begins with recognition of a protein as unfolded. Chaperones and foldases, including BiP, calnexin/calreticulin, and PDI, and specialized lectins that recognize demannosylated N-linked oligosaccharides such as EDEM1-3 and the Yos9p·Hrd3p·Hrd1p ER membrane mannose lectin-ubiquitin ligase complex make this decision (Fig. 2) (Meusser et al. 2005). The roles of ER luminal HSP70 and HSP90 chaperones in selecting proteins for degradation is less well understood, but their cytosolic counterparts are known to interact with co-chaperones that target interacting clients to degradation (Alberti et al. 2003). A similar selection mechanism may operate in the ER. These degradation-promoting co-chaperones should be less abundant or display lower affinity interactions with their chaperone ATPases to allow for productive protein folding. The unfolded protein is transported to a retrotranslocation pore in the ER membrane. For many ERAD substrates this seems to be the SEC61 translocation channel (Fig. 4a), but alternative channels formed by Derlin proteins, or composite channels consisting of subunits of the SEC61 channel and Derlin proteins may also be involved. Valosin-containing protein (VCP)/p97/Cdc48p extracts proteins from the ER and the ER membrane (Meusser et al. 2005). In the cytosol the protein is deglycosylated and polyubiquitinated to direct it to the 26S proteasome. Cytosolic HSP70s and HSP90s interact with these doomed proteins to keep them in a soluble form to prevent their aggregation and precipitation. Ubiquitination is a three step process (Fig. 4a). An ubiquitin-activating enzyme (E1) adenylates the ubiquitin C-terminus, covalently coupling ubiuquitin via a thioester to a cysteine residue in the activating enzyme. Ubiquitin-conjugating enzymes (E2 s) and ubiquitin ligases (E3 s) transfer activated Ub to the target protein via a thioester relay. Accessory proteins, such as HERP, recruit proteasomes to the ER membrane (Schröder 2008a, Strudwick and Schröder 2007).
Fig. 4.
ERAD (a) and autophagy (b). Formation of the phagophore is inhibited by TOR signaling and stimulated by a class III, wortmannin and 3-methyladenine sensitive PI3K. Abbreviations: PPi—pyrophosphate, Ub—ubiquitin. Numbers in circles represent ATG proteins. Reprinted in modified form from Fig. 2 published in Schröder (2008a), copyright 2007, Birkhäuser Basel, with kind permission of Springer Science and Business
A second degradative process recently implicated in clearance of aggregated ER luminal proteins is autophagy (Fig. 4b; Perlmutter 2006). In autophagy a membrane structure called the preautophagosome, phagophore or isolation membrane is formed from an unknown membrane source and engulfs bulk cytosol and whole organelles (Klionsky 2005; Tanida et al. 2004). Vesicle maturation involves two ubiquitination-like protein modification systems, which decorate the membranes of preautophagosomes with two ubiquitin-like proteins, Atg8p/LC3-I, and a tetrameric ATG12·ATG5·ATG16 complex. Lipidated Atg8p/LC3-I (called LC3-II) is also found on the membranes of mature autophagosomes. Mature autophagosomes fuse with lysosomes or vacuoles in which their content is degraded by digestive lysosomal or vacuolar enzymes.
The unfolded protein response
The UPR is a signaling network that adjusts the capacity of the ER-luminal protein folding machinery to the folding demand imposed on this machinery by coordinating the expression of many, if not all, genes encoding components of this machinery, but also induces immune, inflammatory, and apoptotic responses to ER stress [see (Bernales et al. 2006; Marciniak and Ron 2006; Ron and Walter 2007; Schröder 2006; Schröder 2008a, b; Schröder and Kaufman 2005a, b; Schröder and Kaufman 2006) for recent reviews]. In the following sections we summarize signal transduction mechanisms employed by the UPR and physiological response to ER stress activated by the UPR.
Signal transduction mechanisms in the UPR
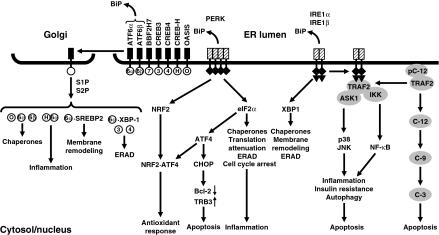
The ER membrane of higher eukaryotes harbors at least three transmembrane proteins activated by ER stress, basic leucine zipper (bZIP) transcription factors synthesized as type II transmembrane proteins such as ATF6α and ATF6β, the protein kinase PERK/PEK/EIF2AK3, and the protein kinase-endoribonucleases IRE1α and IRE1β (Fig. 5). Additional type II transmembrane bZIP transcription factors resident in the ER membrane are CREB3, CREB4, CREB-H, BBF2, and OASIS. BiP is associated with all stress sensors in unstressed cells and released from their ER luminal domains of these stress sensors upon exposure of cells to ER stress, which coincides with their activation.
Fig. 5.
Principal signal transduction pathways in the mammalian UPR. Abbreviations: C-3, caspase 3; C-9, caspase 9; C-12, caspase 12; pC-12, procaspase 12. Reprinted in modified form from Fig. 6 published in Schröder (2008a), copyright 2007, Birkhäuser Basel, with kind permission of Springer Science and Business
ATF6 translocates to the Golgi upon release of BiP. In the Golgi sequential action by two proteases, site-specific proteases 1 and 2 (S1P and S2P), releases the cytosolic domain of ATF6 encompassing its bZIP and transcriptional activation domains (Fig. 5). This ATF6 fragment translocates to the nucleus to induce expression of chaperone genes. ATF6 activates target genes in concert with nuclear factor Y. S1P and S2P also activate the other type II transmembrane bZIP transcription factors. OASIS contributes to activation of the BiP gene, and ATF6 CREB-H heterodimers activate transcription of genes encoding acute phase proteins.
PERK oligomerizes and activates its protein kinase domain when BiP is released from its ER luminal stress sensing domain. Two substrates for the protein kinase activity of PERK are known, the bZIP transcription factor NRF2 and eukaryotic translation initiation factor 2α (eIF2α) (Figs. 5 and 6). In unstressed cells NRF2 is held in the cytosol in an inactive complex with the cytoskeletal anchor protein KEAP1. Phosphorylation of NRF2 by PERK disrupts this complex and triggers relocalization of NRF2 to the nucleus. A NRF2·ATF4 heterodimer activates an antioxidant response (Cullinan and Diehl 2006). Phosphorylation of eIF2α by PERK attenuates general cap-dependent translation by inhibiting loading of the translation initiation factor eIF2 with GTP, and formation of the 43 S preinitiation complex consisting of the 40S ribosomal subunit, GTP-bound eIF2, and methionyl methionine initiator tRNA. One consequence of translational arrest is clearance of short-lived proteins from the cell. Loss of D-type cyclins arrests the cell cycle in G1. Loss of inhibitors of nuclear factor κB (NF-κB) (IκBs) activates NF-κB. Translational arrest in the UPR is transient. Even during translational arrest some mRNAs escape translational inhibition. Translation of mRNAs encoding short upstream open reading frames is stimulated when phosphorylation of eIF2α is increased (Fig. 6; Strudwick and Schröder 2007). An example for such an mRNA is ATF4 mRNA which encodes a bZIP transcription factor. ATF4 activates an antioxidant response (Harding et al. 2003) and induces the inhibitor of mRNA 5′ cap-binding protein 4E-BP1 (Yamaguchi et al. 2008), which contributes to translational arrest during ER stress. The other two escape mechanisms are leaky scanning and internal initiation via internal ribosomal entry sites (Strudwick and Schröder 2007). Only a few mRNAs are known whose translation is stimulated in the UPR. These are the mRNAs encoding ATF4, ATF5 (Zhou et al. 2008), BBF2 (Kondo et al. 2007), and cellular inhibitor of apoptosis 1 (cIAP1)/human inhibitor of apoptosis 2 (hIAP2) (Warnakulasuriyarachchi et al. 2004; Yoshimura et al. 2008). Translational attenuation by PERK is temporary. PERK signaling is attenuated by GADD34, a regulatory subunit of protein phosphatase 1 (PP1). GADD34 is activated late in the ER stress response and directs PP1 toward eIF2α to limit the time window of translational arrest. gadd34−/− cells fail to induce ERO1α and WOLFRAMIN 1, two genes whose protein products have well established functions in protein folding in the ER (Marciniak et al. 2004), and are protected from cell death triggered by ER stress (Zinszner et al. 1998). These observations suggest that recovery from translational arrest restores secretory capacity to cells at the price of increased susceptibility to ER stress and subsequent cell death.
Fig. 6.
Signal transduction by PERK. Abbreviation: m7G—7-methylguanosine. Reprinted from Current Molecular Medicine, copyright 2006, with permission from Bentham Science Publishers
Activation of IRE1 induces non-spliceosomal splicing of mRNAs encoding the bZIP transcription factors Hac1p in Saccharomyces cerevisiae and XBP-1 in metazoans (Fig. 5). IRE1 itself provides the endoribonuclease activity to cleave the exon-intron junctions in these mRNAs. The exons are ligated by an RNA ligase, tRNA ligase in yeast, and an unknown ligase activity in mammalian cells. Both transcription factors activate expression of genes encoding ER-resident molecular chaperones, protein foldases, and genes involved in ERAD, and phospholipid biosynthesis. In mammals, phosphorylated IRE1 assembles a pro-inflammatory and pro-apoptotic signaling complex. Phosphorylated IRE1 sequesters tumor necrosis factor receptor associated factor 2 (TRAF2) from pro-caspase 12 (Yoneda et al. 2001), resulting in clustering and activation of this caspase and activation of a caspase cascade (Fig. 5). TRAF2 recruits apoptosis signal-regulating kinase 1 (ASK1) and IκB kinase (IKK) to IRE1 (Hu et al. 2006; Nishitoh et al. 2002) to activate these protein kinases. ASK1 activates two mitogen-activated protein (MAP) kinase modules leading to activation of the MAP kinase p38 and jun N-terminal kinases (JNK) (Fig. 5). These kinases regulate immune, inflammatory, and apoptotic responses (Roux and Blenis 2004, Schröder 2008a). Association of the pro-apoptotic BCL-2 family proteins BAX and BAK with the cytosolic portion of IRE1 is required for efferent IRE1 signaling, i.e. XBP-1 mRNA splicing and JNK1 activation (Hetz et al. 2006).
Physiological responses regulated by the UPR
Adaptive responses induced by the UPR increase the protein folding capacity of the ER or decrease the protein folding demand imposed on this organelle (Bernales et al. 2006, Ron and Walter 2007; Schröder 2008a). These responses include induction of molecular chaperones and foldases (Dorner et al. 1992) and upregulation of phospholipid synthesis to increase the size of the ER (Sriburi et al. 2004). To decrease the unfolded protein concentration in the ER, the UPR attenuates synthesis of secretory proteins by inhibiting general translation and transcription of genes encoding secretory proteins (Al-Sheikh et al. 2004; Pakula et al. 2003), upregulates ERAD (Friedlander et al. 2000; Yoshida et al. 2003) and autophagy (Ogata et al. 2006; Sakaki et al. 2008), selectively degrades mRNAs encoding secretory proteins (Hollien and Weissman 2006) and proteins that are in the process of translocating into the ER (Fisher et al. 1997; Gusarova et al. 2001). ROS accumulation in ER-stressed cells is countered by an antioxidant response mounted by the transcription factors NRF2 and NF-κB (Cullinan and Diehl 2006; Pham et al. 2004). If these actions fail to restore ER homeostasis the UPR induces apoptosis (Schröder 2008a; Xu et al. 2005). Activation of immune and inflammatory responses by the UPR may contribute to combating infections associated with ER stress, removal of dead cells from tissues and organs, and serve as an organismal alarm signal.
Induction of molecular chaperones and foldases
ATF6α stimulates transcription of genes encoding ER chaperones and foldases by binding to activating transcription factor (ATF)/cAMP response elements (CRE) and the ER stress response elements I and II (ERSE-I and ERSE-II) in the promoters of these genes (Lee et al. 2003; Wu et al. 2007; Yamamoto et al. 2007). ATF6α·ATF6β heterodimers inhibit transcription of the BiP gene (Thuerauf et al. 2007). The other arms of the UPR are also required for full induction of ER chaperone genes. BiP induction is decreased in cells carrying a S51A mutation in eIF2α (Scheuner et al. 2001). XBP-1 is required for induction of several DnaJ co-chaperones including p58IPK, HEDJ, and MDG1 (Lee et al. 2003). Overlap of XBP-1 and ATF6α targets suggests that these two transcription factors fulfill partially redundant functions (Lee et al. 2003). AT6α·XBP-1 heterodimers stimulate transcription of genes involved in ERAD (Wu et al. 2007, Yamamoto et al. 2007). ATF6α activation precedes activation of XBP-1 (Yoshida et al. 2003), resulting in a phase shift from a folding-only to a folding and degradation response. OASIS and XBP-1 contribute to induction of genes encoding ER chaperones and foldases through ERSE-I sites (Lee et al. 2003; Strudwick and Schröder 2007).
Stimulation of phospholipid synthesis
The ER expands in ER-stressed cells (Dorner et al. 1989; Wiest et al. 1990), but in contrast to professional secretory cells the stressed ER is often dilated and does no longer form characteristic cytosolic reticular networks. ATF6 and XBP-1 have been linked to regulation of ER size and lipid synthesis. Cell size, organelle content, total protein synthesis, mitochondrial mass and function, and ribosome number increased in mature B cells overexpressing spliced XBP-1. Knock-down of XBP-1 slightly diminished the ER (Shaffer et al. 2004). XBP-1 induces synthesis of phosphatidylcholine, and increases activity of the CDP-choline pathway leading to an increased surface area and volume of the rough ER in NIH-3T3 cells (Sriburi et al. 2004). A heterodimeric complex of ATF6 and sterol response element (SRE)-binding protein 2 recruits histone deacetylase 1 to the SRE in the low-density lipoprotein receptor (LDLR) promoter (Zeng et al. 2004) to repress cholesterol synthesis. Inhibition of cholesterol synthesis by ATF6 increases the fluidity of the ER membrane to preserve the function of SERCA Ca2+ pumps (Li et al. 2004) and to conserve carbon units for gluconeogenesis (Zeng et al. 2004).
ERAD and autophagy
The UPR induces proteins involved in different steps of ERAD. Spliced XBP-1 induces EDEM, Derlin-1 and Derlin-2, and HERP (Yoshida et al. 2003). ATF6α also induces HERP and Derlin-3 (Wu et al. 2007), and ATF4 contributes to induction of HERP. Thus, all three branches of the UPR contribute to stimulation of ERAD to enhance the clearance of misfolded proteins and decreasing the protein load in the ER during ER stress. The overlap of ERAD targets for ATF6α and XBP-1 can be explained by action of an ATF6α·XBP-1 heterodimer on the promoters of these genes (Yamamoto et al. 2007). In yeast, the UPR also induces the ubiquitin-conjugating enzyme Ubc7p and its membrane anchor Cue1p and the ubiquitin ligase Hrd1p involved in ERAD (Friedlander et al. 2000).
The UPR also stimulates autophagy (Ogata et al. 2006). In ire1α−/− cells or cells treated with JNK inhibitors formation of autophagosomes in response to ER stress was inhibited showing that the IRE1-JNK pathway is required for ER stress-induced autophagy (Ding et al. 2007; Ogata et al. 2006). Polyglutamine-induced conversion of LC3-I to LC3-II is inhibited in cells homozygous for the S51A-eIF2α allele or expressing dominant-negative PERK (Kouroku et al. 2007). Activation of protein kinase C θ, independent of any of the known ER stress sensors, is also required for stimulation of autophagy in ER-stressed cells (Sakaki et al. 2008). Genetic abrogation of autophagy sensitized cells to ER stress, suggesting that autophagy plays an important role in survival of ER stress (Ogata et al. 2006), possibly through its involvement in clearance of polyubiquitinated proteins (Ding et al. 2007).
Two additional degradative mechanisms targeting translocating polypeptide chains and their mRNAs contribute to decreasing the unfolded protein load of the stressed ER. In Drosophila Schneider S2 cells, IRE1 selectively degrades mRNAs associated with ER membrane-bound polyribosomes (Hollien and Weissman 2006). The IRE1 consensus cleavage site in these mRNAs is markedly different from the IRE1 cleavage site in HAC1 or XBP-1 mRNA, but bears some similarity to the cleavage site of IRE1β in 28 S rRNA (Iwawaki et al. 2001). These observations, together with different substrate specificities of the two human IRE1 paralogs (Imagawa et al. 2008) suggest that substrate specificity of IRE1 can be altered by covalent post-translational modification or by recruitment of another RNase to IRE1.
The second mechanism, termed pre-emptive quality control (pQC) (Kang et al. 2006), extracts polypeptide chains whose translocation is stalled from the translocation channel and targets them to the proteasome. Cytosolic HSP70s and HSP90s are required for co-translocational degradation of apolipoprotein B100 (Gusarova et al. 2001), suggesting that a ratcheting mechanism similar to the mechanism employed to translocate polypeptide chains into the ER extracts pQC substrates from the translocation channel. The ribosome and the translocon form a tight junction that blocks access of probes applied at its cytosolic side to the translocating polypeptide chain. Less efficient signal peptides present on pQC substrates weaken this junction (Kang et al. 2006). Thus, a stochastic sampling mechanism may operate in which gated access of cytosolic chaperones to a polypeptide chain engaged with the translocon generates a signal for extraction and proteasomal destruction. Slowly translocating polypeptide chains or polypeptide chains stalled at the translocon interact longer with the translocon, which increases their chances to become a pQC substrate.
Attenuation of protein synthesis
At least four mechanisms attenuate secretory protein synthesis and decrease the folding demand imposed on the ER in the UPR. The most prominent and best characterized of these mechanisms is phosphorylation of eIF2α by activated PERK which inhibits cap-dependent translation. Induction of 4E-BP1 by ATF4 and subsequent inhibition of the 5′-cap binding protein eIF-4E by 4E-BP1 contributes to translational inhibition (Yamaguchi et al. 2008). Transcription of genes encoding secretory proteins is inhibited in lower eukaryotes such as yeasts (Kimata et al. 2006), filamentous fungi (Al-Sheikh et al. 2004; Pakula et al. 2003), and plants (Martínez and Chrispeels 2003). The forth mechanism is attenuation of metabolic activity, which also decreases ROS in ER stressed cells. JNK activation via the IRE1-TRAF2-ASK1 arm leads to serine phosphorylation of insulin receptor substrates (IRS) 1 and 2 (Özcan et al. 2004). IRS serine phosphorylation inhibits IRS phosphorylation at tyrosine residues by the insulin receptor [reviewed in (Draznin 2006; Saltiel and Kahn 2001; Shulman 1999)], promotes IRS-1 degradation (Shah et al. 2004), and inhibits insulin signaling, a condition referred to as insulin resistance. IRS tyrosine phosphorylation triggers binding of SH-2 domain containing proteins, including phosphatidylinositol (PI) 3-kinase (PI3K), to IRS proteins and SH-2 domain-containing (SHC) proteins and their relocalization to the plasma membrane. There PI3K phosphorylates PI, PI-4-phosphate and PI-4,5-bisphosphate, resulting in accumulation of PI-3,4-bisphosphate and PI-3,4,5-trisphosphate in the plasma membrane, and tethering of phosphoinositide-dependent kinases (PDKs) 1 and 2 and several isoforms of protein kinase B (PKB/AKT) to the plasma membrane (White 2002). Induction of TRB3 by CHOP (Ohoka et al. 2005) and inhibition of AKT by TRB3 (Du et al. 2003) contributes to inhibition of insulin signaling. Activated PKBs control many cellular events that stimulate growth, including d-glucose transport, protein, and glycogen synthesis. PKB and PDKs activate the protein kinase mTOR which stimulates translation by phosphorylating the protein kinase p70S6K and the translation initiation inhibitor 4E-BP1 to stimulate translation. SHC proteins activate mitogenic signaling via RAS, RAF, and the MAP kinases ERK1 and ERK2 to stimulate cell growth and division. Mitogenic signaling is largely intact in cells suffering from insulin resistance. Constitutive active mTOR signaling in MEFs defective in the mTOR inhibitory TSC1 TSC2 complex activates the UPR (Ozcan et al. 2008), possibly because of unregulated translation in these cells.
Antioxidant response
ROS accumulate in ER-stressed perk−/− cells (Harding et al. 2003) and in ER-stressed wild type (WT) yeast cells (Haynes et al. 2004). ire1Δ and hac1Δ strains are protected from ROS accumulation (Haynes et al. 2004). Knock-down of ero-1 in Caenorhabditis elegans by RNA interference (RNAi) decreased ROS accumulation (Harding et al. 2003), suggesting that elevation of ERO1 levels by the UPR offsets the cellular redox balance. In yeast, mitochondria are a second source of ROS in ER stressed cells (Haynes et al. 2004). ROS activate redox-sensitive transcription factors such as NF-κB, heat shock transcription factor 1, and NRF2 (Veal et al. 2007). Oxidation of catalytic cysteines in JNK phosphatases activates JNKs (Veal et al. 2007) and may contribute to the development of insulin resistance in ER stressed cells. NF-κB protects cells from tumor necrosis factor (TNF)-stimulated ROS accumulation through induction of ferritin heavy chain (Pham et al. 2004). MEFs deleted for the p65 subunit of NF-κB accumulate ROS during ER stress (Mauro et al. 2006), suggesting that NF-κB has an antioxidant function in the UPR. NRF2 resides within the cytosol of unstressed cells in association with the cytoskeletal anchor protein KEAP1 (Cullinan and Diehl 2006). Phosphorylation of NRF2 by PERK disrupts the NRF2·KEAP1 complex. Oxidation of cysteine residues in KEAP1 by ROS may also contribute to NRF2 activation in ER stressed cells. Upon release from KEAP1 NRF2 translocates to the nucleus to activate transcription of antioxidant genes, genes involved in protein folding and degradation and immune responses, and genes promoting cell growth and survival (Cullinan and Diehl 2006).
Apoptotic response to ER stress
All arms of the UPR stimulate apoptosis via activation of CHOP. chop−/− cells are resistant to ER stress-induced apoptosis (Zinszner et al. 1998). ATF6 and XBP-1 contribute to induction of CHOP. CHOP represses expression of antiapoptotic BCL-2 (McCullough et al. 2001), induces expression of proapoptotic TRB3 (Ohoka et al. 2005), and of GADD34 and ERO1-Lα (Marciniak et al. 2004). However, ATF4 is the most potent activator of CHOP in the UPR (Scheuner et al. 2001). Activation of JNK by IRE1 in the UPR requires the MAP kinase kinase kinase (MAPKKK) ASK1 and promotes apoptosis (Nishitoh et al. 2002, Yokouchi et al. 2008). JNK exerts its proapoptotic effect through phosphorylating and activating proapoptotic BH3-only proteins such as BMF and BIM (Fig. 7a) (Schröder 2008a). Consistent with a proapoptotic role for JNK in the ER stress response is that selective activation of XBP-1 splicing, but not of JNKs, increased survival and growth of cells exposed to drugs that induce ER stress (Lin et al. 2007). traf2−/− and p65−/− cells were more sensitive to ER stress than WT cells (Mauro et al. 2006). Activation of NF-κB by IRE1 through IKK (Hu et al. 2006) inhibits JNK through suppression of ROS (Pham et al. 2004) and induction of GADD45β which interferes with activation of the JNK kinase MKK7 (Papa et al. 2004). NF-κB induces cIAPs to suppress apoptosis (Wajant and Scheurich 2001). It is more difficult to understand why traf2−/− cells, in contrast to ask1−/− cells for example, are more sensitive to ER stress (Mauro et al. 2006). Both, TRAF2 and ASK1, are required for activation of p38, but TRAF2, through interaction with IRE1α mediates activation of IKK and NF-κB, which suppresses JNK-induced apoptosis. TNF-α, a type II transmembrane protein of the plasma membrane, is proteolytically released to the extracellular space and activates TNF-R signaling (Hu et al. 2006). Association of TRAF2 with TNF-R is required for the cytoprotective arm of TNF-R signaling through activation of IKK, NF-κB, and IAPs (Wajant and Scheurich 2001). Intact caspase activation by TNF-R in traf2−/− cells may be responsible for increased cell death of these cells in response to ER stress (Fig. 8).
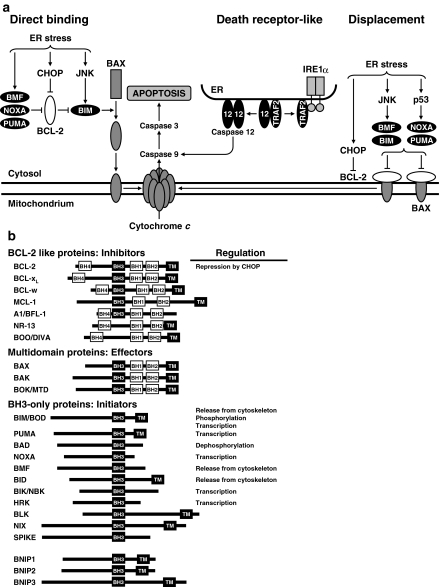
Fig. 7.
Induction of apoptosis in the UPR. (a) Principal pathways according to the direct binding and the displacement models. In the direct binding model (left) proapoptotic BCL-2 proteins such as BIM associate with and activate BAK/BAX, leading to BAK/BAX oligomerization and insertion into the mitochondrial membrane, cytochrome c efflux, and activation of caspase 9. In the displacement model antiapoptotic BCL-2 proteins associate with and inactivate BAK/BAX. Proapoptotic BCL-2 family proteins, whose cytosolic concentration increases in response to intracellular damage, bind to and sequester antiapoptotic BCL-2 proteins from BAK/BAX, leading to BAK/BAX activation (Häcker and Weber 2007; Karst and Li 2007). Reprinted from Fig. 8 published in Schröder (2008a), copyright 2007, Birkhäuser Basel, with kind permission of Springer Science and Business. (b) Anti- and proapoptotic BCL-2 proteins and their regulation in the ER stress response. BH1, BH2, BH3, BH4, and transmembrane (TM) domains are represented by rectangles
Fig. 8.
Autocrine TNF signaling. Membrane-bound TNF (mTNF) is released from the plasma membrane by the protease TACE. Soluble (sTNF) or membrane-anchored TNF interacts with receptors of the TNF receptor (TNF-R) family. Interaction with death domain (DD)-containing receptors such as FAS, TRAIL-R1, and TRAIL-R2 recruits FADD and caspase-8 to the receptor and activates caspase-8. Other DD-containing receptors such as TNF-R1, DR3, and DR6 activate caspase-8 in a complex with TRADD. Conformational changes in the cytosolic domains of TNF-R1 induced by TNF binding release the inhibitory protein SODD from the cytosolic effector domains of the receptor. Binding of TRAF2 to the receptor TRADD complex either recruits cIAP to inhibit caspase-8 activation, or recruits IKK kinase and MAPKKKs such as ASK1 to activate NF-κB and the MAPK p38 and JNK. TRAF2-associated proteins such as RIP contribute to modulation of the outcome of TNF-R1 signaling. TNF-Rs with a cytosolic TRAF-interacting motif (TIM) domain directly associate with TRAF2 to activate IKK, p38, and JNK. TNF-R2 is preferentially activated by mTNF. Decoy receptors lack cytosolic effector domains and inhibit TNF signaling (Dempsey et al. 2003; Wajant and Scheurich 2001). Abbreviation: DED—Death effector domain
IRE1 directly activates procaspase-12 by sequestering TRAF2 from procaspase-12 resulting in clustering and activation of caspase-12 at the ER membrane (Nakagawa et al. 2000; Yoneda et al. 2001). Caspase-12 initiates a caspase cascade leading to activation of the executioner caspase, caspase-3. caspase-12−/− MEFs are protected from ER stress induced cell death (Nakagawa et al. 2000). Human cells do not carry a functional CASPASE-12 gene (Xu et al. 2005). In these cells caspase-4 may substitute for caspase-12. A role for caspase-4 in ER stress induced apoptosis is supported by localization of caspase-4 to the ER, its cleavage upon ER stress, and increased survival of ER stress by caspase-4−/− cells. Transcriptional induction of the proapoptotic BH3-only proteins BIM, PUMA, and NOXA, release of BIM and BMF from inactive cytoskeletal pools, and repression of transcription of antiapoptotic BCL-2 by CHOP shifts the balance of pro- and antiapoptotic BCL-2 proteins toward antiapoptotic BCL-2 proteins and triggers apoptosis (Fig. 7b; Schröder 2008a).
Activation of immune and inflammatory responses
Immune and inflammatory signaling is activated by all three arms of the UPR. Proteolytic release of TNF-α from the plasma membrane to the extracellular space activates TNF-R signaling (Hu et al. 2006), leading to activation of NF-κB, and of the MAP kinases p38 and JNK (Fig. 8). NF-κB activates transcription of inflammatory genes such as TNF-α, TNF-β, interleukin (IL)-2, IL-6, and IL-8 (Baeuerle and Henkel 1994, Hu et al. 2006) and acute phase genes including serum amyloid A precursor, and complement factors B and C4 (Baeuerle and Henkel 1994). Activated JNK phosphorylates and potentiates the transcription factor AP-1, which induces expression of inflammatory cytokines including TNF-α, IL-6, IL-8, and MCP-1 (Gargalovic et al. 2006). A CREB-H ATF6 heterodimer induces inflammatory response genes such as serum amyloid P-component and C-reactive protein (Zhang et al. 2006).
Cell engineering of the early secretory pathway
Reports indicating that inefficient folding of heterologous proteins in the ER results in their intracellular aggregation (Hasemann and Capra 1990; Hsu and Betenbaugh 1997; Hsu et al. 1996; Hsu et al. 1994; Schröder et al. 2002) and association with BiP (Dorner et al. 1987; Hurtley et al. 1989; Jarvis and Summers 1989; Kaufman et al. 1988; Machamer et al. 1990; Suzuki et al. 1991), and, as a consequence, dilation of the ER (Dorner et al. 1989; Gennaro et al. 1991) has triggered interest in engineering of ER resident chaperone systems to boost the folding capacity of the ER of mammalian host cells through overexpression of chaperones and PDIs. These results have by and large yielded small if any increases in cell specific productivities. One possible explanation for these results is that overexpression of one chaperone may not be sufficient increase the folding capacity of the whole folding machinery in the ER and that coordinated elevation of expression of several chaperones and foldases may be necessary to obtain higher and more reproducible increases in cell specific productivities. Engineering of UPR signaling, for example through overexpression of one or a few of the transcription factors activated by the UPR provides an attractive avenue to achieve this goal and has attract considerable attention in the recent past. Results from the engineering of chaperone systems in insect and mammalian cells and of the studies reporting engineering of UPR signaling are reviewed in the following two chapters.
Engineering of the ER-resident protein folding machinery
Modeling of the BiP ATPase cycle predicts a modest increase in secreted IgG levels in BiP overexpressing cells (Whiteley et al. 1997). Such an increase has been observed in some studies (Hsu and Betenbaugh 1997; Whiteley et al. 1997). Other studies reported no effect or even a decrease in heterologous protein secretion (Dorner and Kaufman 1994; Dorner et al. 1988; Dorner et al. 1992; Hsu et al. 1994; Lambert and Merten 1997). Modeling also predicted 2-4 fold increases in secreted IgG levels when the ATPase cycle is slowed down, i.e. in the absence of ATP or the presence of non-hydrolyzable ATP analogs (Whiteley et al. 1997). A plausible explanation for this counterintuitive result is that if ATP-binding and hydrolysis are slowed down, a larger proportion of BiP molecules will exist in the ADP-bound state that has higher affinity for unfolded substrates, thus preventing their non-productive aggregation. Slowing down of the BiP ATPase cycle is a likely consequence of BiP overexpression, as its co-chaperones or ATP itself may become limiting. The affinity of HSP70 chaperones for ADP is ~6 times higher than for ATP. Thus, BiP exchanges ADP for ATP very slowly without catalysis. So why then is this predicted increase in recombinant protein secretion not consistently seen in BiP overexpressing cells? HSP70 chaperones, including BiP, do not work in isolation, but in networks with other HSP70 chaperones (Morano 2007). BiP and Lhs1p coordinate their folding efforts (Steel et al. 2004). Stalled ATPase cycles may be a degradation signal. Elevated BiP levels may increase the prevalence of interactions with degradation-promoting co-chaperones or protein complexes. Increased folding capacity of the BiP ATPase cycle may stall the GRP94 or calnexin-calreticulin chaperone machineries. BiP overexpression attenuates the UPR (Dorner et al. 1992; Kohno et al. 1993), suggesting that BiP levels in WT and BiP overexpressing cells expressing a heterologous protein may not be different. Selective overexpression of BiP in cells experiencing ER stress may blunt induction of other chaperones, thereby causing an imbalance in ER luminal chaperone activities, a situation which may favor degradation. BiP overexpression induces ER-derived BiP-containing vesicles in the cytosol (van der Heide et al. 2002) or nucleus (Morris et al. 1997). These vesicles may be similar to ‘BiP bodies’ observed in yeast in which ER-to-Golgi transport is blocked (Nishikawa et al. 1994) and function in normal cells as cargo selection sites for ER-to-Golgi traffic (Nishikawa et al. 1994) or sites that target proteins for degradation (van der Heide et al. 2002).
Increasing expression levels of calnexin/calreticulin improved secretion of heterologous proteins in insect cells and Chinese hamster ovary (CHO) cells (Chung et al. 2004; Kato et al. 2005). These results are difficult to reconcile with overexpressed calnexin/calreticulin functioning in the calnexin cycle, because expression of all cycle constituents should be maintained at a ratio optimal for maximum cycle capacity. Therefore, increased levels of one cycle constituent should not improve cycle performance. Attractive alternative explanations are that the extra amount of calnexin or calreticulin functions as a chaperone holdase which buffers against an increased unfolded protein load, or that these extra amounts form inactive complexes with lectins such as EDEM1-3 which normally target unfolded proteins to ERAD.
Engineering of PDI expression levels gave a similar picture as described for BiP. Secretion of some heterologous proteins was increased (Kato et al. 2005; Lambert and Merten 1997), but secretion of an equal number of heterologous proteins was not affected (Kitchin and Flickinger 1995; Mohan et al. 2007). PDI may enhance secretion via increased rates of disulfide bond formation and isomerization, or because of its chaperone function. Overexpression of catalytically inactive PDI improved secretion of some heterologous proteins (Hayano et al. 1995), and likewise, overexpression of PDI also improved secretion of heterologous proteins containing no disulfide bonds (Smith et al. 2004), suggesting that PDI overexpression exploits its chaperone function and not its oxidoreductase or isomerase functions. This interpretation is more consistent with the concept that enzymes function at substoichiometric levels compared to their substrates and that efficient chaperone function requires at least equimolar amounts of chaperone and substrate.
Engineering of the UPR
Induction of BiP in several expression systems (Alete et al. 2005; Dorner et al. 1989; Downham et al. 1996; Jones et al. 2005; Miura et al. 2001; Smales et al. 2004; Watowich et al. 1991), of NF-κB in human IL-6 expressing CHO cells (Teruya et al. 2005), and of genes involved in the ER stress response including BiP, GRP94, calnexin, and HERP in human bone morphogenic protein 2 expressing CHO DUKX cells (Doolan et al. 2008) demonstrates that the UPR is activated in many heterologous expression systems. Cooperativity of molecular chaperones and foldases may necessitate increasing expression of several chaperones simultaneously to reproducibly increase productivities. This goal may be reached through constitutive activation of the UPR. Therefore, strong rationales exist to engineer the UPR to boost cell specific productivities. UPR and chaperone engineering may be of greater importance for slowly folding or unstable proteins as these proteins tend to induce the UPR more efficiently (Gething et al. 1986; Watowich et al. 1991). In mammalian cells, engineering of UPR signaling is complicated by existence of several signal transduction pathways (Fig. 5), extensive crosstalk between these pathways, and simultaneous activation of responses beneficial (induction of ER-resident chaperones and expansion of the ER) and detrimental (ERAD, attenuation of protein synthesis, apoptosis, and inflammation) to recombinant protein production. Activation of inflammatory signaling (Fig. 8) as a consequence of UPR activation in ER-stressed production cells or cells engineered for constitutive UPR activity is a concern with regard to product quality, and requires further investigation.
Boosting the beneficial aspects of the UPR, i.e. through overexpression of transcription factors such as ATF6 or XBP-1 has received some attention in the recent past. Overexpression of XBP-1 in mammalian cells increased secretion of interferon γ, erythropoietin, a human monoclonal antibody, secreted alkaline phosphatase, and human vascular endothelial growth factor 121 (Ku et al. 2008; Tigges and Fussenegger 2006). These results may reflect dissection of UPR signaling beneficial for protein secretion from detrimental activities, such as translational attenuation by PERK and induction of apoptosis via PERK and JNK activation. However, these results are not generally applicable. Expression of an humanized anti-CD154 antibody in CHO DG44 cells was only modestly enhanced by individual or combined overexpression of the cytosolic ATF6 fragment, XBP-1, or a fragment of eIF2α carrying the S51A point mutation (Ailor and Reff 2005). Expression of human antithrombin III or granulocyte-macrophage colony-stimulating factor in CHO DXB11 cells was not enhanced by spliced XBP-1 (Ohya et al. 2007). XBP-1 was spliced in antithrombin III expressing CHO DXB11 cells even before introduction of spliced XBP-1 cDNA (Ohya et al. 2007). This result illustrates that potential improvements achievable by engineering of the UPR may be masked by UPR activation in production cell lines. Induction of ERO1-Lα and ERO1-Lβ by XBP-1 elevates ROS (Harding et al. 2003; Haynes et al. 2004; Yokouchi et al. 2008). ROS may damage the heterologous protein product. NF-κB, but not BiP, activation in CHO cells expressing human IL-6 suggests that these cells suffer from oxidative stress (Teruya et al. 2005).
Equally attractive are attempts to abrogate responses detrimental for protein production activated by the UPR. Overexpression of S51A-eIF2α in CHO cells blocks translational attenuation mediated by eIF2α phosphorylation and increased expression of firefly luciferase ~3-fold (Underhill et al. 2003). Guanine nucleotide exchange is catalyzed by eIF2B, which is ~10 times less abundant in cells than eIF2α (Kaufman 2004). A potential limitation of this approach is that only small increases in eIF2α phosphorylation trigger complete translational arrest. Overexpression of ATF4 in antithrombin III-expressing CHO DXB11 cells decreased phosphorylation of eIF2α and improved secretion of antithrombin III (Ohya et al. 2007). ATF4 expression induced CHOP ~2 fold, but did not decrease cell viability. GADD34 expression, an ATF4 target that directs PP1 toward eIF2α, was not significantly increased (Ohya et al. 2007). Manipulation of CHOP expression in NS0 cells did not alter survival of ER stress (Cudna and Dickson 2006), again showing that CHOP is not sufficient to trigger apoptosis in production cells.
Conclusions
In several mammalian expression systems, folding and maturation of the recombinant protein in the ER is the rate- and yield-limiting step for its production and activation of the UPR has been observed. Thus, clear rationales exist to engineer the UPR. Currently, only limited information is available on the effect of UPR engineering on cell specific productivities. However, this data already provides a mixed picture. Some studies have reported improvements, other studies reported no improvements or even decreases in cell specific productivities, a situation which is reminiscent to engineering of chaperone systems. Improvements obtained through engineering of the UPR and of chaperone systems may be masked by endogenous activation of the UPR in expression cell lines (Ohya et al. 2007). Another important aspect of UPR engineering will be to selectively activate those responses beneficial to protein production, while responses detrimental to protein production are not elevated or even abolished. Future engineering of the UPR will hopefully include these considerations, and lead to a more consistent outcome of UPR and chaperone system engineering, an achievement critical for routine chaperone and UPR engineering in commercial settings.
Acknowledgements
We apologize to all whose work could not be cited because of space limitations. This work was supported by funding from the Biotechnology and Biological Sciences Research Council (C513418/1, D01588X/1, E006035/1), the European Commission (HEALTH-F7-2007-201608), and the Wellcome Trust (079821) to M.S.
Abbreviations
- 4E-BP1
4E-Binding protein 1
- ADP
Adenosine diphosphate
- AKT
AKR/J Mice transforming retroviral oncogene
- AP-1
Activation protein 1
- ASK1
Apoptosis signal-regulating kinase 1
- Asn
L-Asparagine
- ATF4
Activating transcription factor 4
- ATF5
Activating transcription factor 5
- ATF6α
Activating transcription factor 6α
- ATF6β
Activating transcription factor 6β
- ATG5
Autophagy-related 5
- Atg8p
Autophagy-related 8 protein
- ATG12
Autophagy-related 12
- ATG16
Autophagy-related 16
- ATP
Adenosine triphosphate
- BAK
BCL-2 Homologous antagonist/killer
- BAP
BiP-Associated protein
- BAX
BCL-2-Associated X protein
- BBF2
Box B-binding factor 2
- BCL-2
B Cell leukemia/lymphoma 2
- BH1
BCL-2 Homology domain 1
- BH2
BCL-2 Homology domain 2
- BH3
BCL-2 Homology domain 3
- BH4
BCL-2 Homology domain 4
- BIM
BCL-2 Interacting mediator of cell death
- BiP
Heavy chain binding protein
- BMF
BCL-2 Modifying factor
- bZIP
Basic leucine zipper
- cAMP
Cyclic adenosine monophosphate
- CD154
Cluster of differentiation 154
- Cdc48p
Cell division cycle 48 protein
- CDP
Cytidine diphosphate
- CHO
Chinese hamster ovary
- CHOP
CCAAT/Enhancer-binding protein (C/EBP) homologous protein
- cIAP1
Cellular inhibitor of apoptosis 1
- CNC
Cap and collar
- CoA
Coenzyme A
- COPII
Coat Protein II
- CRE
cAMP Response element
- CREB3
CRE-Binding protein 3
- CREB4
CRE-Binding protein 4
- CREB-H
CREB Homolog
- Cue1p
Coupling of ubiquitin conjugation to ER degradation 1 protein
- DD
Death domain
- DED
Death effector domain
- DnaJ
2′-Deoxyribonucleic acid chain elongation J
- DR3
Death receptor 3
- DR6
Death receptor 6
- DTT
1,4-DL-Dithiothreitol
- E1
Ubiquitin-activating enzyme
- E2
Factor eluted by DTT (ubiquitin-conjugating enzyme)
- E3
Eluted with high salt or high pH (ubiquitin ligase)
- ECH
Erythroid cell-derived protein with CNC homology
- EDEM1
ER Degradation enhancer, mannosidase α-like 1
- EDEM2
ER Degradation enhancer, mannosidase α-like 2
- EDEM3
ER Degradation enhancer, mannosidase α-like 3
- eIF2α
Eukaryotic translation initiation factor 2α
- eIF2B
Eukaryotic translation initiation factor 2B
- EIF2AK3
eIF2α Kinase 3
- eIF-4E
Eukaryotic translation initiation factor 4E
- ER
Endoplasmic reticulum
- ERAD
ER Associated protein degradation
- ERdj1
ER DnaJ Protein 1
- ERdj2
ER DnaJ Protein 2
- ERdj3
ER DnaJ Protein 3
- ERdj4
ER DnaJ Protein 4
- ERdj5
ER DnaJ Protein 5
- ERGIC-53
ER Golgi intermediate compartment protein of 53 kDa
- ERK1
Extracellular signal regulated kinase 1
- ERK2
Extracellular signal regulated kinase 2
- ERO1-Lα
Ero1p-like α
- ERO1-Lβ
Ero1p-like β
- Ero1p
ER Oxidation 1 protein
- ERp57
ER Protein of 57 kDa
- ERSE-I
ER Stress response element I
- ERSE-II
ER Stress response element II
- Erv2p
Essential for respiration and viability 2 protein
- FAD
Flavine adenine dinucleotide
- FADD
FAS-Associated protein with a novel DD
- GADD34
Growth arrest and DNA damage gene 34
- GADD45β
Growth arrest and DNA damage gene 45β
- Glc
d-Glucose
- α-Glc I
α-Glucosidase I
- α-Glc II
α-Glucosidase II
- GlcNAC
2-N-Acetyl-d-glucosamine
- GRP78
Glucose-regulated protein of 78 kDa
- GRP94
Glucose-regulated protein of 94 kDa
- GRP170
Glucose-regulated protein of 170 kDa
- GrpE
Growth after phage induction E
- Hac1p
Homologous to ATF/CREB1 1 protein
- HEDJ
Human ER-associated DnaJ
- HERP
Hyperhomocysteinemia-induced ER stress-responsive protein
- hIAP2
Human inhibitor of apoptosis 2
- HMG
3-Hydroxy-3-methylglutarate
- Hrd1p
HMG-CoA reductase degradation 1 protein
- Hrd3p
HMG-CoA Reductase degradation 3 protein
- HSP40
Heat shock protein of 40 kDa
- HSP70
Heat shock protein of 70 kDa
- HSP90
Heat shock protein of 90 kDa
- IAP
Inhibitor of apoptosis
- IgG
Immunoglobulin G
- IκB
Inhibitor of NF-κB
- IKK
IκB Kinase
- IL-2
Interleukin 2
- IL-6
Interleukin 6
- IL-8
Interleukin 8
- IRE1α
Inositol requiring 1α
- IRE1β
Inositol requiring 1β
- IRS-1
Insulin receptor substrate 1
- IRS-2
Insulin receptor substrate 2
- JNK
JUN N-Terminal kinase
- JUN
Ju-nana
- KEAP1
Kelch-like ECH-associated protein 1
- LC3
Microtubule-associated protein 1 light chain 3
- LDLR
Low density lipoprotein receptor
- Lhs1p
Luminal HSP70 1 protein
- m7G
7-Methylguanosine
- Man
D-Mannose
- MAP
Mitogen-activated protein
- MAPKKK
MAP Kinase kinase kinase
- MCP-1
Monocyte chemoattractant protein 1
- MDG1
Microvascular differentiation gene 1
- MEF
Mouse embryonic fibroblast
- MKK7
Mitogen-activated protein kinase kinase 7
- mRNA
Messenger RNA
- mTNF
Membrane-bound TNF
- MTJ1
Murine tumor cell DnaJ-like protein 1
- mTOR
Mammalian TOR
- NADPH
Reduced nicotine adenine dinucleotide phosphate
- NF-κB
Nuclear factor κB
- NOXA
NADPH Oxidase activator
- NRF2
Nuclear factor erythroid 2-related factor 2
- NS0
Non-secreting myeloma cell
- OASIS
Old astrocyte specifically induced substance
- ORP150
150 kDa Oxygen-regulated protein
- p38
Protein of 38 kDa
- p58IPK
58 kDa Inhibitor of PKR
- p65
65 kDa Protein
- p70S6K
70,000 Mr 40 S Ribosomal protein S6 kinase
- p97
Protein of 97 kDa
- PDI
Protein disulfide isomerase
- PDK
Phosphoinositide-dependent kinase 2
- PEK
Pancreatic eIF2α kinase
- PERK
PKR-Like ER kinase
- Pi
Inorganic phosphate (HPO42−)
- PI
Phosphatidylinositol
- PI3K
PI 3-Kinase
- PKB
Protein kinase B
- PKR
Double-stranded RNA-activated protein kinase
- PP1
Protein phosphatase 1
- PPi
Pyrophosphate (HP2O73−)
- PPI
Cis–trans peptidyl prolyl isomerase
- pQC
Preemptive quality control
- PUMA
p53 Upregulated modulator of apoptosis
- Q6
Quiescin 6
- QSCN6
Quiescin 6
- QSCN6L1
Quiescin 6-like 1
- QSOX1
Quiescin Q6 sulfhydryl oxidase 1
- QSOX2
Quiescin Q6 sulfhydryl oxidase 2
- RAS
Rat sarcoma
- RIP
Receptor interacting protein
- RNA
Ribonucleic acid
- RNAi
RNA Interference
- ROS
Reactive oxygen species
- rRNA
Ribosomal RNA
- S1P
Site 1 protease
- S2P
Site 2 protease
- SEC61
Secretory 61
- SEC63
Secretory 63
- Ser
l-Serine
- SERCA
Sarcoplasmic or endoplasmic reticulum Ca2+ ATPase
- SH-2
Src Homology 2 domain
- SHC
SH-2 Domain containing
- SIL1
Suppressor of IRE1/LHS1 synthetic lethality 1
- SODD
Silencer of death domains
- SOXN
Neuroblastoma-derived sulfhydryl oxidase
- Src
Sarcoma formation
- SRE
Sterol response element
- sTNF
Soluble TNF
- TACE
TNF-α Converting enzyme
- Thr
l-Threonine
- TIM
TRAF-Interacting motif
- TNF-α
Tumor necrosis factor α
- TNF-R
TNF-α receptor
- TOR
Target of rapamycin
- TRADD
TNF-R-Associated via DD
- TRAF2
TNF-R Associated factor 2
- TRAIL-R1
TNF-Related apoptosis-inducing ligand receptor 1
- TRAIL-R2
TNF-Related apoptosis-inducing ligand receptor 2
- TRB3
Tribbles homolog 3
- tRNA
Transfer RNA
- TSC1
Tuberous sclerosis complex 1
- TSC2
Tuberous sclerosis complex 2
- Ub
Ubiquitin
- Ubc7p
Ubiquitin-conjugating enzyme 7 protein
- UDP
Uridine diphosphate
- UGGT
UDP-Glucose:glycoprotein glucosyltransferase
- UPR
Unfolded protein response
- VCP
Valosin-containing protein
- WT
Wild type
- XBP-1
X-Box-binding protein 1
- Yos9p
Yeast osteosarcoma 9 homolog protein
References
- Ailor EN, Reff ME (2005) Method to increase protein production in culture. US Patent 2005/106,222, 9 May 2005
- Al-Sheikh H, Watson AJ, Lacey GA, Punt PJ, MacKenzie DA, Jeenes DJ et al (2004) Endoplasmic reticulum stress leads to the selective transcriptional downregulation of the glucoamylase gene in Aspergillus niger. Mol Microbiol 53:1731–1742. doi :10.1111/j.1365-2958.2004.04236.x [DOI] [PubMed]
- Alberti S, Esser C, Höhfeld J (2003) BAG-1—a nucleotide exchange factor of Hsc70 with multiple cellular functions. Cell Stress Chaperones 8:225–231. doi :10.1379/1466-1268(2003)008<0225:BNEFOH>2.0.CO;2 [DOI] [PMC free article] [PubMed]
- Alete DE, Racher AJ, Birch JR, Stansfield SH, James DC, Smales CM (2005) Proteomic analysis of enriched microsomal fractions from GS-NS0 murine myeloma cells with varying secreted recombinant monoclonal antibody productivities. Proteomics 5:4689–4704. doi:10.1002/pmic.200500019 [DOI] [PubMed]
- Baeuerle PA, Henkel T (1994) Function and activation of NF-κB in the immune system. Annu Rev Immunol 12:141–179. doi:10.1146/annurev.iy.12.040194.001041 [DOI] [PubMed]
- Barnes LM, Bentley CM, Dickson AJ (2004) Molecular definition of predictive indicators of stable protein expression in recombinant NS0 myeloma cells. Biotechnol Bioeng 85:115–121. doi:10.1002/bit.10893 [DOI] [PubMed]
- Bernales S, Papa FR, Walter P (2006) Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol 22:487–508. doi:10.1146/annurev.cellbio.21.122303.120200 [DOI] [PubMed]
- Biswas C, Ostrovsky O, Makarewich CA, Wanderling S, Gidalevitz T, Argon Y (2007) The peptide binding activity of GRP94 is regulated by calcium. Biochem J 405:233–241. doi:10.1042/BJ20061867 [DOI] [PMC free article] [PubMed]
- Bonifacino JS, Glick BS (2004) The mechanisms of vesicle budding and fusion. Cell 116:153–166. doi:10.1016/S0092-8674(03)01079-1 [DOI] [PubMed]
- Chung JY, Lim SW, Hong YJ, Hwang SO, Lee GM (2004) Effect of doxycycline-regulated calnexin and calreticulin expression on specific thrombopoietin productivity of recombinant Chinese hamster ovary cells. Biotechnol Bioeng 85:539–546. doi:10.1002/bit.10919 [DOI] [PubMed]
- Cudna RE, Dickson AJ (2006) Engineering responsiveness to cell culture stresses: Growth arrest and DNA damage gene 153 (GADD153) and the unfolded protein response (UPR) in NS0 myeloma cells. Biotechnol Bioeng 94:514–521. doi:10.1002/bit.20861 [DOI] [PubMed]
- Cullinan SB, Diehl JA (2006) Coordination of ER and oxidative stress signaling: The PERK/Nrf2 signaling pathway. Int J Biochem Cell Biol 38:317–332. doi:10.1016/j.biocel.2005.09.018 [DOI] [PubMed]
- de StGroth SF, Scheidegger D (1980) Production of monoclonal antibodies: strategy and tactics. J Immunol Methods 35:1–21. doi:10.1016/0022-1759(80)90146-5 [DOI] [PubMed]
- Dejgaard S, Nicolay J, Taheri M, Thomas DY, Bergeron JJ (2004) The ER glycoprotein quality control system. Curr Issues Mol Biol 6:29–42 [PubMed]
- Dempsey PW, Doyle SE, He JQ, Cheng G (2003) The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev 14:193–209. doi:10.1016/S1359-6101(03)00021-2 [DOI] [PubMed]
- Ding W-X, Ni H-M, Gao W, Hou Y-F, Melan MA, Chen X et al (2007) Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem 282:4702–4710. doi:10.1074/jbc.M609267200 [DOI] [PubMed]
- Dinnis DM, James DC (2005) Engineering mammalian cell factories for improved recombinant monoclonal antibody production: lessons from nature? Biotechnol Bioeng 91:180–189. doi:10.1002/bit.20499 [DOI] [PubMed]
- Dollins DE, Warren JJ, Immormino RM, Gewirth DT (2007) Structures of GRP94-nucleotide complexes reveal mechanistic differences between the hsp90 chaperones. Mol Cell 28:41–56. doi:10.1016/j.molcel.2007.08.024 [DOI] [PMC free article] [PubMed]
- Doolan P, Melville M, Gammell P, Sinacore M, Meleady P, McCarthy K et al (2008) Transcriptional profiling of gene expression changes in a PACE-transfected CHO DUKX cell line secreting high levels of rhBMP–2. Mol Biotechnol 39:187–199. doi:10.1007/s12033-008-9039-6 [DOI] [PubMed]
- Dorner AJ, Kaufman RJ (1994) The levels of endoplasmic reticulum proteins and ATP affect folding and secretion of selective proteins. Biologicals 22:103–112. doi:10.1006/biol.1994.1016 [DOI] [PubMed]
- Dorner AJ, Bole DG, Kaufman RJ (1987) The relationship of N-linked glycosylation and heavy chain-binding protein association with the secretion of glycoproteins. J Cell Biol 105:2665–2674. doi:10.1083/jcb.105.6.2665 [DOI] [PMC free article] [PubMed]
- Dorner AJ, Krane MG, Kaufman RJ (1988) Reduction of endogenous GRP78 levels improves secretion of a heterologous protein in CHO cells. Mol Cell Biol 8:4063–4070 [DOI] [PMC free article] [PubMed]
- Dorner AJ, Wasley LC, Kaufman RJ (1989) Increased synthesis of secreted proteins induces expression of glucose-regulated proteins in butyrate-treated Chinese hamster ovary cells. J Biol Chem 264:20602–20607 [PubMed]
- Dorner AJ, Wasley LC, Kaufman RJ (1992) Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J 11:1563–1571 [DOI] [PMC free article] [PubMed]
- Downham MR, Farrell WE, Jenkins HA (1996) Endoplasmic reticulum protein expression in recombinant NS0 myelomas grown in batch culture. Biotechnol Bioeng 51:691–696. doi :10.1002/(SICI)1097-0290(19960920)51:6<691::AID-BIT7>3.0.CO;2-C [DOI] [PubMed]
- Draznin B (2006) Molecular mechanisms of insulin resistance: serine phosphorylation of insulin receptor substrate-1 and increased expression of p85alpha: the two sides of a coin. Diabetes 55:2392–2397. doi:10.2337/db06-0391 [DOI] [PubMed]
- Du K, Herzig S, Kulkarni RN, Montminy M (2003) TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science 300:1574–1577. doi:10.1126/science.1079817 [DOI] [PubMed]
- Ellgaard L, Ruddock LW (2005) The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep 6:28–32. doi:10.1038/sj.embor.7400311 [DOI] [PMC free article] [PubMed]
- Fann CH, Guarna MM, Kilburn DG, Piret JM (1999) Relationship between recombinant activated protein C secretion rates and mRNA levels in baby hamster kidney cells. Biotechnol Bioeng 63:464–472. doi : 10.1002/(SICI)1097-0290(19990520)63:4<464::AID-BIT10>3.0.CO;2-H [DOI] [PubMed]
- Fisher EA, Zhou M, Mitchell DM, Wu X, Omura S, Wang H et al (1997) The degradation of apolipoprotein B100 is mediated by the ubiquitin-proteasome pathway and involves heat shock protein 70. J Biol Chem 272:20427–20434. doi:10.1074/jbc.272.33.20427 [DOI] [PubMed]
- Frey S, Leskovar A, Reinstein J, Buchner J (2007) The ATPase cycle of the endoplasmic chaperone Grp94. J Biol Chem 282:35612–35620. doi:10.1074/jbc.M704647200 [DOI] [PubMed]
- Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T (2000) A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol 2:379–384. doi:10.1038/35017001 [DOI] [PubMed]
- Gargalovic PS, Gharavi NM, Clark MJ, Pagnon J, Yang WP, He A et al (2006) The unfolded protein response is an important regulator of inflammatory genes in endothelial cells. Arterioscler Thromb Vasc Biol 26:2490–2496. doi:10.1161/01.ATV.0000242903.41158.a1 [DOI] [PubMed]
- Gennaro DE, Hoffstein ST, Marks G, Ramos L, Oka MS, Reff ME et al (1991) Quantitative immunocytochemical staining for recombinant tissue-type plasminogen activator in transfected Chinese hamster ovary cells. Proc Soc Exp Biol Med 198:591–598 [DOI] [PubMed]
- Gething M-J, McCammon K, Sambrook J (1986) Expression of wild-type and mutant forms of influenza hemagglutinin: the role of folding in intracellular transport. Cell 46:939–950. doi:10.1016/0092-8674(86)90076-0 [DOI] [PubMed]
- Gusarova V, Caplan AJ, Brodsky JL, Fisher EA (2001) Apoprotein B degradation is promoted by the molecular chaperones hsp90 and hsp70. J Biol Chem 276:24891–24900. doi:10.1074/jbc.M100633200 [DOI] [PubMed]
- Häcker G, Weber A (2007) BH3-only proteins trigger cytochrome c release, but how? Arch Biochem Biophys 462:150–155. doi:10.1016/j.abb.2006.12.022 [DOI] [PubMed]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M et al (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11:619–633. doi:10.1016/S1097-2765(03)00105-9 [DOI] [PubMed]
- Hasemann CA, Capra JD (1990) High-level production of a functional immunoglobulin heterodimer in a baculovirus expression system. Proc Natl Acad Sci USA 87:3942–3946. doi:10.1073/pnas.87.10.3942 [DOI] [PMC free article] [PubMed]
- Hayano T, Hirose M, Kikuchi M (1995) Protein disulfide isomerase mutant lacking its isomerase activity accelerates protein folding in the cell. FEBS Lett 377:505–511. doi:10.1016/0014-5793(95)01410-1 [DOI] [PubMed]
- Haynes CM, Titus EA, Cooper AA (2004) Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol Cell 15:767–776. doi:10.1016/j.molcel.2004.08.025 [DOI] [PubMed]
- Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B et al (2006) Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1α. Science 312:572–576. doi:10.1126/science.1123480 [DOI] [PubMed]
- Hippenmeyer P, Highkin M (1993) High level, stable production of recombinant proteins in mammalian cell culture using the herpesvirus VP16 transactivator. Biotechnology (N Y) 11:1037–1041. doi:10.1038/nbt0993-1037 [DOI] [PubMed]
- Hollien J, Weissman JS (2006) Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313:104–107. doi:10.1126/science.1129631 [DOI] [PubMed]
- Hsu T-A, Betenbaugh MJ (1997) Coexpression of molecular chaperone BiP improves immunoglobulin solubility and IgG secretion from Trichoplusia ni insect cells. Biotechnol Prog 13:96–104. doi:10.1021/bp960088d [DOI] [PubMed]
- Hsu T-A, Watson S, Eiden JJ, Betenbaugh MJ (1996) Rescue of immunoglobulins from insolubility is facilitated by PDI in the baculovirus expression system. Protein Expr Purif 7:281–288. doi:10.1006/prep. 1996.0040 [DOI] [PubMed]
- Hsu TA, Eiden JJ, Bourgarel P, Meo T, Betenbaugh MJ (1994) Effects of co-expressing chaperone BiP on functional antibody production in the baculovirus system. Protein Expr Purif 5:595–603. doi:10.1006/prep. 1994.1082 [DOI] [PubMed]
- Hu P, Han Z, Couvillon AD, Kaufman RJ, Exton JH (2006) Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1α-mediated NF-κB activation and down-regulation of TRAF2 expression. Mol Cell Biol 26:3071–3084. doi:10.1128/MCB.26.8.3071-3084.2006 [DOI] [PMC free article] [PubMed]
- Hurtley SM, Bole DG, Hoover-Litty H, Helenius A, Copeland CS (1989) Interactions of misfolded influenza virus hemagglutinin with binding protein (BiP). J Cell Biol 108:2117–2126. doi:10.1083/jcb.108.6.2117 [DOI] [PMC free article] [PubMed]
- Imagawa Y, Hosoda A, Sasaka S-i, Tsuru A, Kohno K (2008) RNase domains determine the functional difference between IRE1α and IRE1β. FEBS Lett 582:656–660. doi:10.1016/j.febslet.2008.01.038 [DOI] [PubMed]
- Iwawaki T, Hosoda A, Okuda T, Kamigori Y, Nomura-Furuwatari C, Kimata Y et al (2001) Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat Cell Biol 3:158–164. doi:10.1038/35055065 [DOI] [PubMed]
- Jarvis DL, Summers MD (1989) Glycosylation and secretion of human tissue plasminogen activator in recombinant baculovirus-infected insect cells. Mol Cell Biol 9:214–223 [DOI] [PMC free article] [PubMed]
- Jeon HK, Chang KH, Kim KI, Chung IS (2003) Functional expression of recombinant tumstatin in stably transformed Drosophila melanogaster S2 cells. Biotechnol Lett 25:185–189. doi:10.1023/A:1022330429508 [DOI] [PubMed]
- Jones J, Nivitchanyong T, Giblin C, Ciccarone V, Judd D, Gorfien S et al (2005) Optimization of tetracycline-responsive recombinant protein production and effect on cell growth and ER stress in mammalian cells. Biotechnol Bioeng 91:722–732. doi:10.1002/bit.20566 [DOI] [PubMed]
- Kang S-W, Rane NS, Kim SJ, Garrison JL, Taunton J, Hegde RS (2006) Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell 127:999–1013. doi:10.1016/j.cell.2006.10.032 [DOI] [PMC free article] [PubMed]
- Karst AM, Li G (2007) BH3-only proteins in tumorigenesis and malignant melanoma. Cell Mol Life Sci 64:318–330. doi:10.1007/s00018-006-6364-4 [DOI] [PMC free article] [PubMed]
- Kato T, Murata T, Usui T, Park EY (2005) Improvement of the production of GFPuv-β1, 3-N-acetylglucosaminyltransferase 2 fusion protein using a molecular chaperone-assisted insect-cell-based expression system. Biotechnol Bioeng 89:424–433. doi:10.1002/bit.20362 [DOI] [PubMed]
- Kaufman RJ (2004) Regulation of mRNA translation by protein folding in the endoplasmic reticulum. Trends Biochem Sci 29:152–158. doi:10.1016/j.tibs.2004.01.004 [DOI] [PubMed]
- Kaufman RJ, Wasley LC, Dorner AJ (1988) Synthesis, processing, and secretion of recombinant human factor VIII expressed in mammalian cells. J Biol Chem 263:6352–6362 [PubMed]
- Kimata Y, Ishiwata-Kimata Y, Yamada S, Kohno K (2006) Yeast unfolded protein response pathway regulates expression of genes for anti-oxidative stress and for cell surface proteins. Genes Cells 11:59–69. doi:10.1111/j.1365-2443.2005.00921.x [DOI] [PubMed]
- Kitchin K, Flickinger MC (1995) Alteration of hybridoma viability and antibody secretion in transfectomas with inducible overexpression of protein disulfide isomerase. Biotechnol Prog 11:565–574. doi:10.1021/bp00035a011 [DOI] [PubMed]
- Klionsky DJ (2005) The molecular machinery of autophagy: unanswered questions. J Cell Sci 118:7–18. doi:10.1242/jcs.01620 [DOI] [PMC free article] [PubMed]
- Kohno K, Normington K, Sambrook J, Gething MJ, Mori K (1993) The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol 13:877–890 [DOI] [PMC free article] [PubMed]
- Kondo S, Saito A, Hino SI, Murakami T, Ogata M, Kanemoto S et al (2007) BBF2H7, a novel transmembrane bZIP transcription factor, is a new type of ER stress transducer. Mol Cell Biol 27:1716–1729. doi:10.1128/MCB.01552-06 [DOI] [PMC free article] [PubMed]
- Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H et al (2007) ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ 14:230–239. doi:10.1038/sj.cdd.4401984 [DOI] [PubMed]
- Ku SCY, Ng DTW, Yap MGS, Chao S-H (2008) Effects of overexpression of X-box binding protein 1 on recombinant protein production in Chinese hamster ovary and NS0 myeloma cells. Biotechnol Bioeng 99:155–164. doi:10.1002/bit.21562 [DOI] [PubMed]
- Lambert N, Merten OW (1997) Effect of serum-free and serum-containing medium on cellular levels of ER-based proteins in various mouse hybridoma cell lines. Biotechnol Bioeng 54:165–180. doi : 10.1002/(SICI)1097-0290(19970420)54:2<165::AID-BIT8>3.0.CO;2-J [DOI] [PubMed]
- Lederkremer GZ, Glickman MH (2005) A window of opportunity: timing protein degradation by trimming of sugars and ubiquitins. Trends Biochem Sci 30:297–303. doi:10.1016/j.tibs.2005.04.010 [DOI] [PubMed]
- Lee AH, Iwakoshi NN, Glimcher LH (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23:7448–7459. doi:10.1128/MCB.23.21.7448-7459.2003 [DOI] [PMC free article] [PubMed]
- Lee JM, Chang KH, Park JH, Lee YH, Chung IS (2001) Production of recombinant endostatin from stably transformed Trichoplusia ni BTI Tn 5B1-4 cells. Biotechnol Lett 23:1931–1936. doi:10.1023/A:1013726113708
- Li Y, Ge M, Ciani L, Kuriakose G, Westover EJ, Dura M et al (2004) Enrichment of endoplasmic reticulum with cholesterol inhibits sarcoplasmic-endoplasmic reticulum calcium ATPase-2b activity in parallel with increased order of membrane lipids: implications for depletion of endoplasmic reticulum calcium stores and apoptosis in cholesterol-loaded macrophages. J Biol Chem 279:37030–37039. doi:10.1074/jbc.M405195200 [DOI] [PubMed]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B et al (2007) IRE1 signaling affects cell fate during the unfolded protein response. Science 318:944–949. doi:10.1126/science.1146361 [DOI] [PMC free article] [PubMed]
- Machamer CE, Doms RW, Bole DG, Helenius A, Rose JK (1990) Heavy chain binding protein recognizes incompletely disulfide-bonded forms of vesicular stomatitis virus G protein. J Biol Chem 265:6879–6883 [PubMed]
- Marciniak SJ, Ron D (2006) Endoplasmic reticulum stress signaling in disease. Physiol Rev 86:1133–1149. doi:10.1152/physrev.00015.2006 [DOI] [PubMed]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R et al (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 18:3066–3077. doi:10.1101/gad.1250704 [DOI] [PMC free article] [PubMed]
- Martínez IM, Chrispeels MJ (2003) Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15:561–576. doi:10.1105/tpc.007609 [DOI] [PMC free article] [PubMed]
- Mauro C, Crescenzi E, De Mattia R, Pacifico F, Mellone S, Salzano S et al (2006) Central role of the scaffold protein tumor necrosis factor receptor-associated factor 2 in regulating endoplasmic reticulum stress-induced apoptosis. J Biol Chem 281:2631–2638. doi:10.1074/jbc.M502181200 [DOI] [PubMed]
- McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ (2001) Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol 21:1249–1259. doi:10.1128/MCB.21.4.1249-1259.2001 [DOI] [PMC free article] [PubMed]
- Meusser B, Hirsch C, Jarosch E, Sommer T (2005) ERAD: the long road to destruction. Nat Cell Biol 7:766–772. doi:10.1038/ncb0805-766 [DOI] [PubMed]
- Miura T, Katakura Y, Seto P, Zhang YP, Teruya K, Nishimura E et al (2001) Availability of oncogene activated production system for mass production of light chain of human antibody in CHO cells. Cytotechnology 35:9–16. doi:10.1023/A:1008179919857 [DOI] [PMC free article] [PubMed]
- Mohan C, Park SH, Chung JY, Lee GM (2007) Effect of doxycycline-regulated protein disulfide isomerase expression on the specific productivity of recombinant CHO cells: thrombopoietin and antibody. Biotechnol Bioeng 98:611–615. doi:10.1002/bit.21453 [DOI] [PubMed]
- Morano KA (2007) New tricks for an old dog: the evolving world of Hsp70. Ann N Y Acad Sci 1113:1–14. doi:10.1196/annals.1391.018 [DOI] [PubMed]
- Moremen KW, Molinari M (2006) N-linked glycan recognition and processing: the molecular basis of endoplasmic reticulum quality control. Curr Opin Struct Biol 16:592–599. doi:10.1016/j.sbi.2006.08.005 [DOI] [PMC free article] [PubMed]
- Morris JA, Dorner AJ, Edwards CA, Hendershot LM, Kaufman RJ (1997) Immunoglobulin binding protein (BiP) function is required to protect cells from endoplasmic reticulum stress but is not required for the secretion of selective proteins. J Biol Chem 272:4327–4334. doi:10.1074/jbc.272.7.4327 [DOI] [PubMed]
- Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA et al (2000) Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature 403:98–103. doi:10.1038/47513 [DOI] [PubMed]
- Nichols WC, Seligsohn U, Zivelin A, Terry VH, Hertel CE, Wheatley MA et al (1998) Mutations in the ER-Golgi intermediate compartment protein ERGIC-53 cause combined deficiency of coagulation factors V and VIII. Cell 93:61–70. doi:10.1016/S0092-8674(00)81146-0 [DOI] [PubMed]
- Nishikawa S, Hirata A, Nakano A (1994) Inhibition of endoplasmic reticulum (ER)-to-Golgi transport induces relocalization of binding protein (BiP) within the ER to form the BiP bodies. Mol Biol Cell 5:1129–1143 [DOI] [PMC free article] [PubMed]
- Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K et al (2002) ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev 16:1345–1355. doi:10.1101/gad.992302 [DOI] [PMC free article] [PubMed]
- Ogata M, Hino SI, Saito A, Morikawa K, Kondo S, Kanemoto S et al (2006) Autophagy is activated for cell survival after ER stress. Mol Cell Biol 26:9220–9231. doi:10.1128/MCB.01453-06 [DOI] [PMC free article] [PubMed]
- Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H (2005) TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J 24:1243–1255. doi:10.1038/sj.emboj.7600596 [DOI] [PMC free article] [PubMed]
- Ohya T, Hayashi T, Kiyama E, Nishii H, Miki H, Kobayashi K et al (2007) Improved production of recombinant human antithrombin III in Chinese hamster ovary cells by ATF4 overexpression. Biotechnol Bioeng 100:317–324. doi:10.1002/bit.21758 [DOI] [PubMed]
- Özcan U, Cao Q, Yilmaz E, Lee A-H, Iwakoshi NN, Ozdelen E et al (2004) Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 306:457–461. doi:10.1126/science.1103160 [DOI] [PubMed]
- Ozcan U, Ozcan L, Yilmaz E, Düvel K, Sahin M, Manning BD et al (2008) Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell 29:541–551. doi:10.1016/j.molcel.2007.12.023 [DOI] [PMC free article] [PubMed]
- Pakula TM, Laxell M, Huuskonen A, Uusitalo J, Saloheimo M, Penttilä M (2003) The effects of drugs inhibiting protein secretion in the filamentous fungus Trichoderma reesei. Evidence for down-regulation of genes that encode secreted proteins in the stressed cells. J Biol Chem 278:45011–45020. doi:10.1074/jbc.M302372200 [DOI] [PubMed]
- Papa S, Zazzeroni F, Bubici C, Jayawardena S, Alvarez K, Matsuda S et al (2004) Gadd45β mediates the NF-κB suppression of JNK signalling by targeting MKK7/JNKK2. Nat Cell Biol 6:146–153. doi:10.1038/ncb1093 [DOI] [PubMed]
- Parekh R, Forrester K, Wittrup D (1995) Multicopy overexpression of bovine pancreatic trypsin inhibitor saturates the protein folding and secretory capacity of Saccharomyces cerevisiae. Protein Expr Purif 6:537–545. doi:10.1006/prep. 1995.1071 [DOI] [PubMed]
- Park JH, Lee JM, Chung IS (1999) Production of recombinant endostatin from stably transformed Drosophila melanogaster S2 cells. Biotechnol Lett 21:729–733. doi:10.1023/A:1005510821928
- Pendse GJ, Karkare S, Bailey JE (1992) Effect of cloned gene dosage on cell growth and hepatitis B surface antigen synthesis and secretion in recombinant CHO cells. Biotechnol Bioeng 40:119–129. doi:10.1002/bit.260400117 [DOI] [PubMed]
- Perlmutter DH (2006) The role of autophagy in alpha–1-antitrypsin deficiency: a specific cellular response in genetic diseases associated with aggregation-prone proteins. Autophagy 2:258–263 [DOI] [PubMed]
- Pham CG, Bubici C, Zazzeroni F, Papa S, Jones J, Alvarez K et al (2004) Ferritin heavy chain upregulation by NF-κB inhibits TNFα-induced apoptosis by suppressing reactive oxygen species. Cell 119:529–542. doi:10.1016/j.cell.2004.10.017 [DOI] [PubMed]
- Randall TD, Parkhouse RM, Corley RB (1992) J chain synthesis and secretion of hexameric IgM is differentially regulated by lipopolysaccharide and interleukin 5. Proc Natl Acad Sci USA 89:962–966. doi:10.1073/pnas.89.3.962 [DOI] [PMC free article] [PubMed]
- Randow F, Seed B (2001) Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat Cell Biol 3:891–896. doi:10.1038/ncb1001-891 [DOI] [PubMed]
- Robinson DK, Memmert KW (1991) Kinetics of recombinant immunoglobulin production by mammalian-cells in continuous culture. Biotechnol Bioeng 38:972–976. doi:10.1002/bit.260380903 [DOI] [PubMed]
- Ron D, Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8:519–529. doi:10.1038/nrm2199 [DOI] [PubMed]
- Rosser MF, Trotta BM, Marshall MR, Berwin B, Nicchitta CV (2004) Adenosine nucleotides and the regulation of GRP94-client protein interactions. Biochemistry 43:8835–8845. doi:10.1021/bi049539q [DOI] [PubMed]
- Roux PP, Blenis J (2004) ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev 68:320–344. doi:10.1128/MMBR.68.2.320-344.2004 [DOI] [PMC free article] [PubMed]
- Sakaki K, Wu J, Kaufman RJ (2008) Protein kinase Cθ is required for autophagy in response to stress in the endoplasmic reticulum. J Biol Chem 283:15370–15380. doi:10.1074/jbc.M710209200 [DOI] [PMC free article] [PubMed]
- Saltiel AR, Kahn CR (2001) Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414:799–806. doi:10.1038/414799a [DOI] [PubMed]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P et al (2001) Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell 7:1165–1176. doi:10.1016/S1097-2765(01)00265-9 [DOI] [PubMed]
- Schröder M (2006) The unfolded protein response. Mol Biotechnol 34:279–290. doi:10.1385/MB:34:2:279 [DOI] [PubMed]
- Schröder M (2008a) Endoplasmic reticulum stress responses. Cell Mol Life Sci 65:862–894. doi:10.1007/s00018-007-7383-5 [DOI] [PMC free article] [PubMed]
- Schröder M (2008b) Engineering eukaryotic protein factories. Biotechnol Lett 30:187–196. doi:10.1007/s10529-007-9524-1 [DOI] [PMC free article] [PubMed]
- Schröder M, Friedl P (1997) Overexpression of recombinant human antithrombin III in Chinese hamster ovary cells results in malformation and decreased secretion of the recombinant protein. Biotechnol Bioeng 53:547–559. doi :10.1002/(SICI)1097-0290(19970320)53:6<547::AID-BIT2>3.0.CO;2-M [DOI] [PubMed]
- Schröder M, Kaufman RJ (2005a) ER stress and the unfolded protein response. Mutat Res 569:29–63. doi:10.1016/j.mrfmmm.2004.06.056 [DOI] [PubMed]
- Schröder M, Kaufman RJ (2005b) The mammalian unfolded protein response. Annu Rev Biochem 74:739–789. doi:10.1146/annurev.biochem.73.011303.074134 [DOI] [PubMed]
- Schröder M, Kaufman RJ (2006) Divergent roles of IRE1α and PERK in the unfolded protein response. Curr Mol Med 6:5–36. doi:10.2174/156652406775574569 [DOI] [PubMed]
- Schröder M, Körner C, Friedl P (1999) Quantitative analysis of transcription and translation in gene amplified Chinese hamster ovary cells on the basis of a kinetic model. Cytotechnology 29:93–102. doi:10.1023/A:1008077603328 [DOI] [PMC free article] [PubMed]
- Schröder M, Schäfer R, Friedl P (2002) Induction of protein aggregation in an early secretory compartment by elevation of expression level. Biotechnol Bioeng 78:131–140. doi:10.1002/bit.10206 [DOI] [PubMed]
- Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee A-H, Qian SB, Zhao H et al (2004) XBP1, downstream of Blimp–1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21:81–93. doi:10.1016/j.immuni.2004.06.010 [DOI] [PubMed]
- Shah OJ, Wang Z, Hunter T (2004) Inappropriate activation of the TSC/Rheb/mTOR/S6 K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol 14:1650–1656. doi:10.1016/j.cub.2004.08.026 [DOI] [PubMed]
- Shulman GI (1999) Cellular mechanisms of insulin resistance in humans. Am J Cardiol 84:3J–10J. doi:10.1016/S0002-9149(99)00350-1 [DOI] [PubMed]
- Smales CM, Dinnis DM, Stansfield SH, Alete D, Sage EA, Birch JR et al (2004) Comparative proteomic analysis of GS-NS0 murine myeloma cell lines with varying recombinant monoclonal antibody production rate. Biotechnol Bioeng 88:474–488. doi:10.1002/bit.20272 [DOI] [PubMed]
- Smith JD, Tang BC, Robinson AS (2004) Protein disulfide isomerase, but not binding protein, overexpression enhances secretion of a non-disulfide-bonded protein in yeast. Biotechnol Bioeng 85:340–350. doi:10.1002/bit.10853 [DOI] [PubMed]
- Sriburi R, Jackowski S, Mori K, Brewer JW (2004) XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol 167:35–41. doi:10.1083/jcb.200406136 [DOI] [PMC free article] [PubMed]
- Steel GJ, Fullerton DM, Tyson JR, Stirling CJ (2004) Coordinated activation of Hsp70 chaperones. Science 303:98–101. doi:10.1126/science.1092287 [DOI] [PubMed]
- Strudwick N, Schröder M (2007) The unfolded protein response. In: Al-Rubeai M, Fussenegger M (eds) Systems biology. Cell engineering, vol 5. Springer Verlag, Dordrecht, pp 69–157
- Suzuki CK, Bonifacino JS, Lin AY, Davis MM, Klausner RD (1991) Regulating the retention of T-cell receptor α chain variants within the endoplasmic reticulum: Ca2+-dependent association with BiP. J Cell Biol 114:189–205. doi:10.1083/jcb.114.2.189 [DOI] [PMC free article] [PubMed]
- Tanida I, Ueno T, Kominami E (2004) LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol 36:2503–2518. doi:10.1016/j.biocel.2004.05.009 [DOI] [PMC free article] [PubMed]
- Teruya K, Daimon Y, Dong XY, Katakura Y, Miura T, Ichikawa A et al (2005) An approach to further enhance the cellular productivity of exogenous protein hyper-producing Chinese hamster ovary (CHO) cells. Cytotechnology 47:29–36. doi:10.1007/s10616-005-3765-4 [DOI] [PMC free article] [PubMed]
- Thorpe C, Coppock DL (2007) Generating disulfides in multicellular organisms: emerging roles for a new flavoprotein family. J Biol Chem 282:13929–13933. doi:10.1074/jbc.R600037200 [DOI] [PubMed]
- Thuerauf DJ, Marcinko M, Belmont PJ, Glembotski CC (2007) Effects of the isoform-specific characteristics of ATF6α and ATF6β on endoplasmic reticulum stress response gene expression and cell viability. J Biol Chem 282:22865–22878. doi:10.1074/jbc.M701213200 [DOI] [PubMed]
- Tigges M, Fussenegger M (2006) Xbp1-based engineering of secretory capacity enhances the productivity of Chinese hamster ovary cells. Metab Eng 8:264–272. doi:10.1016/j.ymben.2006.01.006 [DOI] [PubMed]
- Tu BP, Weissman JS (2002) The FAD- and O2-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol Cell 10:983–994. doi:10.1016/S1097-2765(02)00696-2 [DOI] [PubMed]
- Tu BP, Weissman JS (2004) Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol 164:341–346. doi:10.1083/jcb.200311055 [DOI] [PMC free article] [PubMed]
- Underhill MF, Coley C, Birch JR, Findlay A, Kallmeier R, Proud CG et al (2003) Engineering mRNA translation initiation to enhance transient gene expression in chinese hamster ovary cells. Biotechnol Prog 19:121–129. doi:10.1021/bp025560b [DOI] [PubMed]
- van Anken E, Romijn EP, Maggioni C, Mezghrani A, Sitia R, Braakman I et al (2003) Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity 18:243–253. doi:10.1016/S1074-7613(03)00024-4 [DOI] [PubMed]
- van der Heide M, Hollenberg CP, van der Klei IJ, Veenhuis M (2002) Overproduction of BiP negatively affects the secretion of Aspergillus niger glucose oxidase by the yeast Hansenula polymorpha. Appl Microbiol Biotechnol 58:487–494. doi:10.1007/s00253-001-0907-2 [DOI] [PubMed]
- Veal EA, Day AM, Morgan BA (2007) Hydrogen peroxide sensing and signaling. Mol Cell 26:1–14. doi:10.1016/j.molcel.2007.03.016 [DOI] [PubMed]
- Wajant H, Scheurich P (2001) Tumor necrosis factor receptor-associated factor (TRAF) 2 and its role in TNF signaling. Int J Biochem Cell Biol 33:19–32. doi:10.1016/S1357-2725(00)00064-9 [DOI] [PubMed]
- Warnakulasuriyarachchi D, Cerquozzi S, Cheung HH, Holcik M (2004) Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J Biol Chem 279:17148–17157. doi:10.1074/jbc.M308737200 [DOI] [PubMed]
- Watowich SS, Morimoto RI, Lamb RA (1991) Flux of the paramyxovirus hemagglutinin-neuraminidase glycoprotein through the endoplasmic reticulum activates transcription of the GRP78-BiP gene. J Virol 65:3590–3597 [DOI] [PMC free article] [PubMed]
- Welker E, Wedemeyer WJ, Narayan M, Scheraga HA (2001) Coupling of conformational folding and disulfide-bond reactions in oxidative folding of proteins. Biochemistry 40:9059–9064. doi:10.1021/bi010409g [DOI] [PubMed]
- Werner RG (2004) Economic aspects of commercial manufacture of biopharmaceuticals. J Biotechnol 113:171–182. doi:10.1016/j.jbiotec.2004.04.036 [DOI] [PubMed]
- White MF (2002) IRS proteins and the common path to diabetes. Am J Physiol Endocrinol Metab 283:E413–E422. doi:10.1152/ajpendo.00514.2001 [DOI] [PubMed]
- Whiteley EM, Hsu T-A, Betenbaugh MJ (1997) Modeling assembly, aggregation, and chaperoning of immunoglobulin G production in insect cells. Biotechnol Bioeng 56:106–116. doi :10.1002/(SICI)1097-0290(19971005)56:1<106::AID-BIT12>3.0.CO;2-I [DOI] [PubMed]
- Wiest DL, Burkhardt JK, Hester S, Hortsch M, Meyer DI, Argon Y (1990) Membrane biogenesis during B cell differentiation: most endoplasmic reticulum proteins are expressed coordinately. J Cell Biol 110:1501–1511. doi:10.1083/jcb.110.5.1501 [DOI] [PMC free article] [PubMed]
- Williams DB (2006) Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J Cell Sci 119:615–623. doi:10.1242/jcs.02856 [DOI] [PubMed]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J et al (2007) ATF6α optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell 13:351–364. doi:10.1016/j.devcel.2007.07.005 [DOI] [PubMed]
- Wurm FM (2004) Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol 22:1393–1398. doi:10.1038/nbt1026 [DOI] [PubMed]
- Xu C, Bailly-Maitre B, Reed JC (2005) Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest 115:2656–2664. doi:10.1172/JCI26373 [DOI] [PMC free article] [PubMed]
- Xu X, Azakami H, Kato A (2004) P-domain and lectin site are involved in the chaperone function of Saccharomyces cerevisiae calnexin homologue. FEBS Lett 570:155–160. doi:10.1016/j.febslet.2004.06.039 [DOI] [PubMed]
- Yamaguchi S, Ishihara H, Yamada T, Tamura A, Usui M, Tominaga R et al (2008) ATF4-mediated induction of 4E-BP1 contributes to pancreatic β cell survival under endoplasmic reticulum stress. Cell Metab 7:269–276. doi:10.1016/j.cmet.2008.01.008 [DOI] [PubMed]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H et al (2007) Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev Cell 13:365–376. doi:10.1016/j.devcel.2007.07.018 [DOI] [PubMed]
- Yokouchi M, Hiramatsu N, Hayakawa K, Okamura M, Du S, Kasai A et al (2008) Involvement of selective reactive oxygen species upstream of proapoptotic branches of unfolded protein response. J Biol Chem 283:4252–4260. doi:10.1074/jbc.M705951200 [DOI] [PubMed]
- Yoneda T, Imaizumi K, Oono K, Yui D, Gomi F, Katayama T et al (2001) Activation of caspase-12, an endoplastic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the ER stress. J Biol Chem 276:13935–13940. doi:10.1074/jbc.M010677200 [DOI] [PubMed]
- Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K (2003) A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell 4:265–271. doi:10.1016/S1534-5807(03)00022-4 [DOI] [PubMed]
- Yoshimura FK, Luo X, Zhao X, Gerard HC, Hudson AP (2008) Up-regulation of a cellular protein at the translational level by a retrovirus. Proc Natl Acad Sci USA 105:5543–5548. doi:10.1073/pnas.0710526105 [DOI] [PMC free article] [PubMed]
- Zang M, Trautmann H, Gandor C, Messi F, Asselbergs F, Leist C et al (1995) Production of recombinant proteins in Chinese hamster ovary cells using a protein-free cell culture medium. Biotechnology (N Y) 13:389–392. doi:10.1038/nbt0495-389 [DOI] [PubMed]
- Zeng L, Lu M, Mori K, Luo S, Lee AS, Zhu Y et al (2004) ATF6 modulates SREBP2-mediated lipogenesis. EMBO J 23:950–958. doi:10.1038/sj.emboj.7600106 [DOI] [PMC free article] [PubMed]
- Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ (2005) The unfolded protein response sensor IRE1α is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest 115:268–281 [DOI] [PMC free article] [PubMed]
- Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT et al (2006) Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124:587–599. doi:10.1016/j.cell.2005.11.040 [DOI] [PubMed]
- Zhou D, Pallam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC (2008) Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem 283:7064–7073. doi:10.1074/jbc.M708530200 [DOI] [PubMed]
- Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H et al (1998) CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev 12:982–995. doi:10.1101/gad.12.7.982 [DOI] [PMC free article] [PubMed]