Abstract
Optimization of cell culture media based on statistical experimental design methodology is a widely used approach for improving cultivation conditions. We applied this methodology to refine the composition of an established culture medium for growth of a human hepatoma cell line, C3A. A selection of growth factors and nutrient supplements were systematically screened according to standard design of experiments (DoE) procedures. The results of the screening indicated that the medium additives hepatocyte growth factor, oncostatin M, and fibroblast growth factor 4 significantly influenced the metabolic activities of the C3A cell line. Surface response methodology revealed that the optimum levels for these factors were 30 ng/ml for hepatocyte growth factor and 35 ng/ml for oncostatin M. Additional experiments on primary human hepatocyte cultures showed high variance in metabolic activities between cells from different individuals, making determination of optimal levels of factors more difficult. Still, it was possible to conclude that hepatocyte growth factor, epidermal growth factor, and oncostatin M had decisive effects on the metabolic functions of primary human hepatocytes.
Keywords: Design of experiments, Bioreactor, C3A, Primary human hepatocytes, Growth factors, Culture medium optimization
Introduction
Cultures of human hepatic cells and liver cell lines are used in a wide range of applications in cell biology and medical research. Despite this, culture media protocols quite often follow traditional recipes or ad hoc experimental observations without being systematically refined for the particular experimental purpose. This often leads to a culturing system with suboptimal control of the cellular mechanisms for cell maintenance and propagation, which limits experimental success.
This situation is particularly remarkable since research on hepatocyte cell biology has over the years uncovered the biological role of several extracellular growth and differentiation factors (Okita 2002). Of particular interest and relevance among these are (1) the hepatocyte growth factor (HGF), due to its ability to promote DNA synthesis (Gomez-Lechon et al. 1995; Strain et al. 1991); (2) albumin, which is involved in controlling osmotic conditions; (3) oncostatin M (OSM), due to its stimulation of fibroblasts and Kaposi cells (Zarling et al. 1986; Linsley et al. 1989); (4) fibroblast growth factors (FGF), which activate tissue repair; and (5) epidermal growth factor (EGF), due to its ability to support DNA synthesis in combination with insulin and glucagon (Block et al. 1996). In order to attain optimal effects during cellular and metabolic experimentation and processing, it is particularly important to target the intracellular concentration levels of these protein factors. In addition to these factors, small extracellular molecules, including carbohydrates, lipids, vitamins, metal ions, and numerous anionic molecules, exert supplementary effects on the metabolism. The intracellular concentrations of all these factors are directly or indirectly affected by the extracellular medium levels, by their transport into the cytoplasm, and by their conversion inside the cell. In consequence, the composition of the culture medium is one of the experimenter’s main options for controlling intracellular processes and the functional activities of the cell. Thus, it is of great interest to optimize these medium factor levels in a coherent and justified way.
Modern statistical methods provide powerful techniques for evaluation of factor effects based on defined system responses for any experimental system (Montgomery 2005). These techniques, commonly referred to as factorial design or design of experiments (DoE), are well suited for screening and optimization of medium factors which are critical for improving cell maintenance and growth and for controlling specific cellular functions such as metabolic activity and growth rate. Previously, DoE has been applied widely in numerous studies of media optimization for bacterial, fungal, and mammalian cell cultures (Mandenius and Brundin 2008). Of particular interest are experiences from screening of medium factors in human cell lines, for example in studies of recombinant protein production in animal cell cultures (Deshpande et al. 2004), and for embryonic stem cell differentiation (Chang and Zandstra 2004).
The common DoE procedure permits a preliminary screening of between five and ten medium factors in a limited number of experiments, a so-called reduced factorial design experimental protocol on two levels of concentrations (Montgomery 2005). In this process, several medium factors are simultaneously compared and their effects are observed and ranked based on analyzed properties or parameters, normally defined as response variables. The response variable or variables should be as relevant as possible to the purpose of the medium design. For example, if efficient cell propagation is the primary aim, cell counts would be the appropriate response; if a certain metabolic state of the culture is the aim, characteristic metabolites should be analyzed and used as response variables. Once the response variables have been determined, statistical performance parameters are generated from subsequent computations based on the DoE procedure, and then used to assess the relevance of the observed effects. Through this, the medium factors are ranked in relation to their influence, and then the most effective factors are selected and further tested in additional experiments. Finally, the results from these experiments are analyzed in a regression model to determine optimal levels of the medium factors.
In this paper, we describe the application of this methodology to evaluate the effect of a number of critical medium factors used in culture media for human hepatocytes. The goal was to optimize the medium composition with respect to metabolic activity of the hepatic cells. Factors investigated included HGF, OSM, FGF4, EGF, albumin, nicotinamide (NicA), and dexamethasone (DexM). The chosen basal medium was a liver cell medium, Heparmed 142, which was developed specifically for serum-free high-density cultures of human hepatocytes (Gerlach et al. 2003; Zeilinger et al. 2004). Experiments were performed using the human hepatoma cell line, C3A, a derivative of the HepG2 cell line (Elkayam et al. 2006; Wang et al. 1998). The highest-ranked media factors from the initial DoE screening were further analyzed in a multiple linear regression model to determine their optimal levels. We also made preliminary comparisons of the results obtained in C3A cell cultures with those obtained in primary human hepatic cells; however, due to the limited availability of primary human cells, these comparisons were restricted to cells from two individuals.
Experimental methods
Cell materials and culture procedures
The hepatoma cell line C3A (CRL-10741, ATCC, Manassas, VA, USA) was expanded at 37°C and 5% CO2 in 175 cm2 cell culture flasks (FalconTM, BD Biosciences, USA) coated with 1 mg/ml collagen A (Biochrom AG, Berlin, Germany). Cultures were maintained in William’s E medium with addition of l-glutamine (2 mmol/l), insulin (20 IU/l), transferrin (5 mg/l), glucagon (3 μg/l), penicillin (100,000 U/l), and streptomycin (100 mg/l), all from Biochrom AG. For passaging, cells were detached enzymatically by incubation with 0.05%/0.02% (w/v) Trypsin/EDTA.
Primary human hepatocytes were gained from two individual donors after partial resection of the liver, with the approval of the local ethical committee and the written agreement of the patients. Cell isolation was performed by collagenase digestion as described previously (Nussler et al. 2008). The cells had a viability of 74 and 86% respectively, as determined by their capacity for trypan blue exclusion. To support cell adhesion after isolation, cells were cultured in the presence of fetal calf serum (FCS, Biochrom AG) during the first days of culture.
For the experiments, C3A cells or freshly isolated primary human liver cells were seeded onto collagen-coated six-well plates (FalconTM, 9.6 cm2/well) at a density of 1.5 × 106 cells/well. A modification of the William’s E medium, developed for serum-free culture of primary hepatocytes at high densities, was used (Heparmed 142 medium, Biochrom AG). This medium is characterized by enrichment of amino acids, monosaccharides, free fatty acids, and trace elements. To adapt the cells to serum-free conditions, the concentration of FCS was stepwise reduced from 10% at day 0 to 5% at day 1 and 0% at day 2. This was achieved by replacing 50% (day 2) or 100% (day 3) of the medium volume with serum free medium containing the investigated medium factors at set concentrations. Afterwards, the medium was exchanged every 2 days up to day 6 with fresh serum free medium with added factors. Samples were collected and analyzed as described below.
Medium factors investigated
Human growth factor (HGF), oncostatin M (OSM), and fibroblast growth factor 4 (FGF4) were obtained from PeproTech, Inc. (Rocky Hill, NJ, USA). Human serum albumin (HSA) was obtained from Baxter Healthcare Corp., Round Lake, IL, USA. Epidermal growth factor (EGF), nicotinamide (NicA), and dexamethasone (DexM) were obtained from Biochrom AG. All substances were added to the medium at two different concentrations (see Table 1).
Table 1.
Factors, ranges, and response variables
| Factor variables | Conc. range (g/ml) | Response variables |
|---|---|---|
| Human serum albumin | 0–6.0 × 10−3 | |
| Oncostatin M | 0–5.0 × 10−8 | |
| Fibroblast growth factor-4 | 0–5.0 × 10−8 | |
| Hepatocyte growth factor | 0–5.0 × 10−8 | |
| Epidermal growth factor | 0–1.0 × 10−7 | |
| Nicotinamide | 1.0–10.0 × 10−5 | |
| Dexamethasone | 0–7.4 × 10−6 | |
| Glucose | ||
| Lactate | ||
| Urea | ||
| LDH |
Analytical methods
The concentrations of the response variables—glucose, urea, lactate, and lactate dehydrogenase (LDH)—in the culture supernatants were measured on days 0, 1, 2, 4, and 6 using automated clinical chemistry analyzers (Roche Diagnostics, Heidelberg, Germany). Analyses were performed using ready-to-use enzymatic test kits (Gluco-quantR, Urea/BUN, lactate, LDH optimized) provided by Roche Diagnostics.
Experimental design
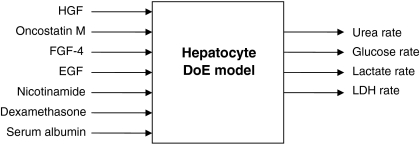
A two-level fractional factorial design was used to screen the effect on human liver cell cultures of the seven selected factors added to the culture medium. Fourteen experiments were used in a resolution III design (i.e. a design where the main factor effects are confounded with two-factor and higher-order interactions) including three center points (see Table 2). The experimental order was randomised in order to minimize unpredictable errors. Samples for analysis and computation were collected simultaneously (day 4 or day 6). Each experiment was carried out in triplicate, performed in separate wells and averaged. The design and the analysis of experiments were performed using the software package Modde™ (Umetrics AB, Umeå, Sweden) (Fig. 1).
Table 2.
Screening design and responses at day 6
| Exp. no. | Factor variables | Response variables | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HSA (mg/ml) | HGF (ng/ml) | OSM (ng/ml) | DexM (mg/l) | FGF4 (ng/ml) | EGF (ng/ml) | NicA (mg/l) | Urea (mg/l) | Lac (mg/l) | LDH (U/l) | |
| 1 | 0 | 0 | 0 | 7.4 | 50 | 100 | 10 | 6.3 | 133 | 35.0 |
| 2 | 6 | 0 | 0 | 0 | 0 | 100 | 100 | 9.0 | 135 | 147 |
| 3 | 0 | 50 | 0 | 0 | 50 | 0 | 100 | 7.6 | 110 | 22.3 |
| 4 | 6 | 50 | 0 | 7.4 | 0 | 0 | 10 | 6.3 | 149 | 23.0 |
| 5 | 0 | 0 | 50 | 7.4 | 0 | 0 | 100 | 8.0 | 132 | 8.0 |
| 6 | 6 | 0 | 50 | 0 | 50 | 0 | 10 | 9.6 | 143 | 27.6 |
| 7 | 0 | 50 | 50 | 0 | 0 | 100 | 10 | 8.0 | 137 | 27.0 |
| 8 | 6 | 50 | 50 | 7.4 | 50 | 100 | 100 | 6.3 | 87.0 | 25.7 |
| 9 | 3 | 25 | 25 | 3.7 | 25 | 50 | 55 | 6.7 | 147 | 24.7 |
| 10 | 3 | 25 | 25 | 3.7 | 25 | 50 | 55 | 8.0 | 149 | 22.7 |
| 11 | 3 | 25 | 25 | 3.7 | 25 | 50 | 55 | 8.3 | 139 | 30.0 |
Fig. 1.
The general outline of the experimental design
On the basis of the results from these analyses, factors were ranked and the three highest ranked factors were subsequently investigated by response surface modeling for further optimization. A two-level full factorial design based on a central composite design (CCD) was used. The optimization required 17 experimental runs including three center points (see Table 4). In the optimization part, all experiments were carried out in duplicate using different wells, and were repeated twice with cells from separate batches. One extra control run with all factors kept at low levels was performed between the two repeats in order to exclude systematic errors from the model.
Table 4.
Media optimization design
| Exp. no. | Factors | Response variables | Comment | |||||
|---|---|---|---|---|---|---|---|---|
| HGF (ng/ml) | OSM (ng/ml) | FGF4 (ng/ml) | Urea (mg/l) | Lac (mg/l) | LDH (U/l) | Alb (μg/ml) | ||
| 1 | 0 | 0 | 0 | 8.0 | 121 | 34.0 | 14.1 | |
| 2 | 40 | 0 | 0 | 11.0 | 123 | 44.0 | 12.0 | |
| 3 | 0 | 50 | 0 | 14.0 | 202 | 66.0 | 18.4 | |
| 4 | 40 | 50 | 0 | 14.0 | 135 | 86.0 | 7.13 | |
| 5 | 0 | 0 | 40 | 7.0 | 76.6 | 28.0 | 17.8 | |
| 6 | 40 | 0 | 40 | 8.0 | 134 | 21.0 | 9.8 | |
| 7 | 0 | 50 | 40 | 11.0 | 177 | 44.0 | 9.2 | |
| 8 | 40 | 50 | 40 | 14.0 | 130 | 127 | 2.6 | |
| 9 | 0 | 25 | 20 | 10.0 | 176 | 42.0 | 7.0 | |
| 10 | 40 | 25 | 20 | 13.0 | 140 | 95.0 | 5.3 | |
| 11 | 20 | 0 | 20 | 9.0 | 139 | 28.0 | 14.6 | |
| 12 | 20 | 50 | 20 | 9.0 | 185 | 43.0 | 5.8 | |
| 13 | 20 | 25 | 0 | 12.0 | 180 | 60.0 | 4.9 | |
| 14 | 20 | 25 | 40 | 9.0 | 186 | 47.0 | 4.9 | |
| 15 | 20 | 25 | 20 | 14.0 | 132 | 74.0 | 6.9 | Center point |
| 16 | 20 | 25 | 20 | 10.0 | 191 | 51.0 | 4.7 | Center point |
| 17 | 20 | 25 | 20 | 11.0 | 175 | 50.0 | 6.0 | Center point |
| 18 | 0 | 0 | 0 | 8.0 | 123 | 32.0 | 18.3 | Blocking exp |
Results and discussion
The typical way of carrying out a DoE plan for cell culture media optimization is to select a realistic number of factors, all of which are considered important for supporting the desired properties of the particular cell type under investigation. These factors are generally tested in a limited number of combinations at two levels of concentration (Mandenius and Brundin 2008). The outcome of the cultivation is judged by relevant responses for the specific cell type, reflecting functional, behavioral, or quality characteristics. The responses must be qualified for quantitative determination. This part of the DoE procedure is referred to as screening; its aim is to select the most influential factor among those tested. Generally, two or three medium factors are identified by this method, and these are then further investigated by complementary testing at the same or modified levels of concentrations. The analytical data obtained are used to set up a refined multiple linear regression (MLR) model which results in a mapping of the dependency of the factors versus the chosen response(s). A response surface plot depicts the optima for the factors.
In this study, the DoE procedure was applied in designing a culture medium with an optimal composition of factors for human hepatocyte cultivation. A well known hepatocyte model, the human hepatoma cell line C3A, was used. The medium factors selected for testing were HSA, OSM, human FGF4, human HGF, human EGF, NicA, and DexM. This selection was based on previous experience in the laboratory and on published results showing a supporting effect of these factors on liver cell maintenance and growth (Gomez-Lechon et al. 1995; Strain et al. 1991; Zarling et al. 1986; Linsley et al. 1989; Block et al. 1996). The lower and higher concentrations for the seven factors were typical levels as reported in the literature or as used in previous experiments (Table 1).
Crucial quality criteria for evaluation of liver cell cultures in vitro are cell integrity and hepatocyte-specific metabolic activities. In the case of a liver cell line, one obvious quality characteristic is cell growth. Considering these aspects, concentrations and rates of formation or depletion of urea, glucose, lactate, and lactate dehydrogenase (LDH) were selected as responses to be analyzed. Urea, being the main product of nitrogen elimination in the urea cycle in hepatocytes as well as a product of urea synthesis, was considered as a useful quality indicator for primary human hepatocytes in vitro (Pless et al. 2006). The cellular uptake of the carbon source, glucose in this case, is associated with most energy-requiring processes, including protein synthesis or metabolic activity. Glucose consumption is also grossly correlated with cell proliferation. Lactate, being the product of anaerobic glycolysis, is generally correlated with glucose consumption in primary cells and cell lines in vitro. Finally, LDH release allows the estimation of cell membrane integrity and indicates the degree of cell damage.
If a DoE screening is to be successful, it is vital to carefully select the ranges of concentrations for the factors studied; the concentration values should cover a sufficiently broad range from a minimal to a maximal level where effects are seen. In the design of a culture medium, the values are normally the initial concentrations of the components. Here, one should also take into account potential interactions with the other factors. Based on previous experience and literature data, the ranges of relevance for the above factors were set as given in Table 1.
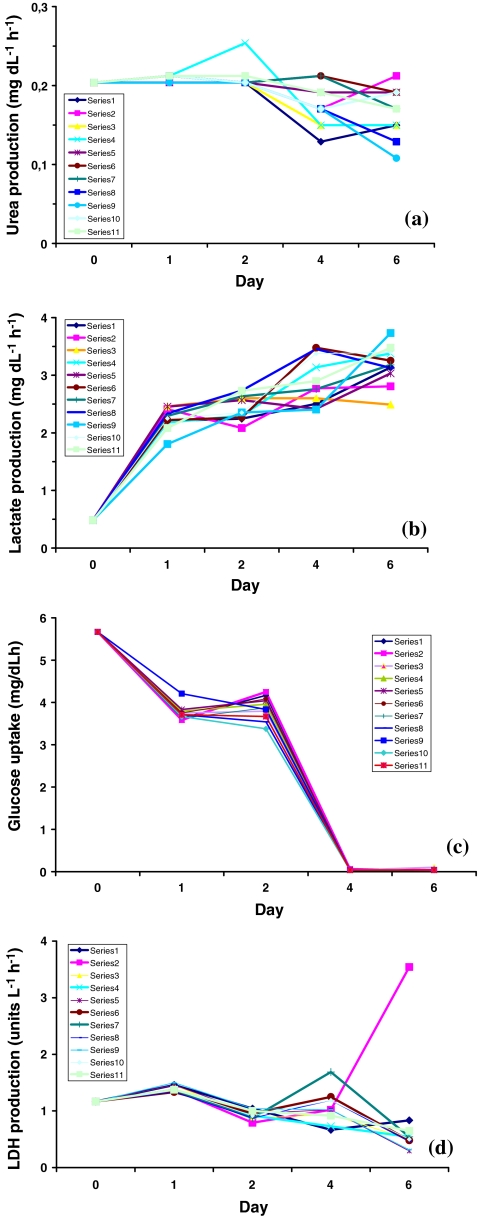
The selected factors were combined in a DoE plan according to Table 2. The plan followed a two-level reduced factorial design comprising 11 experiments. Cells were cultured in triplicate over 7 days, and samples from culture supernatants were taken on days 0, 1, 2, 4, and 6 for determination of the response variables. Figure 2 shows the glucose, lactate, urea, and LDH graphs for the combinations of factors in Table 2. The responses are expressed as rates of consumption (glucose), excretion (lactate, urea), or enzyme release (LDH) per hour during the medium incubation periods.
Fig. 2.
Cultivation diagrams comparing (a) urea production, (b) glucose uptake, (c) lactate production, and (d) lactate dehydrogenase release from the C3A cells at 0, 1, 2, 4, and 6 days. Each diagram depicts 11 graphs representing the experimental runs
The data in Fig. 2 show that at day 6 the four variables had reached steady-state levels. These levels were taken as representative parameters for the physiological state of the hepatocytes in plate culture. It is obvious that glucose was consumed within 4 days with concurrent lactate formation. Glucose consumption appeared to be quite similar for all factor level combinations, while lactate showed slightly larger variations during the sampling interval. Urea was formed more or less in parallel to lactate but varied more significantly due to factor levels. LDH release was low up to day 6 except for two factor combinations. From the graphs shown in Fig. 2, we concluded that day 6 was representative of factor effects on the response variables, and these data were used in DoE analysis of the screening.
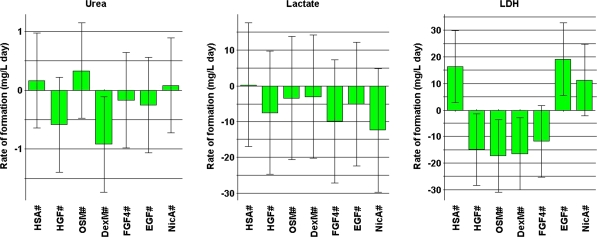
A basic DoE evaluation of the effect of the factors was carried out using the average of the triplicate values. As expected from the graphs above, some factors proved to be more influential than others. Figure 3 shows the effect of the factors on the responses. It is evident that HGF and DexM had the strongest effects on urea production; NicA and FGF4 had the strongest effects on lactate decrease; and OSM, DexM, and HGF had the strongest effects on LDH release. It should be noted that the error of predication in most of the observations was high, a situation not uncommon for any cell culture system. Still, the modeled data allow an undisputable ranking of media component effects.
Fig. 3.
Histograms showing the coefficients of the screening experiments for the response variables urea, lactate, and LDH for each of the seven medium factors
The performance parameters of the regression analysis showed that urea, lactate, and LDH had a good fit to the data of the experiments carried out, though the validity and predictability of the model were high only for urea (Table 3).
Table 3.
Multiple linear regression performance of the screening experiments
| Parameter | Urea | Lactate | LDH |
|---|---|---|---|
| R2 | 0.88 | 0.79 | 0.96 |
| Q2 | 0.85 | −0.19 | −0.20 |
| Model validity | 0.99 | 0.20 | 0.16 |
| Reproducibility | 0.40 | 0.92 | 0.98 |
There was a large degree of variation in the performance parameters; the goodness of fit (R2) varied between 0.79 and 0.99, while the goodness of prediction (Q2) was 0.83 for urea but as low as 0.2 for lactate and LDH. Still, the reproducibility for lactate and LDH was high (for definition of parameters, see e.g. Mandenius and Brundin 2008, or Montgomery 2005).
Despite the low values of some of the model performance parameters, the screening of the C3A cell line gave a clear indication that OSM, HGF, and FGF4 were significant parameters for further optimization.
These three factors were used in a new model using a face-centred central composite design, one of the two commonly used DoE methods. The experimental plan was a full factorial design with 35 experiments including three center points. The experiments were run in two sets in order to increase the statistical relevance (Table 4). Samples were collected on day 4 in order to shorten the experimental plan.
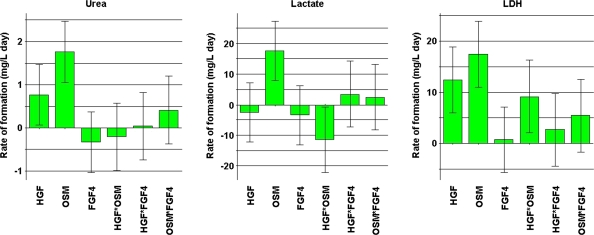
Histograms of the response coefficients for urea, lactate, and LDH are shown in Fig. 4. OSM and HGF had significant effects on urea production. The model also exhibited a slightly positive interaction between OSM and FGF4 in terms of urea production by the C3A cells. In addition, OSM stimulated lactate production, though it also seemed to trigger LDH release, as did HGF. The histograms also show interactive effects of HGF and OSM and of FGF4 and OSM on LDH release. In this part of the study, the errors of prediction of the modeled data were significant.
Fig. 4.
Histograms showing the coefficients of the optimization experiments for the response variables urea, lactate, and LDH for each of the three selected medium factors including interactions
Quality and validity performance parameters of the model are given in Table 5. The fit of data varied between 0.62 and 0.73, which is a slightly lower range than given above for the previous model. However, the validity values lay between 0.68 and 0.97, indicating a definite improvement of reliability.
Table 5.
Model parameters for the optimization experiments
| Model performance | Urea | Lactate | LDH |
|---|---|---|---|
| R2 | 0.63 | 0.62 | 0.73 |
| Q2 | 0.18 | 0.22 | 0.38 |
| Model validity | 0.76 | 0.97 | 0.68 |
| Reproducibility | 0.52 | 0.40 | 0.65 |
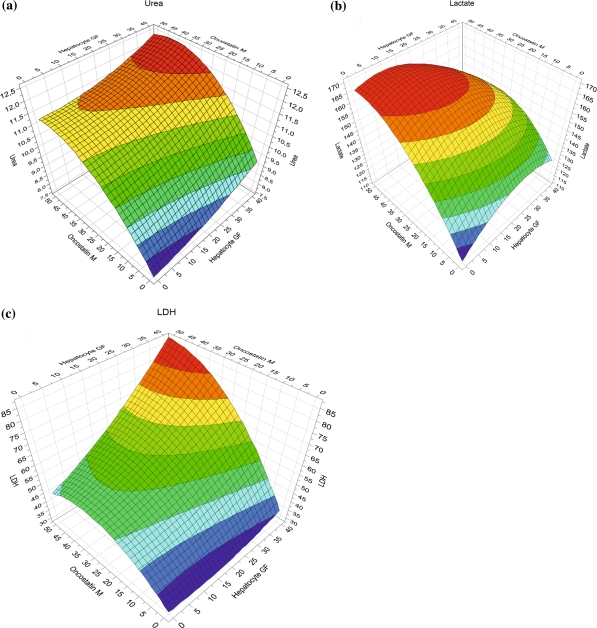
The model allows the generation of response surfaces where optima are distinctly visualized (Fig. 5). In the response surfaces shown, the FGF4 concentration was held at a constant level of 20 ng/ml. Figure 5a shows the result of the model’s prediction for urea formation; an optimum would be reached at a concentration of 30 ng/ml of HGF and 35 ng/ml of OSM. For achieving an optimum lactate production, 25 ng/ml of HGF and 30 ng/ml of OSM (Fig. 5b) were the most effective concentrations. Higher concentrations of these factors (40 ng/ml of HGF and 50 ng/ml of OSM) resulted in a significant increase in LDH release.
Fig. 5.
Response surface graphs for optimization of (a) urea, (b) lactate and (c) LDH at a constant FGF4 concentration of 20 μg/ml
This experimental design was also run with primary human hepatocytes, in order to compare the results with the above-described results from using a cell line. Primary human cells are not widely available, and so the material was restricted to two individual donor cell preparations. The same DoE procedure was employed for each donor cell preparation. Although the results deviated slightly from those found for the cell line, HGF, EGF, and OSM all had decisive effects on the metabolic functions of primary human hepatocytes. HGF proved to be the most significant factor for enhancing urea formation, leading to the highest urea production rate on day 6 for both cell preparations. OSM, however, had a decreasing effect on urea production. This was not consistent with the data obtained with the C3A cell line. However, the results from primary hepatocytes exhibited larger variations between different cell preparations than did samples from the C3A cell line. One major reason for the observed variations in primary human hepatocytes from different donors is the effect of donor-specific factors such as age, medical history, and individual genetic factors. Furthermore, the cell isolation process could also influence the cell metabolism independently of the initial condition of the organ. These variations are not likely to occur when stable cell lines are used.
To obtain more robust data on the effect of the culture medium composition on the in vitro function and quality of primary human hepatocytes, it would be necessary to conduct greater numbers of experiments and to take organ/donor-specific factors into consideration, particularly any potential injury of the donor liver. Such studies could also be helpful in revealing possible correlations between the condition of the organ and the effect of medium factors on isolated cells from individual livers.
Conclusions
This study demonstrates the utility of DoE for screening of factors in the design and optimization of culture media for hepatic cell cultures. A relatively modest number of experiments generated detailed and precise information about a particular cell line’s sensitivity towards already-established growth and differentiation factors. The model visualizes medium requirement deviations between what may appear to be similar cell types/lines but were in fact phenotypic variations, and could motivate slightly modified culture protocols. Of the seven factors investigated, HGF and OSM were identified as the most effective in improving the cell metabolism in vitro in human hepatic cells, The concentrations of these factors were an important determinant for their effect.
The study presupposes that steady states are reached and that media components are present from the beginning. This need not be the case in the design of experiments; factors may be added at different time points in order to achieve a specific cellular effect, thereby refining the cultivation protocol provided the protocol is blocked to these time points.
Hepatoma cell line data showed to be more reliable than primary hepatocyte data. This is not surprising, since primary cell populations are generally less homogeneous. The responses also differed between primary hepatocytes and C3A cells, a result in line with the altered metabolic capacities of the cell line due to transformation of the cells.
In conclusion, the results show the value of using the DoE as a tool for systematic evaluation of medium factors and for ranking their influence in a time-efficient manner.
Acknowledgments
The study was performed within the EU FP6 STREP-project Vitrocellomics. The work on primary hepatocytes was partly funded by the Federal Ministry of Education and Research (BMBF, FKZ 01GG0732 and 01GG0731 to A.N.). The authors would like to thank Dr. Ursula Müller-Vieira, Pharmacelsus GmbH, Saarbrücken, Germany, for valuable contributions.
Abbreviations
- DexM
Dexamethasone
- DoE
Design of experiments
- EGF
Epidermal growth factor
- FGF4
Fibroblast growth factor 4
- HGF
Hepatocyte growth factor
- HSA
Human serum albumin
- LDH
Lactate dehydrogenase
- NicA
Nicotinamide
- OSM
Oncostatin M
References
- Block GD, Locker J, Bowen WC, Petersen BE, Katyal S, Strom SC, Riley T, Howard TA, Michalopoulos GK (1996) Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGFα in a chemically defined medium. J Cell Biol 132:1133–1149 [DOI] [PMC free article] [PubMed]
- Chang KH, Zandstra PW (2004) Quantitative screening of embryonic stem cell differentiation: endoderm formation as a model. Biotechnol Bioeng 88:287–298 [DOI] [PubMed]
- Deshpande RR, Wittmann C, Heinzle E (2004) Microplates with integrated oxygen sensing for medium optimization in animal cell culture. Cytotechnology 46:1–8 [DOI] [PMC free article] [PubMed]
- Elkayam T, Amitay-Shaprut S, Dvir-Ginzburg M, Hartel T, Cohen S (2006) Enhancing the drug metabolism activities of C3A, a human hepatocyte cell line, by tissue engineering within alginate scaffolds. Tissue Eng 12:1357–1368 [DOI] [PubMed]
- Gerlach JC, Mutig K, Sauer IM, Schrade P, Efimova E, Mieder T, Naumann G, Grunwald A, Pless G, Mas A, Bachmann S, Neuhaus P, Zeilinger K (2003) Use of primary human liver cells originating from discarded grafts in a bioreactor for liver support therapy and the prospects of culturing adult liver stem cells in bioreactors: a morphologic study. Transplantation 76:781–786 [DOI] [PubMed]
- Gomez-Lechon M, Castell J, Guillén I, O’Connor E, Nakamura T, Fabra R, Trullenque R (1995) Effects of hepatocyte growth factor on growth and metabolism of human hepatocytes in primary culture. Hepatology 21:1248–1254 [DOI] [PubMed]
- Linsley PS, Bolton-Hanson M, Horn D, Malik N, Kallestad JC, Ochs V, Zarling JM, Shoyab M (1989) Identification and characterization of cellular receptors for the growth regulator oncostatin M. J Biol Chem 264:4282–4289 [PubMed]
- Mandenius CF, Brundin A (2008) Bioprocess optimization using design-of-experiments methodology. Biotechnol Prog (in press) [DOI] [PubMed]
- Montgomery DC (2005) Design and analysis of experiments, 6th edn. Wiley, USA
- Nussler AK, Nussler NC, Merk V, Brulport M, Schormann W, Hengstler JG (2008) In: Santin M (ed) Regenerative medicine today: the holy grail of hepatocyte culturing and therapeutic use. Springer, University of Brighton (in press)
- Okita K (ed) (2002) Growth, proliferation, and apoptosis of hepatocytes. Springer-Verlag, New York
- Pless G, Steffen I, Zeilinger K, Sauer IM, Katenz E, Kehr DC, Roth S, Mieder T, Schwartlander R, Muller C, Wegner B, Hout MS, Gerlach JC (2006) Evaluation of primary human liver cells in bioreactor cultures for extracorporeal liver support on the basis of urea production. Artif Organs 30:686–694 [DOI] [PubMed]
- Strain AJ, Ismail T, Tsubouchi H, Arakaki N, Hishida T, Kitamura N, Daikuhara Y, McMaster P (1991) Native and recombinant human hepatocyte growth factors are highly potent promoters of DNA synthesis in both human and rat hepatocytes. J Clin Invest 87:1853–1857 [DOI] [PMC free article] [PubMed]
- Wang L, Sun J, Li L, Mears D, Horvat M, Sheil AG (1998) Comparison of porcine hepatocytes with human hepatoma (C3A) cells for use in a bioartificial liver support system. Cell Transplant 7:459–468 [DOI] [PubMed]
- Zarling J, Hanson M, Lioubin M, Todaro G (1986) Oncostatin M: a growth regulator produced by differentiated histiocytic lymphoma cells. Proc Natl Acad Sci USA 83:9739–9743 [DOI] [PMC free article] [PubMed]
- Zeilinger K, Holland G, Sauer IM, Efimova E, Kardassis D, Obermayer N, Liu M, Neuhaus P, Gerlach JC (2004) Time course of primary liver cell reorganization in three-dimensional high-density bioreactors for extracorporeal liver support: an immunohistochemical and ultrastructural study. Tissue Eng 10:1113–1124 [DOI] [PubMed]