Abstract
Ascl1 (previously Mash1) is a bHLH transcription factor essential for neuronal differentiation and specification in the nervous system. Although it has been studied for its role in several neural lineages, the full complement of lineages arising from Ascl1 progenitor cells remains unknown. Using an inducible Cre-flox genetic fate mapping strategy, Ascl1 lineages were determined throughout the brain. Ascl1 is present in proliferating progenitor cells but these cells are actively differentiating as evidenced by rapid migration out of germinal zones. Ascl1 lineage cells contribute to distinct cell types in each major brain division: the forebrain including the cerebral cortex, olfactory bulb, hippocampus, striatum, hypothalamus, and thalamic nuclei, the midbrain including superior and inferior colliculi, and the hindbrain including Purkinje and deep cerebellar nuclei cells and cells in the trigeminal sensory system. Ascl1 progenitor cells at early stages in each CNS region become neurons, and at late stages they become oligodendrocytes. In conclusion, Ascl1-expressing progenitor cells in the brain give rise to multiple, but not all, neuronal subtypes and oligodendrocytes depending on the temporal and spatial context, consistent with a broad role in neural differentiation with some subtype specification.
Keywords: Mash1, Ascl1, bHLH transcription factor, genetic fate mapping, cerebellum, brainstem, brain development
INTRODUCTION
In the mammalian central nervous system (CNS), distinct types of neurons are assembled into elaborately interconnected circuits that process complex neural functions. To obtain this refined CNS architecture, diverse populations of neurons must be generated that migrate to specific positions in precise temporal order during embryogenesis. The molecular mechanisms regulating each developmental step in this process are not completely understood. Basic helix-loop-helix (bHLH) transcription factors play a central role in generating neuronal diversity by regulating subtype specification as well as differentiation (Bertrand et al., 2002). Ascl1 (previously Mash1) is a neural bHLH transcription factor restricted to proliferative zones in the developing brain and spinal cord in a spatially specific manner. Multiple studies of mouse embryos lacking Ascl1 suggest that Ascl1 is a neuronal differentiation factor required in diverse but specific neuronal subtypes in the developing central and peripheral nervous systems (Fode et al., 2000; Helms et al., 2005; Horton et al., 1999; Nakada et al., 2004; Pattyn et al., 2004; Perez et al., 1999). Neuronal lineages disrupted in the Ascl1 mutant include interneurons in dorsal spinal cord and telencephalon, both glutamatergic and GABAergic neurons in the mesencephalon, olfactory sensory epithelium, and neurons in the autonomic nervous system in the periphery.
Within a specific neural region, Ascl1 can also influence neuronal subtype specification (Fode et al., 2000; Helms et al., 2005; Nakada et al., 2004; Parras et al., 2002). In dorsal spinal cord, mouse embryos null for Ascl1 lose dI3 and dI5 neurons, whereas overexpression of Ascl1 in the chick neural tube leads to an increase in these specific neuronal populations (Nakada et al., 2004). In neural crest derivatives, Ascl1 is required for autonomic neurons but not sensory neurons (Perez et al., 1999). In the ventral spinal cord, Ascl1 functions in balance with Notch signaling at the choice point where the V2 interneuron population is split into two subpopulations, V2a and V2b (Del Barrio et al., 2007; Peng et al., 2007). Furthermore, Ascl1 is not restricted to neuronal lineages but is also present in progenitors to oligodendrocytes, but not astrocytes, in the spinal cord and adult brain (Battiste et al., 2007; Kim et al., 2007).
A complete fate map of Ascl1 derived lineages, particularly in the brain, has not been described largely due to the transient nature of Ascl1 expression that disappears as the cells exit the cell cycle and migrate extensively during development. This hurdle has been overcome by utilizing Cre recombinase in an in vivo genetic fate mapping strategy to define mature CNS regions receiving contribution from progenitor cells that have transiently expressed Ascl1. This strategy was used to demonstrate that in spinal cord development, Ascl1 progenitors from embryonic day 10.5 (E10.5)–E12.5 give rise to dorsal horn interneurons whereas after E15.5 Ascl1 progenitors give rise to oligodendrocytes (Battiste et al., 2007). In the adult brain, Ascl1 was also found to be present in neuronal progenitors in the dentate gyrus of the hippocampus and the rostral migratory stream in the forebrain, and in oligodendrocyte precursors in white and gray matter (Kim et al., 2007). Consistent with this lineage analysis, forced expression of Ascl1 in neural progenitor cultures biased differentiation of the cells to neurons and oligodendrocytes at the expense of astrocytes (Gokhan et al., 2005; Sugimori et al., 2007).
Here we use the in vivo genetic fate mapping strategy to identify brain regions containing neurons and oligodendrocytes originating from Ascl1 expressing progenitor cells from different embryonic stages. We find that Ascl1-expressing cells are generated continuously throughout embryogenesis from as early as E9.5. As in the spinal cord, the Ascl1 lineage includes both neurons and oligodendrocytes, but not astrocytes. We demonstrate Ascl1 is present in neural progenitor cells in each major brain division. In the cerebellum, depending on the embryonic stage, Ascl1 defined lineages give rise to GABAergic neurons in deep cerebellar nuclei and Purkinje cells. Ascl1 lineages contribute to the trigeminal sensory system from the midbrain to the caudal medulla. Multiple telencephalic regions contain Ascl1 lineage cells including subsets of neurons in the striatum, olfactory bulb, amygdala and piriform cortex. Neurons in the neocortex arise from Ascl1 progenitors present at late embryonic stages. This study correlates the temporal and spatial origin of Ascl1 expressing progenitor cells and their final phenotypes throughout the brain. The diverse identity of the neurons is consistent with Ascl1 requiring additional molecular components for its specification function.
RESULTS AND DISCUSSION
Ascl1 lineage cells identify discrete populations of neurons as well as oligodendrocytes throughout the brain
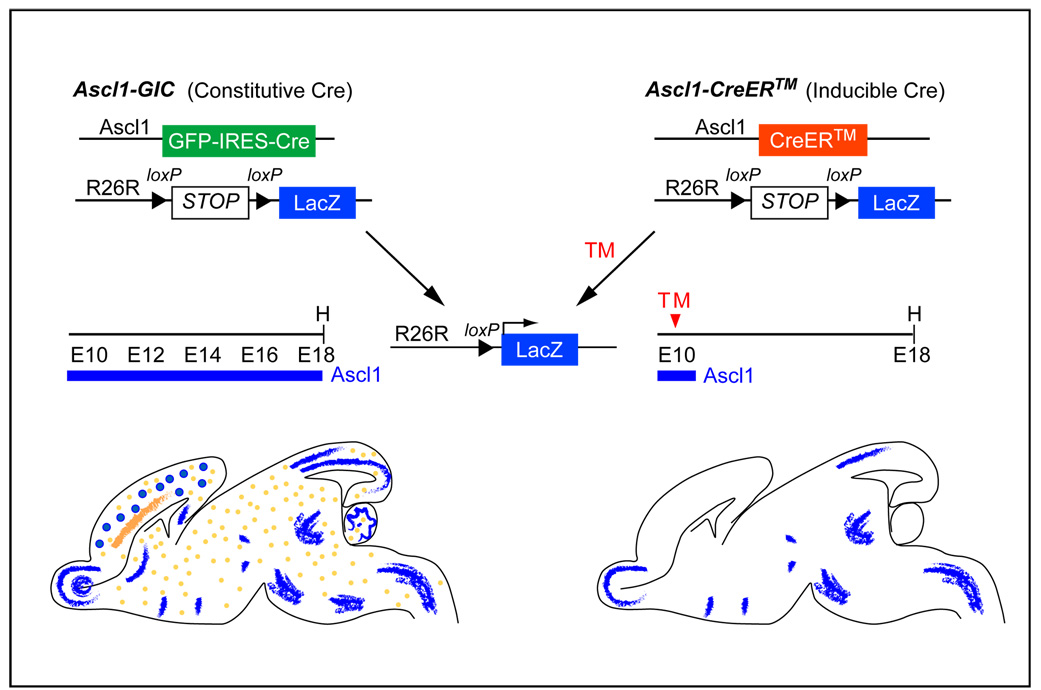
In order to trace the fate of the transiently expressing Ascl1 progenitor cells into the mature brain, we used two BAC (bacterial artificial chromosome) transgenic mouse strains that have been previously described (Battiste et al., 2007; Helms et al., 2005) (Fig. 1). One strain, Ascl1-GIC, expresses constitutively active Cre recombinase in an Ascl1 pattern (Helms et al., 2005). We crossed Ascl1-GIC with R26R-stop-lacZ or R26R-stop-YFP Cre reporter mice to permanently label Ascl1 lineages, since Cre recombinase will excise the stop sequence upstream of LacZ or YFP (Soriano, 1999; Srinivas et al., 2001). Thus, at any given stage, β-gal or YFP positive cells are a cumulative representation of Ascl1 lineages up to that stage. In contrast, the second transgenic strain, Ascl1-CreER™, expresses an inducible Cre recombinase in the Ascl1 pattern, providing temporal control in labeling the Ascl1 lineages. Cre recombination is detectable within 6 hours following tamoxifen treatment, and it persists for approximately 24 h (Hayashi and McMahon, 2002), thus, only the progenitor cells expressing Ascl1 in a restricted time window will be labeled. Together, these mouse strains, combined with neuroanatomical analyses and immunofluorescence with cell-type specific markers, have allowed us to characterize the fate of Ascl1-expressing cells as they progress through development and settle in the mature brain (Fig. 1).
Figure 1. Diagram of Ascl1 transgenic mice and the fate-mapping strategy.
Two transgenic mouse models were generated with a BAC containing Ascl1 >200 kb flanking non-coding sequence. Ascl1-GIC replaces the Ascl1 coding sequence with GFP-IRES-Cre (Helms et al., 2005). This strain will reveal an accumulation of Ascl1 lineage cells when crossed with a Cre reporter mouse strain such as R26R-stop-lacZ (Soriano, 1999). Ascl1-CreER™ replaces the Ascl1 coding sequence with an inducible Cre (Battiste et al., 2007). Only Ascl1 lineage cells originating at the time of tamoxifen (TM) treatment will be detected when crossed with R26R-stop-lacZ. Sagittal views of mouse brains from each paradigm are diagrammed to highlight that the inducible Cre will reveal only a subset of the Ascl1 lineage. Blue represents neurons and orange represents oligodendrocytes E, embryonic stage; H, harvest age.
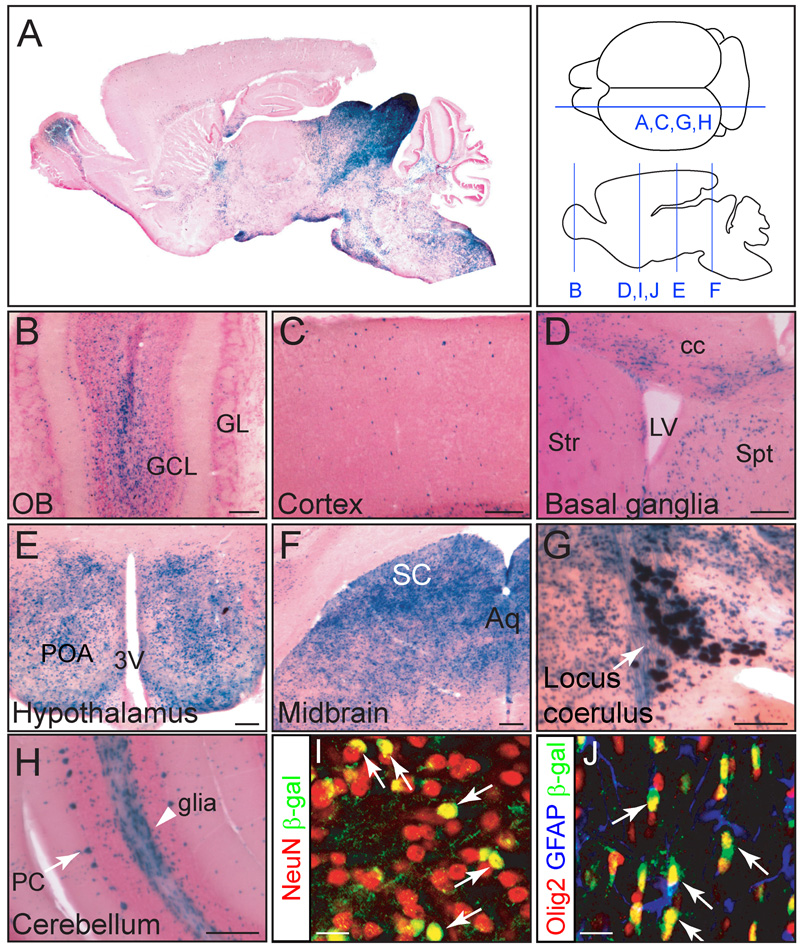
An Ascl1-GIC;R26R-stop-lacZ mouse brain was harvested at postnatal day 30 (P30). An X-gal stained parasagittal section illustrates that Ascl1 lineage cells make extensive but specific contributions to each major subregion in the brain (Fig. 2A). Higher magnification images reveal this specificity (Fig. 2B–H). X-gal stained cells were detected primarily in the granule cell layer in the olfactory bulb (Fig. 2B), in cells dispersed throughout the cortex (Fig. 2C), in the striatum, septum and corpus callosum (Fig. 2D), and in presumptive noradrenergic cells in the locus coerulus (Fig. 2G). These results are consistent with previous studies on Ascl1 function in these lineages (Casarosa et al., 1999; Hirsch et al., 1998; Horton et al., 1999; Kim et al., 2007; Long et al., 2007; Marin et al., 2000). Furthermore, there were X-gal stained cells enriched in other brain regions including the preoptic area of the hypothalamus (Fig. 2E), the superior colliculus in the midbrain (Fig. 2F), and Purkinje cells in the cerebellum (Fig. 2H). Many X-gal stained cells were also found in white matter tracts, regions containing a high percentage of oligodendrocytes (Fig. 2D,H). The Ascl1 lineage cells were identified as neurons and oligodendrocytes since they co-label with the neuronal marker NeuN (Fig. 2I) and the oligodendrocyte marker Olig2 but not the astrocyte marker GFAP (Fig. 2J). This latter finding is consistent with the previous studies defining Ascl1 lineages in neurons and oligodendrocytes of the spinal cord and adult brain (Battiste et al., 2007; Kim et al., 2007; Parras et al., 2007).
Figure 2. Ascl1-expressing cells give rise to discrete cell populations in brain.
(A–H) X-gal staining of P30 brains from Ascl1-GIC;R26R-stop-lacZ transgenic mice including (A) whole brain, (B) olfactory bulb, (C) cerebral cortex, (D) striatum, (E) preoptic area of the hypothalamus, (F) dorsal midbrain, (G) locus coerulus, and (H) cerebellum. (I, J) Immunofluorescence for β-galactosidase in the striatum (I) or the anterior commissure (J). β-gal cells co-express neuronal marker NeuN (I, arrows) or the oligodendrocyte marker Olig2 (J, arrows, but not astrocytes (GFAP). Diagram depicts sectioning plane for each panel. 3V, third ventricle; Aq, aquaduct; cc, corpus callosum; GCL, granule cell layer; GL, glomerular layer; LV, lateral ventricle; OB, olfactory bulb; PC, Purkinje cells; POA, preoptic area; SC, superior colliculus; Spt, septum; Str, striatum. Scale bars = 200µm.
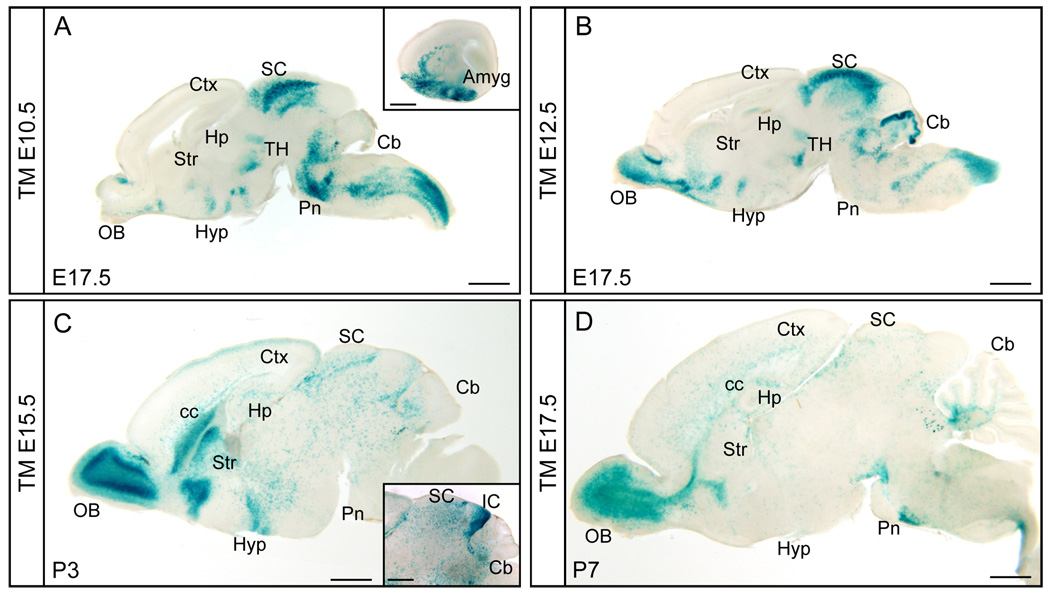
Neurogenesis and gliogenesis are temporally dynamic. To reveal the fate of Ascl1 expressing cells from different stages of embryonic development, we utilized the tamoxifen-inducible Cre line, Ascl1-CreER™. Ascl1-CreER™;R26R-stop-lacZ embryos received tamoxifen by administering the drug to pregnant mice twice with a 6 hour interval on a given embryonic day. The CreER™ recombinase is transiently activated and as the tamoxifen is cleared, the CreER™ returns to the cytoplasm within 24–48 hours (Hayashi and McMahon, 2002). Embryonic or neonatal brains were harvested 7–8 days after tamoxifen administration, sectioned, and X-gal stained. An overview of Ascl1 lineage cells labeled as a consequence of tamoxifen administration at E10.5, E12.5, E15.5, and E17.5 demonstrate these cells distribute with distinctive patterns throughout the brain depending on the embryonic stage labeled (Fig. 3).
Figure 3. Temporal specific fate maps of the Ascl1 lineage in brain.
Sagittal views of X-gal staining of Ascl1-CreER™;R26R-stop-lacZ brains at indicated stages treated with tamoxifen (TM) at embryonic stages indicated. Insets in A and C show amygdala and superior/inferior colliculus from different parasagittal axis sections respectively. Amyg, amygdala; Cb, cerebellum; cc, corpus callosum; Ctx, cortex; Hp, hippocampus; Hyp, hypothalamus; IC, inferior colliculus; OB, olfactory bulb; Pn, Pons; SC, superior colliculus; Str, striatum; TH, thalamus. Scale bars= 1000µm.
Tamoxifen administration at E10.5 resulted in X-gal positive cells that populate ventral regions of the forebrain such as the preoptic area (POA) in the hypothalamus, and amygdala (Fig. 3A and inset). The brain regions that had the highest contribution of Ascl1 lineage cells from this stage include the superior colliculus of the dorsal midbrain, and subdomains within the brainstem (Fig. 3A). With administration of tamoxifen 48 hours later at E12.5, the Ascl1 lineage shifts and populates additional forebrain regions including the olfactory bulb, striatum, and hippocampus (Fig. 3B). The superior colliculus was still strongly labeled, but in the hindbrain a strong contribution of cells was now found in the cerebellum. With tamoxifen at E15.5, Ascl1 lineage cells have dramatically decreased in brainstem regions but they continue to contribute to the olfactory bulb, striatum, POA, and hypothalamus in the forebrain (Fig. 3C). Also at this time, many scattered X-gal stained cells were detected in the cerebral cortex. These cells are likely inhibitory interneurons, consistent with the known birthdates of these cells in the ventral telencephalon migrating to the cortex (Miller, 1985; Miller and Nowakowski, 1988). Notably, oligodendrocytes begin to appear as illustrated by the dispersed X-gal stained cells found throughout the brain and the intensive staining in the corpus callosum (Fig. 3C). And finally by E17.5 tamoxifen administration, the Ascl1 lineage has largely shifted to oligodendrocytes rather than neurons. The identity of the dispersed cells as oligodendrocytes was confirmed by immunofluorescence staining with the marker Olig2 or Sox10 (data not shown). Continued Ascl1 lineage contribution to olfactory bulb neurons at this stage is a notable exception (Fig. 3D). A detailed examination of Ascl1 lineage cells contribution to major brain regions is provided in the following sections. In evaluating the following data, it is important to note that this in vivo genetic fate mapping paradigm is not 100% efficient and likely depends on the level of Cre expression and efficiency of recombination of the Cre-reporter alleles. Because of this limitation, we believe we are preferentially marking the lineage of the highest Ascl1 expressing cells.
Ascl1 lineage in the telencephalon
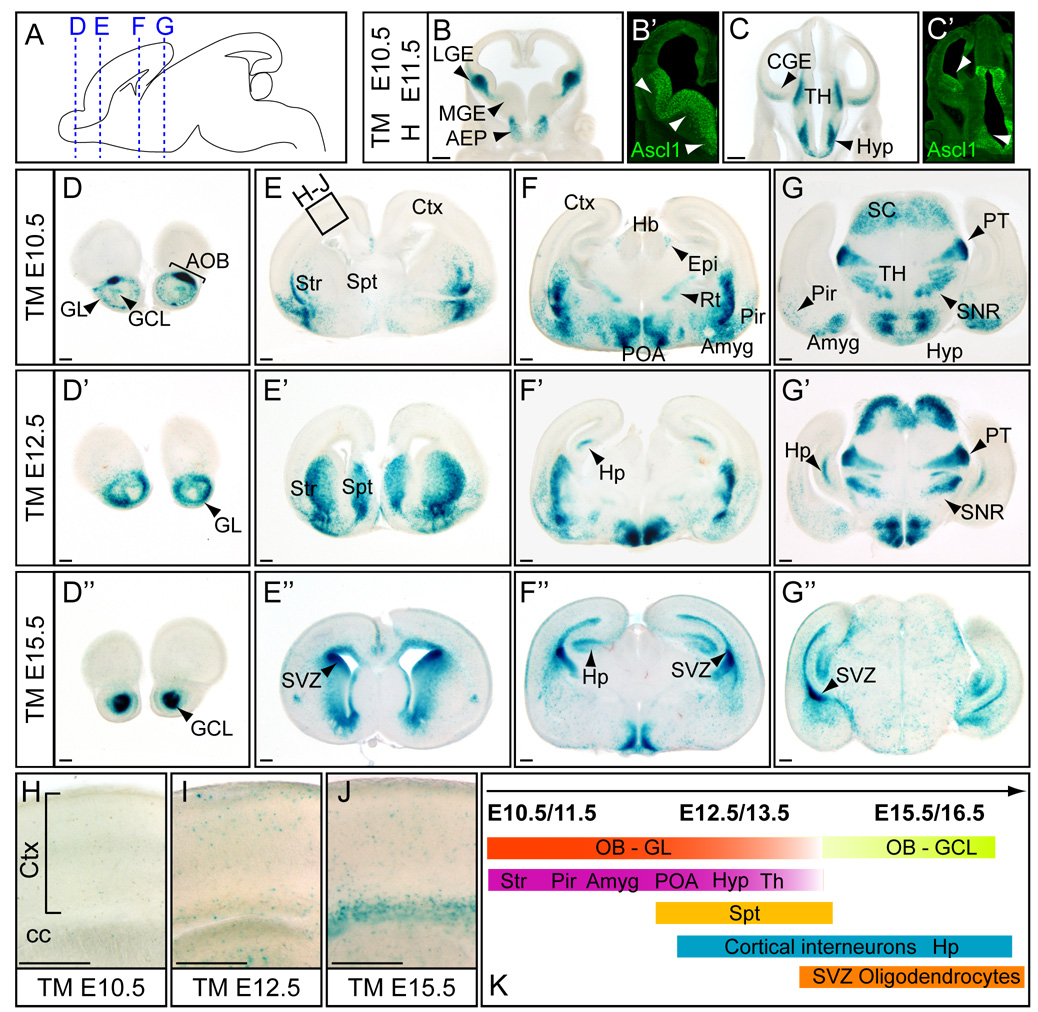
The telencephalon gives rise to diverse brain structures such as the cortex and basal ganglia that contain diverse cell types including pyramidal neurons, interneurons and glia (Corbin et al., 2001; Marin and Rubenstein, 2001; Molyneaux et al., 2007). Neural bHLH transcription factors Neurog1 and Neurog2 (previously Ngn1 and Ngn2) and Ascl1 are expressed in the telencephalic germinal zones and are important in determining distinct neuronal cell types: glutamatergic (Neurog1 and Neurog2) versus GABAergic (Ascl1) (Fode et al., 2000). Briefly, Neurog2 expressing progenitors in dorsal telencephalon give rise to glutamatergic pyramidal neurons in the cerebral cortex via radial migration (Schuurmans et al., 2004). In contrast, Ascl1 expressing progenitors originate in the ganglionic eminences in the ventral telencephalon (Casarosa et al., 1999; Horton et al., 1999). Some of these progenitors migrate tangentially to generate GABAergic interneurons in the neocortex or the olfactory bulb (Fode et al., 2000; Long et al., 2007; Parras et al., 2004). Other Ascl1 progenitors migrate to contribute to interneurons in the striatum (Marin et al., 2000). Among the three major proliferative zones expressing Ascl1 in the ventral telencephalon: the medial (MGE), lateral (LGE) and caudal (CGE) (Fig. 4B’,C’) (Casarosa et al., 1999; Horton et al., 1999), X-gal staining in Ascl1-CreER™ embryos predominantly marks LGE or CGE and not the MGE (Fig. 4B–C). Therefore, the Ascl1 lineages mapped here largely represent the cells originally from the LGE or CGE.
Figure 4. Ascl1 lineage in the forebrain.
(A) Schematic of an E18.5 sagittal brain where dotted lines indicate coronal planes used for panels D–G”. (B–C) X-gal stained coronal sections of telencephalon in Ascl1-CreER™;R26R-stop-lacZ embryos treated at E10.5 and harvested 24 hr later at E11.5 showing the cells of origin marked by Ascl1-CreER™ compared to the endogenous expression patterns of Ascl1 (B’,C’). Ascl1-CreER™;R26R-stop-lacZ embryos exposed to tamoxifen at E10.5 (D–G), E11.5 (D’–G’), and E15.5 (D”–G”) and harvested at E18.5. (D-D”) (H–J) Magnified views (see box in E) to show Ascl1 lineage cells in the cortex and the corpus callosum after E12.5. (K) Summary of the temporal specific generation of discrete Ascl1 lineage cells in forebrain. AEP, anterior entopeduncular area; AOB, accessory olfactory bulb; Amyg, amygdala; cc, corpus callosum; CGE, caudal ganglionic eminence; Ctx, cortex; Epi, epithalamus; GCL, granule cell layer; GL, glomerular layer; Hb, habernula; Hyp, hypothalamus; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; Pir, piriform cortex; POA, preoptic area; PT, pretectal nucleus; Rt, reticular thalamic nucleus; Sc, superior colliculus; SNR, subtrantia nigra (reticular part); Spt, septum; Str, striatum; SVZ, subventricular zone; TH, thalamus. Scale bars = 300µm.
To map the Ascl1 lineages in the telencephalon at different stages of embryonic development, tamoxifen was administered to pregnant females at day 10.5 post coitum (10.5 dpc), 12.5 dpc or 15.5 dpc and Ascl1-CreER™;R26R-stop-lacZ embryos were harvested at E18.5. At each stage, Ascl1 lineage cells contribute to neurons in the olfactory bulb consistent with the LGE origin of these cells (Marin and Rubenstein, 2001). There is a temporal shift with Ascl1 lineage cells contributing to the glomerular cell layers more intensely than the core granule cell layers when tamoxifen was administered at E10.5 or E12.5 (Fig. 4D–D’). In contrast, after E15.5, the Ascl1 progenitors are fated to the granule cell layers (Fig. 3C and Fig. 4D”). The generation of olfactory bulb interneurons from the Ascl1 lineage continues at E17.5 (Fig. 3D) and postnatally into the adult brain (Kim et al., 2007; Parras et al., 2004). In addition to the main olfactory bulb, the Ascl1 lineage also contributes to the accessory olfactory bulb (AOB) mainly from progenitors expressing Ascl1 prior to E12.5 (Fig. 4D).
For the ventral telencephalon, E10.5 and E12.5 Ascl1 lineage cells contribute to the striatum, piriform cortex and amygloid nucleus (Fig. 4E–G). Two days later, E12.5 Ascl1 lineage cells continue to contribute to these regions but now also include the septum (Fig. 4E’–G’). It is known that cortical interneurons arise from the ganglion eminences and migrate tangentially into the cortex (Corbin et al., 2001; Marin and Rubenstein, 2001). Starting after E12.5 tamoxifen administration, we begin to detect X-gal labeled cells in the cortex consistent with the appearance of these interneurons (Fig. 4I). The Ascl1 lineages also contribute to hippocampal neurons after E12.5 and continue through E15.5 (Fig. 4F’–G”). Furthermore, as predicted from the Ascl1 expression pattern and Ascl1 mutant phenotypes, Ascl1 lineage cells did not obviously contribute to glutamatergic projection neurons in the cortex (Fode et al., 2000).
A dramatic shift in cell-types derived from Ascl1 cells occurs after E15.5. At this later embryonic stage, Ascl1 progenitors largely become oligodendrocytes and populate white matter tracts such as the corpus callosum (Fig. 4J) as well as being scattered throughout the gray matter (Fig. 4E”–G”). There is a high density of Ascl1 lineage cells surrounding and emanating from the SVZ of the lateral ventricles. Ascl1 continues to be expressed in the SVZ and in oligodendrocyte progenitors in the adult brain (Kim et al., 2007).
Ascl1 lineage in the diencephalon
Ascl1 expression is detected in distinct progenitor populations in the diencephalon, including the hypothalamus, prethalamus, thalamus, and pretectum (Fig. 4C–C’) (Horton et al., 1999; Vue et al., 2007). Tamoxifen administration at E10.5 in Ascl1-CreER™;R26R-stop-lacZ embryos marks distinct progenitor zones in the diencephalon and allows us to follow the Ascl1 lineage in this region (Fig. 4C). Notably, most thalamic nuclei projecting to the cortex are not Ascl1 derived lineages, consistent with a recent report (Fig. 4F–G’) (Vue et al., 2007). Instead, Ascl1 lineage cells are found in many other nuclei, such as the pretectal nuclei, reticular thalamic nucleus, a cluster of cells lateral to the habenular nucleus and substantia nigra (reticular part) (Fig. 4F–G’). The identity of the pretectal nuclei was verified by lack of Sox2 labeling (Vue et al., 2007; data not shown). Labeling at E15.5 no longer detects these nuclei; instead, scattered X-gal positive cells are found all over the diencephalon, representing oligodendrocytes (Fig. 4G” and data not shown).
Ascl1 lineage cells in the dorsal midbrain show radial migration and sequentially become neurons in superior and inferior colliculi
The superior and inferior colliculi of the midbrain hold many histological parallels with the cerebral cortex. For example, both structures are multilayered, and neurogenesis contributes to each layer in an inside-out manner (Altman and Bayer, 1981a, b; McConnell, 1995). However, neurogenic mechanisms in dorsal midbrain development are less well-studied compared to that in the cerebral cortex. Recent studies characterized the progenitor domains defined by patterns of transcription factors in the developing mesencephalon (Nakatani et al., 2007). Ascl1 expression is detected in all progenitor domains, suggesting Ascl1 derived progeny give rise to diverse neurons rather than a restricted neuronal subtype. Loss of Ascl1 function results in disruption of GABAergic neuron differentiation in this region (Miyoshi et al., 2004). However, the contribution of Ascl1+ progenitors to other neuronal fates has not been investigated. Here we examined the developmental dynamics of Ascl1-defined progenitors for cellular lamination, migration and neuronal subtype.
Administration of tamoxifen at 11.5 dpc to pregnant female mice carrying Ascl1-CreER™;R26R-stop-lacZ embryos and analysis of embryos 24 hours later demonstrates the origin of these cells in the progenitor domains in the mesencephalon (Fig. 5A™A”). Analysis of tamoxifen-treated embryos 6–7 days later reveals some Ascl1 lineage cells migrate in radial arrays (Fig. 5B arrows, E) while others are more scattered and are consistent with tangential migration seen in the telencephalon (Fig. 5B arrowheads). To investigate the identity of the Ascl1 derived neurons, we used the POU homeodomain factor, Brn3a, recently shown to be a pan-glutamatergic neuronal marker (Nakatani et al., 2007). Whereas all marked cells are neurons (Fig. 5C), they distribute between Brn3a+ and Brn3a− populations, suggesting Ascl1+ progenitors not only give rise to GABAergic neurons as previously reported in dorsal midbrain (Miyoshi et al., 2004), but also to glutamatergic neurons (Fig. 5D). With these fate mapping studies, we cannot determine whether cells undergoing different migration patterns distribute between GABA and glutamatergic populations with specificity as has been demonstrated in the developing telencephalon. However, it has been reported that in the developing midbrain, the GABAergic neurons undergo a tangential migration while the glutamatergic neurons undergo a radial migration similar to the migratory pattern known for the cortex (Tan et al., 2002).
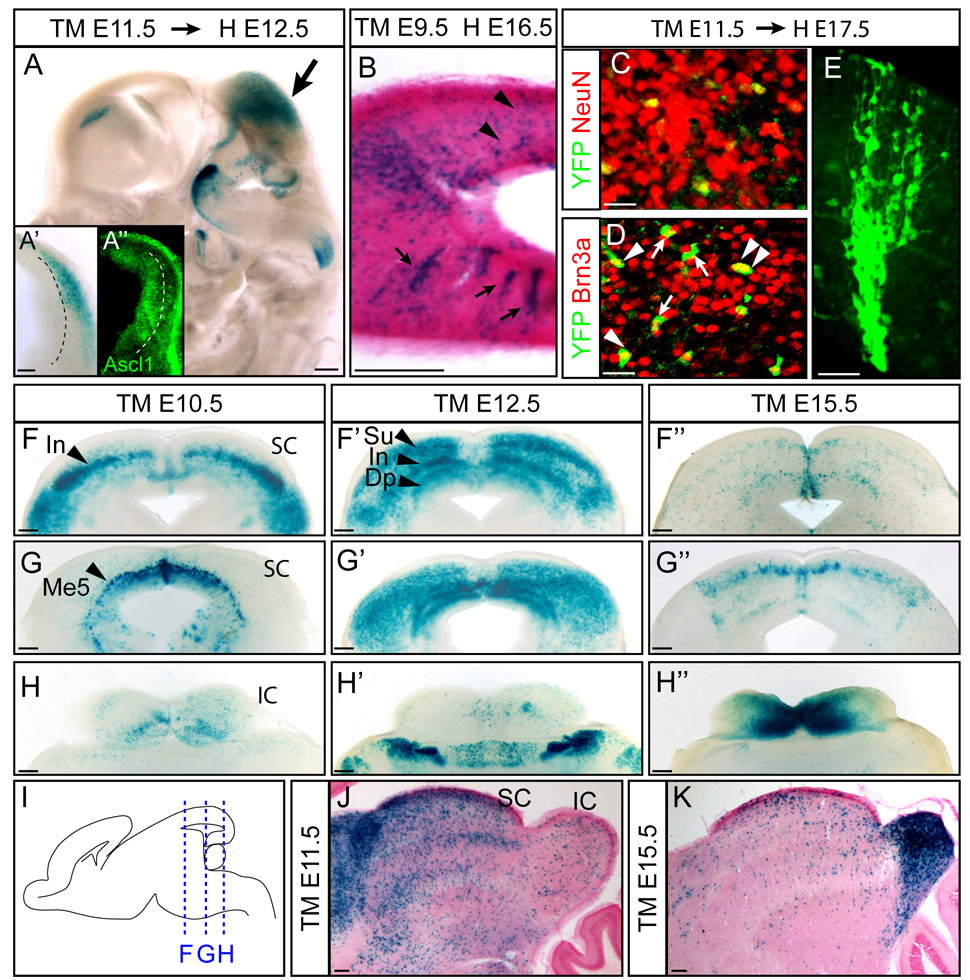
Figure 5. Ascl1 lineage in the dorsal midbrain.
(A) β-gal expression in dorsal mesencephalon (arrow) of Ascl1-CreER™;R26R-stop-lacZ embryos tamoxifen-induced (TM) at E11.5 and harvested (H) at E12.5. Higher magnification of a coronal view shows β-gal present in cells adjacent to the ventricular zone (A’). In contrast, endogenous Ascl1 expression is restricted to the ventricular zone (A”). (B) Ascl1 lineage cells show both scattered (arrowheads) and radially (arrows) arrayed expression patterns. (C–E) In Ascl1-CreER™;R26R-stop-YFP embryos, YFP+ Ascl1 lineage cells are neurons (NeuN+) and some express the glutamatergic marker Brn3a (arrowheads indicate co-expressing cells, arrows indicate no co-expression). (F–H) X-gal stained coronal sections of superior and inferior colliculi from rostal to caudal (from F to H). Ascl1-CreER™;R26R-stop-lacZ embryos were exposed to tamoxifen as indicated and harvested at E18.5. (I) Schematic view of sectioning plane for panels in (F–H”). (J–K) P30 brains of Ascl1-CreER™;R26R-stop-lacZ showed sequential contribution of the Ascl1 lineage to superior and inferior colliculi from early versus late embryogenic stages. IC, inferior colliculus; Me5, mesencephalic trigeminal nucleus; SC, superior colliculus. Scale bars = 20µm for C–E, 200µm for A–B, F–K.
Within the mesencephalon, the superior and inferior colliculi comprise two main functional domains: mutisensory/visual and auditory processing, respectively (Dean et al., 1989). To determine how Ascl1+ cells contribute to these functional domains, we administered tamoxifen at 10.5, 12.5 or 15.5 dpc and analyzed embryos at E18.5. The superior colliculus contains cells derived from progenitor cells expressing Ascl1 during early stages of neurogenesis (~E10.5/E12.5) (Fig. 5F–G”, J). E10.5 Ascl1 lineage cells contribute to the intermediate layer, whereas E12.5 Ascl1 lineage cells populate both the deep and the superficial superior collicular neurons (Fig. 5F–F’ arrows). In contrast, the inferior colliculus contains cells largely derived from progenitor cells expressing Ascl1 at late embryonic stages (E15.5) (Fig. 5H”). This temporal pattern was also illustrated in sagittal sections (Fig. 5J,K). The sequential generation of superior and inferior colliculi is consistent with early birth dating studies using 3H-thymidine (Altman and Bayer, 1981a, b). In addition, early born Ascl1 lineage cells also contribute to the mesencephalic trigeminal nucleus (Me5), the only primary sensory neuronal population in the CNS (Fig. 5F’, arrowhead) (Louvi et al., 2007). In conclusion, Ascl1 lineage contribution to mesencephalon derived brain regions appears broader than other brain regions and encompasses multiple cell types with sequential contribution to rostral then caudal midbrain regions.
Ascl1 lineage cells in the cerebellum
The identification of specific neuronal subtypes that make up the cerebellum, their connections and their development has been extensively studied (Hatten et al., 1997; Wang and Zoghbi, 2001). Two bHLH transcription factors, Atoh1 (previously Math1) and Ptf1a, are major players in the generation of the glutamatergic granule neurons and GABAergic interneurons, respectively (Ben-Arie et al., 1997; Hoshino et al., 2005). Atoh1 is in the upper rhombic lip, a germinal epithelium of the dorsal interface around the fourth ventricle (Fig. 6B). Progenitor cells specified by Atoh1 migrate to the nuclear transitory zone (NTZ) where they express Lhx2/9 (Fig. 6C) and comprise the granule cell progenitors. In contrast, Ptf1a is expressed in the ventricular zone of the cerebeller anlage (Fig. 6D). As these cells mature, they migrate to the cortical transitory zone (CTZ), express Lhx1/5, and contribute to all GABAergic neurons including Purkinje cells in the cerebellum (Chizhikov et al., 2006; Glasgow et al., 2005; Hoshino et al., 2005). Ascl1 is also in the ventricular zone of embryonic cerebellar anlage in an overlapping pattern with Ptf1a (Fig. 6B–D). At E12.5, Ascl1 and Ptf1a are in the ventricular zone where GABAergic neurons are generated, but are excluded from the rhombic lip where the glutamatergic granular cells originate from Atoh1 progenitors (Fig. 6B, D). Ascl1 positive cells do not express Lhx1/5, markers of the postmitotic neurons in the CTZ, suggesting Ascl1 expression is transient in this lineage similar to its expression characteristics in other regions (Fig. 6C).
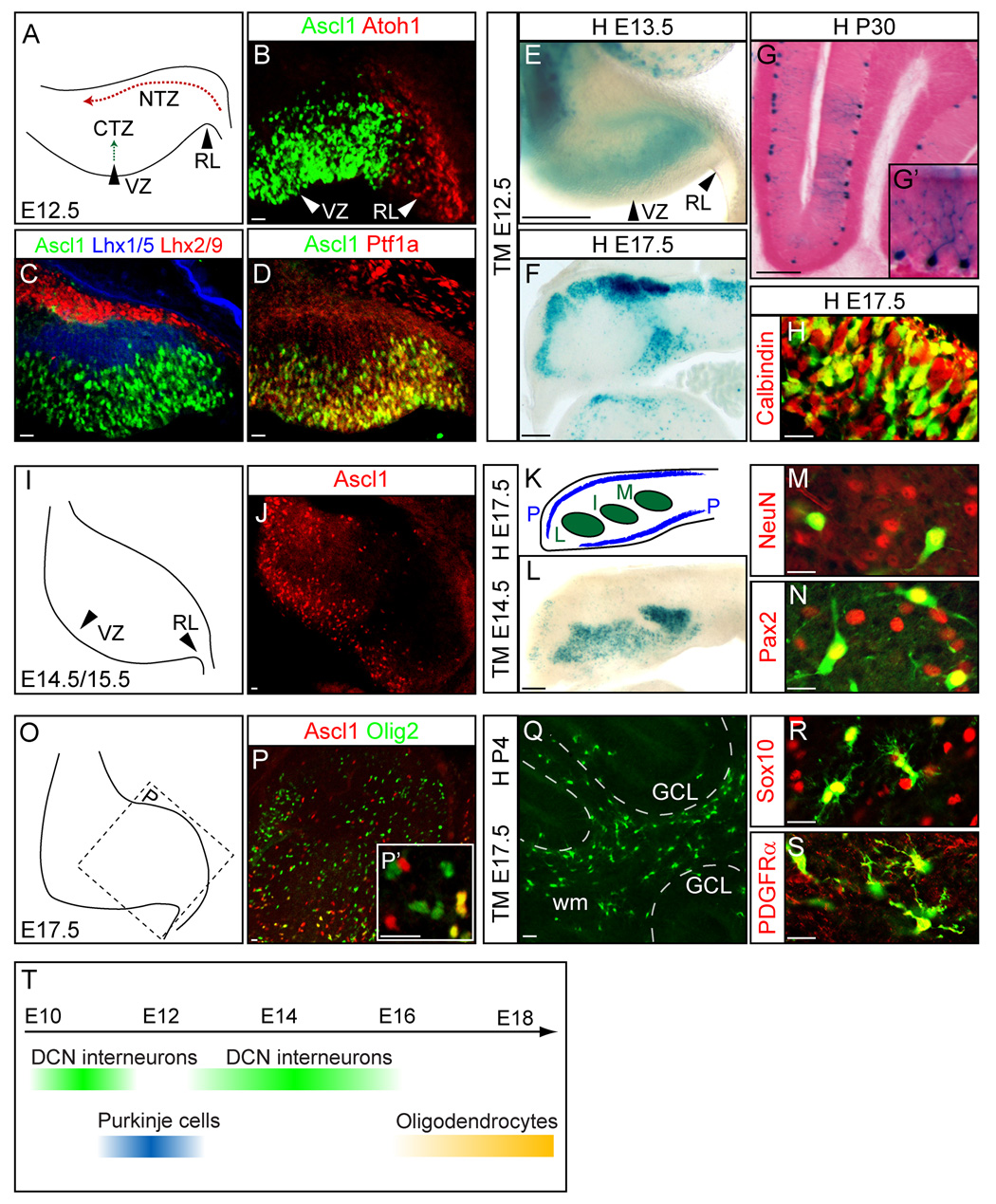
Figure 6. The Ascl1 lineage generates Purkinje cells, deep cerebellar nuclei interneurons and oligodendrocytes at specific stages in the cerebellum.
(A) Diagram depicting E12.5 cerebellum primodium for panels (B–D). Progenitors from the ventricular zone migrate to the cortical transitory zone (CTZ), whereas progenitors from the rhombic lip migrate to the nuclear transitory zone (NTZ). (B–D) Ascl1 is in the ventricular zone (VZ) distinct from Atoh1 in rhombic lip (RL). (C) Transient expression of Ascl1 is not overlapped by CTZ marker Lhx1/5 or NTZ marker Lhx2/9. (D) Ascl1 and Ptf1a are co-expressed in VZ. (E–H) Ascl1-CreER™;R26Rstop-lacZ embryos exposed to tamoxifen at E12.5 and harvested at E13.5 (E), at E17.5 (F,H) or P30 (G). The identity of Purkinje cells are confirmed with a magnified view showing the morphology of Purkinje cells (G’) at P30 or with co-expression of Purkinje cell marker Calbindin (H). (I) Diagram depicting E14.5/E15.5 cerebellum primodium. (J) Ascl1 expression in the VZ of the E15.5 cerebellum. (K) Coronal section of cerebellum with Purkinje cell layer (P) and three deep cerebellar nuclei (L,I,M) at E17.5. (L) β-gal+ Ascl1 lineage cells marked at E14.5 populate all three DCN nuclei, and co-label with neuronal marker NeuN (M) and interneuron marker Pax2 (N). (O) Diagram depicting E17.5 cerebellum primodium. (P) Scattered expression of Ascl1 progenitors in E17.5 cerebellum co-express oligodendrocyte marker Olig2 (P’). (Q–S) Ascl1 progenitors marked at E17.5 and detected at P4 in Ascl1-CreER™;R26R-stop-YFP cerebellum shows most of the Ascl1 lineage in white matter (Q) and co-express Sox10 (R) and PDGFRα (S). (T) Summary diagram depicting temporal specific contribution of Ascl1 lineage cells in cerebellum. CTZ, cortical transitory zone; DCN, deep cerebellar nucleus; GCL, granule cell layer; I, interposed DCN; L, lateral DCN; M, medial DCN; NTZ, nuclear transitory zone; P, Purkinje cells; RL, rhombic lip; VZ, ventricular zone; wm, white matter. Scale bars = 20µm for B–D, H, J, M–S and 200µm for E–F, L.
To determine the fate of the Ascl1 lineage cells, tamoxifen was administered at different times during gestation to pregnant females carrying Ascl1-CreER™;R26R-stop-lacZ embryos. Analysis of Ascl1-CreER™;R26R-stop-lacZ embryos 24 hours after tamoxifen administration at E12.5 illustrates the origin of these cells in the ventricular zone (Fig. 6E). The fate of these progenitors at E17.5 was identified based on location and co-expression of the Purkinje cell marker, calbindin (Fig. 6F, H). Furthermore, when brains were harvested at P30, the X-gal stained cells had the distinct morphology of Purkinje cells (Fig. 6G–G’).
Ascl1 expression in the ventricular zone of the cerebellar primodium starts as early as E10.5 and continues until E15.5 (Fig. 6B–C,J and data not shown). With tamoxifen administration earlier than E11.5 or later than E13.5, Ascl1 lineage cells preferentially became neurons in deep cerebellar nuclei (DCN) rather than Purkinje cells. Ascl1-CreER™;R26R-stop-lacZ E17.5 brains exposed to tamoxifen at E14.5 revealed Ascl1 lineage cells contributing extensively to all three nuclei (medial, interposed, and lateral) (Fig. 6L) and were identified as neurons using co-labeling with NeuN, a pan-neuronal marker (Fig. 6M). DCN contain both glutamatergic and GABAergic neurons. The Ascl1 lineage is restricted to a GABAergic fate determined by expression of Pax2, a GABAergic marker, but not the glutamatergic marker Tbr1 (Fink et al., 2006; Maricich and Herrup, 1999) (Fig. 6N and data not shown). This is consistent with Ascl1 expression restricted to the VZ, whereas the glutamatergic DCN neurons are derived from the rhombic lip cells (Fig. 6B) (Fink et al., 2006; Machold and Fishell, 2005).
Just as in other brain regions, Ascl1 expression in the cerebellum continues through late embryonic and prenatal stages. However, at these late stages, its expression is no longer restricted to the ventricular zone, but rather is detected throughout the cerebellum (Fig. 6P). A subset of these cells co-express the oligodendrocyte lineage marker Olig2, suggesting this population comprises glia progenitors rather than neuronal progenitors (Fig. 6P–P’). Exposure of E17.5 Ascl1-CreER™;R26R-stop-YFP embryos to tamoxifen resulted in Ascl1 lineage cells mainly localized to white matter by P4 (Fig. 6Q). Few YFP expressing cells were located in the Purkinje cell layer or in deep cerebellar nuclei. The identity of these cells as oligodendrocytes was confirmed by co-labeling with Sox10 or PDGFRα (Fig. 6M–N). No overlap was detected with the astrocyte marker GFAP or BLBP, confirming Ascl1 expressing progenitors in the cerebellum do not give rise to astrocytes (data not shown).
Thus, Ascl1 is transient in progenitors to multiple discrete lineages in the cerebellum with a defined temporal sequence that follows the birthdate of these cells (Altman and Bayer, 1985a, b, c; Leto et al., 2006). In contrast to the midbrain, the Ascl1 lineage neurons in the cerebellum appear to be restricted to a GABA neurotransmitter phenotype and include Purkinje cells and DCN neurons. Co-expression of Ascl1 and Ptf1a in the cerebellum ventricular zone is consistent with this restriction to the GABAergic fate since Ptf1a is required for GABAergic neurons in both cerebellum and dorsal spinal cord (Glasgow et al., 2005; Hoshino et al., 2005). As with other brain regions, at late embryonic stages, Ascl1 progenitors no longer give rise to neurons but rather to oligodendrocytes (Fig. 6Q–S and data not shown).
Ascl1 lineages give rise to the trigeminal brainstem nuclei
The lower rhombic lip in the hindbrain is a germinal zone located between the dorsal midbrain and the spinal cord. Progenitor cells from this region migrate extensively to contribute to distinct brainstem nuclei (Rodriguez and Dymecki, 2000). Recent studies have defined rhombic lip progenitor domains by expression of transcription factors patterned along the dorsoventral axis (Fig. 7J) (Landsberg et al., 2005; Sieber et al., 2007; Yamada et al., 2007). For example, it has been shown that progenitors expressing the bHLH transcription factor Atoh1 (dA1) are fated to populate nuclei sending mossy fibers that connect to cerebellar granule cells (Landsberg et al., 2005). Progenitors expressing Ptf1a (dA4) are fated to populate nuclei with climbing fibers that connect to Purkinje cells (Yamada et al., 2007). And finally, the homeodomain factor Lbx1 marks progenitors that contribute to somatosensory and viscerosensory relay neurons (dB1, dB3, dB4) in the brainstem (Sieber et al., 2007).
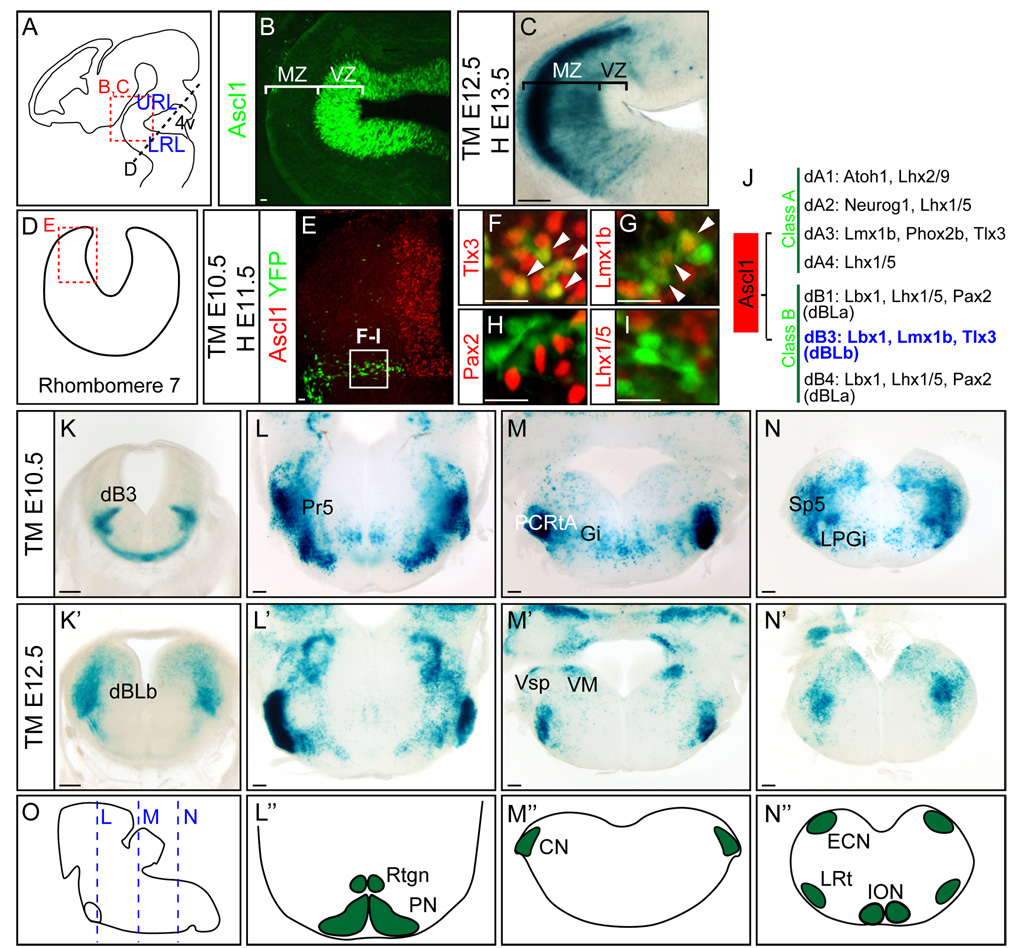
Figure 7. Ascl1 lineage in the trigeminal sensory system in the brainstem.
(A) Diagram of an E12.5 embryo depicting section planes in hindbrain regions shown in (B–D). (B) Ascl1 is restricted to the ventricular zone around the fourth ventricle whereas β-gal marked Ascl1 lineage cells from E12.5 in Ascl1-CreER™;R26R-stop-lacZ embryos are found in the mantle zone by E13.5. (E–I) YFP+ Ascl1 lineage cells in Ascl1-CreER™;R26R-stop-YFP treated with tamoxifen at E10.5 and harvested at E11.5 are dB3 neurons as indicated by co-expression with Tlx3 (H) and Lmx1b (G), but not Pax2 (H) or Lhx1/5 (I). (J) Summary of neuronal subtypes and transcription factors for early (dA1–dB4) and late (dBLa and dBLb) neurogenesis in rhombomere 7 (Sieber et al., 2007). (K–N’) In Ascl1-CreER™;R26R-stop-lacZ embryos, tamoxifen was administered at E10.5 (K–N) and E12.5 (K’–N’), and harvested 24 hours later (K, K’) or at E18.5 (L–N’). (O) Diagram depicting E18.5 embryo brainstem with section plane used in panels (L–N’) shown. (L”–N”) Diagrams of coronal sections of hindbrain showing precerebellar nuclei. 4v, fourth ventricle; CN, cochlear nucleus; ECN, external cuneate nucleus; Gi, gigantocellular reticular nucleus; ION, inferior olive nucleus; LPGi, lateral paragigantocellular nucleus; LRL, lower rhombic lip; LRt, lateral reticular nucleus; MZ, mantle zone; Sp5, spinal trigeminal nucleus; PCRtA, parvicellular reticular nucleus, alpha part; PN, pontine nuclei; Pr5, the primary sensory trigeminal nucleus; Rtgn, reticulotegmental nucleus; URL, upper rhombic lip; VM, medial vestibular nucleus; Vsp, spinal vestibular nucleus; VZ, ventricular zone. Scale bars = 20µm for B, E–I, 200µm for C, K–N’.
Ascl1 is present broadly from dA3 to dB3 throughout rhombomeres 2–7 (Fig. 7E) (Sieber et al., 2007). Similar to neurogenesis in spinal cord, Ascl1 progenitors are generated in two distinct neurogenic phases; early in a stripe-like pattern (E10/11) or late in a salt-and-pepper like pattern (E12/13) (Sieber et al., 2007). Analysis of Ascl1-CreER™;R26R-stop-lacZ embryos harvested 24 hours after E12.5 tamoxifen induction shows rapid differentiation and migration of Ascl1 progenitors to the mantle zone, compared to the restricted expression of Ascl1 in the ventricular zone (Fig. 7B–C). Although Ascl1 is present broadly in the VZ of the lower rhombic lip, in Ascl1-CreER™ embryos, CreER™ is restricted to the dB3 population prior to E12.5 (Fig. 7E and K). This is evident by YFP from Ascl1-CreER™;R26R-stop-YFP overlap with Tlx3 and Lmx1b, but not Pax2 and Lhx1/5 after tamoxifen administration E10.5 and analysis of E11.5 embryos (Fig. 7F–I). Furthermore, after E12.5, the dBLb subset of the Ascl1 lineage is labeled (Fig. 7K’ and data not shown). Detection of only a subset of the Ascl1 lineage in the brainstem in this transgenic strain is consistent with results seen in the spinal cord where dI3 and dI5 population but not dI4 were preferentially marked (Battiste et al., 2007). This may reflect differences in level of expression of Ascl1 in these lineages. Nevertheless, the Ascl1 lineages mapped here represent the fate of progenitors from dB3 and dBLb.
Ascl1-CreER™R26R-stop-lacZ embryos were exposed to tamoxifen at either E10.5 or E12.5, and the Ascl1 lineages in brainstem nuclei were examined at E17.5/E18.5. Ascl1 lineage cells contribute to the trigeminal sensory nuclear complex ranging from the pons into the caudal medulla (Qian et al., 2002). Ascl1 dB3 and dBLb progenitors populate the spinal trigeminal nucleus (Sp5) consistent with previous studies of Lbx1 (Fig. 7L–N’) (Sieber et al., 2007). In addition, Ascl1 lineage cells include the primary sensory nucleus (Pr5), and other associated nuclei such as the parvicellular reticular nucleus (PCRtA), and the gigantocellular nucleus (Gi) (Fig. L–N’). Notably, since the mesencephalic trigeminal nucleus (Me5) in dorsal midbrain is also derived from the Ascl1 lineage, Ascl1 progenitors appear to preferentially contribute a functional network for the somatosensory relay system (Fig. 5F’). In contrast, Ascl1 progenitors in dB3 and dBLb are not fated to the six precerebellar nuclei (pontine gray, reticulotegmental, vestibular, lateral reticular, external cuneate, and inferior olivary nuclei) (Fig. L–N”). Thus, Ascl1-CreER™ transgenic mice have allowed us to determine the fate of a subpopulation of the Ascl1 progenitors in the developing brainstem.
Concluding Remarks
In this study, we describe the temporal fate map for Ascl1 lineages throughout the brain. A summary of the CNS structures that receive contribution from Ascl1 lineage cells is provided in Table 1. Neuronal progenitors marked by CreER™ in Ascl1-CreER™ are dynamic in that they are transitioning from proliferating progenitors to differentiating neurons. This is inferred from experiments where embryos were harvested within 24 hours of induction by tamoxifen and lineage marked cells were already found lateral to progenitor domains and express markers of differentiating markers. This is similar to what was seen in embryonic spinal cord and adult neurogenesis (Battiste et al., 2007; Kim et al., 2007), and is consistent with the interpretation that cells with high Ascl1 levels, as would be preferentially labeled in this paradigm, are differentiating. Indeed, overexpression of Ascl1 in chick neural tube induces progenitors to rapidly exit the cell cycle, move out of the VZ and express markers of neuronal differentiation (Nakada et al., 2004).
Table 1. Neural derivatives of Ascl1 lineage in CNS.
Neuron Oligodendrocyte but not Astrocyte (Battiste 2007, Parras 2007, Sugimori 2007&2008)
| Anatomical Structure | Cell Types/Nuclei | References | |
|---|---|---|---|
| Forebrain | Olfactory Bulb :(MOB and AOB) | ND | This study, Long 2007, Parras 2004 |
| Cortex | Interneurons in granule cell and glomerular layer | This study, Casarosa 1999 | |
| Basal Ganglia (Striatum, Amygdala) | Cortical interneurons | This study, Marin 2000 | |
| Hippocampus | Cholinergic or CR+ Interneurons | This study, Pleasure 2000 | |
| Preoptic Area | Pyramidal and granule neurons | This study | |
| Midbrain | Superior/Inferior Colliculus | Both glutamateric and GABAergic neurons | This study, Miyoshi 2004, Nakatani 2007 |
| Thalamus | Thalamic nuclei (Epi, Pre, Rt) | This study, Tuttle 2000, Vue 2007 | |
| Hypothalamus | POMC neurons | Mcnay 2006 | |
| Trigeminal sensory system | Me5 | This study | |
| Hindbrain | Cerebellum | Deep cerebellar nuclei interneurons, Purkinje cells | This study |
| Trigeminal sensory system | Spinal trigeminal nucleus (Sp5), associated nuclei (Pr, Gi, PCRtA) | This study, Qian 2002 | |
| Locus coeruleus | Noradrenergic neurons | This study, Hirsch 1998 | |
| Dorsal raphe nucleus | Serotonergic neurons | Pattyn 2004 | |
| Spinal Cord | dI3,5, v2, dILA&B neurons | Helms 2005, Li 2005, Wildner 2006 | |
AOB, accessory olfactory bulb; CR, calrethnin; Epi, epithalamus; Gi, gigantocellular reticular nucleus; Me5, mesencephalic trigeminal nucleus; MOB, main olfactory bulb; ND, not determined; PCRtA, parvicellular reticular nucleus; Pr, pontine reticular nucleus; Pre, pretectal thalamus; Rt, reticular thalamus.
Ascl1 progenitors give rise to diverse neuronal subtypes including GABAergic (Fode et al., 2000; Horton et al., 1999), glutamatergic (Helms et al., 2005), serotonergic (Pattyn et al., 2004), noradrenergic (Hirsch et al., 1998) and acetylcholinergic (Marin et al., 2000). This is in contrast to other transcription factor marked populations such as Atoh1 and Tlx3 which appear restricted to glutamatergic neurons, and Pax2 which is restricted to GABAergic lineages. Ascl1 does appear to function in neuronal specification but this is region dependent. For example, in early dorsal spinal cord, Ascl1 is required for dI3 and dI5 and overexpression results in excess dI3 and dI5 (Helms et al., 2005). In the second round of neurogenesis in the dorsal spinal cord, Ascl1 is important for generating normal numbers of the GABAergic dILA neurons (Mizuguchi et al., 2006; Wildner et al., 2006). Identifying interacting factors that work with Ascl1 will be important for understanding how it functions to generate neuronal diversity.
Ascl1 is present in progenitors to oligodendrocytes in the embryonic spinal cord, and in embryonic and adult brain. However, not all oligodendrocytes arise from Ascl1 expressing cells since the earliest oligodendrocytes arise in the ventral neural tube from non-Ascl1 cells (Lu et al., 2002; Sugimori et al., 2008; Zhou and Anderson, 2002). However, in each brain region examined, the late stage expression of Ascl1 is preferentially marking oligodendrocyte progenitors, not neuronal progenitors or astrocytes. The function of Ascl1 in oligodendrocyte development is just beginning to be uncovered. Oligodendrocytes in Ascl1 mutant mice fail to express the full complement of known markers such as Nkx2.2 (Sugimori et al., 2008).
To interpret in vivo lineage studies using inducible Cre recombinase in transgenic mice, it is important to understand the limitations of this paradigm. First, there is always the caveat that expression from the transgene is not 100% reliable due to position effects or lack of regulatory information. The BAC used to generate the transgenic mice includes over 100 kb of sequence both 5’ and 3’ flanking the Ascl1 coding region, increasing the likelihood that important transcription control regions are included. However, we know some Ascl1 lineages are not detected such as cells derived from the MGE in the telencephalon, and a subset of neurons in the spinal cord and brainstem. Second, the induction of Cre is not expected to be 100% efficient in deleting the STOP sequence from the reporter so it is not expected that 100% of the cells derived from Ascl1 will be marked. However, in a majority of cases, expression from this BAC transgenic line appears to reflect endogenous Ascl1 expression, and thus, identifying X-gal or YFP marked cells in a specific brain region provides strong support that these cells are derived from Ascl1-expressing progenitor cells.
EXPERIMENTAL METHODS
Transgenic mice and Tamoxifen injection
Ascl1-GIC, Ascl1-CreER™, R26R-stop-lacZ, and R26R-stop-YFP mice have been previously described. Briefly, Ascl1-GIC and Ascl1-CreER™ mice are BAC transgenic mice where GFP-IRES-Cre or CreER™ replaces the Ascl1 coding region, respectively (Battiste et al., 2007; Helms et al., 2005). R26R-stop-YFP (or lacZ) mice are Cre recombinase reporter mice (Soriano, 1999; Srinivas et al., 2001).
Tamoxifen induction of Cre recombinase was accomplished by intraperitoneal injection of pregnant females at a given day post coitum (dpc) twice with 6 hr interval (noon and 6 pm) with 50–75 mg/kg tamoxifen (Sigma, T55648) in sunflower oil. Embryonic or adult brains were harvested at the times specified after tamoxifen treatment.
X-gal staining and immunofluorescence staining
For X-gal staining, whole embryos (E9.5–13.5) or dissected brains (E15.5-P3) were fixed by immersion in 4% formaldehyde for 1–2 hours at room temperature. After washing in phosphate buffer, the tissues were incubated overnight in X-gal staining solution (1mg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal), 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 2mM MgCl2 in PBS and 0.02% NP-40). Tissues were washed in phosphate buffer, post-fixed with 4% formaldehyde overnight at 4 °C, and vibratome sectioned at 200 µm. Adult brains were dissected from the skull after animals were anesthetized with Avertin and perfused with 4% formaldehyde transcardially. The brains were post-fixed with 4% formaldehyde overnight at 4 °C, rinsed in phosphate buffer, and vibratome sectioned sagittally at 200 µm. Sections were mounted on slides and X-gal stained. Tissue sections were photographed using an Olympus SZX12 or Zeiss Discovery V12 microscope.
For immunofluorescence, tissues were fixed as above except for the late gestation embryos or neonates, which were ex-sanguinated with cold PBS before brains were removed and immersion fixed. The fixed tissues were rinsed in PBS, cryoprotected in 30% sucrose overnight at 4 °C, embedded frozen in OCT and cryosectioned at 30–50 µm. For immunofluorescence staining, free floating sections or sections mounted on slides were incubated in the appropriate dilution of primary antibody in PBS/3% donkey serum/0.2% NP-40, followed by appropriate secondary antibody conjugated with Alexa Fluor 488, 594, or 647 (Molecular Probes). Mouse monoclonal antibodies used were: GFAP (1:400, Sigma-Aldrich, G3893), NeuN (1:1000, Chemicon, MAB377), PDGFRα (1:200, BD Biosciences), Lhx1/5 (1:100, Developmental Studies Hybridoma Bank, 4F2), and APC (1:100, Oncogene Sciences, clone CC-1). Rabbit polyclonal antibodies used were: GFP (1:500, Molecular Probes, A6455), Sox2 (1:3000, Chemicon), Calbindin (1:1000, Swant), TH (1:2500, Chemicon), Atoh1 (1:100, (Helms and Johnson, 1998)), Lhx2/9 (1:8000, a gift from T. Jessell), Pax2 (1:500, Zymed), and Olig2 (1:2000, Chemicon). Chick GFP (1:500, Aves lab), goat β-gal (1:500, Biogenesis), and guinea pig Sox10 (1:2000, gift M. Wegner) were also used. Rabbit anti-Ptf1a (1:10,000), guinea pig anti-Brn3a (1:10,000) and anti-Ascl1 antibody (1:10,000) were generated for this study using bacterially produced recombinant GST-fusion proteins as antigens (GST-Ptf1a plasmid provided by H. Edlund, Umea University, Sweden, and GST-Brn3a from E. Turner, Univ. of California, San Diego). Fluorescence imaging was carried out on a BioRad MRC 1024 confocal microscope. For each experiment multiple sections from at least 3 different animals were analyzed.
Neuroanatomical analyses
Nomenclature of anatomical structures for the adult brain was assigned using The Mouse Brain in Stereotaxic Coordinates (Paxinos and Franklin, 2001) as a reference guide. For analysis of the embryonic brain, The Atlas of Mouse Development (Kaufman, 1992), Atlas of Prenatal Rat Brain Development (Altman and Bayer, 1995), and Chemoarchitectonic Atlas of the Developing Mouse Brain (Jacobowitz and Abbott, 1997) were used as guides.
ACKNOWLEDGEMENTS
We are grateful for the outstanding technical assistance from T. Savage, J. Dumas, and Dr. K. Hori with special technical tips provided by Dr. R. Storm. We thank Dr. S. Dymecki for providing expertise in confirming anatomical structures in the brainstem. We also appreciate the generous gifts from Drs. T. Jessell for the anti-Lhx2/9 antibody, M. Wegner for the anti-Sox10 antibody, H. Edlund for the GST-Ptf1a expression construct, and E. Turner for the GST-Brn3a expression construct, and the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa for providing monoclonal antibodies. This work was funded by NIH NS32817 to JEJ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Altman J, Bayer SA. Time of origin of neurons of the rat inferior colliculus and the relations between cytogenesis and tonotopic order in the auditory pathway. Experimental brain research. Experimentelle Hirnforschung. 1981a;42:411–423. doi: 10.1007/BF00237506. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Time of origin of neurons of the rat superior colliculus in relation to other components of the visual and visuomotor pathways. Experimental brain research. Experimentelle Hirnforschung. 1981b;42:424–434. doi: 10.1007/BF00237507. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Embryonic development of the rat cerebellum. II. Translocation and regional distribution of the deep neurons. The Journal of comparative neurology. 1985a;231:27–41. doi: 10.1002/cne.902310104. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Embryonic development of the rat cerebellum. III. Regional differences in the time of origin, migration, and settling of Purkinje cells. The Journal of comparative neurology. 1985b;231:42–65. doi: 10.1002/cne.902310105. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Embryonic development of the rat cerebellum: deliniation of the cerebellar primordium and early cell movements. J. Comp. Neurol. 1985c;231:1–26. doi: 10.1002/cne.902310103. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Atlas of prenatal rat brain development. Boca Raton: CRC Press; 1995. [Google Scholar]
- Battiste J, Helms AW, Kim EJ, Savage TK, Lagace DC, Mandyam CD, Eisch AJ, Miyoshi G, Johnson JE. Ascl1 defines sequentially generated lineage-restricted neuronal and oligodendrocyte precursor cells in the spinal cord. Development. 2007;134:285–293. doi: 10.1242/dev.02727. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat. Rev. Neuroscience. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- Chizhikov VV, Lindgren AG, Currle DS, Rose MF, Monuki ES, Millen KJ. The roof plate regulates cerebellar cell-type specification and proliferation. Development. 2006;133:2793–2804. doi: 10.1242/dev.02441. [DOI] [PubMed] [Google Scholar]
- Corbin JG, Nery S, Fishell G. Telencephalic cells take a tangent: non-radial migration in the mammalian forebrain. Nature neuroscience. 2001;4 Suppl:1177–1182. doi: 10.1038/nn749. [DOI] [PubMed] [Google Scholar]
- Dean P, Redgrave P, Westby GW. Event or emergency? Two response systems in the mammalian superior colliculus. Trends in neurosciences. 1989;12:137–147. doi: 10.1016/0166-2236(89)90052-0. [DOI] [PubMed] [Google Scholar]
- Del Barrio MG, Taveira-Marques R, Muroyama Y, Yuk DI, Li S, Wines-Samuelson M, Shen J, Smith HK, Xiang M, Rowitch D, Richardson WD. A regulatory network involving Foxn4, Mash1 and delta-like 4/Notch1 generates V2a and V2b spinal interneurons from a common progenitor pool. Development. 2007;134:3427–3436. doi: 10.1242/dev.005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink AJ, Englund C, Daza RA, Pham D, Lau C, Nivison M, Kowalczyk T, Hevner RF. Development of the deep cerebellar nuclei: transcription factors and cell migration from the rhombic lip. J Neurosci. 2006;26:3066–3076. doi: 10.1523/JNEUROSCI.5203-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Glasgow SM, Henke RM, Macdonald RJ, Wright CV, Johnson JE. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132:5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- Gokhan S, Marin-Husstege M, Yung SY, Fontanez D, Casaccia-Bonnefil P, Mehler MF. Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression. J Neurosci. 2005;25:8311–8321. doi: 10.1523/JNEUROSCI.1850-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME, Alder J, Zimmerman K, Heintz N. Genes involved in cerebellar cell specification and differentiation. Current opinion in neurobiology. 1997;7:40–47. doi: 10.1016/s0959-4388(97)80118-3. [DOI] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Helms AW, Battiste J, Henke RM, Nakada Y, Simplicio N, Guillemot F, Johnson JE. Sequential roles for Mash1 and Ngn2 in the generation of dorsal spinal cord interneurons. Development. 2005;132:2709–2719. doi: 10.1242/dev.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms AW, Johnson JE. Progenitors of dorsal commissural interneurons are defined by MATH1 expression. Development. 1998;125:919–925. doi: 10.1242/dev.125.5.919. [DOI] [PubMed] [Google Scholar]
- Hirsch MR, Tiveron MC, Guillemot F, Brunet JF, Goridis C. Control of noradrenergic differentiation and Phox2a expression by MASH1 in the central and peripheral nervous system. Development. 1998;125:599–608. doi: 10.1242/dev.125.4.599. [DOI] [PubMed] [Google Scholar]
- Horton S, Meredith A, Richardson JA, Johnson JE. Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor MASH1. Mol. Cell. Neurosci. 1999;14:355–369. doi: 10.1006/mcne.1999.0791. [DOI] [PubMed] [Google Scholar]
- Hoshino M, MNakamura S, Mori K, Kawauchi T, Terao M, Nishimura Y, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, Terashima T, Wright CV, Kawaguchi Y, Nakao K, Nabeshima Y. Ptf1a, a bHLH Transcriptional Gene, Defines GABAergic Neuronal Fates in Cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Jacobowitz DM, Abbott LC. Chemoarchitectonic atlas of the developing mouse brain. Boca Raton, FL: CRC Press; 1997. [Google Scholar]
- Kaufman MH. The atlas of mouse development. San Diego: Academic Press; 1992. [Google Scholar]
- Kim EJ, Leung CT, Reed RR, Johnson JE. In vivo analysis of Ascl1 defined progenitors reveals distinct developmental dynamics during adult neurogenesis and gliogenesis. J Neurosci. 2007;27:12764–12774. doi: 10.1523/JNEUROSCI.3178-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg RL, Awatramani RB, Hunter NL, Farago AF, DiPietrantonio HJ, Rodriguez CI, Dymecki SM. Hindbrain rhombic lip is comprised of discrete progenitor cell populations allocated by Pax6. Neuron. 2005;48:933–947. doi: 10.1016/j.neuron.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Leto K, Carletti B, Williams IM, Magrassi L, Rossi F. Different types of cerebellar GABAergic interneurons originate from a common pool of multipotent progenitor cells. J Neurosci. 2006;26:11682–11694. doi: 10.1523/JNEUROSCI.3656-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Garel S, Alvarez-Dolado M, Yoshikawa K, Osumi N, Alvarez-Buylla A, Rubenstein JL. Dlx-dependent and -independent regulation of olfactory bulb interneuron differentiation. J Neurosci. 2007;27:3230–3243. doi: 10.1523/JNEUROSCI.5265-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Yoshida M, Grove EA. The derivatives of the Wnt3a lineage in the central nervous system. The Journal of comparative neurology. 2007;504:550–569. doi: 10.1002/cne.21461. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Maricich SM, Herrup K. Pax-2 expression defines a subset of GABAergic interneurons and their precursors in the developing murine cerebellum. Journal of neurobiology. 1999;41:281–294. doi: 10.1002/(sici)1097-4695(19991105)41:2<281::aid-neu10>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Rubenstein JL. A long, remarkable journey: tangential migration in the telencephalon. Nature reviews. 2001;2:780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- McConnell SK. Constructing the cerebral cortex: neurogenesis and fate determination. Neuron. 1995;15:761–768. doi: 10.1016/0896-6273(95)90168-x. [DOI] [PubMed] [Google Scholar]
- Miller MW. Cogeneration of retrogradely labeled corticocortical projection and GABA-immunoreactive local circuit neurons in cerebral cortex. Brain research. 1985;355:187–192. doi: 10.1016/0165-3806(85)90040-9. [DOI] [PubMed] [Google Scholar]
- Miller MW, Nowakowski RS. Use of bromodeoxyuridine-immunohistochemistry to examine the proliferation, migration and time of origin of cells in the central nervous system. Brain research. 1988;457:44–52. doi: 10.1016/0006-8993(88)90055-8. [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Bessho Y, Yamada S, Kageyama R. Identification of a novel basic helix-loop-helix gene, Heslike, and its role in GABAergic neurogenesis. J Neurosci. 2004;24:3672–3682. doi: 10.1523/JNEUROSCI.5327-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi R, Kriks S, Cordes R, Gossler A, Ma Q, Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nature neuroscience. 2006;9:770–778. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nature reviews. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Nakada Y, Hunsaker TL, Henke RM, Johnson JE. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus cell-type specification. Development. 2004;131:1319–1330. doi: 10.1242/dev.01008. [DOI] [PubMed] [Google Scholar]
- Nakatani T, Minaki Y, Kumai M, Ono Y. Helt determines GABAergic over glutamatergic neuronal fate by repressing Ngn genes in the developing mesencephalon. Development. 2007;134:2783–2793. doi: 10.1242/dev.02870. [DOI] [PubMed] [Google Scholar]
- Parras CM, Galli R, Britz O, Soares S, Galichet C, Battiste J, Johnson JE, Nakafuku M, Vescovi A, Guillemot F. Mash1 specifies neurons and oligodendrocytes in the postnatal brain. The EMBO journal. 2004;23:4495–4505. doi: 10.1038/sj.emboj.7600447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras CM, Hunt C, Sugimori M, Nakafuku M, Rowitch D, Guillemot F. The proneural gene Mash1 specifies an early population of telencephalic oligodendrocytes. J Neurosci. 2007;27:4233–4242. doi: 10.1523/JNEUROSCI.0126-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras CM, Schuurmans C, Scardigli R, Kim J, Anderson DJ, Guillemot F. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes & Development. 2002;16:324–338. doi: 10.1101/gad.940902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Simplicio N, van Doorninck JH, Goridis C, Guillemot F, Brunet JF. Ascl1/Mash1 is required for the development of central serotonergic neurons. Nature neuroscience. 2004;7:589–595. doi: 10.1038/nn1247. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. Second ed. San Diego: Academic Press; 2001. [Google Scholar]
- Peng CY, Yajima H, Burns CE, Zon LI, Sisodia SS, Pfaff SL, Sharma K. Notch and MAML signaling drives Scl-dependent interneuron diversity in the spinal cord. Neuron. 2007;53:813–827. doi: 10.1016/j.neuron.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez SE, Rebelo S, Anderson DJ. Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Development. 1999;126:1715–1728. doi: 10.1242/dev.126.8.1715. [DOI] [PubMed] [Google Scholar]
- Qian Y, Shirasawa S, Chen C, Cheng L, Ma Q. Proper development of relay somatic sensory neurons and D2/D4 interneurons requires homeobox genes Rnx/Tlx3and Tlx1. Genes & Development. 2002;16:1220–1233. doi: 10.1101/gad.982802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CI, Dymecki SM. Origin of the precerebellar system. Neuron. 2000;27:475–486. doi: 10.1016/s0896-6273(00)00059-3. [DOI] [PubMed] [Google Scholar]
- Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin LM, Seibt J, Tang H, Cunningham JM, Dyck R, Walsh C, Campbell K, Polleux F, Guillemot F. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. The EMBO journal. 2004;23:2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber MA, Storm R, Martinez-de-la-Torre M, Muller T, Wende H, Reuter K, Vasyutina E, Birchmeier C. Lbx1 acts as a selector gene in the fate determination of somatosensory and viscerosensory relay neurons in the hindbrain. J Neurosci. 2007;27:4902–4909. doi: 10.1523/JNEUROSCI.0717-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genetics. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimori M, Nagao M, Bertrand N, Parras CM, Guillemot F, Nakafuku M. Combinatorial actions of patterning and HLH transcription factors in the spatiotemporal control of neurogenesis and gliogenesis in the developing spinal cord. Development. 2007;134:1617–1629. doi: 10.1242/dev.001255. [DOI] [PubMed] [Google Scholar]
- Sugimori M, Nagao M, Parras CM, Nakatani H, Lebel M, Guillemot F, Nakafuku M. Ascl1 is required for oligodendrocyte development in the spinal cord. Development. 2008;135:1271–1281. doi: 10.1242/dev.015370. [DOI] [PubMed] [Google Scholar]
- Tan SS, Valcanis H, Kalloniatis M, Harvey A. Cellular dispersion patterns and phenotypes in the developing mouse superior colliculus. Dev Biol. 2002;241:117–131. doi: 10.1006/dbio.2001.0505. [DOI] [PubMed] [Google Scholar]
- Vue TY, Aaker J, Taniguchi A, Kazemzadeh C, Skidmore JM, Martin DM, Martin JF, Treier M, Nakagawa Y. Characterization of progenitor domains in the developing mouse thalamus. The Journal of comparative neurology. 2007;505:73–91. doi: 10.1002/cne.21467. [DOI] [PubMed] [Google Scholar]
- Wang VY, Zoghbi HY. Genetic regulation of cerebellar development. Nature reviews. 2001;2:484–491. doi: 10.1038/35081558. [DOI] [PubMed] [Google Scholar]
- Wildner H, Muller T, Cho SH, Brohl D, Cepko CL, Guillemot F, Birchmeier C. dILA neurons in the dorsal spinal cord are the product of terminal and non-terminal asymmetric progenitor cell divisions, and require Mash1 for their development. Development. 2006;133:2105–2113. doi: 10.1242/dev.02345. [DOI] [PubMed] [Google Scholar]
- Yamada M, Terao M, Terashima T, Fujiyama T, Kawaguchi Y, Nabeshima Y, Hoshino M. Origin of climbing fiber neurons and their developmental dependence on Ptf1a. J Neurosci. 2007;27:10924–10934. doi: 10.1523/JNEUROSCI.1423-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]