Abstract
Thioredoxin reductase and thioredoxin constitute the cellular thioredoxin system, which provides reducing equivalents to numerous intracellular target disulfides. Mammalian thioredoxin reductase contains the rare amino acid selenocysteine. Known as the “21st” amino acid, selenocysteine is inserted into proteins by recoding UGA stop codons. Some model eukaryotic organisms lack the ability to insert selenocysteine, and prokaryotes have a different recoding apparatus than do eukaryotes, thus making heterologous expression of mammalian selenoproteins difficult. Here, we present a semisynthetic method for preparing mammalian thioredoxin reductase. This method produces the first 487 amino acids of mouse thioredoxin reductase-3 as an intein fusion protein in Escherichia coli cells. The missing C-terminal tripeptide containing selenocysteine is then ligated to the thioester-tagged protein by expressed protein ligation. The semisynthetic version of thioredoxin reductase that we produce in this manner has kcat values ranging from 1500 min−1 to 2220 min−1 towards thioredoxin and has strong peroxidase activity, indicating a functional form of the enzyme. We produced the semisynthetic thioredoxin reductase with a total yield of 24 mg from 6 liters of E. coli culture (4 mg/L). This method allows production of a fully functional, semisynthetic selenoenzyme that is amenable to structure-function studies. A second semisynthetic system is also reported that makes use of peptide complementation to produce a partially active enzyme. The results of our peptide complementation studies reveal that a tetrapeptide that cannot ligate to the enzyme (Ac-Gly-Cys-Sec-Gly) can form a non-covalent complex with the truncated enzyme to form a weak complex. This non-covalent peptide/enzyme complex has 350–500 fold lower activity than the semisynthetic enzyme produced by peptide ligation.
Mammalian thioredoxin reductase (TR1) is a pyridine nucleotide-disulfide oxidoreductase that functions in vivo to reduce the catalytic disulfide bond of thioredoxin. Thioredoxin reductase, together with thioredoxin (Trx) and Trx peroxidase, make up the thioredoxin system. This system is essential for cell viability, providing essential reducing equivalents to a number of important cellular targets such as ribonucleotide reductase (1–3).
Mammalian TRs contain the rare amino acid selenocysteine (Sec), the so-called “21st” amino acid in protein (4–5). This amino acid has been found in 25 human proteins and is present in all three domains of life (6). The codon that directs incorporation of Sec into a protein chain is the UGA codon, which normally serves as a termination signal. In the cases in which UGA directs the incorporation of Sec, a specific stem-loop structure is present in the mRNA. This RNA structure, known as a selenocysteine insertion sequence (SECIS) element, is responsible for the recoding of the UGA codon as a sense codon for Sec by recruiting protein factors necessary to bind the specialized tRNA(Ser)(Sec) needed to direct incorporation of Sec (7–9). In prokaryotes such as E. coli, the SECIS element is proximal to the UGA codon, whereas in eukaryotes the SECIS element is located in the 3′ untranslated region of the mRNA (10–12). In addition, there are sequence and structural differences between the two types of SECIS elements (13). These distinctions make heterologous expression of mammalian selenoproteins in E. coli highly problematic. There have been attempts at engineering eukaryotic mRNAs to contain a prokaryotic SECIS element such that Sec can be incorporated into a heterologously expressed protein in E. coli cells (14–15). This method is difficult to implement since prokaryotic SECIS elements also contain coding information for amino acids. Thus, a eukaryotic selenoprotein expressed in E. coli would be mutated because the SECIS structure, as part of the mRNA, must be maintained to decode the UGA codon as Sec. Such an approach has been used for glutathione-S-transferase, in which the catalytic Cys residue was replaced by a Sec residue by engineering the mRNA to contain a SECIS element. The result was a protein containing not only the desired Sec residue, but also a mutation in the region downstream of Sec because of the requirement of the in-frame SECIS element (14). In addition, 85% of the produced protein was present in the truncated form that terminated at the UGA codon, despite the use of a specialized bacterial strain to produce the needed accessory proteins for the biosynthesis of selenocysteine and selenocysteine-tRNA.
However in TR, the placement of Sec as the penultimate amino acid makes it possible to produce a recombinant TR in E. coli by fusing a bacterial SECIS element to the 3′ end of the gene. Such a strategy was used to successfully produce a recombinant TR by Arnér and coworkers (16). This same group was able to optimize conditions for production of TR in E. coli such that 40 mg of rat TR1 containing 50% selenocysteine was produced per liter of bacterial culture (17). One complication of this method is reported by Sandman & Noren, who demonstrated that the efficiency of Sec insertion is influenced by the nucleotide immediately downstream of the UGA codon (18). The results showed that in E. coli, tryptophan (Trp) insertion competes with Sec insertion when a purine base is immediately downstream of the UGA codon. This is the situation that exists in mammalian TR’s with the conserved peptide sequence of Gly-Cys-Sec-Gly (glycine begins with the purine guanine). Thus, TR produced by this method would contain some tryptophan contamination as the penultimate amino acid. This problem unique to bacterial expression of mammalian TR, was partially overcome by Arnér and coworkers, who developed a “Sel-tag” system to further purify TR. This system uses the high affinity of a phenylarsine oxide ligand for the reduced form of the GCUG tetrapeptide to purify the recombinant TR containing selenocysteine. This method enriches the selenium content of the TR preparation produced in E. coli (19).
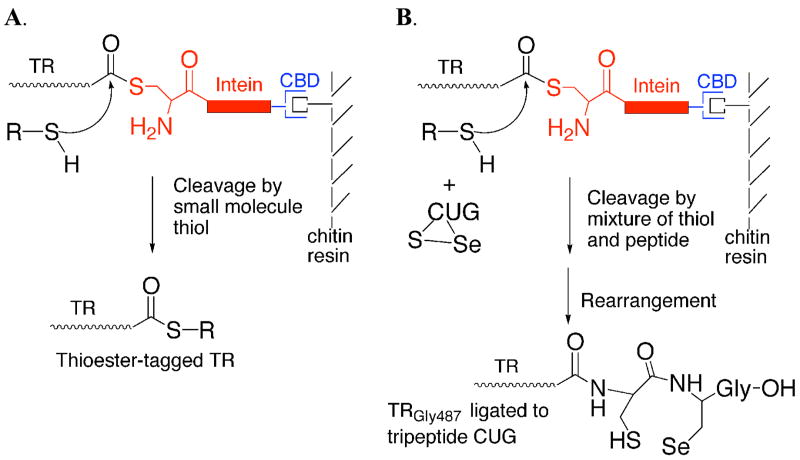
Here, we report the development of an alternative method for producing an active mammalian TR containing selenocysteine. This method uses intein-mediated peptide ligation to create a semisynthetic thioredoxin reductase enzyme. Thioredoxin reductase contains selenocysteine in a C-terminal tripeptide of sequence Cys-Sec-Gly, where the Sec residue is the penultimate amino acid (20). The location of the Sec residue in the C-terminus makes TR an ideal candidate for semisynthesis. Our strategy for producing a semisynthetic TR is shown in Figure 1.
Figure 1.
(A) Thiol-mediated cleavage of mTR-3-intein fusion construct by addition of exogenous thiol. The junction between mTR-3 and the intein exists in equilibrium between amide and thioester forms. Addition of exogenous thiol causes cleavage of the target protein from the thioester form of the fusion protein. (B) Mechanism of incorporation of tripeptide CUG to mTR-3-intein fusion protein. The ligation is done by adding oxidized tripeptide to the cleavage buffer containing 70 mM thiol. The excess thiol both reduces the tripeptide and causes cleavage of mTR-3 from the intein fusion. The reduced peptide can then attack the thioester-tagged protein and become stably ligated to the protein because of the presence of the N-terminal amino group which enables rapid rearrangement to the amide form of the protein.
The semisynthetic system reported in this paper will allow molecular details of the catalytic cycle to be studied that are not possible with biophysical techniques such as X-ray crystallography. For example, the crystal structure of a cysteine mutant form of rat TR1 has been reported (21). However, this structure reported electron density for the enzyme containing a reduced GCCG motif at the C-terminus. No information is available about the 8-membered ring structure that results as the enzyme cycles between reduced and oxidized forms. The crystal structure of the mouse TR3 (mTR-3) enzyme has also been recently reported to 2.6 Å resolution (22). However, no electron density could be observed for the last four residues of mTR-3 because of very high conformational flexibility of this C-terminal tail. As a result, the redox-active tetrapeptide had to be modeled into the active site. Without empirical data about the conformation of this tetrapeptide (and the resulting 8-membered ring in the oxidized state), no firm conclusions can be drawn about catalytic details of the enzyme mechanism. While the method developed by Arnér and coworkers clearly allows for production of the recombinant TR enzyme in high yield, our semisynthetic approach allows for structure-function analysis of the C-terminus to be performed by NMR spectroscopy with site-specific spectroscopic probes such as 77Se, 13C, and 15N. A number of other modifications to include non-standard amino acids would also be possible and are unique to this system. Thus the system reported here will allow for the elucidation of mechanistic details that are currently not known and could not be obtained by currently available methods.
In the course of developing this method, we also discovered that we could create a semisynthetic TR by using a technique called “peptide complementation”. In such a system, a truncated enzyme missing part of its active site, can be restored to full or partial activity by exogenous addition of a synthetic peptide that “complements” the active site with the missing amino acids. The most prominent example of peptide complementation is the S-protein/S-peptide system developed by Richards (23). In this system, RNase A can be cleaved between Ala20 and Ser21 by subtilisin to produce two peptide fragments. Separately, these two peptides have no ribonuclease activity. When combined together in a non-covalent complex, the two peptide segments combine to form an enzyme with 100% activity. This type of peptide or fragment complementation system was the first system in which “mutants” of the wild type enzyme could be tested in an in vitro assay system, as it allowed for a number of amino acids to be substituted for in the wild type enzyme by direct replacement of the amino acid via peptide synthesis (24). A number of other examples of peptide complementation are known in the literature including S. aureus nuclease, soybean trypsin inhibitor, and thioredoxin (25–27). A review detailing the historical development of peptide complementation as “semisynthesis” has been published by Offord2 (28).
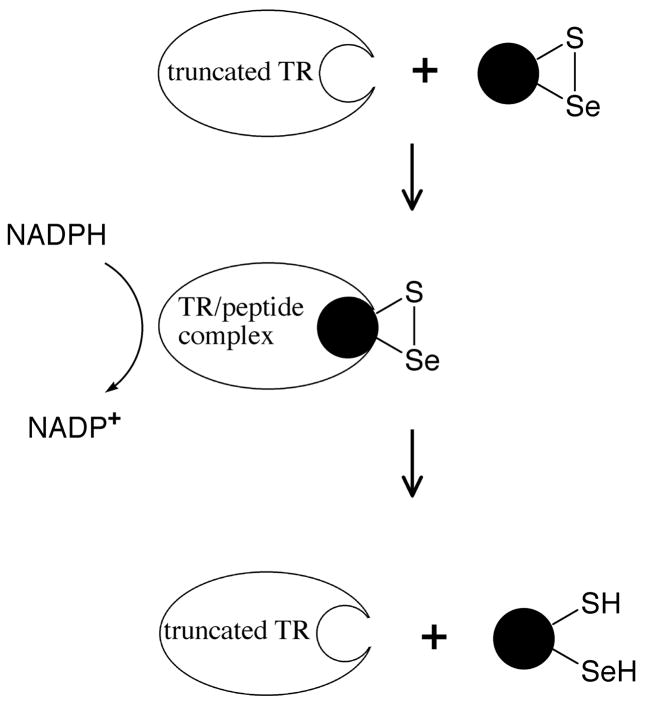
In our peptide complementation system, a synthetic peptide containing Sec (but which cannot be ligated to the thioester-tagged TR) is added to a truncated form of the enzyme to restore partial activity. This type of approach is shown in Figure 2. As detailed herein, the enzyme produced from the peptide complementation approach has much lower activity than the semisynthetic enzyme in which successful ligation of tripeptide Cys-Sec-Gly has occurred.
Figure 2.
Peptide complementation to create a transient-semisynthetic TR. Oxidized peptides containing selenocysteine can serve as substrates for the truncated enzyme because they fit into the active site of the enzyme and are able to be reduced by the N-terminal dithiol (Figure 6). This system is unlike the S-protein/S-peptide system because the peptide binds weakly to the active site. For simplicity, the N-terminal redox-active disulfide and the other subunit of the TR dimer is not shown.
MATERIALS AND METHODS
Materials
Solvents for peptide synthesis were purchased from EMD Biosciences (San Diego, CA). Fmoc amino acids were purchased from Synpep Corp (Dublin, CA). Resins for solid-phase synthesis were purchased from Novabiochem (San Diego, CA). All other chemicals were purchased from either Sigma-Aldrich or Fisher Scientific. Restriction endonucleases Nco I, Sap I, Hind III, Eco RI, Vent DNA Polymerase, T4 DNA Ligase and reaction buffers were all supplied by New England Biolabs (Beverley, MA). Recombinant E. coli thioredoxin was purchased from American Diagnostica (Greenwich, CT). Enzyme kinetics were performed on a Cary50 UV/Vis Spectrophotometer.
Peptide Synthesis
Fmoc-selenocysteine(pMob)-OH was synthesized as reported previously (29–30). This derivative of Sec was then used to make tripeptide Cys-Sec-Gly (CUG)3 or tetrapeptide Ac-GCUG using solid phase peptide synthesis as described below. The tripeptide was synthesized on a 0.25-mmol scale (three batches were combined for a total scale of 0.75 mmol). The peptide was synthesized using a glass vessel that was shaken by a Model 75 Burrell wrist action shaker. Each batch used 1.39 g of Fmoc-Gly-PEG-PS resin (Applied Biosystems, 0.18 g/mol). Deprotection of the Nα-Fmoc protecting group was carried out using three 10-minute agitations with 20% piperdine/80% DMF, fully covering the resin and rinsing between each deblock addition twice with DMF. Success of the deblocking was monitored qualitatively using a ninhydrin test (31). Couplings were performed using a 4-fold excess of Fmoc-amino acid, HOBt, HATU, and trimethylpyridine (TMP) (used as the base). Preactivation of the amino acid was not performed in order to minimize the racemization of L-cysteine and L-selenocysteine. These coupling conditions are essentially the same as those of Barany and coworkers (32), which were likewise used to minimize racemization of cysteine. The activated amino acid was added directly to the deblocked resin and couplings were judged complete by ninhydrin testing after allowing the reaction to proceed for 1 h. The removal of the final Nα-Fmoc protecting group completed the peptide synthesis. The final yield of the crude, Sec(Mob) protected tripeptide was 235 mg (70% yield). Tetrapeptide Ac-GCUG was synthesized in the same manner except for the addition of an acetylation step. The cleavage of the peptide off of the resin support, along with deprotection of the side-chain protecting groups, was done in a 2 h reaction using 95% TFA/2.5% EDT/2.5% water. The cleavage filtrate was pipetted into cold, anhydrous diethyl ether, where the peptide was observed to precipitate. Centrifugation at 3000 rpm for 10 min pelleted the peptide, which was followed by 2 additional wash/precipitation cycles. Peptides were dissolved in a minimal amount of water and then lyophilized. The p-methoxybenzyl group of selenocysteine was then removed by dissolving the lyophilized peptides in a cleavage cocktail containing m-cresol/thioanisole/TFA/TMSOTf in a ratio of 50:120:690:194 for 1 h at room temperature as previously described (33). The peptides were then precipitated again in cold ether and lyophilized. The tripeptide was used without further purification while peptide Ac-GCUG was purified using preparative HPLC.
Mutagenesis and Transformation
DNA primers were purchased from Integrated DNA Technologies Inc. (Coralville, IA) for the mutation of the Sec residue (encoded by TGA) at position 489 to cysteine (encoded by TGT). The template used was plasmid pTR3, which contained the full-length sequence of the mouse thioredoxin reductase-3 gene (accession #AF171053). Mutagenesis was performed by PCR on the GeneAmp PCR System 2400 by Perkin Elmer Life Sciences Inc. (Boston, MA) using Vent DNA polymerase. The PCR product was purified using the QIAquick Purification Kit from Qiagen (Valencia, CA) according to product instructions. The PCR product containing the mutated sequence was inserted into plasmid pTYB3 (New England Biolabs) by restriction digest with Nco I and Sap I for 2 h at 37 °C followed by purification with a Qiagen spin-column, and ligation with T4 DNA Ligase at 16 °C for 16 h. The ligase was heat inactivated at 70 °C for 30 min, followed by incubation at 37 °C with EcoR I. EcoR I only cuts the plasmid that has not been ligated with the PCR insert, thus enriching the pool of positive clones. The resulting plasmid (pTYB3TR) containing the gene for ampicillin resistance and a T7 promoter, was used to produce the thioredoxin reductase-intein-chitin binding domain fusion protein in E coli ER2566 cells.
E. coli DH5α cells, purchased from Stratagene (La Jolla, CA), were transformed with pTYB3TR by addition of 100 ng of plasmid to 200 μL of cells and incubation on ice for 30 min. The cells were heat shocked at 37 °C for 2.5 min and placed back on ice for 5 min. A 1-mL aliquot of LB broth was added to the cells and the mixture incubated at 37 °C for 60 min. The cells were then plated onto LB-ampicillin agar and incubated overnight. Colonies were then picked and used to inoculate 3-mL of 2xYT media containing 0.2 mg/mL ampicillin and grown in a shaking incubator for 24 h at 37 °C. The plasmid was purified from culture using the Perfectprep Plasmid Mini Kit from Eppendorf (Hamburg, Germany). Transformants were screened by restriction analysis and then analyzed by agarose gel electrophoresis. Positive clones were then sequenced to verify the complete coding sequence of mTR-3.
Thioredoxin Reductase Expression and Cleavage from Chitin Resin
The full-length U489C mutant as well as a truncated version of mTR-3 that was missing the C-terminal tripeptide CUG, were expressed in E. coli ER2566 cells (New England Biolabs). The cell culture (6 L) contained 0.2 mg/mL ampicillin, and cells were grown at 25 °C until an O.D. of 0.6 at 600 nm was achieved. Expression was then induced for 6 h by the addition of IPTG to a final concentration of 0.5 mM. Cells were harvested by centrifugation and frozen at −20 °C overnight. The cell pellet was homogenized with 300 mL of buffer containing 150 mM sodium chloride (NaCl) in 50 mM 3-(N-Morpholino) Propane-Sulfonic Acid (MOPS) at pH 7.0 and probe-sonicated on ice for 20 min. The suspension was then centrifuged at 9000 rpm in a Beckman J21B preparative centrifuge (JA-14 rotor) Beckman Coulter (Fullerton, CA).
The supernatant was gravity-loaded onto a column containing chitin resin (New England Biolabs) with a bed volume of 50 mL, and the column washed with MOPS/NaCl until the absorbance at 280 nm of the eluant dropped to below 0.1. For the truncated product and the full-length U489C mutant, the column was then equilibrated with cleavage buffer of composition 50 mM MOPS, 150 mM NaCl, and 70 mM N-methyl-mercaptoacetamide (NMA) or βME at pH 7.2. The resin-bound TR-intein fusion protein was then incubated with this buffer at 4 °C overnight.
Ligation of CUG peptide to Truncated Construct
The ligation of the synthetic CUG peptide to the truncated enzyme was performed with concomitant cleavage from the chitin resin. The resin-bound truncated construct was cleaved from the chitin resin with cleavage buffer containing 12 mM peptide (305 μmol − 100 mg) added directly to the chitin column. Based on typical yields of protein, this represents a 3000 to 1 ratio of peptide to protein. For a more optimized procedure with respect to ligation efficiency, the same amount of peptide (in cleavage buffer) was added to the chitin column in which only half the total amount of protein (supernatant from 3 L of cell culture instead of 6 L) was loaded onto the column.
Final Purification of Enzymes
The protein was batch eluted from the chitin resin with 1–3 column volumes of the cleavage buffer, depending on preparation scale. The ligated product was dialyzed extensively with the MOPS/NaCl buffer at pH 7.0 with a molecular weight cutoff of 6000 daltons for removal of any unligated peptide. A separate control experiment was performed in which the truncated form of mTR-3 was isolated in the same manner as the semisynthetic TR. Each step in the purification of the truncated mTR-3 was the same except that tripeptide CUG was not added to the chitin column.
The pooled protein fractions were prepared for the next step in the purification by the addition of Tris to 50 mM and ammonium sulfate to 1.0 M, and the pH was adjusted to 7.5. Samples were then gravity-loaded onto a column containing Fast Flow 6 Low Substitution Phenyl Sepharose from Pharmacia-Amersham Biosciences (Uppsala, Sweden) with a bed volume of 60 mL. Protein was eluted from the column with a 400-mL gradient of ammonium sulfate (1.0 M- 0 M) with collection of 4-mL fractions. Fractions were analyzed by determining A280 and further analyzed by 12% SDS-PAGE. Fractions containing mTR-3 were pooled, dialyzed against 10 mM potassium phosphate/10 mM NaCl at pH 7.8, and gravity-loaded onto DEAE Sephacel from Sigma Diagnostics (St. Louis, MO) with a bed volume of 60 mL. The protein was eluted with a 400-mL linear gradient of NaCl (10–300 mM), collecting 4-mL fractions. Fractions containing mTR-3 were verified by SDS-PAGE, pooled, dialyzed against 100 mM potassium phosphate pH 7.4/150 mM NaCl, and concentrated using Centriprep 10 concentrators from Millipore (Bedford, MA). The purification process was monitored on 12% Tris-Glycine SDS-PAGE with Novex pre-cast gels from Invitrogen (Carlsbad, CA) and stained as previously described (34).
Production of Recombinant TR in E. coli cells
Recombinant mTR3 (Turanov and Gladyshev, manuscript in preparation) was prepared to serve as control for the semisynthetic mTR3. Briefly, bacterial expression construct pET-28a-mTR3 was coexpressed in E. coli BL21(DE3) cells with the pSUABC plasmid (17) in the presence of 2 μM sodium selenite. The mTR3 construct contained an E. coli fdhH SECIS element immediately downstream of the stop signal and coded for an N-terminal His-tag. Cells were grown overnight at 30 °C and harvested by centrifugation. To purify TR3, bacterial cells were sonicated in the solution containing 50 mM sodium phosphate, 150 mM NaCl, pH 8.0, and Protease Inhibitor Cocktail EDTA-free (Roche). The soluble fraction was then fractionated on a TALONTM column (BD Biosciences Clontech) using a linear gradient of 20–200 mM imidazole. Fractions containing mTR3 were dialyzed against PBS, pH 7.3, and further isolated using an ADP-Sepharose (Amersham) column. The bound proteins were eluted with 1 M NaCl in PBS, TR3-containnig fractions were dialyzed, and the N-terminal His-tag was removed using His-tag Cleavage Kit (Novagen).
Determination of Selenium Content of the Semisynthetic TR
Semisynthetic TR was analyzed by inductively coupled plasma-mass spectrometry (ICP-MS) to determine the selenium content of the enzyme. The TR samples were in phosphate-buffered saline, and the corresponding buffer was also analyzed in parallel as a control. The selenium content (in ppm) was then recalculated to the molar amount of the analyzed protein.
Enzyme Activity Assays
The activity of the recombinant protein was evaluated as reduction of thioredoxin with NADPH and monitored by decreasing absorbance at 340 nm as previously described (35). Activity was also evaluated by reduction of hydrogen peroxide in the presence of NADPH and monitored at 340 nm (35). Data were linear over a 2-min range after collection on a Cary 50 spectrophotometer from Varian (Walnut Creek, CA). Peptide Ac-GCUG was also assayed as a substrate under similar conditions as the thioredoxin assay, but in addition, the assay was performed in the absence of thioredoxin and insulin, as well as several permutations as indicated in the Results. Kinetic constants were determined from a plot of vo vs. substrate concentration and curve fitting the data to the Michaelis-Menten equation.
“Off-Resin” Ligation Efficiency
The cell extract from 6 L of E. coli cells was loaded onto a chitin-agarose column as described above. After extensive washing with column buffer, the resin was treated with cleavage buffer to elute the protein. The eluate was concentrated as before and then dialyzed against 50 mM MOPS buffer, pH 7.0 with 150 mM NaCl to remove excess NMA. The concentrated protein sample was then divided into two aliquots. The tripeptide CUG was added to each aliquot at a concentration of 5 mM. One aliquot received NMA while a second aliquot (which served as a negative control) received only buffer. The samples were allowed to incubate at room temperature for 4 h, after which they were dialyzed against 50 mM potassium phosphate buffer, pH 7.4, containing 150 mM NaCl and 1 mM EDTA to remove excess peptide and reagent. After dialysis the samples were assayed using 90 μM thioredoxin to determine a specific activity.
Comparison of the Activity of Semisynthetic TR to Recombinant TR
Specific catalytic activities were determined using the insulin assay as described previously (36). Briefly, 500 μL reaction mixtures containing 3 μM semisynthetic TR or the corresponding mouse recombinant TR3 were used. As substrates, 5 μM E. coli Trx or recombinant rat mitochondrial Trx2 were used. Progress in the reaction was followed by the decrease in absorbance at 340 nm, resulting from the oxidation of NADPH.
Mass Determination of Truncated and Semisynthetic TR’s
The enzymes were concentrated to 1 mg/mL in 10 mM ammonium bicarbonate buffer pH 8.0. The protein samples were then spotted on a 100-well plate at various dilutions. A solution of sinnapinic acid at 10 mg/mL was overlaid onto the protein spots and the samples were left to air dry. A Voyager-DETM MALDI-TOF mass spectrometer in the linear mode was then used to analyze the samples. The instrument was externally calibrated with BSA.
Peptide Mass Mapping of Tryptic Fragments
Purified thioredoxin reductase (truncated or semisynthetic) at a concentration of 0.3 mg/mL, was digested using 10 μg/mL sequencing grade trypsin (Promega) in 150 mM ammonium bicarbonate at 37 °C for 16 h. The digested sample was stored frozen until mass spectrometric analysis. Samples for MALDI-TOF were prepared by serial dilution in 10 mg/mL matrix, 2,5-Dihydroxybenzoic Acid (DHB) or α-Cyano-4-Hydroxycinnamic Acid (CHCA), with 50% acetonitrile/0.05% TFA as a diluent. Samples were applied to the MALDI plate as 1-μL spots and dried under vacuum.
A Voyager-DETM PRO Workstation (Applied Biosystems) was used to analyze MALDI-TOF samples. Positive ion spectra (mass + H+) were collected in reflector mode with an accelerating voltage of 20,000 and a delayed extraction time of 175 nsec. Data was collected as the accumulated spectra containing a minimum of 10 spectra at 50 laser shots per spectrum. Monoisotopic masses were corrected by external-calibration using Applied Biosystems Calibration Mixture 2; Angiotensin I (1296.6853), ACTH clip 1–17 (2093.0867), clip 18–39 (2465.1989) and clip 7–38 (3657.9294) prepared in the equivalent matrix. The resulting (mass + H+) data were compared to a theoretical digest generated using the ExPASy Peptide Mass Proteomics tool.
Digested samples were also analyzed by LC/MS using a Thermo-Finnigan LCQ Deca XP Plus liquid chromatograph ion trap mass spectrometry with peaks subjected to MS/MS fragmentation sequencing by collision-induced dissociation. Full MS of parent ions and MS/MS fragmentation data were processed using the Turbo-SEQUEST database for peptide identification. The (mass + H+) of the semisynthetic C-terminal tryptic fragment (SGLEPTVTGCUG), molecular formula Se1C44H73N12O18S1 representing the oxidized form, was compared to the theoretical isotope pattern generated using MS-Isotope from USCF ProteinProspector v4.0.5.
RESULTS
Expression and Purification
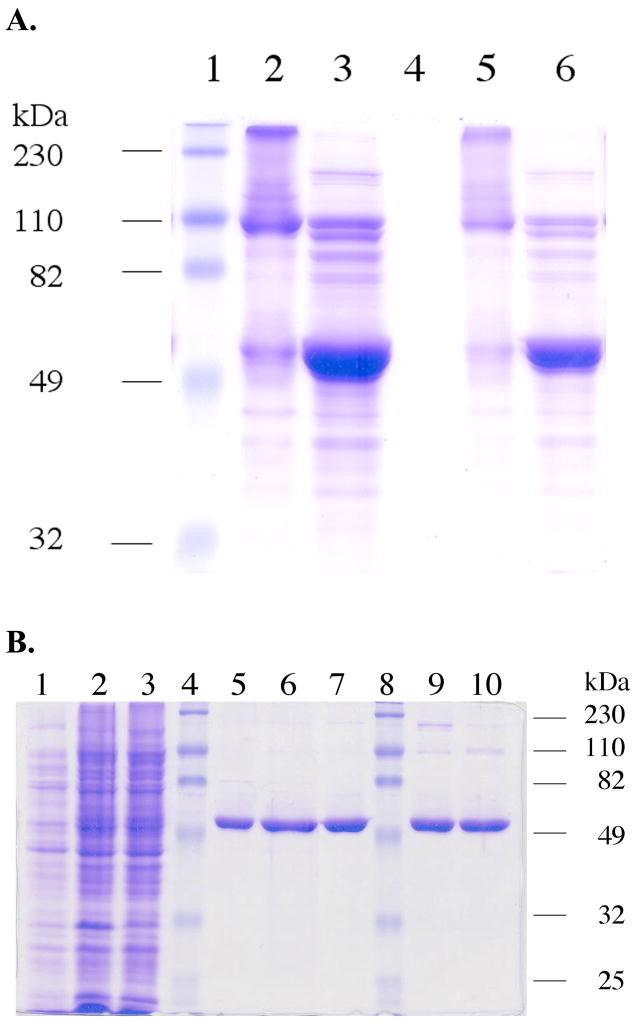
We have constructed a fusion protein that consists of mouse thioredoxin reductase-3, intein, and chitin binding domain. The chitin binding domain is a small peptide that has high affinity for chitin-agarose, and this resin provides a high level of purification of the fusion protein. The fusion construct is made such that glycine 487 of mTR-3 is the amino acid at the junction between mTR-3 and the intein. Addition of a cleavage buffer containing thiol (NMA, βME, or DTT), causes elution of the thioester-tagged mTR-3 from the affinity column. Figure 3A shows the effect of the addition of a thiol-containing buffer to the isolated fusion protein. In this experiment, the fusion protein was first isolated in an impure form on a DEAE anion exchange column (the cell extract was directly applied to the DEAE column for this experiment without any other chromatographic steps). The fusion protein appears as a band near 110 kDa in the 12% SDS-PAGE gel. Upon addition of thiol-containing buffer, the fusion protein is cleaved to liberate the thioester-tagged mTR-3 from the VMA1 intein-chitin binding domain fusion as shown in Figure 3A. Figure 3B shows a 12% SDS-PAGE gel of pooled protein fractions from each step in the purification process. As the chitin resin is an affinity resin, the purity of the eluted mTR-3 (either truncated or semisynthetic) is very good with only minor contaminants showing. These contaminants are removed after the phenyl sepharose and DEAE chromatographic steps as shown in Figure 3B. The purity of the truncated mTR-3 is such that large crystals can be grown from the purified enzyme (data not shown). A preliminary crystal structure of our truncated enzyme has been reported (37). The purity of the semisynthetic enzyme is similar to the truncated enzyme as shown in Figure 3B. The purity of the recombinant mTR-3 (with engineered SECIS element) is >95% as judged by SDS-PAGE (see Figure 1 in the Supporting Information).
Figure 3.
(A) A 12% SDS-PAGE gel showing the effect of addition of thiol to the TR-intein fusion protein. The fusion protein was isolated in a semi-pure form by DEAE ion exchange chromatography as the first step instead of the chitin affinity resin. Fractions containing the fusion protein were then pooled. The protein sample was then loaded onto the 12% gel with or without 50 mM DTT. Lane 1: molecular weight markers. Lane 2: (10 μL loading) TR-intein fusion protein without addition of DTT. The fusion protein appears as a band near 110 kDa. Some hydrolysis of the fusion protein is apparent. Very high molecular weight bands are also present, most likely due to disulfide cross-linking in the absence of reducing agent. Lane 3: (10 μL loading) TR-intein fusion protein sample treated with 50 mM DTT. A very large band at 55 kDa now appears. Lane 4: Blank. Lane 5: the sample as in lane 2 except that 5 μL was loaded. Lane 6: same as lane 3 except that 5 μL was loaded.
(B) 12% SDS-PAGE characterization of recombinant mouse thioredoxin reductase 3. Lane 1: Cell culture pre-induction. Lane 2: IPTG-induced cell culture. Lane 3: Supernatant of the cell lysate. Lane 4 and 8: Molecular weight markers. Lane 5: Truncated TR cleaved from the intein on-resin by 2-mercaptoethanol and eluted from the chitin resin. Lane 6: Truncated TR eluted from the phenyl sepharose column. Lane 7: Truncated TR after elution from the DEAE column. Lane 9: The purified semisynthetic TR after elution from the DEAE column. Lane 10: Purified truncated TR cleaved from the intein on-resin by the amino acid cysteine.
Selenium Content of Semisynthetic Enzyme
Selenium content of the semisynthetic enzyme (using our “normal” protocol), determined by ICP-MS, was found to be 0.63 mol of selenium per mol of enzyme. The selenium content of the semisynthetic enzyme using our optimized protocol was found to be 0.91 mol of selenium per mol of enzyme. The selenium content of the recombinant TR3 prepared by fusing a bacterial SECIS element to the 3′ end of the gene had a selenium content in the range of 0.4 – 0.5 mol of selenium per mole of enzyme depending on the preparation.
Peptide Complementation: Peptides as Substrates for the Truncated Enzyme
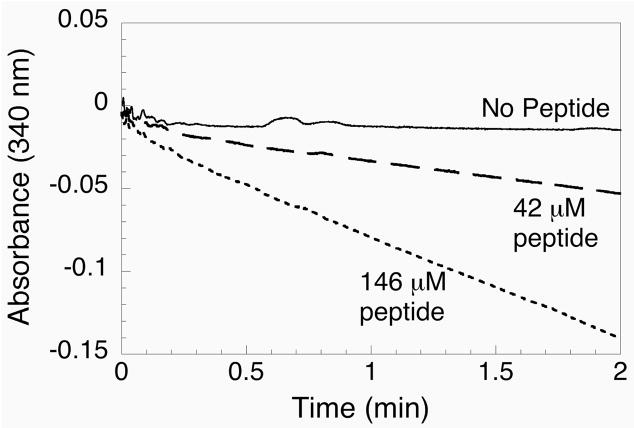
Our first attempt at developing a semisynthetic system for producing TR was inspired by the S-protein/S-peptide system developed by Richards (23). A peptide complementation approach was first tried because only 3 amino acids out of a total of 490 are missing from the intact enzyme. Peptide Ac-GCUG was synthesized and tested for its ability to “complement” the truncated enzyme missing tripeptide CUG. This tetrapeptide is N-acylated and begins with a glycine residue and as such, cannot ligate to the thioester-tagged enzyme produced from the intein fusion. Figure 4 shows the results of titrating increasing amounts of Ac-GCUG peptide into the reaction mixture containing mTR-3, NADPH, thioredoxin, and insulin. The results from this titration show that NADPH was clearly consumed upon addition of high concentrations of the peptide.
Figure 4.
Assay of peptide complementation experiment using truncated mTR-3 and synthetic peptide Ac-GCUG. The consumption of NADPH is monitored at 340 nm. When no peptide is added to the cuvette, no decrease in absorbance is observed. However, as increasing amounts of peptide are added to the assay, a progressive consumption of NADPH is observed.
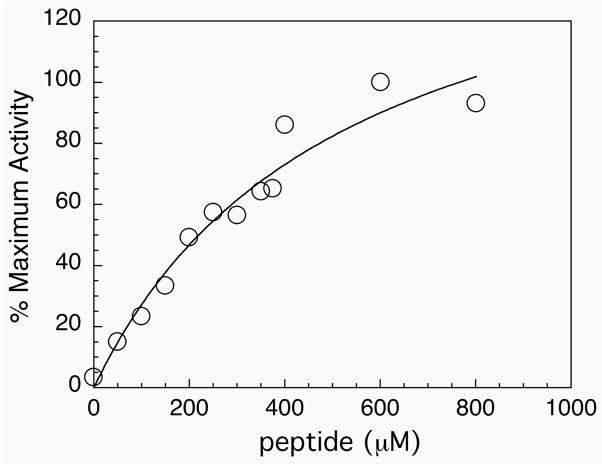
Further variations of this assay were performed in which either thioredoxin, insulin, or both were missing from the assay. The results of these experiments are summarized in Table 1 and demonstrate that the selenium-containing peptide was required for NADPH consumption. Thioredoxin is the natural substrate for the full-length selenium-containing enzyme (38). It is reduced during the course of the reaction, and insulin in turn is reduced by thioredoxin. The function of the insulin in the assay is to provide a source of oxidant for thioredoxin so that the concentration of the oxidized thioredoxin remains high during the course of the assay. When thioredoxin was absent, the oxidized form of the peptide was reduced by the truncated form of the enzyme, resulting in the consumption of NADPH. The results in Table 1 show that more NADPH is consumed when both thioredoxin and insulin are present in the assay. Thioredoxin may be reduced by the transiently formed enzyme/peptide complex, or thioredoxin may get reduced by the resulting reduced peptide. These results demonstrate that the peptide is primarily acting as a substrate for the truncated enzyme, rather than forming a reconstituted enzyme as in the S-protein/S-peptide complex. The acylated peptide cannot ligate to the thioester-tagged enzyme and forms a weak complex with the truncated TR. The relative weakness of this complex was illustrated by titrating the Ac-GCUG peptide as a function of NADPH consumption and plotting the per cent activity vs. peptide concentration as shown in Figure 5. The truncated enzyme becomes saturated with Ac-GCUG tetrapeptide at concentrations above 1 mM and an estimation of the Kd for the complex is ~0.5 mM. Thus, this small peptide forms a very weak association with the truncated enzyme. These results can be put into perspective by examining the mechanism of reduction of thioredoxin as shown in Figure 6. The enzyme transfers electrons from NADPH to the C-terminus of the enzyme containing the selenenylsulfide group. The free selenol then attacks the disulfide bond of thioredoxin. If this peptide is missing from the reaction, then no reduction can occur. However, if oxidized peptide is provided to the enzyme that is missing this peptide, then the oxidized peptide becomes a substrate for the enzyme and NADPH is consumed, which is what is observed experimentally.
Table 1.
a Summary of TR/Ac-GCUG peptide complementation kinetics
| b Enzyme | 100 μM peptide | 160 μM insulin | 3 μM Trx | c Rate (nmoles NADPH/min) |
|---|---|---|---|---|
| TRGly487 | + | + | + | 4.20 ± 0.60 |
| TRGly487 | + | − | − | 2.25 ± 0.21 |
| TRGly487 | + | − | + | 1.96 ± 0.086 |
| TRGly487 | + | + | − | 2.60 ± 0.062 |
Enzyme assays were performed in a volume of 600 μL containing 100 mM potassium phosphate buffer, pH 7.0, 1 mM EDTA, with 8.4 μM enzyme. Other assay components are as indicated in the table.
Indicates that the truncated form of the enzyme ends at residue 487
The rate is calculated by monitoring the loss of NADPH at 340 nm.
Figure 5.
Saturation curve for the addition of tetrapeptide Ac-GCUG to the truncated enzyme. The enzyme becomes saturated at concentrations above 1 mM, thus the association between the peptide and truncated enzyme is weak.
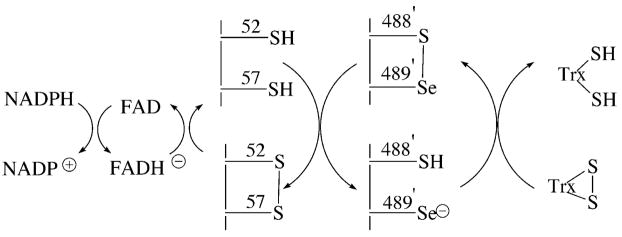
Figure 6.
Proposed reaction mechanism for catalysis by mouse thioredoxin reductase-3. The “prime” designation for residues 488 and 489 indicate that they are present on the other subunit of the TR dimer.
Activity of the Semisynthetic Enzyme – Activity Towards Thioredoxin
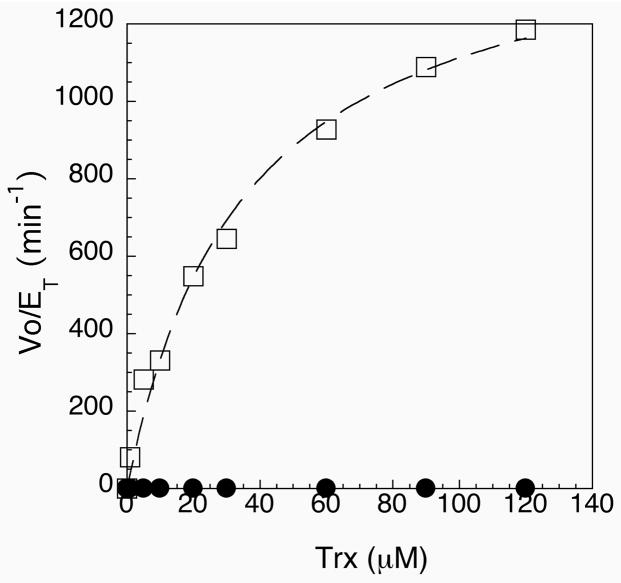
The peptide complementation system described above led to the development of a semisynthetic enzyme that covalently links the tripeptide containing selenocysteine to the main body of the enzyme through a peptide linkage. The semisynthetic enzyme could be constructed by addition of peptide CUG to the cleavage buffer, as described above. Since this peptide has an N-terminal cysteine residue (and is not N-acylated), the sulfhydryl group of the cysteine residue can attack the C-terminal thioester group of the enzyme. Rearrangement then occurs to the amide form (39). In order to test whether the peptide becomes truly incorporated into the enzyme, we tested the ability of the semisynthetic enzyme to reduce thioredoxin. The presence of a Sec residue has been shown to be necessary to reduce E. coli thioredoxin with high efficiency (35). The semisynthetic enzyme at a concentration of 2 nM showed excellent activity towards E. coli thioredoxin in comparison to both the truncated form and a mutant form of the enzyme in which a cysteine residue replaces Sec489 as part of a full-length construct. A Michaelis-Menten plot for our semisynthetic construct in comparison to the Sec489Cys mutant is shown in Figure 7, and a summary of our kinetic data is listed in Table 2. As shown in Figure 7, the Sec489Cys mutant has much lower activity, as would be expected and as was reported previously (35). In contrast to the cysteine mutant enzyme, our semisynthetic enzyme shows very high activity, with values of kcat ranging from 1500 min−1 to 2220 min−1, depending on the preparation (see also Figure 2 in the Supporting Information). This high activity strongly indicates that the C-terminal tripeptide was successfully ligated to the thioester-tagged enzyme. The enzyme preparation with a higher kcat value resulted from using a higher peptide/protein ratio during the ligation and resulted with an enzyme with a higher selenium content (91% Se). The enzyme with a higher Se content also gave a lower yield of enzyme, 9 mg from 3 L of cell culture vs. 24 mg from 6 L of cell culture for the enzyme with a kcat of 1500 min−1 (63% Se). The truncated enzyme (ending with amino acid 487 and missing the tripeptide) has no detectable activity towards oxidized thioredoxin.
Figure 7.
Michaelis-Menten plot of Vo/ET vs. thioredoxin concentration for semisynthetic enzyme (open squares) and the Sec489Cys mutant (closed circles). Both curves could be fitted to a hyperbolic plot. Values of kcat and Km were obtained from the curve fit and are listed in Table 2.
Table 2.
Activity of Semisynthetic and Mutant Enzymes Towards Thioredoxin
| Enzyme | kcat (min−1) | Km (μM) |
|---|---|---|
| a TR-Gly487 | No activity | No activity |
| b TRSec489Cys | 1.2 ± 0.11 | 6.6 ± 1.8 |
| TR-semisynthetic (63% Se) | 1500 ± 81 | 35 ± 5 |
| c TR-semisynthetic (91% Se) | 2220 ± 78 | 67.6 ± 6 |
| d rat TR1 | 2500 | 3.3 |
The truncated enzyme ends at glycine 487 and is missing the C-terminal tripeptide.
The full-length mutant in which cysteine replaces the catalytic selenocysteine residue.
Data from optimized procedure using a higher ratio of peptide to protein as described under Methods.
Taken from (35).
Peroxidase Activity
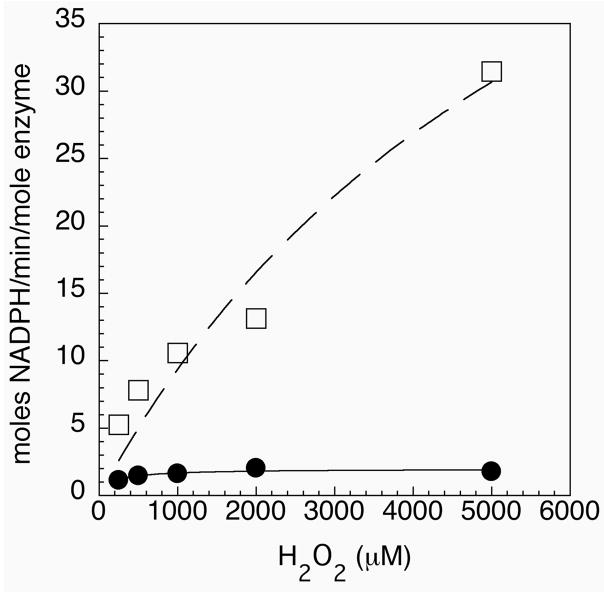
Mammalian thioredoxin reductases also show hydrogen peroxidase activity. This activity is a characteristic feature of the mammalian enzymes because non-selenium containing TR’s have been shown to have little, if any, hydrogen peroxidase activity (35). Figure 8 shows the results of our hydrogen peroxidase activity assays with our semisynthetic enzyme and the Sec489Cys mutant. The mutant enzyme has barely detectable hydrogen peroxidase activity, while the semisynthetic enzyme shows high peroxidase activity with a kcat of 71 min−1 (± 7) and a Km of 6.6 mM (± 0.50). This rate was achieved with only 2 nM of semisynthetic enzyme present in the assay. The kinetic data derived from Figure 8 compares favorably to the data reported by Zhong and Holmgren (35) for reduction of H2O2 by human placental TR1. The high peroxidase activity of our enzyme provides additional strong support for the incorporation of the selenocysteine-containing tripeptide by our semisynthetic approach.
Figure 8.
Peroxidase activity of semisynthetic enzyme (open squares) and the Sec489Cys mutant (closed circles). Activity is reported as moles of NADPH oxidized per minute per mole of enzyme.
Ligation Efficiency for “off-resin” Ligations
As shown in Figure 1, our method of making a semisynthetic TR makes use of the addition of the oxidized tripeptide to a thiol containing cleavage buffer, which is then added directly to the resin-bound protein. The thiol in the cleavage buffer can either liberate the protein from the resin to produce a thioester-tagged protein or it can reduce the oxidized tripeptide, which in turn could directly attack the resin-bound protein and become incorporated as part of the polypeptide chain. Alternatively, the liberated thioester-tagged protein can undergo a thioester exchange reaction with the reduced tripeptide and then be incorporated into the polypeptide via a stable amide bond. All three pathways lead to the eventual incorporation of the tripeptide into the larger polypeptide of mTR-3. The key to ligating this tripeptide to the larger polypeptide of mTR-3 is keeping this tripeptide reduced by using a large excess of thiol in the cleavage buffer. The thiol in the cleavage buffer, functions to affect cleavage of the resin-bound protein and also to keep the tripeptide reduced so that it can ligate to the liberated thioester-tagged protein.
We next compared the efficiency of the ligation when the peptide is added directly to a slurry of resin-bound TR and cleavage buffer as compared to adding the peptide to a semi-purified, concentrated form of the thioester-tagged TR “off-resin.” Concentrated thioester-tagged protein was either treated with oxidized peptide or oxidized peptide plus 70 mM NMA. After dialysis to remove excess peptide and reagents, the resulting enzymes were assayed for thioredoxin reductase activity. The results are summarized in Table 3 and clearly show that the efficiency of ligation is much higher when the peptide is added directly to the slurry of resin-bound TR and cleavage buffer. One reason for this decline in efficiency when the ligation is done “off-resin” is that the lifetime of the thioester-tagged TR may be short in this protein context. Hydrolysis of the thioester would yield the C-terminal carboxylate form of the protein, which is unproductive with respect to ligation. Second, the reduced tripeptide can directly attack the resin-bound TR-intein fusion protein with concomitant ligation. This process should be kinetically faster than when an intermediary thiol attacks first.
Table 3.
a Comparison of methods used to construct a semisynthetic TR.
| Chitin Resin | Reductant | TR concentration in assay [nM] | mol NADPH/min/mol TR |
|---|---|---|---|
| On | NMA | 2 | 1008 |
| Off | NMA | 100 | 48 |
| Off | none | 100 | 8 |
The enzyme was assayed using 90 μM thioredoxin as substrate. All other assay conditions were the same as those described in Methods.
Comparison of the Activity of Semisynthetic TR to Recombinant TR
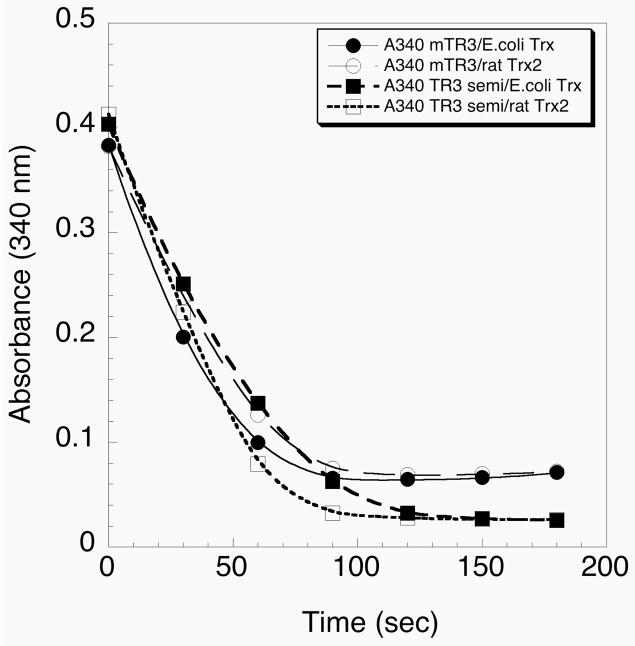
The activity of the semisynthetic enzyme was compared to that of the recombinant mTR-3. The latter protein was expressed in E. coli and contained 0.5 equivalents of selenium (Turanov and Gladyshev, manuscript in preparation). The recombinant TR enzyme is made by fusing a bacterial SECIS element to the 3′ end of the TR3 gene. This allows for heterologous expression of the recombinant enzyme in E. coli. The results in Figure 9 show that the recombinant mTR-3 and the semisynthetic mTR-3 have similar activities with both E. coli Trx and recombinant rat Trx2. These data further verify that the semisynthesis method resulted in a functional TR.
Figure 9.
Comparison of the activities of semisynthetic TR and recombinant TR. The activity of the recombinant TR produced in E. coli by using an engineered SECIS element is represented by the circles. The activity of the semisynthetic TR is represented by the squares. E. coli thioredoxin (closed symbol) and rat thioredoxin-2 (open symbol) were used as substrates.
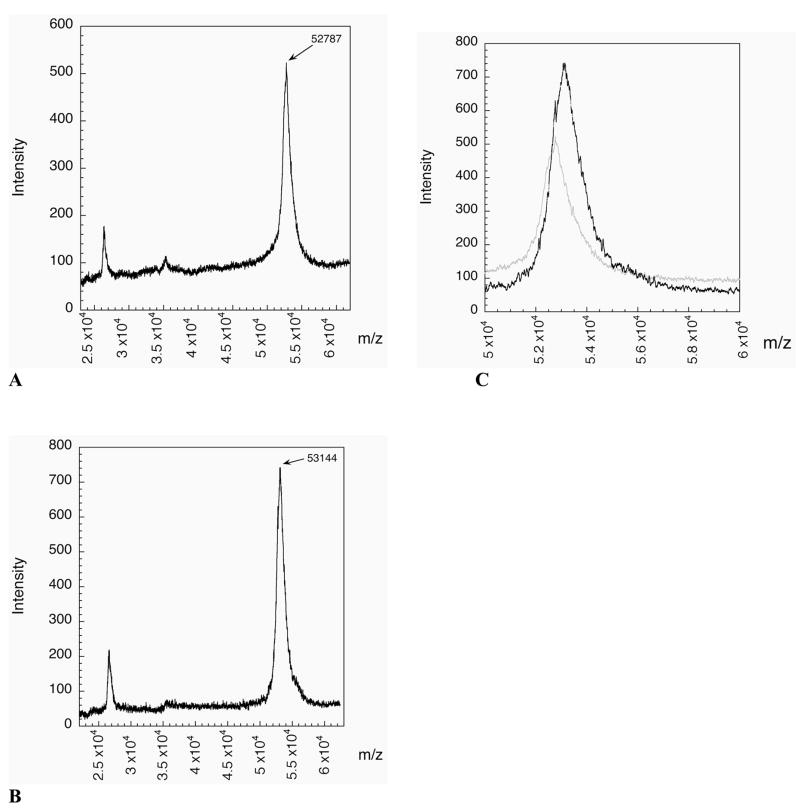
Mass Determination of Truncated and Semisynthetic TR’s
The mass spectrographs of the truncated and semisynthetic TR’s are shown in Figures 10A and 10B, respectively. The predicted masses of the truncated and semisynthetic enzymes are 52,857 (carboxylate form) and 53,164 daltons, respectively. The experimentally observed masses were 52,787 and 53,144 daltons, respectively, and are in good agreement with the expected values. An overlay of the two spectra (Figure 10C) shows a clear mass increase due to the presence of the ligated peptide. The experimental mass increase of the semisynthetic enzyme is 357 daltons, which is very close to the expected mass increase of 329 daltons.
Figure 10.
(A) MALDI-MS of truncated mTR-3 missing the C-terminal tripeptide. The average mass for the carboxylic acid form of the enzyme is 52,857 and the mass of the thioester-tagged enzyme is 52,944. The mass difference between the two forms is 87 daltons which makes identifying the exact form of the enzyme impossible at the resolution of the spectrometer (1/100). The observed mass of 52,787 is closer to the carboxylate form of the enzyme. (B) MALDI-MS of the semisynthetic form of mTR-3. The observed mass of 53,144 is in good agreement with the predicted mass of 53,164 (oxidized form). The increase in mass between the two forms is 357 daltons, in good agreement with the expected mass increase of 329 daltons. (C) Overlay of the two spectra. The semisynthetic spectrum is in black and the truncated spectrum is in light gray. The difference in mass is clearly observable. The semisynthetic spectrum has a shoulder which lines up with the peak of the truncated spectrum. The peak width of the semisynthetic enzyme shows that it is a mixture of the two forms.
Peptide Mass Mapping of Tryptic Fragments
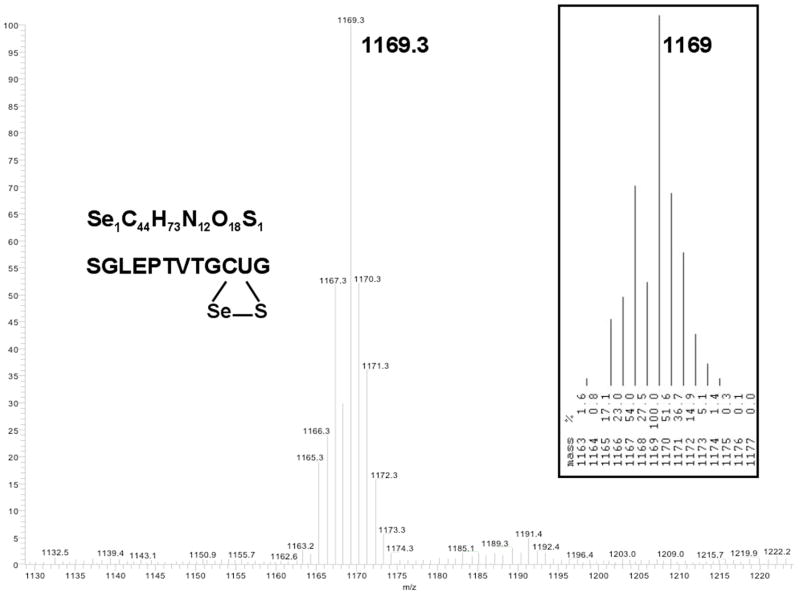
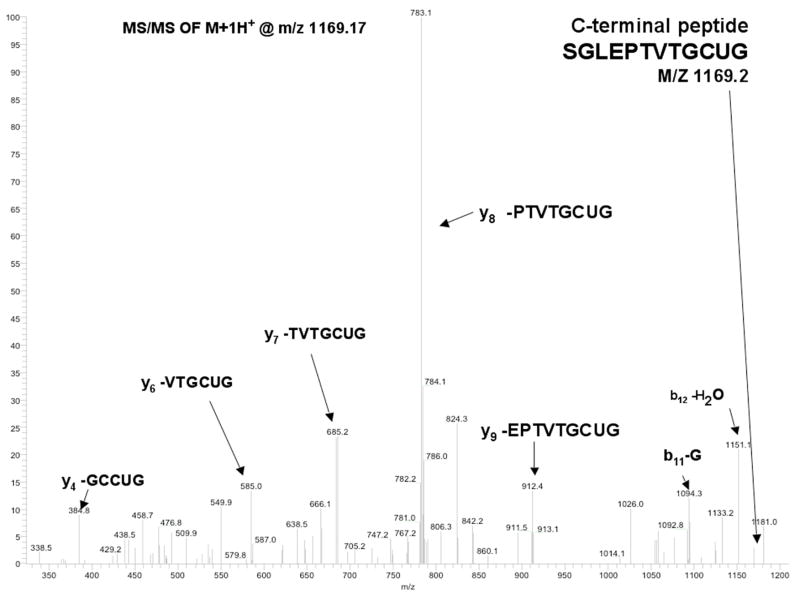
In order to unambiguously demonstrate that our peptide had been ligated to the C-terminus of mTR-3, we undertook a peptide mass mapping experiment. In this experiment, both the truncated and semisynthetic enzymes were digested with trypsin and then the peptide fragments were analyzed by both MALDI-TOF MS and ESI-MS. A summary of all of the identified peptides is available in the Supporting Information (Table 1). Approximately 70% of the total peptides were identified by mass mapping. Figure 11 shows a peptide with sequence of SGLEPTVTGCUG found in the ESI-MS experiment, which has a m/z of 1169.3 daltons (calculated m/z of 1169.4). The mass of this peptide corresponds exactly to the mass of the C-terminal peptide containing selenocysteine. Further proof of the identity of this peptide is provided by a MS-MS experiment shown in Figure 12. In this experiment, the peptide is fragmented by collision induced dissociation (CID) and the resulting ions unambiguously identify the sequence of the peptide as the peptide corresponding to the C-terminus.
Figure 11.
ESI-MS of peptide of sequence SGLEPTVTGCUG. The peptide corresponds to the C-terminal peptide of mTR-3 containing the ligated tripeptide CUG. The inset at right is the predicted isotope pattern for this peptide containing selenium. The mass of the peptide corresponds to the oxidized form as would be expected for a peptide containing selenocysteine.
Figure 12.
Legend: MS/MS of peptide SGLEPTVTGCUG. The peptide was fragmented by CID. Ions produced by fragmentation are labeled in the figure with the corresponding peptide sequence.
DISCUSSION
Enzymologists attempting to study eukaryotic selenocysteine-containing enzymes have been hampered by the lack of a heterologous system for producing selenoenzymes in large amounts for biophysical studies. The lack of a recombinant system also means that construction of mutants for structure-function studies is difficult or not possible. We have presented here a simple system for the construction of a semisynthetic selenocysteine-containing thioredoxin reductase.
Our method divides the enzyme into two modules, a non-selenium containing module produced as an intein-fusion protein in E. coli cells and a selenium-containing module produced as a synthetic tripeptide. The two modules are then joined together using expressed peptide ligation (29–30) which is becoming an increasingly common methodology for producing recombinant proteins with unusual features, non-standard amino acids, or that are difficult to purify from natural sources (40).
Expressed protein ligation (EPL) can be used to incorporate selenocysteine into a protein in several ways. First, selenocysteine can take the place of an N-terminal cysteine residue and be used directly at the ligation junction. There are several examples in the literature using this approach (29, 41–42). A second derivation of EPL used here incorporates selenocysteine away from the site of ligation. Thioredoxin reductase is a good candidate to be engineered by EPL because the selenocysteine residue is the penultimate amino acid in the polypeptide chain. Moreover, there is a conveniently located cysteine residue adjacent to the selenocysteine residue to utilize in the ligation reaction. Lastly, the peptide is fortuitously short in sequence.
Although there are some technical challenges to synthesizing a peptide containing selenocysteine such as relatively high expense and complex deprotection schemes (43–44), there are also challenges to making selenoproteins using genetic manipulation. Arnér and coworkers have developed a system for producing mammalian thioredoxin reductase by splicing a prokaryotic SECIS element to the 3′ end of the TR gene (16). This method also requires a special strain that increases the expression of the SelA, SelB, and SelC gene products needed to overproduce the accessory proteins for the selenocysteine incorporation machinery. In using EPL to produce thioredoxin reductase the challenges are chemical, while in using genetic engineering, the challenges are biological. Although our yields are more modest (4 mg/L of cell culture) than those reported by Arnér and colleagues (20 mg/L of cell culture), the two methods were applied to two different thioredoxin reductases (TR3 in our study and TR1 in the previous study); hence a direct comparison of the two methods cannot be made here.
The reason that mammalian thioredoxin reductase can be made both by recombinant techniques as developed by Arnér and colleagues, or by EPL as reported here, is that the Sec residue is very close to the C-terminus (the penultimate residue). Of the 25 proteins containing Sec in the human proteome (6), several of these proteins would be amenable to recombinant production using the engineered SECIS element technique. These are selenoproteins designated as K, O and S (in addition to three TRs) (6). In these proteins, the Sec residue is either one or two residues removed from the C-terminus. For expression of these proteins, a bacterial SECIS element can be placed at the 3′ end of the gene as was done for TR1. These same proteins could also be produced using EPL. In addition, EPL should be capable of the production of selenoproteins I and R, because in these proteins Sec is near the C-terminus and would require the synthesis of peptides of length 11 and 22 residues respectively for use by EPL (well within the reach of peptide technology). Several more selenoproteins of the human proteome could be synthesized by joining two or three peptide segments together using the native chemical ligation technique (total chemical protein synthesis). These selenoproteins are designated as W, H, and M and contain 87, 122, and 145 amino acid residues, respectively. These proteins are thus small enough to be made by chemical means.
The decision to produce a selenoprotein by using an engineered SECIS or by EPL will depend on factors such as yield, post-translational modifications of the protein, solubility of the expressed protein in the heterologous host, and technical expertise (chemical or biological as mentioned above) with the chosen system. EPL provides important advantages in protein engineering experiments, as labeled amino acids and non-standard amino acids can be incorporated into the protein to test structure-function relationships as discussed below.
In the case of TR, spectroscopic labels can be incorporated directly into the tripeptide. For example, a 13C-labeled cysteine residue could be incorporated in the tripeptide used in the ligation reaction. The result would be a semisynthetic enzyme that incorporated a 13C-label on a single atom out of thousands of atoms in the enzyme. This could then be used to determine the pKa of Cys488. Likewise 77Se could be used in the synthesis of the tripeptide and the resulting semisynthetic enzyme would have a unique label to do 77Se NMR experiments. A second use of our semisynthetic system would be in its use in incorporating non-standard amino acids at the C-terminus to explore structure-function relationships. The crystal structure of the cysteine mutant form of the cytosolic rat enzyme revealed that the C-terminal carboxyl group is involved in a salt bridge with Lys29 (21). This interaction could be probed by synthesizing a tripeptide that was amidated at the C-terminus (CONH2 vs COO-) so that this interaction would be disrupted. This modification is impossible using standard protein engineering techniques.
There are multiple lines of evidence that we were able to create a semisynthetic TR. (i) The thioredoxin reductase activity of our enzyme was very high, with a kcat of either 1500 min−1 or 2220 min−1 depending on the preparation. This compares very favorably to the value of 2500 min−1 reported for rat TR1 (35). The Km for thioredoxin of our semisynthetic enzyme is in the range of 35–70 μM. This is 10–20 fold higher than the Km reported for rat TR1, but the two enzymes have only 56% sequence identity so some difference in Km is to be expected (the difference is between the cytosolic form of the rat enzyme vs. the mitochondrial form of the mouse enzyme). (ii) The semisynthetic enzyme shows high hydrogen peroxidase activity. This activity is evident in Figure 8, which shows the peroxidase activity of our semisynthetic enzyme and that of our Sec489Cys mutant, which has barely detectable peroxidase activity. The peroxidase activity of the enzyme is indicative of the presence of selenium because selenium is strongly nucleophilic towards the oxygen-oxygen bond, whereas sulfur is much less so (45–46). (iii) Comparison of the activity of the semisynthetic enzyme to that of the recombinant TR3 produced with an engineered SECIS element in Figure 9 shows that the two enzymes produced by two different methods have similar activities. (iv) The increased mass of the semisynthetic enzyme compared to the truncated enzyme in Figure 10C shows that the mass has increased close to the expected (329 daltons). (v) Peptide mass mapping experiments followed by MS/MS demonstrated that the tripeptide becames covalently linked to the C-terminus of the enzyme (Figures 11 and 12). (vi) ICP-MS analysis of our protein samples showed that Se was incorporated into our enzyme at a level of 63–91%.
The overall yield of TR from this method was either 24 mg from 6 L (4 mg/L) of cell culture yielding an enzyme with a kcat of 1500 min−1, or 9 mg of enzyme from 3 L (3 mg/L) of cell culture with a kcat of 2220 min−1 (optimized procedure). In both cases, the final preparations of enzymes are a mixture of the truncated form and the full-length form. The ligation efficiency can be estimated from ICP-MS analysis since the ligated tripeptide is the only source of selenium for the enzyme. Thus, in the enzyme preparation with a kcat of 1500 min−1, the ligation efficiency is 63%, since there was 0.63 mol of Se per mole of enzyme. In the optimized procedure producing an enzyme with a kcat of 2220 min−1, the ligation efficiency was 91%, because there was 0.91 mol of Se per mol of enzyme.
We have also developed a peptide complementation approach to creating a semisynthetic thioredoxin reductase. In this system, the synthetic peptide can fit into the active site of a truncated form of the enzyme to restore partial activity. The peptide complementation approach developed here is not a robust semisynthetic system as the S-peptide/S-protein system is, but rather the results are more indicative of the peptide being a substrate for the truncated form of the enzyme. The data in Table 1 show that all that is required for consumption of NADPH is the presence of oxidized selenium-containing peptide. An increase in activity is observed when either thioredoxin or insulin is added to the assay, but the increase in activity is probably due to both insulin and thioredoxin providing a source of oxidant to maintain a high concentration of oxidized peptide in the assay. A further increase in activity is noted when both insulin and thioredoxin are present in the assay. It is likely that thioredoxin becomes chemically reduced by the reduced form of the selenium-containing peptide, which itself is reduced by the truncated form of the enzyme. However, we cannot state this as a certainty.
It is useful to compare the activities of the two systems. The activity of the non-covalent peptide complementation system is 4.2 nmoles NADPH/min, while the activity of the covalent semisynthetic system is 1500–2220 nmoles of NADPH/min (thioredoxin as substrate). Thus the semisynthetic enzyme has 350–500 fold higher activity than that of the non-covalent complex, further validating the success of the peptide ligation approach.
The more important finding is that oxidized selenocysteine-containing peptides can serve as substrates for the truncated form of the enzyme. During the enzymatic cycle, the enzyme must reduce the resulting 8-membered ring structure that forms between neighboring cysteine and selenocysteine residues. This peptide complementation system can be exploited to examine the kinetics of a single step in the enzyme reaction cycle. This system will be used for future structure-function studies.
Conclusions
We have presented a novel method for constructing a semisynthetic thioredoxin reductase. The semisynthetic system reported here will allow for detailed structure-function studies to be performed in the future.
Supplementary Material
A complete table summarizing the peptide mass mapping experiments is available in the Supporting Information. A Michaelis-Menten plot showing the activity of the optimized preparation of TR towards Trx is also available. Finally, a gel showing the purity of the TR produced using an engineered SECIS element is available in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
The authors wish to acknowledge Ms. Marque St. John and Mr. Brian Lacey for performing some preliminary experiments of peptide complementation with thioredoxin reductase. We wish to thank Dr. Jack Benner of New England Biolabs for performing some of the mass spectrometry experiments. We wish to thank Dr. Chris Noren of New England Biolabs for helpful comments and suggestions with this manuscript. We also wish to thank Dr. Dwight Matthews and Dr. Jack Heim of the University of Vermont Mass Spectrometry Facility for the help in the peptide mass mapping experiments.
Footnotes
These studies were supported by National Institutes of Health Grants GM070742 to RJH, GM044783 to RTR, and GM065204 to VNG.
βME, beta mercaptoethanol; BSA, bovine serum albumin; CID, collision induced dissociation; DEAE, diethylaminoethyl; DCM, dichloromethane; DMF, dimethylformamide; DTNB, 5, 5′-dithiobis(2-nitrobenzoic acid); EDT, ethanedithiol; EPL, expressed protein ligation; ESI-MS, electrospray ionization mass spectrometry; Fmoc, 9-fluorenylmethoxycarbonyl; HATU, N-[(dimethylamino)-1H-1,2,3-triazolo[4,5-b]pyridin-1-yl]methylene]-N-methylmethanaminiumhexafluorophosphate N-oxide; HOBt, 1-hydroxybenzotriazole; ICP-MS, inductively coupled plasma-mass spectrometry; IPTG, isopropyl-β-D-thiogalactopyranoside; MALDI-TOF MS, matrix assisted laser desorption ionization-time of flight mass spectrometry; MOPS, 3-(N-morpholino) propane-sulfonic acid; mTR-3, mouse thioredoxin reductase 3; NADPH, β-nicotinamide adenine dinucleotide phosphate, reduced; NMA, N-methyl-mercaptoacetamide; PEG-PS; poly(ethylene glycol)-polystyrene graft resin support; pMOB, p-methoxybenzyl; Sec, selenocysteine; SECIS, selenocysteine insertion sequence; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; SPPS, solid phase peptide synthesis; Trx, thioredoxin; TR, thioredoxin reductase; TFA, trifluoroacetic acid; TMP, 2,4,6-trimethylpyridine (collidine); TMSOTf, trimethylsilyl trifluoromethanesulfonate; Tris, tris(hydroxymethyl)aminomethane; Trt, trityl.
We use the term “semisynthesis” in its historical context. In the older literature, a non-covalent association between a peptide and protein fragment had been considered to be a semisynthetic enzyme, e.g. the S-peptide/S-protein system. Please see page 22 of the monograph by Offord (28) as well as Chapter 8. More recently, with the development of peptide ligation techniques, the term semisynthesis has been confined to those systems in which a covalent peptide bond is formed between a synthetic peptide and a protein fragment.
U is the one letter abbreviation for the amino acid selenocysteine.
References
- 1.Mustacich D, Powis G. Thioredoxin reductase. Biochem J. 2000;346:1–8. [PMC free article] [PubMed] [Google Scholar]
- 2.Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 3.Arnér ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 4.Atkins JF, Gesteland RF. The twenty-first amino acid. Nature. 2000;407:463–465. doi: 10.1038/35035189. [DOI] [PubMed] [Google Scholar]
- 5.Bock A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 6.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 7.Heider J, Baron C, Bock A. Coding from a distance: dissection of the mRNA determinants required for the incorporation of selenocysteine into protein. EMBO J. 1992;11:3759–3766. doi: 10.1002/j.1460-2075.1992.tb05461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Harney JW, Driscoll DM, Hatfield DL, Berry MJ. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:158–163. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fagegaltier D, Huber N, Yamada K, Mizutani T, Carbon P, Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forchhammer K, Leinfelder W, Bock A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature. 1989;342:453–456. doi: 10.1038/342453a0. [DOI] [PubMed] [Google Scholar]
- 11.Forchhammer K, Rucknagel KP, Bock A. Purification and biochemical characterization of SELB, a translation factor involved in selenoprotein synthesis. J Biol Chem. 1990;265:9346–9350. [PubMed] [Google Scholar]
- 12.Berry MJ, Banu L, Harney JW, Larsen PR. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 1993;12:3315–3322. doi: 10.1002/j.1460-2075.1993.tb06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krol A. Evolutionarily different RNA motifs and RNA-protein complexes to achieve selenoprotein synthesis. Biochimie. 2002;84:765–774. doi: 10.1016/s0300-9084(02)01405-0. [DOI] [PubMed] [Google Scholar]
- 14.Jiang Z, Arnér ES, Mu Y, Johansson L, Shi J, Zhao S, Liu S, Wang R, Zhang T, Yan G, Liu J, Shen J, Luo G. Expression of selenocysteine-containing glutathione S-transferase in Escherichia coli. Biochem Biophys Res Commun. 2004;321:94–101. doi: 10.1016/j.bbrc.2004.06.110. [DOI] [PubMed] [Google Scholar]
- 15.Bar-Noy S, Moscovitz J. Mouse methionine sulfoxide reductase B: effect of selenocysteine incorporation on its activity and expression of the seleno-containing enzyme in bacterial and mammalian cells. Biochem Biophys Res Comm. 2002;297:956–961. doi: 10.1016/s0006-291x(02)02314-8. [DOI] [PubMed] [Google Scholar]
- 16.Arnér ESJ, Sarioglu H, Lottspeich F, Holmgren A, Bock A. High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the SelA, SelB, and SelC genes. J Mol Biol. 1999;292:1003–1016. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- 17.Rengby O, Johansson L, Carlson LA, Serini E, Vlamis-Gardikas A, Karsnas P, Arnér ESJ. Assessment of production conditions for efficient use of Escherichia coli in high-yield heterologous recombinant selenoprotein synthesis. Appl Environ Microbiol. 2004;70:5159–5167. doi: 10.1128/AEM.70.9.5159-5167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandman KE, Noren CJ. The efficiency of Escherichia coli selenocysteine insertion is influenced by the immediate downstream nucleotide. Nucleic Acids Res. 2000;28:755–761. doi: 10.1093/nar/28.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson L, Chen C, Thorell JO, Fredriksson A, Stone-Elander S, Gafvelin G, Arnér ESJ. Exploiting the 21sst amino acid-purifying and labeling proteins by selenolate targeting. Nat Methods. 2004;1:61–66. doi: 10.1038/nmeth707. [DOI] [PubMed] [Google Scholar]
- 20.Gladyshev VN, Jeang KT, Stadtman TC. Selenocysteine identified as the penultimate C-terminal residue in human T-cell thioredoxin reductase corresponds to TGA in the human placental gene. Proc Natl Acad Sci USA. 1996;93:6146–51. doi: 10.1073/pnas.93.12.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandalova T, Zhong L, Lindquist Y, Holmgren A, Schneider G. Three-dimensional structure of a mammalian thioredoxin reductase: Implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc Natl Acad Sci USA. 2001;98:9533–9538. doi: 10.1073/pnas.171178698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biterova EI, Turanov AA, Gladyshev VN, Barycki JJ. Crystal structures of oxidized and reduced mitochondrial thioredoxin reductase provide molecular details of the reaction mechanism. Proc Natl Acad Sci USA. 2005;102:15018–15023. doi: 10.1073/pnas.0504218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards FM, Vithayathil PJ. The preparation of subtilisin-modified ribonuclease and the separation of the peptide and protein components. J Biol Chem. 1959;234:1459–1465. [PubMed] [Google Scholar]
- 24.van Batenburg OD, Voskuyl-Holtkamp I, Schattenkerk C, Hoes K, Kerling KE, Havinga E. The role of the imidazoyl nitrogen atoms of histidine-12 in ribonuclease S. Biochem J. 1977;163:385–387. doi: 10.1042/bj1630385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taniuchi H, Anfinsen CB, Sodja A. Nuclease-T: an active derivative of staphylococcal nuclease composed of two noncovalently bonded peptide fragments. Proc Natl Acad Sci USA. 1967;58:1235–1242. doi: 10.1073/pnas.58.3.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato I, Tominaga N. Soybean trypsin inhibitor-C: An active derivative of soybean trypsin inhibitor composed of two noncovalently bonded peptide fragments. FEBS Lett. 1970;10:313–316. doi: 10.1016/0014-5793(70)80461-6. [DOI] [PubMed] [Google Scholar]
- 27.Holmgren A. Thioredoxin-C’: Reconstitution of an active form of Escherichia coli thioredoxin from two noncovalently linked cyanogens bromide peptide fragments. FEBS Lett. 1972;24:351–354. doi: 10.1016/0014-5793(72)80389-2. [DOI] [PubMed] [Google Scholar]
- 28.Offord RE. Semisynthetic Proteins. John Wiley & Sons Ltd.; New York: 1980. pp. 156–193. [Google Scholar]
- 29.Hondal RJ, Nilsson BL, Raines RT. Selenocysteine in native chemical ligation and expressed protein ligation. J Am Chem Soc. 2001;123:5140–5141. doi: 10.1021/ja005885t. [DOI] [PubMed] [Google Scholar]
- 30.Hondal RJ, Raines RT. Semisynthesis of proteins containing selenocysteine. Methods Enzymol. 2002;347:70–83. doi: 10.1016/s0076-6879(02)47009-7. [DOI] [PubMed] [Google Scholar]
- 31.Muting D, Kaiser E. On quantitative determination of the alpha-amino nitrogen in biological material using the ninhydrin reaction. Hoppe Seylers Z Physiol Chem. 1963;332:276–281. doi: 10.1515/bchm2.1963.332.1.276. [DOI] [PubMed] [Google Scholar]
- 32.Han Y, Albericio F, Barany G. Occurrence and minimization of cysteine racemization during stepwise solid-phase peptide synthesis. J Org Chem. 1997;62:4307–4312. doi: 10.1021/jo9622744. [DOI] [PubMed] [Google Scholar]
- 33.Koide T, Itoh H, Otaka A, Yasui H, Kuroda M, Esaki N, Soda K, Fuji N. Synthetic study on selenocystine-containing peptides. Chem Pharm Bull. 1993;41:5020–506. doi: 10.1248/cpb.41.502. [DOI] [PubMed] [Google Scholar]
- 34.Fairbanks G, Steck TL, Wallach DF. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971;10:2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- 35.Zhong L, Holmgren A. Essential role of selenium in the catalytic activities of mammalian thioredoxin reductase revealed by characterization of recombinant enzymes with selenocysteine mutations. J Biol Chem. 2000;275:18121–18128. doi: 10.1074/jbc.M000690200. [DOI] [PubMed] [Google Scholar]
- 36.Arnér ES, Zhong L, Holmgren A. Preparation and assay of mammalian thioredoxin and thioredoxin reductase. Methods Enzymol. 1999;300:226–239. doi: 10.1016/s0076-6879(99)00129-9. [DOI] [PubMed] [Google Scholar]
- 37.Eckenroth BE, Everse SJ, Adams TE, Hondal RJ. Crystallization of mammalian thioredoxin reductase for structural studies using synthetic active site peptides. FASEB Meeting; April 2–6 (2005); San Diego, CA. Abstract #2600. FASEB Abstracts. [Google Scholar]
- 38.Holmgren A Bovine thioredoxin system. Purification of thioredoxin reductase from calf liver and thymus and studies of its function in disulfide reduction. J Biol Chem. 1977;252:4600–4606. [PubMed] [Google Scholar]
- 39.Noren CJ, Wang J, Perler FB. The currently accepted mechanism of protein splicing. Angew Chem Int Ed. 2000;39:450–466. [PubMed] [Google Scholar]
- 40.Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 41.Quaderer R, Sewing A, Hilvert D. Selenocysteine-mediated native chemical ligation. Helv Chim Acta. 2001;84:1191–1206. [Google Scholar]
- 42.Berry SM, Giesselman MD, Nilges MJ, van der Donk WA, Lu Y. An engineered azurin variant containing a selenocysteine copper ligand. J Am Chem Soc. 2002;124:2084–2085. doi: 10.1021/ja0169163. [DOI] [PubMed] [Google Scholar]
- 43.Gieselman MD, Zhu Y, Zhou H, Galonic D, van der Donk WA. Selenocysteine derivatives for chemoselective ligations. ChemBioChem. 2002;3:709–716. doi: 10.1002/1439-7633(20020802)3:8<709::AID-CBIC709>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 44.Hondal RJ. Incorporation of selenocysteine into proteins using peptide ligation. Prot Pep Lett. 2005;12:757–764. doi: 10.2174/0929866054864319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang SI, Spears CP. Structure-activity studies on organoselenium alkylating agents. J Pharm Sci. 1990;79:57–62. doi: 10.1002/jps.2600790114. [DOI] [PubMed] [Google Scholar]
- 46.Jacob C, Giles GI, Giles NM, Sies H. Sulfur and selenium: The role of oxidation state in protein structure and function. Angew Chem Int Ed. 2003;42:4742–4758. doi: 10.1002/anie.200300573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A complete table summarizing the peptide mass mapping experiments is available in the Supporting Information. A Michaelis-Menten plot showing the activity of the optimized preparation of TR towards Trx is also available. Finally, a gel showing the purity of the TR produced using an engineered SECIS element is available in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.