Abstract
The identification of complex disease susceptibility loci through genome-wide association studies (GWAS) has recently become possible and is now a method of choice for investigating the genetic basis of complex traits. The number of results from such studies is constantly increasing but the challenge lying forward is to identify the biological context in which these statistically significant candidate variants act. Regulatory variation plays an important role in shaping phenotypic differences among individuals and thus is very likely to also influence disease susceptibility. As such, integrating gene expression data and other disease relevant intermediate phenotypes with GWAS results could potentially help prioritize fine-mapping efforts and provide a shortcut to disease biology. Combining these different levels of information in a meaningful way is however not trivial. In the present review, we outline the several approaches that have been explored so far in this sense and their achievements. We also discuss the limitations of the methods and how upcoming technological developments could help circumvent these limitations. Overall, such efforts will be very helpful in understanding initially regulatory effects on disease and disease etiology in general.
INTRODUCTION
The ability of genome-wide association studies (GWAS) to help understand the genetic basis of complex disorders has recently become apparent. Well-documented common human genetic variation maps (e.g. HapMap project) (1), large patient samples with accurately recorded phenotypic information as well as appropriate statistical methods to assess significance (2) and account for potential biases, have all contributed to the current outburst of successful GWAS. Numerous susceptibility variants for a large number of complex diseases have been reported and effectively replicated. A present catalog of published GWAS (http://www.genome.gov/26525384) includes single nucleotide polymorphisms (SNPs) not only associated with major common disorders [Crohn’s disease (3), type 2 diabetes (4), lung cancer (5) etc.] but also with disease-relevant or anthropomorphic quantitative traits [e.g. body mass index (6) or height (7)].
What has not kept the pace however with the capacity to design and perform successful GWAS is our ability to understand how variants discovered via this hypothesis-free approach influence complex traits, In fact, few of the association studies go beyond reporting the most statistically significant hits and if they do, the suggested functionality is typically speculative, based on available annotation of genes in the vicinity of the variants. Since many of the discovered susceptibility polymorphisms fall in non-coding regions and with an increasing number of regulatory variants already implicated in a series of common disorders (8), one conventional approach has been to interrogate disease associated SNPs for associations with differential gene expression. Moffatt et al. (9) found that the same most significant SNPs associated with childhood asthma risk also explain ∼29.5% of the variance in ORMDL3 transcript levels, measured in lymphoblastoid cell lines. While an interesting observation, this still cannot be regarded as convincing evidence for a causal relationship between ORMDL3 and asthma onset. The concurrent progress towards uncovering the genetic basis of regulatory variation (10) has revealed an abundance of expression quantitative trait loci (eQTLs) in the human genome, making an accidental overlap between these and disease signals very likely. Thus, while gene expression is a very informative and immediate ‘DNA phenotype’, integrating expression data and disease studies genetics for an ultimate understanding of disease etiology is not straightforward.
ADVANCES AND CURRENT ISSUES IN EXPRESSION AND DISEASE STUDIES
Power of current eQTL studies
Natural variation in human gene expression has been recently quantified on a genome-wide scale using microarray technologies. Linkage and association studies coupling expression with genetic variability data have started to reveal the genetics underlying part of this variation, including complex allele-specific interactions (11) and its relatively high level of heritability (12–15). Most of the variants discovered with these approaches (a field also called Genetical Genomics) explain variance in transcript levels of nearby genes (so called cis eQTLs) but a few distal acting regulators have also been reported (trans associations). The sample sizes of genome-wide expression association studies have been fairly small though, meaning that the discoveries made so far represent generally large genetic effects [Stranger et al. (12) report an R2 coefficient of determination ranging from 0.27 to almost 1 for the SNP–gene associations detected in the 270 HapMap individuals]. The magnitude of the discovered effects drops when pooling populations together with appropriate corrections, a direct consequence of the increased statistical power due to the larger sample sizes. The importance of appropriate statistical power has been extensively demonstrated in complex disease GWAS, where samples of a few thousand paired cases and controls have become a prerequisite (16). The main reason for this requirement is the fact that the individual contribution of genetic variants towards complex trait determination is known to be small. In fact, all susceptibility alleles discovered so far explain only a small fraction of disease risk, with odds ratios typically in the range of 1.2–1.5 (16,17). Given the marked difference between the magnitudes of detected genetic effects on expression variation and disease predisposition, respectively, it is not surprising that only few instances of overlapping signals have been observed, even when expression in a disease relevant tissue was considered. Small genetic effects on expression variation or complex interactions between regulatory variants with moderate or large effects could become decisive on a permissive environmental background. Current expression analyses are underpowered with respect to these kinds of discoveries; hence whole-genome expression association studies on larger samples would be very desirable. Such efforts are on the way, including the quantification of expression levels in blood cells of 820 HapMap III individuals from eight populations (Barbara Stranger, Stephen Montgomery and Emmanouil Dermitzakis, personal communication). Combined with SNP genotyping data, this resource will give insight into the level of expression differences among populations and generate many additional eQTLs with more subtle effects, some of them potentially related to disease.
Tissue-specific phenotypes
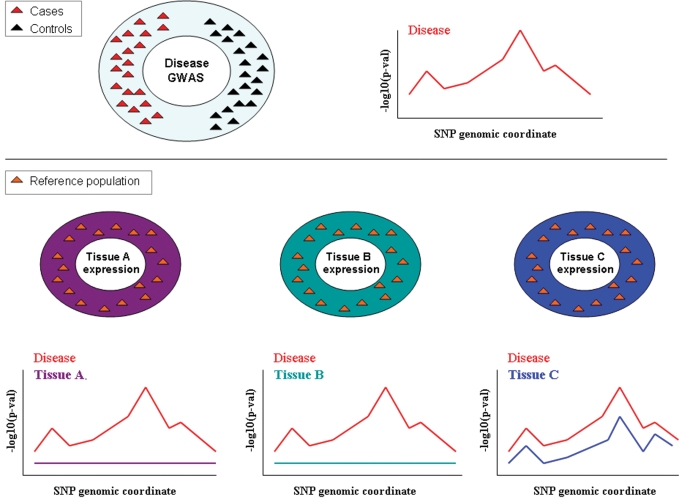
Confined by the availability of human tissue samples, expression experiments have been initially performed in lymphocytes or immortalized lymphoblastoid cell lines. In addition to having differential spatial and temporal expression patterns, some genes are only expressed in specific tissues. Also, many diseases manifest their phenotype in certain tissues exclusively. For these reasons, overlaying expression and disease signals is informative only if expression measurements are carried out in tissue types relevant to disease (Fig. 1). Particularly because our notion of relevance is still subjective in this case, identifying regulatory regions in multiple tissues is imperative for both a better understanding of the regulatory mechanisms in general and also the extent to which they are shared across tissues. Promising advancement in this direction can already be found in the literature. For the first time, genetic variants influencing normal human cortical expression have been reported (18). Myers et al. find little overlap between their set of eQTLs and results from previous studies on blood-derived cells. While differences in genotyping platforms and power can also partially explain this discrepancy, it is very likely that variants discovered here underlie brain-specific control of gene expression. These findings in conjunction with results from GWAS may help uncover the genetic basis of neurological disorders.
Figure 1.
Expression and disease association studies. Current expression studies are performed on a limited number of tissues derived from a small number of individuals compared with disease association studies. The availability of large collections of cell types from a large number of individuals will allow the identification of eQTLs that explain disease effects, such as the one at the top of the panel, when the relevant cell type/tissue is analyzed, as indicated for Tissue C at the bottom of the panel. This may not be successful in less relevant tissues such as in the case of Tissues A and B at the bottom of the panel.
A more comprehensive view on the extent of shared eQTLs among tissues has been presented in a recent study by Schadt et al. (19). The authors have analyzed 400 human liver samples and identified more than 6000 SNP–gene associations. Many of these genes had already been linked to a variety of complex diseases, a fact to be expected given that liver is known to be essential for many human metabolic processes. In addition to data from other disease GWAS, these eQTLs were integrated with gene expression and clinical data from segregating mouse populations in order to build probabilistic gene networks (20,21). This approach has led to the prioritization of susceptibility genes for coronary artery disease, LDL cholesterol levels and type I diabetes (T1D). More importantly, the same expression platform employed for the human liver cohort here had been also used on a set of human blood and adipose tissues in another study (22). This allowed a direct evaluation of the cis eQTL overlap in the three tissues, which amounted to ∼30%. While still a rough estimate, this amount of shared regulatory control makes a good case for interrogating disease susceptibility variants for expression associations in any available data set. Nevertheless, the remaining unshared fraction reflects the expected tissue-specific regulatory activity. These biological processes will have to be studied in appropriate tissues before they become useful for understanding tissue-specific complex disorders. It is of course still unclear what the pattern of diminishing returns is across human tissues and what set of tissues could serve as highly informative in large cohort collections.
Finding the causal variants in expression and disease GWAS
Even once expression data become available for a large variety of human tissues, the ability to pinpoint causal variants predisposing to disease via a regulatory mechanism will still depend on the density at which such variants have been interrogated. Currently, all genotyping platforms survey only a subset of the human sequence variation (23). Typing all common SNPs in the genome (currently estimated around 10 million) has been replaced by the inference of tag SNPs, a subset of approximately 300 000–600 000 markers, depending on the population. This reduction exploits the pervasiveness of linkage disequilibrium (LD) in the human genome (24) and captures most of the common genetic variation in a region of interest. Still, this means that reported susceptibility variants are most probably only tagging the real functional variants and are not causal themselves. Hence, initial discoveries must be followed by fine mapping of the regions harboring the most significant statistical signals (25). So far this has not been thoroughly attempted, primarily because in the absence of other prior biological information, such tasks are financially prohibitive. Next generation sequencing will allow transcriptomic studies that will go deep into the transcript structure and abundance and will resolve issues of alternative splicing. In terms of in-depth interrogation of human genetic variation, a new resource (http://www.1000genomes.org/) promises to facilitate these efforts by generating a human genetic variation map at unprecedented resolution. The 1000 Genomes Project will sequence more than 1000 individuals with the final goal of cataloging almost all variants found at minor allele >1% frequency in human populations. Within genes, sequencing will go even deeper, down to 0.5% frequency. As a result, much like common variation documented by the HapMap has been used so far to perform successful GWAS, this project will further support disease studies aiming at those relatively rare variants. Importantly, it will allow the assessment of the individual impact of rare variants on complex traits, which has not been understood so far.
APPROACHES OF COMBINING EXPRESSION AND DISEASE DATA
Comparison of expression profiles in cases and controls
Gene expression signatures in relation to complex diseases have been investigated well before the era of GWAS. The area of cancer genetics is one such example. Expression profiles of different tumors have been broadly analyzed in order to categorize cancer subtypes and even identify the genes that determine the subtype identification (26). Higher resolution diagnoses followed, as well as the hope that candidate cancer genes could be more easily identified. An obvious approach seemed to be the comparison of gene expression levels in the same tissue from patients and non-diseased individuals and from the differences observed to infer potentially causal pathways and biochemical functions. Given the substantial expression profile changes it causes in the cell, the complexity of genome rearrangements and the somatic nature of some of the effects, cancer is likely to be an exceptionally difficult case, and requires a slightly different treatment so we do not discuss it in this review. Even in other diseases, the daunting task in this type of experiment remains distinguishing causal from reactive effects. Changes at the cellular level will be produced as a result of the disease, and discriminating these from causal changes is extremely challenging. In addition, not all individuals have the same causal pathways so it is not necessarily true that disease and non-disease individuals have clear and well-defined differences in gene expression when compared. Finally, it is essential to derive the genetic dimension of such differences and not simply describe the gene expression differences. For all these reasons, it becomes evident that expression information derived from a reference set of samples rather than patients would be more useful for determining the disease predisposing genetic factors, while diagnosis purposes can still be served by quantifying expression in patient-derived samples. Patient samples and cellular resources will also be very useful in the context of cell-based studies that may investigate the impact of challenging these cells with various agents and candidate drugs.
Individual effect of disease variants on transcription levels
Expression variation in the general population is being actively researched and its genetic determinants uncovered. Without the burden of chasing a reactive effect, another common approach has been to use this data for determining whether disease variants from GWAS are also responsible for natural variation in human transcript levels. The underlying principle is the following: if one allele is more frequent in cases than controls and at the same time it is causal for gene expression effects of a nearby gene, which is itself important for the disease, then it is likely that causality can be established. Few genes have been prioritized by this method (19), but several caveats make it a difficult task. For example, the same best candidate SNP could be significantly associated with discernible expression differences in several nearby genes. Deciding which one among those is the best candidate would not be feasible. Moreover, absence of evidence is not evidence of absence: not finding expression associations with some genes flanking the variant of interest (provided that expression data is available for all of those genes and there is an effect on expression that eventually induces disease) does not mean that they are not potential disease contributors. Plenty of reasons from reduced power in the expression study to interrogation of a cell type irrelevant to disease could explain that.
Lessons from gene networks
The most successful integration of disease and expression data, circumventing most of the previously listed caveats has been achieved by Chen et al. (27). The authors exploit for the first time the complexity of interactions that lead to disease by modeling it as a system. Gene expression networks from liver and adipose tissues of segregating mouse populations were constructed and subsequently, network disruptions caused by DNA variations were observed. This allowed the identification of a particular network (macrophage-enriched metabolic network) enriched for genes correlated to obesity-related phenotypes. Experimental validation of the top candidate genes further supports the validity of this method. A similarly constructed network was also identified in a human blood and adipose tissue cohort (22), overlapping significantly with the mouse derived one. In addition to this discovery, gene network theory applied here demonstrated once more the naivety with which some of the links between expression and disease could be made. Despite the fact that variants associated with obesity traits resided in a region with known Apoa2 eQTLs, a gene involved in metabolic activity, it was other genes in the region that exhibited a significant linkage pattern with the phenotypes of interest. This argues for an independent relationship between Apoa2 expression and the metabolic traits under study, even though the same locus is associated with both. Clearly, the ability to distinguish between such coincidental and actual causal effects is essential. Studies based on the use of networks rather than sets of eQTLs show some theoretical promise since they account for additional complexity, but until we have a good sense of the complexity framework in which we can embed the impact of genetic variation they will lag behind.
Statistical challenges in combining GWAS and eQTL studies
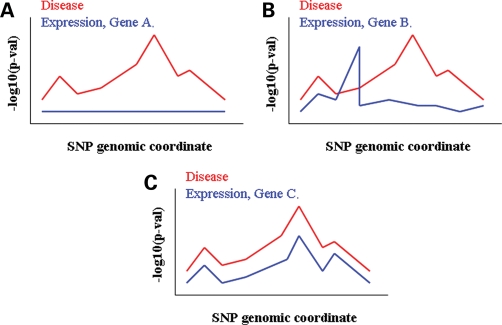
Finally, the ever-increasing technological advancement supporting both expression and disease GWAS in humans, calls for the development of proper statistical tools to combine the two directly. Local genomic regions of interest with association data available from both types of analyses could provide gene prioritization clues, if explored properly. Superimposing patterns of association at commonly tested loci in expression and disease could distinguish between multiple genes affected in cis by the same variants (Fig. 2). This kind of discrimination would not be possible by solely examining the most significant disease variant for effects on expression. However, the same property that makes such correlations sound—LD—also forces the need for proper statistical assessment of the results. All variants in a genomic interval that have been historically co-segregating with the causal variant will be significantly associated with the two phenotypes by chance. On one hand, this allows present association studies to be performed on just subsets of SNPs, but on the other hand, regions of high LD will exhibit tighter relationships between expression and disease patterns. This is not biologically meaningful in terms of disease and as such, correcting out the LD effect is necessary. In addition, as already pointed out, eQTLs are ubiquitously distributed throughout the genome. When analyzing genome-wide expression data in lymphoblastoid cell lines of the HapMap II individuals, Stranger et al. (12) found 4043 genes with at least one eQTL at 0.05 permutation threshold. Considering the 32 996 genome-wide hotspot intervals estimated by McVean et al. (28) from the HapMap data, we can approximate that the probability of finding an eQTL in any hotspot interval in the human genome (and therefore likely in LD with a disease variant in the interval) by chance is ∼0.123. Therefore, irrespective of the LD pattern, the probability of finding a good match between disease and expression signals is high. These numbers will be even higher when studies with larger sample sizes and increased sets of tissues are available where it is possible that every single SNP may be in LD with an eQTL in some population and/or some tissue. In this context, a statistical framework accounting for LD while at the same time evaluating the probability of observing disease and expression correlations by chance would be ideal.
Figure 2.
Local superimposition of whole-genome disease and expression associations. For any given genomic interval, same SNPs will have been independently tested for associations with a disease and transcript levels of a set of genes, respectively. (A) No significant eQTLs detected in the interval for Gene A. (B) Significant eQTLs for Gene B do not also display significant disease associations. (C) The association patterns of disease and Gene C expression fit very well, making it the most likely candidate out of the three for a possible regulatory-mediated disease effect.
CONCLUSIONS AND FUTURE DIRECTIONS
Genetics has played a crucial role in the advancement of medical sciences by facilitating the understanding of human disease. In addition to the early success in elucidating numerous rare monogenic disorders (29), it has also recently become possible to discover genetic markers robustly associated with complex diseases. Being able to overcome the statistical limitation of linkage studies (30), GWAS are now routinely employed for a variety of human conditions. By themselves however, statistically significant signals resulting from such studies still cannot answer important biological questions such as which are the causal variants and how do they lead to disease. Thus, the necessity of incorporating additional information when studying complex disease etiology has become apparent. Motivated by the simultaneous progress in understanding the role of regulatory variants in shaping human phenotypic variation, gene expression has become an obvious candidate as informative intermediate ‘DNA phenotype’. How to best integrate the two levels of complexity is nonetheless far from obvious. Currently, both expression and disease GWAS suffer from critical limitations. Expression studies have been performed on relatively small samples and on a restricted variety of tissue. Also, the transcriptome’s diversity such studies explore is bound to be incomplete, with only few probes presently designed per gene. In terms of the genetic variety underlying phenotypic differences, the picture is also far from complete. Genotyping platforms survey only a subset of the known human genetic variability, so the candidates found are most probably not causal, but just correlated with the real functional variants because of LD. All these caveats explain why a clear link between the genetics of disease and that of gene expression has not been established yet, despite its existence. Fortunately, the incredibly high rate at which technology is advancing will soon eliminate some of these limitations. Next generation sequencing has already made efforts like the 1000 Genomes Project possible, which would have seemed unthinkable few years ago. On the same note, we envisage in the near future a public resource providing complete expression profiles from a multitude of healthy human tissues derived from large samples of individuals. In conjunction with next generation sequencing data which will also greatly improve the resolution of disease GWAS, this resource would be the ideal solution to the present-day problems. In the meantime, alternative methods of integrating genotypic and phenotypic information towards a better understanding of human disease will have to be developed.
FUNDING
A.C.N. and E.T.D. are supported by Wellcome Trust funding.
ACKNOWLEDGEMENTS
We would like to thank Barbara Stranger, Stephen Montgomery, Antigone Dimas, Simon Tavare, Panos Deloukas and Gillean McVean for helpful discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rice T.K., Schork N.J., Rao D.C. Methods for handling multiple testing. Adv. Genet. 2008;60:293–308. doi: 10.1016/S0065-2660(07)00412-9. [DOI] [PubMed] [Google Scholar]
- 3.Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeggini E., Scott L.J., Saxena R., Voight B.F., Marchini J.L., Hu T., de Bakker P.I., Abecasis G.R., Almgren P., Andersen G., et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hung R.J., McKay J.D., Gaborieau V., Boffetta P., Hashibe M., Zaridze D., Mukeria A., Szeszenia-Dabrowska N., Lissowska J., Rudnai P., et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 6.Loos R.J., Lindgren C.M., Li S., Wheeler E., Zhao J.H., Prokopenko I., Inouye M., Freathy R.M., Attwood A.P., Beckmann J.S., et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudbjartsson D.F., Walters G.B., Thorleifsson G., Stefansson H., Halldorsson B.V., Zusmanovich P., Sulem P., Thorlacius S., Gylfason A., Steinberg S., et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 8.Stranger B.E., Dermitzakis E.T. From DNA to RNA to disease and back: the ‘central dogma’ of regulatory disease variation. Hum. Genomics. 2006;2:383–390. doi: 10.1186/1479-7364-2-6-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moffatt M.F., Kabesch M., Liang L., Dixon A.L., Strachan D., Heath S., Depner M., von Berg A., Bufe A., Rietschel E., et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 10.Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao H., Cox D.R., Frazer K.A. Allele-specific KRT1 expression is a complex trait. PLoS Genet. 2006;2:e93. doi: 10.1371/journal.pgen.0020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D., et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 14.Goring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G., et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 15.Cheung V.G., Spielman R.S., Ewens K.G., Weber T.M., Morley M., Burdick J.T. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., Perry J.R., Elliott K.S., Lango H., Rayner N.W., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers A.J., Gibbs J.R., Webster J.A., Rohrer K., Zhao A., Marlowe L., Kaleem M., Leung D., Bryden L., Nath P., et al. A survey of genetic human cortical gene expression. Nat. Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 19.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schadt E.E., Lamb J., Yang X., Zhu J., Edwards S., Guhathakurta D., Sieberts S.K., Monks S., Reitman M., Zhang C., et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J., Wiener M.C., Zhang C., Fridman A., Minch E., Lum P.Y., Sachs J.R., Schadt E.E. Increasing the power to detect causal associations by combining genotypic and expression data in segregating populations. PLoS Comput. Biol. 2007;3:e69. doi: 10.1371/journal.pcbi.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S., et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 23.Hirschhorn J.N., Daly M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 24.Cardon L.R., Abecasis G.R. Using haplotype blocks to map human complex trait loci. Trends Genet. 2003;19:135–140. doi: 10.1016/S0168-9525(03)00022-2. [DOI] [PubMed] [Google Scholar]
- 25.McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P., Hirschhorn J.N. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 26.Khan J., Wei J.S., Ringner M., Saal L.H., Ladanyi M., Westermann F., Berthold F., Schwab M., Antonescu C.R., Peterson C., et al. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat. Med. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., Zhu J., Lum P.Y., Yang X., Pinto S., MacNeil D.J., Zhang C., Lamb J., Edwards S., Sieberts S.K., et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McVean G.A., Myers S.R., Hunt S., Deloukas P., Bentley D.R., Donnelly P. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 29.Jimenez-Sanchez G., Childs B., Valle D. Human disease genes. Nature. 2001;409:853–855. doi: 10.1038/35057050. [DOI] [PubMed] [Google Scholar]
- 30.Risch N., Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]