Abstract
Genome-wide association studies are providing new insights into the genetic basis of metabolic and cardiovascular traits. In the past 3 years, common variants in ∼50 loci have been strongly associated with metabolic and cardiovascular traits. Several of these loci have implicated genes without a previously known connection with metabolism. Further studies will be required to characterize the full impact of these loci on metabolism. Many of the identified loci include multiple independent variants that influence the same metabolic or cardiovascular trait and a few loci harbor independent variants that each influence distinct traits. The total proportion of trait heritability explained by variants identified so far is still modest (typically <10%). Future studies will build on these successes by identifying additional common and rare variants and by determining the functional impact of the underlying alleles and genes.
INTRODUCTION
Metabolic and cardiovascular diseases (CVD) are common, complex traits with a substantial public health burden. Although disease prevalence varies with age, gender and population, the World Health Organization estimates that in 2005, 17.5 million deaths (∼30% of the global total) were due to CVD (1). In many individuals, CVD coexists with a set of metabolic risk factors including diabetes, central obesity, unhealthy lipid profiles, high blood pressure, a prothrombotic state, and a proinflammatory state, each of which is likely regulated by genetic and environmental risk factors (2). There is a strong genetic component to these traits; one recent large-scale study found that on an average, genetic effects explain 25% of the variance for 20 measures of cardiovascular function, 51% for five anthropometric measures, and 40% for 38 blood tests including cholesterol and other metabolic measures (3).
Until recently, the catalog of common genetic variants influencing metabolism and CVD was limited and obtained painfully slowly. Candidate gene studies and linkage analyses identified only modest numbers of susceptibility loci that were consistently replicated in additional large studies. In contrast, genome-wide association (GWA) studies have dramatically increased the number of common variants with confirmed association with metabolic and cardiovascular traits. In this article, we review the recent progress identifying loci through GWA studies and outline the challenges for the future. Given space limitations, our aim is to provide a flavor of the discoveries made to date, and to synthesize important trends and future directions. Thus, we focus on GWA studies of >200 000 single nucleotide polymorphisms (SNPs) that reported at least one SNP exceeding an arbitrary statistical significance threshold of P = 5 × 10−8. We have tried to include independent GWA studies that identified the same locus within ∼3 months of the first report. Of the traits relevant to metabolism and cardiovascular risk, we focus primarily on type 2 diabetes, coronary artery disease (CAD) and myocardial infarction (MI), cholesterol and lipid levels, and obesity-related traits.
TYPE 2 DIABETES
In the past decade, candidate gene studies and linkage analyses provided definitive evidence for common susceptibility variants in at least five loci. Specifically, candidate gene studies based on prior understanding of biological function or of monogenic forms of diabetes identified common variants in the genes encoding peroxisome proliferator-activated receptor-g (PPARG) (4,5), potassium inwardly-rectifying channel, subfamily J, member 11 (KCNJ11) (6), Wolfram syndrome 1 (WFS1) (7), and transcription factor 2, hepatic (TCF2) (8). Linkage analysis and fine mapping identified common type 2 diabetes (T2D)-associated variants within transcription factor 7-like 2 (TCF7L2) (9), a locus not previously suspected to influence T2D or related traits and which remains the most strongly associated T2D susceptibility locus to date (Table 1).
Table 1.
Loci associated with metabolic or cardiovascular traits and identified or confirmed by GWA with significance P < 5 × 10−8
| Trait | Nearby gene(s) | Representative SNP | P-value | OR | 95% CI | Effect | References |
|---|---|---|---|---|---|---|---|
| T2D | TCF7L2 | rs7903146 | 4.6 × 10−49 | 1.37 | 1.32–1.43 | – | (11–14) |
| T2D | CDKN2A-CDKN2B | rs2383208 | 2.7 × 10−15 | 1.20 | 1.15–1.25 | – | (11–14) |
| T2D | FTO | rs8050136 | 1.5 × 10−12 | 1.17 | 1.12–1.22 | – | (11–14) |
| T2D | THADA | rs7578597 | 1.1 × 10−9 | 1.15 | 1.10–1.20 | – | (18) |
| T2D | IGF2BP2 | rs4402960 | 8.9 × 10−15 | 1.14 | 1.10–1.18 | – | (11–14) |
| T2D | KCNJ11 | rs5219 | 5.4 × 10−11 | 1.14 | 1.10–1.19 | – | (11–14) |
| T2D | PPARG | rs1801282 | 2.3 × 10−6 a | 1.14 | 1.08–1.20 | – | (11–14) |
| T2D | CDKAL1 | rs4712523 | 1.6 × 10−12 | 1.13 | 1.09–1.17 | – | (11–14) |
| T2D | HHEX-IDE | rs1111875 | 6.9 × 10−10 | 1.13 | 1.09–1.17 | – | (11–14) |
| T2D | NOTCH2 | rs10923931 | 4.1 × 10−8 | 1.13 | 1.08–1.17 | – | (18) |
| T2D | SLC30A8 | rs13266634 | 2.4 × 10−7 a | 1.12 | 1.07–1.17 | – | (11–14) |
| T2D | CDC123-CAMK1D | rs12779790 | 1.2 × 10−10 | 1.11 | 1.07–1.14 | – | (18) |
| T2D | JAZF1 | rs864745 | 5.0 × 10−14 | 1.10 | 1.07–1.13 | – | (18) |
| T2D | TSPAN8-LGR5 | rs7961581 | 1.1 × 10−9 | 1.09 | 1.06–1.12 | – | (18) |
| T2D | ADAMTS9 | rs4607103 | 1.2 × 10−8 | 1.09 | 1.06–1.12 | – | (18) |
| CAD | CDKN2A-CDKN2B | rs1333049 | 2.9 × 10−19 | 1.36 | 1.27–1.46 | – | (13,22,23,28) |
| CAD | MTHFD1L | rs6922269 | 2.9 × 10−8 | 1.23 | 1.15–1.33 | – | (13,28) |
| CAD | CELSR2-PSRC1-SORT1 | rs599839 | 4.1 × 10−9 | 1.29 | 1.18–1.40 | – | (13,28) |
| HDL-C | CETP | rs3764261 | 2.3 × 10−57 | – | – | 3.47 mg/dl | (29,30,32,33) |
| HDL-C | LPL | rs10503669 | 4.1 × 10−19 | – | – | 2.09 mg/dl | (29,30,32,33) |
| HDL-C | LIPC | rs4775041 | 3.2 × 10−20 | – | – | 1.38 mg/dl | (29,30) |
| HDL-C | LIPG | rs2156552 | 6.4 × 10−12 | – | – | 1.20 mg/dl | (29,30) |
| HDL-C | GALNT2 | rs2144300 | 2.6 × 10−14 | – | – | 1.11 mg/dl | (29,30) |
| HDL-C | ABCA1 | rs4149268 | 1.2 × 10−10 | – | – | 0.82 mg/dl | (29,30,33) |
| HDL-C | MMAB-MVK | rs2338104 | 3.4 × 10−8 | – | – | 0.48 mg/dl | (29) |
| LDL-C | LDLR | rs6511720 | 4.2 × 10−26 | – | – | 9.17 mg/dl | (29–31) |
| LDL-C | APOE-C1-C4 | rs4420638 | 3.0 × 10−43 | – | – | 6.61 mg/dl | (29–32) |
| LDL-C | CELSR2-PSRC1-SORT1 | rs599839 | 6.1 × 10−33 | – | – | 5.48 mg/dl | (29–32) |
| LDL-C | APOB | rs562338 | 5.6 × 10−22 | – | – | 4.89 mg/dl | (29–32) |
| LDL-C | NCAN-CILP2 | rs16996148 | 2.7 × 10−9 | – | – | 3.32 mg/dl | (29,30) |
| LDL-C | PCSK9 | rs11206510 | 3.5 × 10−11 | – | – | 3.04 mg/dl | (29,30) |
| LDL-C | HMGCR | rs12654264 | 1 × 10−20 | – | – | 3.30 mg/dl | (30) |
| Triglycerides | APOA5-A4-C3-A1 | rs12286037 | 1.0 × 10−26 | – | – | 25.82 mg/dl | (29,30,32,33) |
| Triglycerides | LPL | rs10503669 | 3.9 × 10−22 | – | – | 11.57 mg/dl | (29,30,32,33) |
| Triglycerides | GCKR | rs780094 | 6.1 × 10−32 | – | – | 8.59 mg/dl | (29,30,32) |
| Triglycerides | MLXIPL | rs17145738 | 2.0 × 10−12 | – | – | 8.21 mg/dl | (29,30,33) |
| Triglycerides | ANGPTL3 | rs1748195 | 1.7 × 10−10 | – | – | 7.12 mg/dl | (29,30) |
| Triglycerides | TRIB1 | rs17321515 | 7.0 × 10−13 | – | – | 6.42 mg/dl | (29,30) |
| Triglycerides | NCAN-CILP2 | rs16996148 | 2.5 × 10−9 | – | – | 6.10 mg/dl | (29,30) |
| Triglycerides | LIPC | rs4775041 | 1.6 × 10−8 | – | – | 3.62 mg/dl | (29) |
| Triglycerides | GALNT2 | rs4846914 | 7 × 10−15 | – | – | 6.40 mg/dl | (30) |
| BMI | FTO | rs9939609 | 3 × 10−35 | – | – | ∼0.4 kg/m2 | (16,17) |
| BMI/waist circumference | MC4R | rs1778231 | 2.8 × 10−15 | – | – | .05 logBMI Z score |
(40,41) |
| Fat mass | CTNNBL1 | rs6013029 | 5.0 × 10−8 | – | – | 5.96 kg | (42) |
| QT interval | NOS1AP | rs10494366 | <1 × 10−10 b | – | – | 2.45 msb | (44) |
| Fasting glucose | G6PC2 | rs563694 | 6.4 × 10−33 | – | – | 0.01–0.16 mm | (46,47) |
| Serum uric acid | SLC2A9 | rs6855911 | 1.8 × 10−16 | – | – | 0.3 mg/dl | (32,48–50) |
| CRP | APOE | rs769449 | 8.9 × 10−21 | – | – | –0.261c | (51,52) |
| CRP | CRP | rs3091244 | 6.2 × 10−28 | – | – | 0.203c | (51,52) |
| CRP | LEPR | rs1892534 | 6.5 × 10−21 | – | – | −0.170c | (52) |
| CRP | HNF1A | rs7310409 | 6.8 × 10−17 | – | – | −0.154c | (51,52) |
| CRP | GCKR | rs780094 | 6.7 × 10−15 | – | – | 0.140c | (52) |
| CRP | 12q23 | rs10778213 | 1.2 × 10−10 | – | – | –0.115c | (52) |
| CRP | IL6R | rs8192284 | 1.9 × 10−8 | – | – | −0.101c | (52) |
Table includes a single SNP reported at a given locus, with a corresponding significance level and odds ratio (OR) or measure of effect from one of the listed references. Within each trait, loci are sorted by OR or effect size. GWA studies are included as references if they reported a result within a few months of the first report. T2D, type 2 diabetes; CAD, coronary artery disease; HDL-C, high density lipoprotein cholesterol; LDL-C, low density lipoprotein cholesterol; BMI, body mass index; CRP, C-reactive protein.
aSignificance exceeds 5 × 10−8 threshold with additional samples.
bSignificance and average per allele difference from subset of samples in Table 1 of reference (44).
cRegression coefficient.
In 2007, the first GWA studies describing T2D susceptibility loci were reported. Five GWA studies were conducted in populations of European ancestry, with the most significant SNPs followed up by genotyping of additional samples (10–15). These studies replicated evidence for TCF7L2, PPARG and KCNJ11, and identified at least six additional loci (Table 1). Strongly significant variants were identified at solute carrier family 30 (zinc transporter), member 8 (SLC30A8), insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2), fat mass and obesity-associated (FTO), near hematopoietically expressed homeobox and insulin degrading enzyme (HHEX-IDE), CDK5 regulatory subunit-associated protein 1-like 1 (CDKAL1), and distal to the genes cyclin-dependent kinase inhibitors 2A and 2B (CDKN2A-CDKN2B, which encode proteins also known as p16INK4a and p15INK4b). Among the reported SNPs, the only likely functional variant identified is a non-synonymous Arg325Trp substitution in SLC30A8. Although most of these loci implicate pancreatic beta-cell biological processes, the T2D-associated common variants in FTO are also strongly associated with body mass index (BMI) (16,17).
More recently, a large meta-analysis of ∼2.2 million genotyped or imputed SNPs in 10 128 individuals with further genotyping in up to 79 792 individuals identified six additional loci meeting a statistical significance threshold of P = 5 × 10−8 (18). These six signals are located in or near the genes JAZF1, CDC123-CAMK1D, TSPAN8-LGR5, THADA, ADAMTS9 and NOTCH2 (Table 1). The first three of these loci listed are associated with measures of insulin release, again implicating abnormal pancreatic beta-cell function in the pathogenesis of T2D (19). The total number of T2D susceptibility loci is now at least 17, and larger scale meta-analyses currently underway have the potential to identify additional susceptibility variants.
CAD AND MI
Compared with T2D, fewer genetic variants meet our threshold of P = 5 × 10−8 in published GWA studies of CAD and MI. An early study of 65 671 gene-based SNPs in individuals of Japanese ancestry identified association with MI at SNPs in lymphotoxin-A (LTA), including one SNP whose alleles were also associated with LTA transcription level (20), although later studies provided inconsistent replication (21).
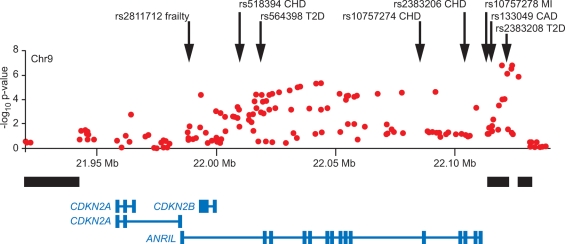
Three recent independent GWA studies of CAD or MI identified strong evidence of association with SNPs near the CDKN2A and CDKN2B genes (13,22,23) (Table 1). These studies reported SNPs in strong linkage disequilibrium (LD) with each other in populations of European ancestry (Fig. 1), and secondary signals in the adjacent LD block were identified as well (23). The variants most strongly associated with CAD are not in strong LD (r2 < .01) with the variants most strongly associated with T2D described earlier, although secondary signals for both traits are in LD with each other (e.g. r2 between secondary coronary heart disease SNP rs518394 and secondary T2D SNP rs564398 = 0.75 in HapMap CEU). As shown in Figure 1, this genomic region also contains SNPs associated with frailty (24) and melanoma (25). In addition to the CDKN2A and CDKN2B genes, which play a role in the cell cycle, this region contains the non-coding antisense RNA named ANRIL, which has been shown to be expressed in tissues involved in atherosclerosis (26,27). A replication study confirmed that the CAD and T2D associations were independent of each other and that these SNPs were not associated with differences in plasma levels of cholesterol, fibrinogen, albumin, uric acid, bilirubin, or homocysteine (27).
Figure 1.
Evidence for association with several traits on chromosome 9p21. Evidence for association with type 2 diabetes (T2D) is shown for single nucleotide polymorphisms (SNPs) evaluated in 10 128 GWA samples (18); P-values are substantially more significant when follow-up samples are included (see Table 1). Arrows indicate the locations of SNPs reported to be associated with frailty, coronary heart disease (CHD), myocardial infarction (MI), and coronary artery disease (CAD). Black bars indicate the locations of recombination hotspots, and the locations of genes and transcripts are indicated at the bottom.
One GWA study identified additional SNPs associated with CAD using replication samples from an MI study (28). One SNP is in the gene for methylenetetrahydrofolate dehydrogenase (NADP+-dependent) 1-like protein (MTHFD1L). The encoded protein is involved in the synthesis of purines and regeneration of methionine, and may influence homocysteine levels. Another locus with strong evidence of association maps near the PSRC1 gene; as described in the next section, the same SNP is also associated with low density lipoprotein cholesterol (LDL-C). Several other additional loci have been reported that do not quite reach the threshold of P = 5 × 10−8. Further functional experiments will be required to discern the molecular mechanisms mediating the impact on CAD and MI of this collection of signals.
CHOLESTEROL LIPOPROTEINS AND LIPIDS
Among the cardiovascular and metabolic traits, analyses of lipoprotein and lipid levels have been among the most successful in terms of the number of loci identified or confirmed by GWA studies (Table 1). An initial GWA study reported an association of the glucokinase regulator (GCKR) with serum triglycerides (12). Two subsequent GWA studies analyzed high density lipoprotein cholesterol (HDL-C), LDL-C, and triglycerides concentrations in 8816 individuals and performed follow-up analysis in independent sample sets (29,30). Together, these studies showed strong support for association with variants in or near GCKR and 11 additional previously reported loci, as well as seven new loci. The previously identified loci included three apolipoprotein genes or gene clusters (APOB, APOE-APOC1-APOC4-APOC2, APOA1-APOC3-APOA4-APOA5); hepatic, endothelial and lipoprotein lipases (LIPC, LIPG, LPL); cholesterol ester transfer protein (CETP); LDL receptor (LDLR); hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR); proprotein convertase subtilisin/kexin type 9 (PCSK9); and ATP-binding cassette, subfamily A, member 1 (ABCA1). For several of these loci, the common variant identified by GWA is in weak LD with previously identified common variants and likely represents a new signal; e.g. at the LIPC locus, a set of common variants ∼50 kb upstream of the gene are strongly associated with HDL-C and appear to be independent of previously identified variants that overlap the transcribed sequence of the gene (29).
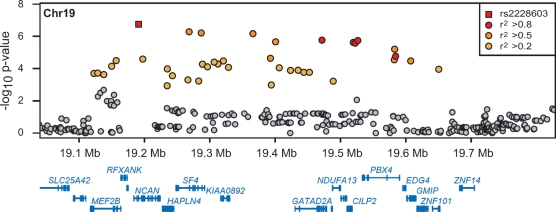
The two studies also reported seven novel loci associated with lipoprotein or lipid levels (29,30). SNPs near the mevalonate kinase (MVK) – methylmalonic aciduria cblB type (MMAB) locus and in the N-acetylgalactosaminyltransferase GALNT2 were found to be associated with HDL-C. SNPs near the SORT1-PSRC1-CELSR2 locus were found to be associated with LDL-C, and two additional studies independently identified this locus (31,32). An SNP at this locus, rs646776, was strongly associated with transcript concentrations of the three neighboring genes and explained 58–86% of the inter-individual variability in their transcript levels (30). Of 11 SNPs at seven loci associated with LDL-C, Willer et al. noted that all of the alleles associated with increased LDL-C were also associated with CAD in a Wellcome Trust Case Control Consortium (WTCCC) sample (13). SNPs primarily associated with triglycerides were identified near the G-protein-coupled-receptor-induced protein tribbles 1 (TRIB1), near angiopoietin-like 3 (ANGPTL3), and in MLX interacting protein-like (MLXIPL). Kooner et al. (33) independently reported MLXIPL association with triglycerides, including a non-synonymous SNP (rs3812316, Gln241His). SNPs at a locus encompassing more than a dozen genes near NCAN were strongly associated with both triglycerides and LDL-C (Fig. 2); the large number of genes, none of which are overwhelming candidates for a role in lipoprotein or lipid biology, highlights the challenge ahead to determine the underlying functional gene. Taken together, the combined effects of these common variants account for only 5–8% of the variation in the three lipid traits, leaving most of the heritability of these traits to be explained.
Figure 2.
Evidence for association with low density lipoprotein cholesterol (LDL-C) at a locus spanning many genes. Evidence for association is shown for evaluated single nucleotide polymorphisms (SNPs) in 8816 GWA samples (29). SNPs are colored according to their degree of linkage disequilibrium with rs2228603, the SNP most strongly associated with LDL-C in this region. The bottom panel depicts the locations of genes. Figure adapted from Willer et al. (29).
OBESITY, BMI, WAIST CIRCUMFERENCE AND FAT MASS
Over the last two decades, candidate gene and linkage studies have reported many loci for obesity and related traits, but few have been reproducibly confirmed. A review of the obesity literature through October 2005 summarized associations with 127 candidate genes and 253 quantitative trait loci for obesity-related phenotypes from linkage studies (34). Recently, common non-synonymous variants in proprotein convertase subtilisin/kexin type 1 (PCSK1) were found to be associated with obesity (P < 8 × 10−8) in adults and children (35). An early GWA report from the Framingham Heart Study offspring study described a locus near the insulin-induced gene 2 (INSIG2) gene associated with BMI (36), and a follow-up study of ∼17 000 individuals in nine cohorts from eight populations across multiple ethnicities confirmed the association in both unrelated and family-based samples, but with a modest effect (37).
In 2007, two independent GWA studies identified obesity-associated variants within the first intron of the FTO gene (16,17). The association between FTO variants and BMI was independently identified in a third study (38) and has since been replicated in many cohorts. FTO encodes a 2-oxoglutarate-dependent nucleic acid demethylase, and the functional link between this methylase (or another nearby gene) and obesity is not yet known (39), emphasizing the benefits of unbiased genome-wide studies to identify susceptibility genes.
Two recent large GWA studies for obesity-related traits identified associated SNPs near the melanocortin-4 receptor (MC4R) gene. A study of BMI in 16 876 samples with follow-up in >60 000 adults and almost 6000 children identified variants >100 kb downstream of MC4R (40), and a study of waist circumference and insulin resistance in 2684 individuals described similar associations (41). MC4R contains uncommon amino acid substitutions Val103Ile and Ile251Leu known previously to influence obesity, but the associations with common variants downstream of MC4R cannot be explained by these substitutions (40). Together FTO and MC4R variants explain only a small portion of inter-individual variation in BMI, ∼1.2 kg/m2 in adults (40).
A third recent GWA study for BMI and fat mass in a sample of 1000 unrelated United States Caucasians with follow-up in >3800 individuals in a French case–control sample identified variants in catenin, beta-like 1 (CTNNBL1) (42). The initial evidence of association (P = 5.0 × 10−8 for fat mass) became weaker after correction for stratification using genomic control (43). Replication in additional cohorts will be important to determine the impact of this locus.
OTHER QUANTITATIVE TRAITS
GWA studies have been reported for several other quantitative traits relevant to metabolic and cardiovascular traits (Table 1). One of the first successes was a multi-stage GWA study of the QT interval, a risk factor for sudden cardiac death that identified association with variants near the NOS1AP gene (44). More recently, a very large number of traits associated with cardiovascular and metabolic traits have been dissected using GWA. For fasting glucose level, common sequence variants in the glucokinase (GCK) promoter were reported to be strongly associated (P ∼ 10−9) based on candidate gene analyses (45), and two GWA studies identified variants in islet-specific glucose-6-phosphatase, catalytic, 2 (G6PC2) (46,47). The first GWAs for serum uric acid levels identified strongly significant variants at solute carrier family 2 (facilitated glucose transporter), member 9 (SLC2A9) (32,48). An additional independent GWA study reported a significantly stronger association between SLC2A9 variants and urate concentration in women compared with men (49), and another independent GWA demonstrated that SLC2A9 can transport uric acid in a Xenopus laevis model (50).
Plasma concentrations of C-reactive protein (CRP) reflect systemic inflammation, which co-occurs with CVD; two GWA studies of plasma CRP level identified seven significant (P < 5 × 10−8) loci (51,52). The most strongly associated loci include the CRP gene itself and the APOE locus, both of which were previously known from candidate gene studies, as well as the leptin receptor (LEPR). In addition, association with CRP is observed for variants associated with other traits or disease, including HNF1 homeobox A (HNF1A), variants of which are known to cause maturity-onset diabetes of the young, and GCKR, associated with triglycerides as described earlier. CRP levels are also associated with the interleukin 6 receptor (IL6R) and a gene desert region on chromosome 12q23.2.
Genes for several additional cardiovascular and metabolic traits from GWA studies have not yet been reported. Notably lacking from the GWA harvest to date are loci for hypertension and systolic and diastolic blood pressure, and very large meta-analyses may be needed to detect reproducibly associated SNPs. The WTCCC reported no results achieving a threshold of 5 × 10−7 for hypertension (13), suggesting that there may be fewer common alleles with large effect on this trait.
CHALLENGES FOR THE FUTURE
Many challenges remain to identify the genetic variants that influence cardiovascular and metabolic traits. Larger sample sizes will be needed to continue to implicate additional loci, and as cohorts are combined, heterogeneity across studies may become a problem. Careful consideration of the potential sources of heterogeneity will be necessary. Many of the GWA studies reported to date are from samples of European ancestry, and it will be important to assess the identified variants in other populations, notably those of African and Asian ancestry, and further loci may be identified by carrying out de novo GWA studies in these populations. Also, the current GWA studies do not thoroughly evaluate structural variants and rare alleles, which are important targets of resequencing; identifying these and also the alleles that form the basis for each initial association are likely to increase the heritability that can be explained.
Further analyses also may lead to additional biological insights. Studies of gene–gene interactions may indicate how combinations of genetic variants impact disease risk, and studies of gene-by-environment interactions, especially interactions with dietary and physical activity factors, may further elucidate the mechanisms of inter-individual trait variability. Many challenges await to identify the functional alleles and genes at the implicated loci, as several loci contain many genes and tens to hundreds of genetic variants associated with a disease or trait. The predictive value of identified variants is currently limited (53), but with resequencing and identification of more loci such predictions could improve. Together, gene and allele identification may eventually have clinical utility.
CONCLUSION
The first few years of GWA studies for cardiovascular and metabolic traits have been filled with many successes. Dozens of genes identified previously by candidate gene or linkage analyses have been convincingly replicated in large studies with genome-wide levels of significance (Table 1). Many new loci have been implicated in disease susceptibility or trait variability, including some obvious likely functional genes and some loci in gene deserts or only near genes without known function. Although loci have been identified with stringent statistical significance, the effect sizes are modest and account for only a small fraction of variability so far.
Several loci exhibit allelic heterogeneity, including both common and rare alleles involved in disease. For example, non-synonymous variants in MC4R are associated with extreme obesity, and common variants >100 kb from the gene are associated with modest changes in BMI and waist circumference. Allelic heterogeneity will increase as loci are resequenced and additional rare alleles are identified. Some loci have been identified as associated with more than one cardiovascular or metabolic trait, such as the sets of SNPs near the genes CDKN2A, CDKN2B and ANRIL that are associated with T2D, CAD, MI, frailty and melanoma. Biological studies will be important to evaluate the shared and unique risk alleles at this and other loci. The abundance of new susceptibility loci is ushering in an exciting new period in complex disease genetics and providing many opportunities for future research.
FUNDING
This work was supported by the National Institutes of Health [DK072193 and DK078150 to K.L.M., DK062370 to M.B., and HG02651 and HL084729 to G.R.A.].
Conflict of Interest statement. The authors declare no conflicts of interest related to this work or publication.
REFERENCES
- 1.World Health Organization. Fact sheet N°317 – Cardiovascular diseases. 2007 http://www.who.int/mediacentre/factsheets/fs317/en/index.html . [Google Scholar]
- 2.NCEP Expert Panel. Third report of the National Cholesterol Education Program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (adult treatment panel III) final report. Circulation. 2002;106:3143–3421. [PubMed] [Google Scholar]
- 3.Pilia G., Chen W.M., Scuteri A., Orru M., Albai G., Dei M., Lai S., Usala G., Lai M., Loi P., et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deeb S.S., Fajas L., Nemoto M., Pihlajamaki J., Mykkanen L., Kuusisto J., Laakso M., Fujimoto W., Auwerx J. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat. Genet. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 5.Altshuler D., Hirschhorn J.N., Klannemark M., Lindgren C.M., Vohl M.C., Nemesh J., Lane C.R., Schaffner S.F., Bolk S., Brewer C., et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 6.Gloyn A.L., Weedon M.N., Owen K.R., Turner M.J., Knight B.A., Hitman G., Walker M., Levy J.C., Sampson M., Halford S., et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52:568–572. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- 7.Sandhu M.S., Weedon M.N., Fawcett K.A., Wasson J., Debenham S.L., Daly A., Lango H., Frayling T.M., Neumann R.J., Sherva R., et al. Common variants in WFS1 confer risk of type 2 diabetes. Nat. Genet. 2007;39:951–953. doi: 10.1038/ng2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winckler W., Weedon M.N., Graham R.R., McCarroll S.A., Purcell S., Almgren P., Tuomi T., Gaudet D., Bostrom K.B., Walker M., et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007;56:685–693. doi: 10.2337/db06-0202. [DOI] [PubMed] [Google Scholar]
- 9.Grant S.F., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A., et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 10.Sladek R., Rocheleau G., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 11.Scott L.J., Mohlke K.L., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diabetes Genetics Initiative. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 13.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeggini E., Weedon M.N., Lindgren C.M., Frayling T.M., Elliott K.S., Lango H., Timpson N.J., Perry J.R., Rayner N.W., Freathy R.M., et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinthorsdottir V., Thorleifsson G., Reynisdottir I., Benediktsson R., Jonsdottir T., Walters G.B., Styrkarsdottir U., Gretarsdottir S., Emilsson V., Ghosh S., et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 16.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., Perry J.R., Elliott K.S., Lango H., Rayner N.W., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scuteri A., Sanna S., Chen W.M., Uda M., Albai G., Strait J., Najjar S., Nagaraja R., Orru M., Usala G., et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeggini E., Scott L.J., Saxena R., Voight B.F., Marchini J.L., Hu T., de Bakker P.I., Abecasis G.R., Almgren P., Andersen G., et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grarup N., Andersen G., Krarup N.T., Albrechtsen A., Schmitz O., Jorgensen T., Borch-Johnsen K., Hansen T., Pedersen O. Association testing of novel type 2 diabetes risk-alleles in the JAZF1, CDC123/CAMK1D, TSPAN8, THADA, ADAMTS9 and NOTCH2 loci with insulin release, insulin sensitivity and obesity in a population-based sample of 4,516 glucose-tolerant middle-aged Danes. Diabetes. 2008;57:2534–2540. doi: 10.2337/db08-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozaki K., Ohnishi Y., Iida A., Sekine A., Yamada R., Tsunoda T., Sato H., Sato H., Hori M., Nakamura Y., et al. Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat. Genet. 2002;32:650–654. doi: 10.1038/ng1047. [DOI] [PubMed] [Google Scholar]
- 21.Clarke R., Xu P., Bennett D., Lewington S., Zondervan K., Parish S., Palmer A., Clark S., Cardon L., Peto R., et al. Lymphotoxin-alpha gene and risk of myocardial infarction in 6,928 cases and 2,712 controls in the ISIS case-control study. PLoS Genet. 2006;2:e107. doi: 10.1371/journal.pgen.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helgadottir A., Thorleifsson G., Manolescu A., Gretarsdottir S., Blondal T., Jonasdottir A., Jonasdottir A., Sigurdsson A., Baker A., Palsson A., et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 23.McPherson R., Pertsemlidis A., Kavaslar N., Stewart A., Roberts R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melzer D., Frayling T.M., Murray A., Hurst A.J., Harries L.W., Song H., Khaw K., Luben R., Surtees P.G., Bandinelli S.S., et al. A common variant of the p16(INK4a) genetic region is associated with physical function in older people. Mech. Ageing Dev. 2007;128:370–377. doi: 10.1016/j.mad.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pho L., Grossman D., Leachman S.A. Melanoma genetics: a review of genetic factors and clinical phenotypes in familial melanoma. Curr. Opin. Oncol. 2006;18:173–179. doi: 10.1097/01.cco.0000208791.22442.09. [DOI] [PubMed] [Google Scholar]
- 26.Pasmant E., Laurendeau I., Heron D., Vidaud M., Vidaud D., Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 27.Broadbent H.M., Peden J.F., Lorkowski S., Goel A., Ongen H., Green F., Clarke R., Collins R., Franzosi M.G., Tognoni G., et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum. Mol. Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 28.Samani N.J., Erdmann J., Hall A.S., Hengstenberg C., Mangino M., Mayer B., Dixon R.J., Meitinger T., Braund P., Wichmann H.E., et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willer C.J., Sanna S., Jackson A.U., Scuteri A., Bonnycastle L.L., Clarke R., Heath S.C., Timpson N.J., Najjar S.S., Stringham H.M., et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N.P., Rieder M.J., Cooper G.M., Roos C., Voight B.F., Havulinna A.S., et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandhu M.S., Waterworth D.M., Debenham S.L., Wheeler E., Papadakis K., Zhao J.H., Song K., Yuan X., Johnson T., Ashford S., et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace C., Newhouse S.J., Braund P., Zhang F., Tobin M., Falchi M., Ahmadi K., Dobson R.J., Marcano A.C., Hajat C., et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am. J. Hum. Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kooner J.S., Chambers J.C., Aguilar-Salinas C.A., Hinds D.A., Hyde C.L., Warnes G.R., Gomez Perez F.J., Frazer K.A., Elliott P., Scott J., et al. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat. Genet. 2008;40:149–151. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- 34.Rankinen T., Zuberi A., Chagnon Y.C., Weisnagel S.J., Argyropoulos G., Walts B., Perusse L., Bouchard C. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 35.Benzinou M., Creemers J.W., Choquet H., Lobbens S., Dina C., Durand E., Guerardel A., Boutin P., Jouret B., Heude B., et al. Common nonsynonymous variants in PCSK1 confer risk of obesity. Nat. Genet. 2008;40:943–945. doi: 10.1038/ng.177. [DOI] [PubMed] [Google Scholar]
- 36.Herbert A., Gerry N.P., McQueen M.B., Heid I.M., Pfeufer A., Illig T., Wichmann H.E., Meitinger T., Hunter D., Hu F.B., et al. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 37.Lyon H.N., Emilsson V., Hinney A., Heid I.M., Lasky-Su J., Zhu X., Thorleifsson G., Gunnarsdottir S., Walters G.B., Thorsteinsdottir U., et al. The association of a SNP upstream of INSIG2 with body mass index is reproduced in several but not all cohorts. PLoS Genet. 2007;3:e61. doi: 10.1371/journal.pgen.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dina C., Meyre D., Gallina S., Durand E., Korner A., Jacobson P., Carlsson L.M., Kiess W., Vatin V., Lecoeur C., et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 39.Gerken T., Girard C.A., Tung Y.C., Webby C.J., Saudek V., Hewitson K.S., Yeo G.S., McDonough M.A., Cunliffe S., McNeill L.A., et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loos R.J., Lindgren C.M., Li S., Wheeler E., Zhao J.H., Prokopenko I., Inouye M., Freathy R.M., Attwood A.P., Beckmann J.S., et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chambers J.C., Elliott P., Zabaneh D., Zhang W., Li Y., Froguel P., Balding D., Scott J., Kooner J.S. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat. Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y.J., Liu X.G., Wang L., Dina C., Yan H., Liu J.F., Levy S., Papasian C.J., Drees B.M., Hamilton J.J., et al. Genome-wide association scans identified CTNNBL1 as a novel gene for obesity. Hum. Mol. Genet. 2008;17:1803–1813. doi: 10.1093/hmg/ddn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 44.Arking D.E., Pfeufer A., Post W., Kao W.H., Newton-Cheh C., Ikeda M., West K., Kashuk C., Akyol M., Perz S., et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat. Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 45.Weedon M.N., Clark V.J., Qian Y., Ben-Shlomo Y., Timpson N., Ebrahim S., Lawlor D.A., Pembrey M.E., Ring S., Wilkin T.J., et al. A common haplotype of the glucokinase gene alters fasting glucose and birth weight: association in six studies and population-genetics analyses. Am. J. Hum. Genet. 2006;79:991–1001. doi: 10.1086/509517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen W.M., Erdos M.R., Jackson A.U., Saxena R., Sanna S., Silver K.D., Timpson N.J., Hansen T., Orru M., Grazia Piras M., et al. Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J. Clin. Invest. 2008;118:2620–2628. doi: 10.1172/JCI34566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bouatia-Naji N., Rocheleau G., Van Lommel L., Lemaire K., Schuit F., Cavalcanti-Proenca C., Marchand M., Hartikainen A.L., Sovio U., De Graeve F., et al. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science. 2008;320:1085–1088. doi: 10.1126/science.1156849. [DOI] [PubMed] [Google Scholar]
- 48.Li S., Sanna S., Maschio A., Busonero F., Usala G., Mulas A., Lai S., Dei M., Orru M., Albai G., et al. The GLUT9 gene is associated with serum uric acid levels in Sardinia and Chianti cohorts. PLoS Genet. 2007;3:e194. doi: 10.1371/journal.pgen.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doring A., Gieger C., Mehta D., Gohlke H., Prokisch H., Coassin S., Fischer G., Henke K., Klopp N., Kronenberg F., et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat. Genet. 2008;40:430–436. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- 50.Vitart V., Rudan I., Hayward C., Gray N.K., Floyd J., Palmer C.N., Knott S.A., Kolcic I., Polasek O., Graessler J., et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat. Genet. 2008;40:437–442. doi: 10.1038/ng.106. [DOI] [PubMed] [Google Scholar]
- 51.Reiner A.P., Barber M.J., Guan Y., Ridker P.M., Lange L.A., Chasman D.I., Walston J.D., Cooper G.M., Jenny N.S., Rieder M.J., et al. Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am. J. Hum. Genet. 2008;82:1193–1201. doi: 10.1016/j.ajhg.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ridker P.M., Pare G., Parker A., Zee R.Y., Danik J.S., Buring J.E., Kwiatkowski D., Cook N.R., Miletich J.P., Chasman D.I. Loci related to metabolic-syndrome pathways including LEPR, HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women’s Genome Health Study. Am. J. Hum. Genet. 2008;82:1185–1192. doi: 10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kathiresan S., Melander O., Anevski D., Guiducci C., Burtt N.P., Roos C., Hirschhorn J.N., Berglund G., Hedblad B., Groop L., et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N. Engl. J. Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]