Abstract
The different gene members of the Egr family of transcriptional regulators have often been considered to have related functions in brain, based on their co-expression in many cell-types and structures, the relatively high homology of the translated proteins and their ability to bind to the same consensus DNA binding sequence. Recent research, however, suggest this might not be the case. In this review, we focus on the current understanding of the functional roles of the different Egr family members in learning and memory. We briefly outline evidence from mutant mice that Egr1 is required specifically for the consolidation of long-term memory, while Egr3 is primarily essential for short-term memory. We also review our own recent findings from newly generated forebrain-specific conditional Egr2 mutant mice, which revealed that Egr2, as opposed to Egr1 and Egr3, is dispensable for several forms of learning and memory and on the contrary can act as an inhibitory constraint for certain cognitive functions. The studies reviewed here highlight the fact that Egr family members may have different, and in certain circumstances antagonistic functions in the adult brain.
Keywords: transcription factor, Egr1, zif268, Egr2, Krox20, learning, memory
Introduction
The formation of long-term memory has been shown to be dependent on the synthesis of new proteins but the specific molecular mechanisms that are essential for learning and memory processes and how particular classes of molecules contribute to these events is not fully understood. Several studies indicate that immediate early genes (IEGs) such as Fos, Jun, or Egr1 may play an important role in learning and memory processes (Bozon et al., 2003b; Dragunow, 1996; Knapska and Kaczmarek, 2004). The IEGs are a class of genes that are rapidly and transiently activated by a variety of signalling cascades and phosphorylation events in a protein synthesis-independent manner in response to neuronal activation. IEGs encode many functionally different products such as secreted proteins, cytoplasmic enzymes and inducible transcription factors that can activate downstream target genes (Herdegen and Leah, 1998). As such, the rapid activation of IEGs encoding transcriptional regulators constitutes an early genomic response in a strategic position in the molecular cascades leading to persistent neuronal modification. To date several lines of evidence suggest that their regulated expression following neuronal activation is a key mechanism for the recruitment of specific gene programs involved in the stabilisation of neural modification required for the laying down of long-term memories.
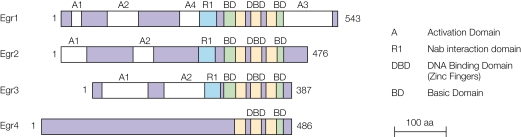
Among the studies of the functional role of inducible transcriptional regulators in synaptic plasticity and memory processes, many have focused the Early Growth Response (Egr) gene family (for reviews, see Abraham et al., 1991; Bozon et al., 2002; Tischmeyer and Grimm, 1999). It was notably demonstrated that a member of Egr family, Egr1 (also known as NGFI-A, Krox-24, Zif268, Tis8 or ZENK), is involved in the maintenance of long-term potentiation (LTP), an activity-dependent form of synaptic plasticity believed to play a crucial role in the formation of memories (Jones et al., 2001). Egr1 is also essential for the consolidation of several forms of long-term memory (Bozon et al., 2002, 2003b; Jones et al., 2001; Malkani et al., 2004) and for reconsolidation of memory after reactivation during retrieval (Bozon et al., 2003a; Lee et al., 2004). In addition, enhanced memory strength was recently reported in transgenic mice with forebrain over-expression of Egr1 (Baumgärtel et al., 2008). While the role of Egr1 in memory is relatively well characterised, that of other members of the Egr family has only recently began to be investigated. The Egr family in fact comprises four members: Egr1, Egr2 (Krox-20), Egr3 (Pilot) and Egr4 (NGFI-C) (Beckmann and Wilce, 1997; O'Donovan et al., 1999; Figure 1). They encode closely related transcription factors that contain three Cys2-Hys2 zinc fingers shared by the four members and that can bind to the same cognate GC-rich consensus DNA binding motif, the Egr response element (ERE). The high homology between the three zinc finger sequences of the four members of the Egr family (Beckmann and Wilce, 1997) suggests that the proteins may well bind to cis-regulatory regions of at least a subset of the same target genes (Chavrier et al., 1990; Lemaire et al., 1990; Swirnoff and Milbrandt, 1995). However, outside their common DNA-binding domain, the sequences of the different Egr members have less homology, suggesting they can be differentially regulated by selective signalling pathways and may therefore serve different functions (Beckmann and Wilce, 1997; Herdegen and Leah, 1998). Nevertheless a few conserved domains have been identified and are depicted in Figure 1. Of particular interest is the interaction domain for the Nab proteins, which is found in Egr1, Egr2 and Egr3. Nab1 and Nab2 have initially been described as repressors of the transcriptional activity of Egr proteins (Russo et al., 1995; Svaren et al., 1996; Swirnoff et al., 1998). However recent data suggest that they can also act as synergistic factors for Egr2 function at least (Desmazières et al., in press; Le et al., 2005). Finally, a detailed structure-function analysis of the Egr2 protein has been performed recently and has revealed that other domains within the N-terminal part of the protein are involved in the activation of specific target genes (Desmazières, Charnay and Gilardi-Hebenstreit, unpublished data). One of these domains is conserved in Egr1 and Egr3.
Figure 1.
Schematic representation of the four members of the Egr family (Beckmann and Wilce, 1997; O'Donovan et al., 2000; Russo et al., 1995). The most closely related members are Egr2 and Egr3, followed by Egr1. Egr4 is much more distant. Several conserved sequences are featured: the zinc fingers that are extremely well conserved between all members; two basic regions flanking the zinc fingers that are conserved between Egr1, Egr2 and Egr3, the C-terminal one being also observed in Egr4; the Nab interaction domain that is present in Egr1, Egr2 and Egr3.
In brain, Egr family members show, in general, a similar regional profile of basal expression (Beckmann and Wilce, 1997). mRNA and proteins are detected in several areas of the neocortex, hippocampus, entorhinal cortex, amygdala, nucleus accumbens, striatum, olfactory bulb and cerebellar cortex, with different levels of expression (reviewed in Beckmann and Wilce, 1997). In the cerebral cortex, the highest levels of Egr1, Egr3 and Egr4 are detected in layers II and IV, whereas Egr2 predominates in layers II and III (Beckmann and Wilce, 1997). In the hippocampus, all Egr family members are highly expressed in CA1-3 pyramidal cells, with negligible basal levels in granule cells of the dentate gyrus (Beckmann and Wilce, 1997; Herdegen et al., 1993; Mack et al., 1990; Yamagata et al., 1994). In addition to their basal expression, the expression of Egr family members can be induced in different brain areas after a variety of neuronal stimulation paradigms such as the induction of LTP or maximal electroconvulsive shocks (MECS) inducing seizure activity, or in behavioural circumstances after exposure to novel environments of specific learning experiences (reviewed in Beckmann and Wilce, 1997; Davis et al., 2003; Knapska and Kaczmarek, 2004).
Functional Role of the Egr Gene Family Members
Egr1 is the most widely studied member of the Egr family in processes of synaptic plasticity and memory. The origin of Egr1 research in this field can be traced back to the discovery of its rapid induction in dentate granule cells following the induction of LTP (Cole et al., 1989; Wisden et al., 1990) and its expression has since been widely used as a marker of neuronal plasticity in a learning context (see Davis et al., 2003; Knapska and Kaczmarek, 2004 for reviews). Using Egr1 mutant mice, it has been demonstrated that Egr1-deficiency results in a complete and specific loss of the maintenance of late-LTP at perforant path-to-dentate granule cell synapses (Jones et al., 2001). In parallel, a unique feature of Egr1-deficient mice is their inability to form long-term memories in a variety of behavioural tasks such as conditioned taste aversion, olfactory discrimination, novel object recognition and spatial navigation, while short-term memory, as well as early-LTP, is intact (Jones et al., 2001). More recently, the behavioural phenotype of Egr3 mutant mice has also been characterised and major differences with the neural and behavioural phenotype of Egr1 mutant mice have been found (Gallitano-Mendel et al., 2007; Li et al., 2007). Egr3 mutant mice were found to have motor abnormalities due to defects in muscle spindles morphogenesis (Tourtellotte and Mildbrandt, 1998), heightened reactivity to stressful stimuli, abnormalities in their adaptation to novel and stressful stimuli, and abnormalities in social interactions (Gallitano-Mendel et al., 2007). In addition, clear short-term memory deficits were identified in contextual fear conditioning and object recognition, with a consequential defect in long-term memory performance, together with compromised early-phase of LTP in area CA1 of the hippocampus (Li et al., 2007). Deficits in NR2B-containing NMDA receptor function and in CA1 hippocampal LTD have also been reported in these mice (Gallitano-Mendel et al., 2007). In contrast to the role of Egr1 and Egr3 in different aspects of synaptic plasticity and memory processes, very little is known about the role of Egr2 and Egr4. To our knowledge, no study has yet examined the functional role of Egr4 in these processes. For Egr2, a specific function has been documented in the regulation of peripheral nerve myelination (Topilko et al., 1994), hindbrain segmentation (Schneider-Maunoury et al., 1993; Swiatek and Gridley, 1993) and endochondreal bone formation (Levi et al., 1996). Examination of its functional role in adult brain, however, has been hampered by the fact that mutant mice carrying two Egr2 null alleles show high perinatal lethality (Decker et al., 2006; Schneider-Maunoury et al., 1993; Swiatek and Gridley, 1993; Topilko et al., 1994), a short lifespan presumably due to defects in hindbrain rythmogenic neural networks controlling respiratory functions (Jacquin et al., 1996).
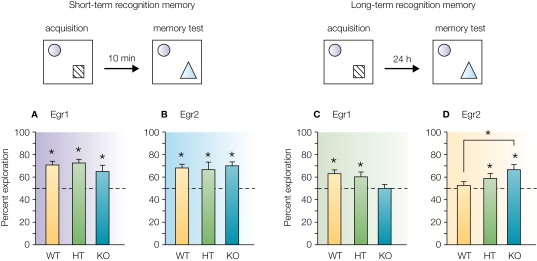
To circumvent perinatal lethality, we recently generated novel Egr2 conditional mutant mice based on a floxed allele allowing postnatal, CaMKII promoter-dependent Cre-mediated excision of Egr2 selectively in adult forebrain neurons (Poirier et al., 2007). In this study, we investigated whether lack of Egr2 in mice results in behavioural deficits similar to those observed in Egr1 or Egr3 mutant mice. Performance of homozygous and heterozygous conditional mutant mice, and of control littermates, was thus investigated in a battery of behavioural tasks to evaluate motor capacity and exploratory behaviour (grid-suspension, traction reflex and rotarod tests, open-field activity), emotional reactivity (elevated-plus maze test), as well as learning and memory abilities in tasks such as object recognition, spatial learning in the water maze, trace fear conditioning and conditioned taste aversion learning. Homozygous and heterozygous conditional Egr2 mutant mice appeared healthy and showed no abnormal spontaneous behaviour or brain structure abnormalities. They showed no sign of anxiety disturbance; nor was their motor activity and coordination impaired. In contrast to our initial expectations, we found that none of the forms of learning examined were impaired in Egr2 deficient mice. Both conditional homozygous and heterozygous transgenic mice demonstrated normal learning and memory performance in spatial navigation, normal associative memory in a conditioned taste aversion paradigm and normal fear learning and memory in a trace fear conditioning paradigm. Surprisingly, however, we found that conditional Egr2 mutant mice had superior implicit motor skill learning abilities in an accelerating rotarod task, and enhanced long-term object recognition memory (Poirier et al., 2007). With the training protocol used to assess recognition memory, control mice could form short-term, but not long-term recognition memory, while conditional Egr2 mice, with identical short-term memory performance to that of their controls, were able to form a long-term memory for objects (Figure 2), suggesting facilitation of consolidation processes. In this task, performance of heterozygous mice was in between that of knockout and wild-type mice, supporting a gene-dosage effect. In summary, our results revealed that forebrain Egr2-deficiency does not result in learning or memory impairments, but on the contrary, facilitates performance in certain tasks.
Figure 2.
Short-term and long-term object recognition memory in Egr1 and Egr2 deficient mice. During acquisition, wild-type (WT) heterozygous (HT) and homozygous (KO) mice were exposed to tow objects and retention was tested 10 min or 24 h later by replacing one familiar object by a novel object. The histograms represent the time spent exploring the novel object during memory retention. (A and B) Ten minutes after acquisition, both Egr1 and Egr2 mutant mice spent significantly more time exploring the novel than the familiar object, showing good short-term recognition memory. (C and D) Long-term recognition memory tested 24 h after acquisition. A clear deficit was observed in Egr1 mutant mice (C). By contrast, Egr2 mutant mice showed better performance than their WT controls (D). In both cases, heterozygous mice with half the complement of the Egr proteins showed intermediate performance, suggesting gene-dosage effect. Modified from Jones et al. (2001) and Poirier et al. (2007).
Clearly, the behavioural phenotype of conditional Egr2 mutant mice appears in sharp contrast to that of Egr1 and Egr3 mutant mice when comparing their performance in similar tasks. Whereas Egr1-deficiency leads to significant memory deficits in conditioned taste aversion, spatial learning and novel object recognition when tested 24 h after acquisition, no specific impairment was found for these forms of memories in Egr2 mutant mice. More surprisingly, as summarized in Figure 2, long-term recognition memory is severely affected in Egr1 mutant mice (Jones et al., 2001), whereas Egr2 mutant mice demonstrated a better memory ability than their controls (Poirier et al., 2007). In comparison, Egr3 mutant mice were shown not only to have deficits in long-term memory in a similar object recognition task, but also compromised short-term memory (Li et al., 2007). These results clearly establish that Erg family members have distinct, and in certain circumstances antagonistic functions in the adult brain.
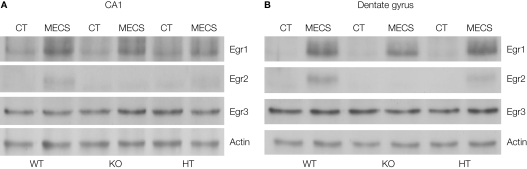
In Egr2-deficient mice, we analysed the basal and stimulation-induced expression of Egr1 and Egr3 proteins in the dentate gyrus and area CA1 of the hippocampus, using maximal electroconvulsive shock (MECS) (Poirier et al., 2007; Figure 3). We detected no over-expression of Egr1 or Egr3 in the mice, suggesting that the absence of memory deficits and the gain of function in certain tasks in Egr2-deficient mice is not due to a compensatory increase in basal or induced expression of Egr1 or Egr3 in these structures.
Figure 3.
Basal and MECS-induced expression of Egr family members in the hippocampus of Egr2 mutant mice. Egr1, Egr2 and Egr3 expression levels were measured by Western immunoblotting from area CA1 and the dentate gyrus of mice from the three genotypes in a control condition (CT) and 2 hours after MECS-induced seizure. Actin was used as a control protein. In the absence of Egr2, basal and MECS-induced expression of Egr1 and Egr3 was not significantly modified. Data from Poirier et al. (2007).
Regulation of Egr Family Members in Synaptic Plasticity and Learning
As with Egr1 and Egr3, the expression of Egr2 is regulated after certain forms of pharmacological or physiological stimuli. Induction of Egr2 mRNA or protein has been observed for example after kainic acid injection (Gass et al., 1994) or seizure activity (Bhat et al., 1992, but see Mack et al., 1992). All three Egr members are rapidly regulated after induction of LTP in the dentate gyrus, although the regulation of Egr2 and Egr3 appears to require stronger LTP-inducing stimuli, but then expression of the proteins lasts longer than that of Egr1 (Cole et al., 1989; Williams et al., 1995; Worley et al., 1993; Yamagata et al., 1994).
In learning, Egr1 mRNA or protein are also rapidly induced in specific brain structures in a variety of tasks such as active avoidance learning (Nikolaev et al., 1992), brightness discrimination (Grimm and Tischmeyer, 1997), visual paired associate learning in monkeys (Okuno and Miyashita, 1996; Tokuyama et al., 2002), song learning in birds (Bolhuis et al., 2000; Jarvis et al., 1995; Mello et al., 1992), learning and/or retrieval of contextual and cued fear learning (Frankland et al., 2004; Hall et al., 2001; Malkani and Rosen, 2000; Weitemier and Ryabinin, 2004) and spatial learning (Guzowski et al., 2001; Maviel et al., 2004). In general, as in synaptic plasticity paradigms, the induction of Egr1 occurs rapidly after learning or exposure to learning-associated cues, and is transient, suggesting a role in the transition from short- to long-term memory.
Much less is known about the regulation of other Egr family members during learning. In single-trial contextual fear conditioning, induction of Egr2, Egr3 and Egr4 has not been clearly observed (Malkani and Rosen, 2000). In a recent study, however, Desteno and Schmauss (2008) showed induction of Egr2 expression in areas of the medial prefrontal cortex of mice performing an attention-set-shifting task, but not in a spatial working memory task. In this study, the authors have detected no induction of Egr1 and Egr3 in the same brain structures in the attention-set-shifting task. These observations suggest that Egr family members may be differentially induced during selective cognitive tasks. Further research is needed however to expand our knowledge of the specific conditions under which distinct Egr family members are activated, and of their localization and kinetics of expression in relation to specific types of learning.
Although a wide range of neurotransmitter systems can induce expression of Egr family members (Beckmann and Wilce for a review), the cell-signalling pathways leading to the regulated expression of Egr members in synaptic plasticity and learning have not been precisely defined. Induction of Egr1 in the hippocampus has been the focus of some investigations and it has been in particular shown that activity-dependent expression of Egr1 is dependent upon NMDA receptor (Cole et al., 1989; Gass et al., 1993; Wisden et al., 1990) and MAPK/ERK activation (Davis et al., 2000), both of which are known to play a key role in synaptic plasticity and learning. Conversely, Egr1 can be negatively regulated by the protein phosphatase calcineurin, a negative regulator of neuronal signalling and of memory formation (Baumgärtel et al., 2008). Much less is known about activity-dependent induction of Egr2 and Egr3 in the brain. Certain evidence suggests that inactivation of the adenosine 2A receptor in mice results in an up-regulation of Egr2 associated with down-regulation of Egr1 in the striatum (Yu et al., 2005), and that mGluR5 antagonists induce down-regulation of Egr2, but not of other Egr members in the rat cortex (Gass and Olive, 2008). In addition both Egr1 and Egr2 have been shown to be regulated by BDNF/TrkB in a CCAAT/enhancer binding proteins (C/EBPα,-β) dependent manner (Calella et al., 2007). To our knowledge, no study has yet examined the signalling cascades involved in the regulated expression of Egr3 in the adult brain. These data reinforce the notion of a differential sensitivity of Egr members activation in response to distinct types of stimuli, however whether this type of differential regulation of the Egr member transcription factors occurs during the consolidation of memory is yet to be known.
Conclusion
Our recent findings provide the first evidence that the absence of Egr2 in forebrain neurons may facilitate certain forms of learning and memory and reinforce the notion that Egr1, Egr2 and Egr3 can have different functions in the adult brain and, in certain circumstances, antagonistic functions. Such antagonistic roles played by Egr members have already been observed during the development of Schwann cell lineage where Egr1 and Egr2 are expressed successively and in a mutually exclusive manner (Topilko et al., 1997) and more recently in the regulation of T-cell function (Collins et al., 2008), suggesting they might compete and possibly repress each other. The molecular basis of these antagonistic actions however has not yet been established. In the adult brain, the mechanisms whereby Egr members could exert antagonistic functions in learning and memory are also not elucidated. One possibility is the control of different sets of downstream targets genes. Egr family members might also regulate some of the same downstream genes, but in opposite manners, due to non-conserved regulatory regions. Although certain Egr direct target genes have been characterized in different tissues such as the pituitary for Egr1 (Lee et al., 1996; Topilko et al., 1998) and hindbrain for Egr2 (Sham et al., 1993), little is known about the Egr direct target genes in the adult brain. In neuronal cells, it has been suggested that Egr1 regulates expression of synapsin I and II and of the cdk5-regulator p35 (Harada et al., 2001; Petersohn et al., 1995; Thiel et al., 1994). The list of putative Egr1 target genes derived from a large-scale profiling study conducted in PC12 cells have also pointed to a major role of Egr1 in controlling various proteins of the proteasome (James et al., 2005). Recently, a proteomic study conducted in Egr1 over-expressing mice revealed changes in the expression of proteins of many functional groups, including transcription factors, DNA and RNA modifying enzymes, components of the proteasomal and ubiquitination machinery, kinases, phosphatases and small GTPases, cytoskeletal proteins and various proteins involved in vesicle recycling and postsynaptic organization (Baumgärtel et al., 2008). Using Egr1-deficient and Egr3-deficient mice, it has also been shown that the plasticity-associated gene Arg3.1/Arc is a direct transcriptional target of Egr1 and Egr3 (Li et al., 2005) and Ehrengruber et al. (2000) using adenoviral gene transfer, identified Egr3 and Nab2 as Egr1 target genes in neuroblastoma cells. Few studies have directly examined potential target genes of other Egr members. During development, Egr2 was shown to activate Hox genes and genes associated with nerve myelination such as myelin basic protein, myelin protein zero and desert hedgehog (Jang et al., 2006; Manzanares et al., 2002; Nagarajan et al., 2001; Nonchev et al., 1996; Seitanidou et al., 1997; Sham et al., 1993; Topilko et al., 1994). In the striatum of adult adenosine 2A receptor knockout mice, Yu et al. (2005) identified a number of up and down-regulated genes, including an increased expression of Egr2 while Egr1 expression was decreased, and found an over-representation of predicted Egr2 binding sites in the promoter regions of the regulated genes. In this study, Egr2 binding to and regulation of the protein phosphatase PP2A was confirmed by chromatin immunoprecipitation.
Despite recognising the same ERE sequence in promoter regions, site recognition involves additional factors that interact specifically with non-conserved regions of Egr proteins, adding a further layer of complexity in target gene regulation. Complex control of Egr-mediated gene regulation is illustrated by studies showing that Egr1 and Egr3 directly bind to, and regulate expression of the low affinity neurotrophin receptor (p75NTR) in Schwann cells by binding preferentially to two distinct Egr binding motifs in the promoter region of the p75NTR gene, with a transactivation ability which varies with Egr1 and Egr3 proteins levels (Gao et al., 2007), highlighting the fact that Egr members may not have entirely redundant functions. While these studies point to the possibility that Egr family members may control distinct gene programs, the precise identity of the regulated genes is still far from reach and would need to be characterized in a learning context to understand mechanistically how different Egr members affect cognitive functions. The rapidly evolving technologies for gene screening and the availability of Egr-specific mutant mice will likely serve as powerful tools to characterize specific gene programs that are under the control of each Egr family member in the brain in vivo.
Finally, the fact that Egr2 acts as an inhibitory constrain on certain cognitive functions may also involve its potency to positively regulate the expression of Nab1 and Nab2 (Desmazières, Charnay and Gilardi-Hebenstreit, unpublished data; Mechta-Grigoriou et al., 2000; Nagarajan et al., 2001). Egr proteins possess a domain for interaction with the Nab1 and Nab2 proteins that were initially described as transcriptional co-repressors (Russo et al., 1995; Svaren et al., 1996; Swirnoff et al., 1998), but are also likely to act in a positive manner in specific cell types (Desmazières et al., 2008). In this respect, it is interesting to note that a recent study identified multiple Egr binding sites on the promoter of Nab2 in a region critical for promoter activity, suggesting that Egr members are direct regulators of Nab2 (Kumbrink et al., 2005). Thus, it is tempting to speculate that the absence of Egr2 in neurons may result in a reduced expression of Nab proteins, and that this would increase or prolong transcriptional activity of other Egr family members or affect other transcriptional regulators possessing Nab-mediated activities, resulting in a facilitation of certain types of memory. To date, we are only beginning to understand the functional roles of the different members of this family of transcriptional regulators in higher cognitive functions. These advances now give us the possibility to define the complex combination of activity-dependent recruitment of the different Egr family members more precisely, and how they interact with each other and with their co-regulators and what specific gene programs they control in specific brain structures during learning and memory.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to François Tronche for the gift of the CaMKII-Cre line, and to Claire Chevalier, Nathalie Samson, Pascale Veyrac and Sandra Vandergeenst for animal care. Our deepest thanks go to Emmanuel Taillebourg who was involved in the initiation of the generation of the conditional mutant.
This work was supported by CNRS, INSERM, and by grants from the French Ministry of Research (ACI-NEURO-NIC-0027) to S.D. and P.C. and from EU Marie Curie Research Training Program (RTN-CT-2003-504231) to S.L. H.C was supported by a fellowship from the French Ministry of Research.
References
- Abraham W. C., Dragunow M., Tate W. P. (1991). The role of immediate early genes in the stabilization of long-term potentiation. Mol. Neurobiol. 5, 297–314 [DOI] [PubMed] [Google Scholar]
- Baumgärtel K., Genoux D., Welzl H., Tweedie-Cullen R. Y., Koshibu K., Livingstone-Zatchej M., Mamie1 C., Mansuy I. M. (2008). Control of the establishment of aversive memory by calcineurin and Zif268. Nature Neurosci. 11, 572–578 10.1038/nn.2113 [DOI] [PubMed] [Google Scholar]
- Beckmann A. M., Wilce P. A. (1997). Egr transcription factors in the nervous system. Neurochem. Int. 31, 477–510 10.1016/S0197-0186(96)00136-2 [DOI] [PubMed] [Google Scholar]
- Bhat R. V., Worley P. F., Cole A. J., Baraban J. M. (1992). Activation of the zinc finger encoding gene krox-20 in adult rat brain: comparison with zif268. Mol. Brain Res. 13, 263–266 10.1016/0169-328X(92)90034-9 [DOI] [PubMed] [Google Scholar]
- Bolhuis J. J., Zijlstra G. G., den Boer-Visser A. M., Van Der Zee E. A. (2000). Localized neuronal activation in the zebra finch brain is related to the strength of song learning. Proc. Natl. Acad. Sci. USA 97, 2282–2285 10.1073/pnas.030539097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozon B., Davis S., Laroche S. (2002). Regulated transcription of the immediate early gene Zif268: mechanisms and gene dosage-dependent function in synaptic plasticity and memory formation. Hippocampus 12, 570–577 10.1002/hipo.10100 [DOI] [PubMed] [Google Scholar]
- Bozon B., Davis S., Laroche S. (2003a). A requirement for the immediate early gene zif268 in reconsolidation of recognition memory after retrieval. Neuron 40, 695–701 10.1016/S0896-6273(03)00674-3 [DOI] [PubMed] [Google Scholar]
- Bozon B., Kelly A., Josselyn S. A., Silva A. J., Davis S., Laroche S. (2003b). MAPK, CREB and zif268 are all required for the consolidation of recognition memory. Phil. Trans. R. Soc. B 358, 805–814 10.1098/rstb.2002.1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calella A. M., Nerlov C., Lopez R. G., Sciarretta C., von Bohlen Und Halbach O., Bereshchenko O., Minichiello L. (2007). Neurotrophin/Trk receptor signaling mediates C/EBPalpha, -beta and NeuroD recruitment to immediate-early gene promoters in neuronal cells and requires C/EBPs to induce immediate-early gene transcription. Neural Develop. 2, 1–21 10.1186/1749-8104-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavrier P., Vesque C., Galliot B., Vigneron M., Dolle P., Duboule D., Charnay P. (1990). The segment-specific gene Krox-20 encodes a transcription factor with binding sites in the promoter region of the Hox-1.4 gene. EMBO J. 9, 1209–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole A. J., Saffen D. W., Baraban J. M., Worley P. F. (1989). Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature 340, 474–476 10.1038/340474a0 [DOI] [PubMed] [Google Scholar]
- Collins S., Lutz M. A., Zarek P. E., Anders R. A., Kersh G. J., Powell J. D. (2008). Opposing regulation of T cell function by Egr-1/NAB2 and Egr-2/Egr-3. Eur. J. Immunol. 38, 528–536 10.1002/eji.200737157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S., Bozon B., Laroche S. (2003). How necessary is the activation of the immediate early gene zif268 in synaptic plasticity and learning? Behav. Brain Res. 142, 17–30 10.1016/S0166-4328(02)00421-7 [DOI] [PubMed] [Google Scholar]
- Davis S., Vanhoutte P., Pages C., Caboche J., Laroche S. (2000). The MAPK/ERK cascade targets both Elk-1 and cAMP response element-binding protein to control long-term potentiation-dependent gene expression in the dentate gyrus in vivo. J. Neurosci. 20, 4563–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker L., Desmarquet-Trin-Dinh C., Taillebourg E., Ghislain J., Vallat J. M., Charnay P. (2006). Peripheral myelin maintenance is a dynamic process requiring constant Krox20 expression. J. Neurosci. 26, 9771–9779 10.1523/JNEUROSCI.0716-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmazières A., Decker L., Vallat J.-M., Charnay P., Gilardi-Hebenstreit P. (2008). Disruption of Krox20-Nab interaction leads to a peripheral neuropathy with biphasic evolution. J. Neurosci. 28, 5891–5900 10.1523/JNEUROSCI.5187-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desteno D. A., Schmauss C. (2008). Induction of early growth response gene 2 expression in the forebrain of mice performing an attention-set-shifting task. Neuroscience 18, 417–428 10.1016/j.neuroscience.2008.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragunow M. (1996). A role for immediate-early transcription factors in learning and memory. Behav. Genet. 26, 293–299 10.1007/BF02359385 [DOI] [PubMed] [Google Scholar]
- Ehrengruber M. U., Muhlebach S. G., Söhrman S., Leutenegger C. M., Lester H. A., Davidson N. (2000). Modulation of early growth response (EGR) transcription factor-dependent gene expression by using recombinant adenovirus. Gene 258, 63–69 10.1016/S0378-1119(00)00445-5 [DOI] [PubMed] [Google Scholar]
- Frankland P. W., Bontempi B., Talton L. E., Kaczmarek L., Silva A. J. (2004). The involvement of the anterior cingulate cortex in remote contextual fear memory. Science 304, 881–883 10.1126/science.1094804 [DOI] [PubMed] [Google Scholar]
- Gallitano-Mendel A., Izumi Y., Tokuda K., Zorumski C. F., Howell M. P., Muglia L. J., Wozniak D. F., Milbrandt J. (2007). The immediate early gene early growth response gene 3 mediates adaptation to stress and novelty. Neuroscience 148, 633–643 10.1016/j.neuroscience.2007.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Daugherty R. L., Tourtellotte W. G. (2007). Regulation of low affinity neurotrophin receptor (p75(NTR)) by early growth response (Egr) transcriptional regulators. Mol. Cell Neurosci. 36, 501–514 10.1016/j.mcn.2007.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass J. T., Olive M. F. (2008). Transcriptional profiling of the rat frontal cortex following administration of the mGlu5 receptor antagonists MPEP and MTEP. Eur. J. Pharmacol. 584, 253–262 10.1016/j.ejphar.2008.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass P., Herdegen T., Bravo R., Kiessling M. (1993). Induction and suppression of immediate early genes in specific rat brain regions by the non-competitive N-methyl-D-aspartate receptor antagonist MK-801. Neuroscience 53, 749–758 10.1016/0306-4522(93)90621-L [DOI] [PubMed] [Google Scholar]
- Gass P., Herdegen T., Bravo R., Kiessling M. (1994). High induction threshold for transcription factor KROX-20 in the rat brain: partial co-expression with heat shock protein 70 following limbic seizures. Mol. Brain Res. 23, 292–298 10.1016/0169-328X(94)90238-0 [DOI] [PubMed] [Google Scholar]
- Grimm R., Tischmeyer W. (1997). Complex patterns of immediate early gene induction in rat brain following brightness discrimination training and pseudotraining. Behav. Brain Res. 84, 109–116 10.1016/S0166-4328(97)83330-X [DOI] [PubMed] [Google Scholar]
- Guzowski J. F., Setlow B., Wagner E. K., McGaugh J. L. (2001). Experience-dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate-early genes Arc, c-fos, and zif268. J. Neurosci. 21, 5089–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Thomas K. L., Everitt B. J. (2001). Cellular imaging of zif268 expression in the hippocampus and amygdala during contextual and cued fear memory retreival: selective activation of hippocampal CA1 neurons during the recall of contextual memories. J. Neurosci. 21, 2186–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T., Morooka T., Ogawa S., Nishida E. (2001). ERK induces p35, a neuron-specific activator of Cdk5, through induction of Egr1. Nat. Cell Biol. 3, 453–459 10.1038/35074516 [DOI] [PubMed] [Google Scholar]
- Herdegen T., Kiessling M., Bele S., Bravo R., Zimmermann M., Gass P. (1993). The KROX-20 transcription factor in the rat central and peripheral nervous systems: novel expression pattern of an immediate early gene-encoding protein. Neuroscience 57, 41–52 10.1016/0306-4522(93)90110-2 [DOI] [PubMed] [Google Scholar]
- Herdegen T., Leah J. D. (1998). Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res. Rev. 28, 370–490 10.1016/S0165-0173(98)00018-6 [DOI] [PubMed] [Google Scholar]
- Jacquin T. D., Borday V., Schneider-Maunoury S., Topilko P., Ghilini G., Kato F., Charnay P., Champagnat J. (1996). Reorganization of pontine rhythmogenic neuronal networks in Krox-20 knockout mice. Neuron 17, 747–758 10.1016/S0896-6273(00)80206-8 [DOI] [PubMed] [Google Scholar]
- James A. B., Conway A. M., Morris B. J. (2005). Genomic profiling of the neuronal target genes of the plasticity-related transcription factor – Zif268. J. Neurochem. 95, 796–810 10.1111/j.1471-4159.2005.03400.x [DOI] [PubMed] [Google Scholar]
- Jang S. W., LeBlanc S. E., Roopra A., Wrabetz L., Svaren J. (2006). In vivo detection of Egr2 binding to target genes during peripheral nerve myelination. J. Neurochem. 98, 1678–1687 10.1111/j.1471-4159.2006.04069.x [DOI] [PubMed] [Google Scholar]
- Jarvis E. D., Mello C. V., Nottebohm F. (1995). Associative learning and stimulus novelty influence the song-induced expression of an immediate early gene in the canary forebrain. Learn. Mem. 2, 62–80 10.1101/lm.2.2.62 [DOI] [PubMed] [Google Scholar]
- Jones M. W., Errington M. L., French P. J., Fine A., Bliss T. V. P., Garel S., Charnay P., Bozon B., Laroche S., Davis S. (2001). A requirement for the immediate early gene Zif268 in the expression of late LTP and the consolidation of long-term memories. Nat. Neurosci. 4, 289–296 10.1038/85138 [DOI] [PubMed] [Google Scholar]
- Knapska E., Kaczmarek L. (2004). A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Prog. Neurobiol. 74, 183–211 10.1016/j.pneurobio.2004.05.007 [DOI] [PubMed] [Google Scholar]
- Kumbrink J., Gerlinger M., Johnson J. P. (2005). Egr-1 induces the expression of its corepressor nab2 by activation of the nab2 promoter thereby establishing a negative feedback loop. J. Biol. Chem. 280, 42785–42793 10.1074/jbc.M511079200 [DOI] [PubMed] [Google Scholar]
- Le N., Nagarajan R., Wang J. Y., Svaren J., LaPash C., Araki T., Schmidt R. E., Milbrandt J. (2005). Nab proteins are essential for peripheral nervous system myelination. Nat. Neurosci. 8, 932–940 [DOI] [PubMed] [Google Scholar]
- Lee J. L., Everitt B. J., Thomas K. L. (2004). Independent cellular processes for hippocampal memory consolidation and reconsolidation. Science 304, 839–843 10.1126/science.1095760 [DOI] [PubMed] [Google Scholar]
- Lee S. L., Sadovsky Y., Swirnoff A. H., Polish J. A., Goda P., Gavrilina G., Milbrandt J. (1996). Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1). Science 273, 1219–1221 10.1126/science.273.5279.1219 [DOI] [PubMed] [Google Scholar]
- Lemaire P., Vesque C., Schmitt J., Stunnenberg H., Frank R., Charnay P. (1990). The serum-inducible mouse gene Krox-24 encodes a sequence-specific transcriptional activator. Mol. Cell. Biol. 10, 3456–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi G., Topilko P., Schneider-Maunoury S., Lasagna M., Mantero S., Cancedda R., Charnay P. (1996). Defective bone formation in Krox-20 mutant mice. Development 122, 113–120 [DOI] [PubMed] [Google Scholar]
- Li L., Carter J., Gao X., Whitehead J., Tourtellotte W. G. (2005). The neuroplasticity-associated arc gene is a direct transcriptional target of early growth response (Egr) transcription factors. Mol. Cell Biol. 25, 10286–10300 10.1128/MCB.25.23.10286-10300.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Yun S. H., Keblesh J., Trommer B. L., Xiong H., Radulovic J., Tourtellotte W. G. (2007). Egr3, a synaptic activity regulated transcription factor that is essential for learning and memory. Mol. Cell Neurosci. 35, 76–88 10.1016/j.mcn.2007.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack K., Day M., Milbrandt J., Gottlieb D. I. (1990). Localization of the NGF1-A protein in the rat brain. Mol. Brain Res. 8, 177–180 10.1016/0169-328X(90)90062-I [DOI] [PubMed] [Google Scholar]
- Mack K. J., Cortner J., Mack P., Farnham P. J. (1992). Krox-20 messenger RNA and protein expression in the adult central nervous system. Mol. Brain Res. 14, 117–123 10.1016/0169-328X(92)90018-7 [DOI] [PubMed] [Google Scholar]
- Malkani S., Rosen J. B. (2000). Specific induction of early growth response gene 1 in the lateral nucleus of the amygdalal following contextual fear conditioning in rats. Neuroscience 97, 693–702 10.1016/S0306-4522(00)00058-0 [DOI] [PubMed] [Google Scholar]
- Malkani S., Wallace K. J., Donley M. P., Rosen J. B. (2004). An egr-1 (zif268) antisense oligodeoxynucleotide infused into the amygdala disrupts fear conditioning. Learn. Mem. 11, 617–624 10.1101/lm.73104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanares M., Nardelli J., Gilardi-Hebenstreit P., Marshall H., Giudicelli F., Martinez-Pastor M. T., Krumlauf R., Charnay P. (2002). Krox20 and kreisler co-operate in the transcriptional control of Hoxb3 hindbrain segmental expression. EMBO J. 21, 365–376 10.1093/emboj/21.3.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maviel T., Durkin T. P., Menzaghi F., Bontempi B. (2004). Sites of neocortical reorganization critical for remote spatial memory. Science 305, 96–99 10.1126/science.1098180 [DOI] [PubMed] [Google Scholar]
- Mechta-Grigoriou F., Garel S., Charnay P. (2000). Nab proteins mediate a negative feedback loop controlling Krox-20 activity in the developing hindbrain. Development 127, 119–128 [DOI] [PubMed] [Google Scholar]
- Mello C. V., Vicario D. S., Clayton D. F. (1992). Song presentation induces gene expression in the songbird forebrain. Proc. Natl. Acad. Sci. USA 89, 6818–6822 10.1073/pnas.89.15.6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R., Svaren J., Le N., Araki T., Watson M., Milbrandt J. (2001). EGR2 mutations in inherited neuropathies dominant-negatively inhibit myelin gene expression. Neuron 30, 355–368 10.1016/S0896-6273(01)00282-3 [DOI] [PubMed] [Google Scholar]
- Nikolaev E., Kaminska B., Tischmeyer W., Matthies H., Kaczmarek L. (1992). Induction of expression of genes encoding transcription factors in the rat brain elicited by behavioral training. Brain Res. Bull. 28, 479–484 10.1016/0361-9230(92)90050-8 [DOI] [PubMed] [Google Scholar]
- Nonchev S., Vesque C., Maconochie M., Seitanidou T., Ariza-McNaughton L., Frain M., Marshall H., Sham M. H., Krumlauf R., Charnay P. (1996). Segmental expression of Hoxa-2 in the hindbrain is directly regulated by Krox-20. Development 122, 543–554 [DOI] [PubMed] [Google Scholar]
- Okuno H., Miyashita Y. (1996). Expression of the transcription factor Zif268 in the temporal cortex of monkeys during visual paired associate learning. Eur. J. Neurosci. 8, 2118–2128 10.1111/j.1460-9568.1996.tb00733.x [DOI] [PubMed] [Google Scholar]
- O'Donovan K. J., Levkovitz Y., Ahn D., Baraban J. M. (2000). Functional comparison of Egr3 transcription factor isoforms: identification of an activation domain in the N-terminal segment absent from Egr3beta, a major isoform expressed in brain. J. Neurochem. 75, 1352–1357 10.1046/j.1471-4159.2000.0751352.x [DOI] [PubMed] [Google Scholar]
- O'Donovan K. J., Tourtellotte W. G., Milbrandt J., Baraban J. M. (1999). The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuronscience. Trends Neurosci. 22, 167–173 10.1016/S0166-2236(98)01343-5 [DOI] [PubMed] [Google Scholar]
- Petersohn D., Schoch S., Brinkmann D. R., Thiel G. (1995). The human synapsin II gene promoter. Possible role for the transcription factor zif268/egr-1, polyoma enhancer activator 3, and AP2. J. Biol. Chem. 270, 24361–24369 10.1074/jbc.270.41.24361 [DOI] [PubMed] [Google Scholar]
- Poirier R., Cheval H., Mailhes C., Charnay P., Davis S., Laroche S. (2007). Paradoxal role of an Egr transcription factor family member, Egr2/Krox20, in learning and memory. Front. Behav. Neurosci. 1, 6 10.3389/neuro.08.006.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo M. W., Sevetson B. R., Milbrandt J. (1995). Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc. Natl. Acad. Sci. USA 92, 6873–6877 10.1073/pnas.92.15.6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Maunoury S., Topilko P., Seitandou T., Levi G., Cohen-Tannoudji M., Pournin S., Babinet C., Charnay P. (1993). Disruption of Krox-20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell 75, 1199–1214 10.1016/0092-8674(93)90329-O [DOI] [PubMed] [Google Scholar]
- Seitanidou T., Schneider-Maunoury S., Desmarquet C., Wilkinson D. G., Charnay P. (1997). Krox-20 is a key regulator of rhombomere-specific gene expression in the developing hindbrain. Mech. Dev. 65, 31–42 10.1016/S0925-4773(97)00051-8 [DOI] [PubMed] [Google Scholar]
- Sham M. H., Vesque C., Nonchev S., Marshall H., Frain M., Das Gupta R., Whiting J., Wilkinson D., Charnay P., Krumlauf R. (1993). The zinc finger gene Krox-20 regulates Hox-B2 (Hox-2.8) during hindbrain segmentation. Cell 72, 183–196 10.1016/0092-8674(93)90659-E [DOI] [PubMed] [Google Scholar]
- Svaren J., Sevetson B. R., Apel E. D., Zimonjic D. B., Popescu N. C., Milbrandt J. (1996). NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol. Cell Biol. 16, 3545–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatek P. J., Gridley T. (1993). Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox-20. Genes Dev. 7, 2071–2084 10.1101/gad.7.11.2071 [DOI] [PubMed] [Google Scholar]
- Swirnoff A. H., Apel E. D., Svaren J., Sevetson B. R., Zimonjic D. B., Popescu N. C., Milbrandt J. (1998). Nab1, a corepressor of NGFI-A (Egr-1), contains an active transcriptional repression domain. Mol. Cell Biol. 18, 512–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swirnoff A. H., Milbrandt J. (1995). DNA-binding specificity of NGF1-A and related zinc finger transcription factors. Mol. Cell Biol. 15, 2275–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G., Schoch S., Petersohn D. (1994). Regulation of synapsin I gene expression by the zinc finger transcription factor zif268/egr-1. J. Biol. Chem. 269, 15294–15301 [PubMed] [Google Scholar]
- Tischmeyer W., Grimm R. (1999). Activation of immediate early genes and memory formation. Cell. Mol. Life Sci. 55, 564–574 10.1007/s000180050315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyama W., Okuno H., Hashimoto T., Li Y. X., Miyashita Y. (2002). Selective zif268 mRNA induction in the perirhinal cortex of macaque monkeys during formation of visual pair-association memory. J. Neurochem. 81, 60–70 10.1046/j.1471-4159.2002.00790.x [DOI] [PubMed] [Google Scholar]
- Topilko P., Levi G., Merlo G., Mantero S., Desmarquet C., Mancardi G., Charnay P. (1997). Differential regulation of the zinc finger genes Krox-20 and Krox-24 (Egr-1) suggests antagonistic roles in Schwann cells. J. Neurosci. Res. 50, 702–712 [DOI] [PubMed] [Google Scholar]
- Topilko P., Schneider-Maunoury S., Levi G., Baron-Van Evercooren A., Chennoufi A. B., Seitanidou T., Babinet C., Charnay P. (1994). Krox-20 controls myelination in the peripheral nervous system. Nature 371, 796–799 10.1038/371796a0 [DOI] [PubMed] [Google Scholar]
- Topilko P., Schneider-Maunoury S., Levi G., Trembleau A., Gourdji D., Driancourt M. A., Rao C. V., Charnay P. (1998). Multiple pituitary and ovarian defects in Krox-24 (NGFIA/Egr-1) targeted mice. Mol. Endocrinol. 12, 107–122 10.1210/me.12.1.107 [DOI] [PubMed] [Google Scholar]
- Tourtellotte W. G., Milbrandt J. (1998). Sensory ataxia and muscle spindle agenesis in mice lacking the transcription factor Egr3. Nat. Genet. 20, 87–91 10.1038/1757 [DOI] [PubMed] [Google Scholar]
- Weitemier A. Z., Ryabinin A. E. (2004). Subregion-specific differences in hippocampal activity between delay and trace fear conditioning: an immunohistochemical analysis. Brain Res. 995, 55–65 10.1016/j.brainres.2003.09.054 [DOI] [PubMed] [Google Scholar]
- Williams J., Dragunow M., Lawlor P. A., Mason S. E., Abraham W. C., Leah J., Bravo R., Demmer J., Tate W. (1995). Krox20 may play a key role in the stabilisation of long-term potentiation. Mol. Brain Res. 28, 87–93 10.1016/0169-328X(94)00187-J [DOI] [PubMed] [Google Scholar]
- Wisden W., Errington M. L., Williams S., Dunnett S. B., Waters C., Hitchcock D., Evan G., Bliss T. V., Hunt S. P. (1990). Differential expression of immediate early genes in the hippocampus and spinal cord. Neuron 4, 603–614 10.1016/0896-6273(90)90118-Y [DOI] [PubMed] [Google Scholar]
- Worley P. F., Bhat R. V., Baraban J. M., Erickson C. A., McNaughton B. L., Barnes C. A. (1993). Thresholds of synaptic activation of transcription factors in hippocampus: correlation with long-term enhancement. J. Neurosci. 13, 4776–4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K., Kaufmann W. E., Lanahan A., Papapavlous M., Barnes C. A., Andreasson K. I., Worley P. F. (1994). Egr3/Pilot, a zinc finger transcription factor, is rapidly regulated by activity in brain neurons and colocalize with Egr-1/zif268. Learn. Mem. 1, 140–152 [PubMed] [Google Scholar]
- Yu L., Haverty P. M., Mariani J., Wang Y., Shen H. Y., Schwarzschild M. A., Weng Z., Chen J. F. (2005). Genetic and pharmacological inactivation of adenosine A2A receptor reveals an Egr-2-mediated transcriptional regulatory network in the mouse striatum. Physiol. Genomics 23, 89–102 10.1152/physiolgenomics.00068.2005 [DOI] [PubMed] [Google Scholar]