Abstract
Four homologs to the Drosophila homeotic gene spalt (sal) exist in both humans and mice (SALL1 to SALL4/Sall1 to Sall4, respectively). Mutations in both SALL1 and SALL4 result in the autosomal-dominant developmental disorders Townes-Brocks and Okihiro syndrome, respectively. In contrast, no human diseases have been associated with SALL2 to date, and Sall2-deficient mice have shown no apparent abnormal phenotype. We generated mice deficient in Sall2 and, contrary to previous reports, 11% of our Sall2-deficient mice showed background-specific neural tube defects, suggesting that Sall2 has a role in neurogenesis. To investigate whether Sall4 may compensate for the absence of Sall2, we generated compound Sall2 knockout/Sall4 genetrap mutant mice. In these mutants, the incidence of neural tube defects was significantly increased. Furthermore, we found a similar phenotype in compound Sall1/4 mutant mice, and in vitro studies showed that SALL1, SALL2, and SALL4 all co-localized in the nucleus. We therefore suggest a fundamental and redundant function of the Sall proteins in murine neurulation, with the heterozygous loss of a particular SALL protein also possibly compensated in humans during development.
The homeotic spalt (sal) gene of Drosophila melanogaster determines the identity of the anterior head and the posterior tail regions in early development.1,2 At later stages, sal is involved in the development of the wing disk, trachea, and sensory organs.3,4,5,6 Humans and mice have four functional sal-related genes SALL1 to SALL4 (Sall1 to Sall4 in mice). The human SALL genes encode transcription factors presenting a characteristic structure of evenly distributed zinc finger domains. Mutations in SALL1 result in Townes-Brocks syndrome (TBS, OMIM 107480), a rare autosomal-dominant malformation syndrome that is characterized by dysplastic ears, preaxial polydactyly and/or triphalangeal thumbs, imperforate anus, renal malformations, and heart anomalies.7,8,9 Mutations in SALL2 are not associated with any disease, however, it has been suggested that SALL2 may act as a tumor suppressor.10,11 Deletion of SALL3 has been implicated in the phenotype of the 18q-deletion syndrome.12 Mutations in SALL4 cause autosomal-dominant Okihiro/Duane-Radial Ray syndrome (DRRS, OMIM 607323), combining radial ray defects and Duane anomaly.13 Additional clinical features such as anal, renal, cardiac, ear and foot malformations, hearing loss, postnatal growth retardation, and facial asymmetry have been documented.
Sall1-null mutant mice die perinatally because of severe kidney dysgenesis or agenesis,14 whereas Sall2-deficient mice were reported to show no apparent phenotype.15 Homozygous Sall4 knockout mice die shortly after implantation, and Sall4 haploinsufficiency results in anorectal and heart anomalies as well as exencephaly.16 Mice lacking both Sall1 and Sall2 show kidney phenotypes comparable to those of Sall1 single knockout mice, suggesting that both genes do not genetically interact in vivo. In contrast, Sall1/4 double heterozygotes exhibit uni- or bilateral renal agenesis, exencephaly, anorectal malformations, and ventricular septum defects with increased incidence in comparison to Sall4 heterozygotes. This argues for a redundancy in the function of these two Sall genes.
The SALL proteins were shown to be important factors in complex regulatory networks in early and late vertebrate embryogenesis. Murine SALL4 is an essential transcription factor for the early development of inner cell mass-derived cell lineages and is required for the maintenance of embryonic stem cell pluripotency.17,18 At later stages, SALL4 controls Fgf10 expression in the limbs and Gja5 expression in the heart.19 In zebrafish, fgf10 expression is indirectly activated by sall1a and sall4 via activation of fgfr24.20 XsalF from Xenopus determines the position of forebrain and midbrain21 and Xsal3, csal3, and sall1a are essential for nephrogenesis.22,23,24
Cranial neurulation begins during gastrulation, when the neuroepithelium is induced to differentiate from the dorsal midline ectoderm to form the neural plate. Subsequently, the neural folds form at the edges of the neural plate, elevate from a semihorizontal to a vertical position, and shift from convex to concave in a complex process that involves midline neuroepithelial bending and expansion of the underlying cranial mesenchyme. Later on, the tips of the neural folds converge along the dorsal midline and fuse to form the neural tube.25,26,27,28 Neural tube defects (NTDs) are a group of heterogeneous and complex congenital anomalies of the central nervous system. Most of the mouse NTD mutants reflect a failure in neural fold elevation, and as a consequence, they present as exencephaly, spina bifida, or rachischisis at later stages. In mice, more than 150 mutations have been reported that cause NTDs, but no clinical or experimental study has provided unequivocal evidence for a definitive role for any of these genes in the causation of NTDs in humans. In addition, considerable variation is seen in the pattern of morphogenesis between inbred mouse strains. This indicates that neurogenesis is a complex process with a complex genetic network.
We have generated Sall2-null mutant mice, and in contrast to previous reports, we present evidence that Sall2 has a role in neurogenesis. The incidence of NTDs in Sall2−/− embryos is drastically increased in Sall2/4 compound mutants. In addition, compound Sall1/4 mutants also display NTDs similar to Sall2/4 mutants, suggesting an in vivo redundant role for the Sall genes during murine neurulation.
Materials and Methods
Generation of Sall2−/− Mice
The targeting vector was generated by incorporating the 5′ BglII/BglII fragment (4.3 kb) and the 3′ BglII/BamHI (3.0 kb) fragment into the pTKneo vector (kindly provided by Nils Brose, MPI für Experimentelle Medizin, Göttingen, Germany). The 5′ fragment was subcloned into pBluescript (Stratagene, La Jolla, CA) and subsequently cloned into a SalI-ClaI site 5′ of the Neor gene. The 3′ fragment was excised from a pBluescript subclone and inserted into a BglII-BamHI site 3′ of the Neor gene. RI cells were electroporated, selected, and clones resistant to G418 were isolated and screened by Southern blot. The DNA isolated from the duplicate plate was digested with EcoRI, electrophoresed, transferred on nitrocellulose membrane (Amersham, Little Chalfont, UK), and hybridized to radioactive probes to confirm the correct homologous recombination. Nine of thirty-six clones were correctly targeted. Clones were then injected into 129SV/J blastocysts. Resulting chimeric animals were bred with C57BL/6 or 129SV/J females. Mutant animals studied were of F2 and later generations. Mice were genotyped by using genomic polymerase chain reaction (PCR). For genotyping we used the primer mix: SAF1: 5′-GGCTCACAACCATCCGTAACA-3′; ExR1 5′-ACACCTGCTCACCTCCATCG-3′; NeoR1 5′-GCGCGAATTCGATGATCCTGAACGGC-3′. The size of the amplicon is 400 bp for the wild-type allele and 620 bp for the Sall2 mutant allele.
Sall2+/− mice on a 129SV/J-DBA/2, 129SV/J-NZW, and 129SV/J-CD1 background were generated by crossing Sall2−/− mice on a 129SV/J background to mice on the inbred DBA/2 and NZW and the outbred CD1 background, respectively. Sall2−/− embryos from F1 intercrosses were analyzed at E12.5 to E14.5. Inbred NZW mice were obtained from The Jackson Laboratory (Bar Harbor, MA), and inbred DBA/2 and outbred CD1 mice were obtained from Charles River (Lyon, France).
Generation of Sall4+/gt Mice
Clone W097E01 (embryonic stem cells 129 × 1/SvJ2) carrying an insertion of the gene trap vector pT1βgeo in intron 1 of the Sall4 gene (Sall4GT(pT1Betageo)1Flo, hereafter termed Sall4gt) was identified within the German Gene Trap Consortium (GGTC; www.genetrap.de).29 Mutant mice were generated after injection into C57BL/6 blastocyst and resulting chimeras were bred to C57BL/6J females. Mice were genotyped using the primer mix: S4F 5′-TGGGGATTCCGGACTTGCTTC-3′; S4R 5′-TTTAAAAGCGGCGCCACTAGAG-3′; pGEOR6 5′-GAGATGGATTGGCAGATGTAGC-3′. The size of the amplicon is 284 bp for the wild-type allele and 234 bp for the Sall4gt mutant allele.
Generation of Sall1+/− Mice
The targeting vector was made by insertion of a 5′ 6.2-kb XhoI/ClaI fragment and a 3′ 1.1-kb XbaI fragment into the pTKneo vector. The construct was linearized with NotI and transfected into R1 embryonic stem cells. Clones resistant to G418 were isolated, screened, and analyzed as described above to confirm homologous recombination. Twenty-seven of forty-three clones were positive. Clones were then injected into 129SV/J blastocysts. Resulting chimeric animals were bred with C57BL/6 females. Mutant animals studied were of F2 and later generations. Mice were genotyped by using genomic PCR. For genotyping of the wild-type allele, we used the primers Ex2 F164 5′-TGATTACACGACATTTCCTACTG-3′ and SA R164 5′-GACGACTCAAGTAAA AAGCACAA-3′ (fragment size 400 bp). For the recombinant allele, PCR was performed with primers Neo F264 5′-GGCTGACCGCTTCCTCGTG-3′ and SA R164 (fragment size 750 bp).
Histological Analysis
Embryos were collected in phosphate-buffered saline (PBS), fixed in Serra (ethanol:37% formaldehyde:acetic acid, 6:3:1), embedded in paraffin, and sectioned to 7 μm. Sections were stained with hematoxylin and eosin (H&E). For cell proliferation, sections were stained with rabbit anti-phospho-histone 3 (H3P) antibody (catalog no. 06-570, Upstate, Chicago, IL). Detection of apoptotic cells in paraffin sections was performed using a rabbit anti-caspase 3-active antibody (catalog no. AF-835; R&D Systems, Minneapolis, MN). The ABC kit (Vector Laboratories, Burlingame, CA) was used following the manufacturer’s protocol. In every section, the number of stained cells was scored and the surface area of neuroepithelium was determined using the AxioVs40 V 4.4.0.0 software (Zeiss, Jena, Germany). Images were captured by a Zeiss Axiocam MRc5 charge-coupled device camera.
In Situ Hybridization Analysis
Whole mount in situ hybridization was performed following a standard procedure,30 with digoxigenin-labeled antisense riboprobes for Sall2, Sall4,31 Fgf8,32 and Msx1.33 For expression analysis of Sall2 a 1082-bp Sall2 BamHI fragment of exon 2 (Figure 1A) containing sequences coding for the third double zinc finger and 3′-UTR was subcloned into pBluescriptKS (Stratagene).
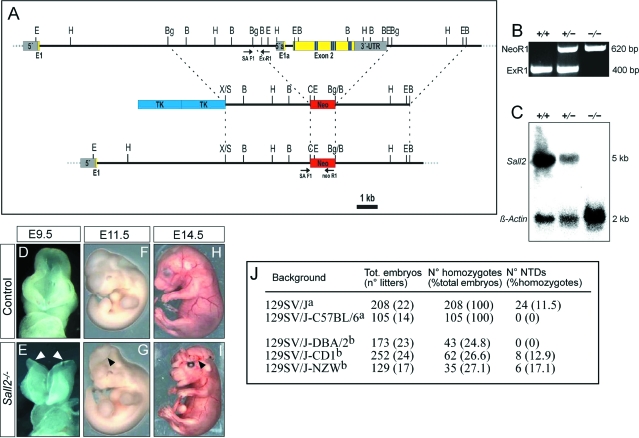
Figure 1.
Generation and analysis of Sall2−/− mice. A: Strategy for inactivation of the Sall2 gene. Exons 1a and 2 were replaced by the neomycin cassette of the pTKneo vector resulting in a recombinant allele. Exons including zinc finger domains (blue stripes), sequencing primers (arrows), neomycin cassette (Neo), and thymidine kinase (Tk) are depicted B: BamHl, Bg: Bglll, C: Clal, E: EcoRl, H: Hindlll, S: Sall, X: Xbal. B: PCR to identify the Sall2 wild-type allele (+) and the Sall2 mutant allele (−). The strategy is indicated in A. We used a primer mix with the forward primer SAF1 and the reverse primers ExR1 (for wild-type allele) and NeoR1 (for Sall2 mutant allele). C: Northern-blot analysis of Sall2-deficient mice. Each lane contains 20 μg of total RNA prepared from kidneys of wild-type mice (+/+), as well as of heterozygous (+/−) and homozygous (−/−) Sall2 knockout mice. The Sall2 probe detects a 5-kb transcript. D–I: Gross morphology of Sall2−/− embryos at different stages. D and E: At E9.5, the neural tube is already closed in control embryos (D), whereas in some Sall2−/− embryos the neural folds remain unfused (E, arrowheads). F–I: At E11.5 and at E14.5 some Sall2−/− embryos show excencephaly (G and I, arrowheads). J: Incidence of NTDs of Sall2−/− embryos on different genetic backgrounds. aFor the 129SVJ and 129SVJ-C57BL/6 backgrounds, embryos analyzed were from homozygous Sall2 mutant intercrosses from F3 and later generations; bfor the other three backgrounds, embryos analyzed were from heterozygous Sall2 mutant F1 intercrosses.
Intracellular Co-Localization Analysis
For co-localization assays, COS-7 and HEK293 cells were grown on coverslips (5 × 104 cells) and transiently transfected with expression plasmids using Lipofectamine (Invitrogen, Carlsbad, CA). After washing with PBS, the cells were treated with methanol for fixation (10 minutes), washed twice with PBS, and equilibrated in IF buffer [10 mmol/L Tris, pH 7.5, 100 mmol/L NaCl, 0,05% (v/v) Tween20, 1% (w/v) bovine serum albumin] for 30 minutes. Cells were incubated with primary antibodies (anti-FLAG; Sigma-Aldrich, St. Louis, MO; and anti-HA; BD Biosciences, Heidelberg, Germany), washed with PBS, incubated with secondary biotinylated antibodies (Vector Laboratories), and signals were detected using the fluorescent avidin kit (Vector Laboratories). Nuclear counterstaining was accomplished by 4,6-diamidino-2-phenylindole staining showing the location of the nuclei for orientation. Finally, coverslips were mounted on glass slides using Slowfade Gold (Invitrogen) and fluorescence signals were visualized by confocal laser-scanning microscopy.
Quantitative Real-Time Reverse Transcriptase (RT)-PCR
For quantitative real-time RT-PCR (qRT-PCR) total RNA was isolated from E8.5 stage embryos and cDNA was prepared by reverse transcription using oligo(dT) primers. The relative expression of the Sall4 exon 1-2 transcript was analyzed using the primers E1RTF1 (5′-GAAGCCCCAGCACATCAAC-3′) and E2RTR2 (5′-CTGAGGCTTCATCGCAGTT-3′). The exon 1a-2 transcript was amplified with the primer pair E1aRTF1 (5′-CGGACTGCGACACACACC-3′) and E2RTR2. The qRT-PCR was performed using the ABI Prism Real-Time Detection System 7900 (Applied Biosystems, Darmstadt, Germany). For the quantification of RT-PCR products, we used the SYBR Green assay system (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. In brief, the RT-PCR reactions were prepared by adding 5 μl of 2× SYBR Green mix, 2.5 μl of primer mix (forward and reverse, 1 pmol/μl), and 2.5 μl of cDNA template. The RT-PCR amplification condition was the following: 94°C 30 seconds, 63°C 30 seconds, 72°C 30 seconds for 45 cycles. Data analysis was done by SDS 2.1 software (Applied Biosystems) using standard curve method and Microsoft Office Excel (Microsoft, Redmond, WA). To verify the specificity and efficiency of real-time analysis, the PCR products were analyzed by monitoring their dissociation curves. The expression levels of investigated transcripts were normalized to β-actin mRNA expression to compensate for difference in sample amounts. The primer sequences used for amplification of the β-actin transcript were actinF (5′-CTTTGCAGCTCCTTCGTTGC-3′) and actinR (5′-ACGATGGAGGGGAATACAGC-3′).
Northern Blot
For expression analysis of Sall2 a 1082-bp Sall2 BamHI fragment34 containing sequences coding for the third double zinc finger and 3′ UTR (exon 2) was subcloned into pBluescriptKS (Stratagene). A 32P-labeled probe of this subclone was generated by use of Rediprime II labeling system (Amersham Biosciences) and hybridized to Northern blots containing RNA of adult murine kidney tissue. RNA extraction, Northern blotting, and hybridization were performed as described.35
Results
Sall2−/− Embryos Show NTDs
We have generated Sall2−/− mice by deletion of exons 1a and 2 via homologous recombination, resulting in a loss of all regions encoding zinc finger domains and loss of the complete coding region for the exon 1a-exon 2 transcript (Figure 1, A–C). Sall2+/− mice developed normally and had no phenotypic abnormalities. When intercrossed to generate homozygous Sall2−/− mutants, again no phenotypic abnormalities were observed on a mixed 129SV/J-C57BL/6 background (105 Sall2−/− embryos analyzed). However, when we analyzed Sall2−/− mice on a 129SV/J background, the litter size was constantly below the strain-specific average. We therefore decided to analyze the phenotypic appearance of the embryos and noted a significant perinatal lethality (data not shown). We observed abnormalities during neural tube development at E9.5 (Figure 1E) and exencephaly at E11.5 in some Sall2−/− embryos (Figure 1G). To quantify the frequency of the phenotype, we collected 208 Sall2−/− embryos at E14.5, and 24 (11.5%) embryos displayed exencephaly (Figure 1I). While this work was in progress, a Sall2−/− mouse was described. Because these mutants developed to term without pathological findings, a dispensable role for Sall2 in murine embryogenesis has been postulated.15 In contrast, our results indicate that Sall2 is required for normal embryonic neural tube development, depending on the genetic background. To confirm this, we generated Sall2−/− mice on three new mixed backgrounds: 129SV/J-DBA/2, 129SV/J-CD1, and 129SV/J-NZW. We found NTDs in homozygous Sall2 mutants on the 129SV/J-CD1 and the 129SV/J-NZW backgrounds at a frequency of 12.9% and 17.1%, respectively, but not on the 129SV/J-DBA/2 background (Figure 1J). This result confirms that Sall2 plays a role during neurulation. It also shows that the frequency of NTDs displayed by Sall2−/− embryos varies depending on the genetic background, indicating that the different genetic constitutions of the various mouse backgrounds modulates the effect of the loss of Sall2.
Early Expression of Sall2 in the Cranial Region
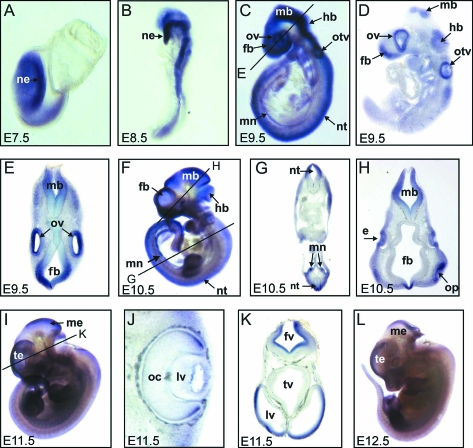
Previous studies have revealed Sall2 expression in the metanephros and the spinal cord at E11.5. At later stages, the expression was localized to the kidney and the brain.15 Because we observed NTDs in Sall2−/− embryos already at E9.5, we decided to study Sall2 expression at early developmental stages. In situ hybridization for Sall2 revealed expression at E7.5 in the neural ectoderm, the mesoderm, and a broad anterior region of the head (Figure 2A). The same expression pattern was observed at E8.5 (Figure 2B). Later, the expression became restricted to distinct structures: strong signals were seen in the forebrain, midbrain, hindbrain, and the mesonephros at E9.5 (Figure 2, C–E) and at E10.5 (Figure 2, F–H). Analysis of the sections indicated that Sall2 is expressed dorsally in the neuroepithelium all along the body axis (Figure 2, D and E). In addition, Sall2 expression could be detected in the otic vesicle at E9.5 (Figure 2, C–E), in the lens placode at E10.5, and in the optic cup at E11.5 (Figure 2, H and J). Starting at E11.5, Sall2 expression in the neural tube decreased but was maintained in the telencephalon and the third ventricle until E12.5 (Figure 2, I, K, and L). This pattern of expression is consistent with a role of Sall2 in neurulation.
Figure 2.
Sall2 expression during murine embryogenesis. A–C, F, I, and L: Whole mount in situ hybridization for Sall2. D, E, G, H, J, and K: Vibratome sections of the stained embryos. e, eye; fb, forebrain; fv, fourth ventricle; hb, hindbrain; lv, lateral ventricle; mb, midbrain; me, mesencephalon; mn, mesonephros; ne, neuroectoderm; nt, neural tube; oc, optic cup; op, olfactory pit; otv, otic vesicle; ov, optic vesicle; te, telencephalon; tv, third ventricle. For the description of the results see the text.
Redundancy between Sall2 and Sall4 in Neural Tube Development
The low penetrance of the cranial abnormalities in the Sall2−/− embryos might be explained by the compensation by other factors. Sall4, a closely related gene, was previously shown to display a similar expression pattern during embryogenesis,31 and we confirmed by in situ hybridization that both genes were simultaneously expressed in the developing neuroectoderm of the cranial region at E8.5 (Figure 3, A and B). To screen for a possible redundant role during neural tube development, we decided to generate compound Sall2/4 mutant mice.
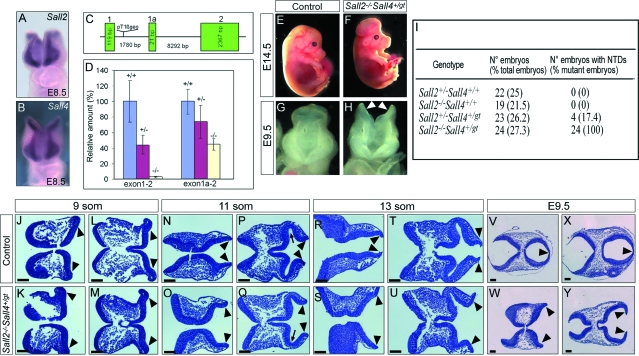
Figure 3.
Analysis of Sall2/4 compound mutants. A and B: In situ hybridization for Sall2 (A) and for Sall4 (B) on mouse embryos revealed that both genes have a similar pattern of expression in the cranial neuroepithelium at E8.5. C: Schematic representation of the integration of the pT1βgeo gene trap vector in the Sall4 locus. Only the first three exons are shown. Integration occurred in intron 1, between exon 1 and the newly identified alternative exon 1a. D: Quantitative RT-PCR of E7.5 wild-type (+/+) and heterozygous (+/−) and homozygous (−/−) Sall4gt embryos for the Sall4 locus. Primers were designed to include either exon 1 and exon 2 (exon1–2) or exon 1a and exon 2 (exon 1a-2). Note that in Sall4gt/gt embryos, exon1–2 transcripts are abolished whereas 45% of exon 1a-2 transcripts are still present when compared to wild-type embryos. E–H: Sall2−/−Sall4+/gt embryos at E14.5 showed exencephaly (F). At E9.5, the forebrain and midbrain neural folds of mutant embryos did not fuse (H, arrowheads). I: Incidence of NTDs in the four mutant genotypes resulting from the cross between Sall2+/−Sall4+/gt mice and Sall2−/− mice. J–Y: Transverse serial sections of controls (J, L, N, P, R, T, V, X) and Sall2−/−Sall4+/gt embryos (K, M, O, Q, S, U, W, Y) stained with H&E demonstrated a failure in elevation and central fusion of the neural folds in the mutants. Arrowheads point to the tips of the neural folds, and arrows (P, Q) point to the optic vesicle. For a detailed explanation see the text. Scale bars = 100 μm.
Heterozygous Sall4 gene trap mutant mice, Sall4+/gt, generated as described in the Materials and Methods, did not reveal any phenotypic abnormalities. In contrast, Sall4gt/gt homozygous embryos displayed a phenotype similar to Sall4-deficient knockout mice,16 but they died at approximately E7.5, 1 day later than reported for homozygous knockouts (unpublished observations). By database searches we identified an EST clone (BY729264) comprising an alternative first exon upstream of exon 2 that contained a different translation initiation site. Comparison of the EST sequence to the Sall4 genomic region identified the localization of the alternative exon closely downstream of the genetrap integration site. The newly identified exon was named exon 1a (Figure 3C). To determine the impact of the integration of the gene trap vector we performed quantitative RT-PCR at E7.5. Our analysis revealed that in Sall4gt homozygous mutants the mRNA expression initiated from exon 1 was completely abolished whereas the expression initiated from exon 1a was reduced to 45% when compared to wild-type embryos (Figure 3D). Therefore, the genetrap integration resulted in a hypomorphic Sall4 mutant allele. Analysis of the cross between Sall2+/−Sall4+/gt mice and Sall2−/− mice revealed that Sall2−/−Sall4+/gt developed NTDs with a full penetrance of the phenotype (24 of 24), whereas the phenotype was only partially penetrant in Sall2/4 compound heterozygous mutant embryos (4 of 23). We found no NTDs in the other genetic combinations (Figure 3I).
To determine when phenotypic differences between Sall2+/−Sall4+/gt and control embryos can first be detected, we examined H&E-stained sections of the cranial region between E8.0 to E8.5 (9 to 13 somite stage). At the 9-somite stage, the neural folds are already prominent in the cephalic region of both control and mutant embryos, lying in a semihorizontal position (Figure 3, J–M). At the 11-somite stage, the neural folds in the cephalic region are elevating and start to become parallel in control embryos (Figure 3N). This bending is also visible in the mutant neural folds, but to a much lesser extent, and the distance between the neural tips is always increased (Figure 3O). In a more rostral location, the development of an optic vesicle is visible in both control and mutant embryos (Figure 3, P and Q; arrow), but the distance between the neural tips remains increased in mutant embryos (Figure 3Q). At the 13-somite stage, in control embryos, the neural folds in the cephalic region are lying parallel, and the neural tips are close to fusing (Figure 3R). In the mutant embryos, the degree of bending of the neural folds is similar to the 11-somite stage, and the neural tips are very far apart (Figure 3S). In a more rostral location, the edges of the neural tube are parallel and very close to each other in control embryos (Figure 3T), whereas they are very separated in the mutant embryos (Figure 3U). At E9.5, the neural tube is already fused in control embryos (Figure 3, V and X), whereas the neural folds remain unfused in the mutants (Figure 3, W and Y). Altogether, these results show that Sall2−/−Sall4+/gt embryos have a failure in neural fold elevation and fusion, which finally leads to the excencephally seen at later stages. They also indicate that Sall2 and Sall4 have a redundant function in neural tube closure.
Increased Apoptosis in Sall2−/−Sall4+/gt Embryos
To test whether a complete gene misregulation may be responsible for the phenotype observed in Sall2−/−Sall4+/gt embryos, we analyzed by in situ hybridization the expression of Fgf8, a marker for the neuroepithelium of the frontonasal region, the midbrain-hindbrain boundary, and the branchial arch,32 and Msx1, that is expressed in the forebrain and the midbrain.33 No difference in expression was seen for these two markers between Sall2−/−Sall4+/gt and wild-type embryos (Figure 4, A–D). Thus, a significant misregulation of these two marker genes seems not to be the origin of the neural tube closure defects seen in the Sall2−/−Sall4+/gt mutant embryos.
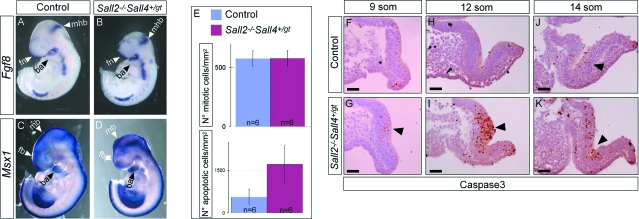
Figure 4.
Increased apoptosis in Sall2−/−Sall4+/gt neural folds. A–D: In situ hybridization for Fgf8 and Msx1 at E9.5 revealed no difference in the pattern of expression of both genes between control (A, C) and Sall2−/−Sall4+/gt embryos (B, D). E: Analysis of cell proliferation and apoptosis of control and Sall2−/−Sall4+/gt mutant embryos at 9 to 13 somite stages. For cell proliferation no statistical difference was found between controls and Sall2−/−Sall4+/gt embryos (n = 6; unpaired t-test, P = 0.9554) (E, top). In contrast, a statistically significant difference was seen in the number of apoptotic cells between control and Sall2−/−Sall4+/gt embryos (n = 6; unpaired t-test, P = 0.0026) (E, bottom). F, H, and J: Analysis of the neuroepithelium of control embryos stained for caspase 3 revealed some positive cells, in particular close to the bending of the neural tube (arrowhead in J). G, I, and K: In the equivalent regions of Sall2−/−Sall4+/gt embryos the number of positive cells was markedly increased. Arrowheads point to the apoptotic cells. ba, branchial arch; fb, forebrain; mhb, midbrain-hindbrain boundary; fn, frontonasal region; mb, midbrain. Scale bars = 50 μm.
To test whether the neuroepithelium of Sall2−/−Sall4+/gt mutants may reveal any difference in cell proliferation or cell death we performed immunohistochemistry for the mitosis marker H3P and for the apoptotic marker caspase 3 at the 9- to 13-somite stage. We found no difference in cell proliferation between control and mutant embryos (Figure 4E, top). In contrast, we found an increased number of apoptotic cells (Figure 4E, bottom). Analysis of the neuroepithelium of control embryos stained for caspase 3 revealed some positive cells, in particular close to the bending of the neural tube (Figure 4, F, H, and J). In the equivalent regions in the mutants the number of positive cells was markedly increased (Figure 4, G, I, and K).
Sall1 and Sall4 Act Redundantly during Murine Neurulation
The murine Sall1 gene is closely related to Sall2 and Sall4 and is also expressed in the neural tube. In compound Sall1/2-null mutants no neural tube abnormalities have been described.15 To screen for a possible redundant role between Sall1 and Sall4 in murine neurulation, we crossed Sall1+/− mice to mice harboring the hypomorphic Sall4gt allele. For this purpose we generated Sall1+/− mice by homologous recombination resulting in a deletion of exons 2 and 3 including all zinc finger domains. Heterozygous Sall1 mutant mice were phenotypically normal and fertile, whereas homozygous Sall1 mutants died perinatally as a result of a bilateral kidney agenesis, as previously described.14
By crossing Sall1+/− and Sall4+/gt mice we obtained 4 Sall1+/−Sall4+/gt mutants of 79 offspring. While this work was in progress, a Sall1/4 compound heterozygous mutant was described. Interestingly, 45% of these Sall1+/−Sall4+/− mutants showed NTDs, but none of them survived after birth.16 We therefore speculate that the remaining expression of our Sall4gt hypomorphic allele (see above) may account for the incomplete penetrance and therefore may rescue a small number of embryos from early lethality.
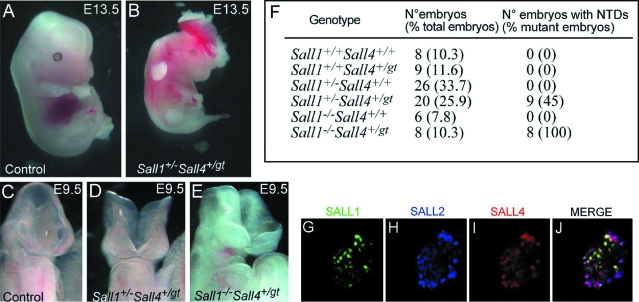
Analysis of the embryos from the cross between Sall1+/−Sall4+/gt mice and Sall1+/− mice revealed that Sall1−/−Sall4+/gt embryos presented with general growth retardation at E10.5 and died at approximately E11.5, probably because of cardiac failure (data not shown); they developed NTDs with a full penetrance of the phenotype (eight of eight) (Figure 5, E and F). In contrast the phenotype was partially penetrant in Sall1+/−Sall4+/gt compound heterozygous mutant embryos (9 of 20) (Figure 5, B, D, and F). We found no NTDs in the other genetic combinations (Figure 5F). In conclusion, our data suggest that the higher the dosage of Sall4 on a Sall1+/− background, or the higher the dosage of Sall1 on a Sall4+/gt background, the less severe is the penetrance of the resulting cranial phenotype. This supports the idea of functional redundancy of both genes during murine neurulation.
Figure 5.
Analysis of Sall1/4 compound mutants. A and B: Gross morphology at E13.5 of a control embryo (A) and a Sall1/4 double heterozygous mutant embryo presenting exencephaly (B). C–E: At E9.5, some of the Sall1/4 double heterozygous embryos displayed defects in neural tube closure (D). All Sall1−/−Sall4+/gt embryos obtained showed a severe phenotype at E9.5 (E). F: Incidence of NTDs in the different genotypes resulting from the cross between Sall1+/−Sall4+/gt mice and Sall1+/− mice. G–J: Intracellular co-localization of SALL1, SALL2, and SALL4. COS-7 cells expressing SALL1-GFP (green), SALL2-Flu (blue), and SALL4-FLAG (red) showed that SALL2 and SALL4 essentially co-localized at the nuclear periphery, whereas SALL1 showed a more homogenous distribution.
It has previously been shown that SALL1 and SALL4 are located in distinct nuclear dots representing heterochromatic structures.36,37 We therefore investigated whether SALL1, SALL2, and SALL4 co-localize in the nucleus. By triple-transfecting COS-7 cells with fusion proteins SALL1-GFP, SALL2-Flu, and SALL4-FLAG we found that SALL2 and SALL4 preferentially co-localize at the nuclear margins, whereas SALL1 is more centrally located. However, all proteins co-localize in dot-like structures in the nucleus, favoring the idea that they may operate as redundant transcription factors (Figure 5, G–J).
Discussion
Previous data suggested a dispensable role for Sall2 in embryogenesis because no human disease is associated with mutations in the SALL2 gene and because Sall2−/− mice do not present any phenotypic abnormalities.15 In contrast, here we present evidence that Sall2 may have a role in neurulation as we found NTDs in Sall2-deficient mice. Our Sall2-null mutant embryos presented this phenotypic abnormality only on a 129SV/J background, but not on a mixed 129SV/J-C57BL/6 background, the background of the Sall2 mutants described by Sato and colleagues.15 To confirm this phenomenon of strain-specific variations in the penetrance of the phenotype, we studied Sall2−/− mice on three other backgrounds, and found that Sall2−/− mice also developed NTDs on the mixed backgrounds 129SV/J-CD1 and 129SV/J-NZW, but not on a mixed 129SV/J-DBA/2 background. These data are consistent with the notion that genetic loci controlling neural tube development are polymorphic between the mouse strains. Indeed, strain-specific variations in the penetrance of NTDs displayed by homozygous null mutants have been frequently reported. One of the factors that may affect these strain-specific differences in NTDs is the position of neural tube closure 2, which varies between mouse strains.38,39 The position of closure 2 is located within the midbrain in the DBA/2 strain, is located more rostrally, at the midbrain-forebrain boundary, in the CD1 strain and is located even more rostrally, in the forebrain, in the NZW strain.38 A rostral location of closure 2 has been proposed to be a risk factor for cranial NTDs. Accordingly, in Sp2H mutants, the frequency of cranial NTDs is reduced after transfer to the DBA/2 background and is increased after transfer to the NZW background.38 Consistent with these data, we found no NTDs in embryos on the mixed 129SV/J-DBA/2 background, but we did observe NTDs in embryos on the 129SV/J-CD1 and 129SV/J-NZW backgrounds. Our data are thus consistent with the notion that a protective role of caudal closure 2 may account for the differences in the penetrance of NTDs seen in our Sall2-deficient mice. As nothing is known up to now where site 2 closure occurs in the 129SV and the C57BL/6 strains, it is not possible to discuss whether or not the absence of NTDs observed in Sall2−/− mutant mice on the mixed 129SV/C57BL/6 background may be because of a caudal site 2 closure protective effect. But one must also consider that we analyzed mixed genetics backgrounds, thus, the shift in the position of closure 2 was not as prominent as on a pure genetic background. So, in addition to the position of closure 2, other genetic factors must be involved in the strain-specific differences observed.
The low incidence of NTDs in Sall2−/− embryos, the similar structure of the SALL proteins, the overlapping expression patterns of Sall1, Sall2, and Sall4 in the cranial region, and the overlapping clinical features of patients with TBS and Okihiro syndrome led us to the hypothesis of a redundant function of the SALL proteins in neurogenesis. We therefore generated Sall2/4 and Sall1/4 compound mutants. Morphological analyses of the embryos revealed that the higher the SALL dosage, the less penetrant were the NTDs. In addition, we showed that SALL1, SALL2, and SALL4 co-localize in the nucleus, and previous GST-pulldown studies have demonstrated that the SALL proteins form homo- and heterodimers.40 These in vivo and in vitro results indicate an essential and redundant role of the murine Sall genes in neurulation.
Histological analysis of our Sall2/4 compound mutants revealed a failure in the elevation of the neural folds in the forebrain and midbrain. In Sall2−/−Sall4+/gt embryos the neural folds remain in a semihorizontal position and they never elevate and fuse. The exencephaly shown by mutant embryos at later stages is probably a direct consequence of the failure in elevation. Taken together, these results indicate that Sall2/4 have an essential role in the process of neural fold elevation during neurogenesis.
Neural tube closure is a complex and multistep process that involves lifting, bending, and fusion of the neural folds. Therefore, cell proliferation and cell death must be tightly regulated during this process. We did not find increased cell proliferation in Sall2−/−Sall4+/gt embryos, but we detected an increased number of apoptotic cells. Little is known about the molecular function of Sall2 and Sall4. Sall2 has been proposed to act in some cell types as a regulator of cell growth and survival,11 and Sall4 has been shown to be a regulator of survival and apoptosis in human leukemic cells.41 It is thus conceivable that both genes may have an anti-apoptotic function also during early neural development. Alternatively, Sall2/4 may control other unknown function(s) during neurulation, and the increased number of apoptotic cells may simply be a consequence of the failure in neural fold elevation.
NTDs are only rarely seen in TBS and Okihiro syndrome. In TBS, two families are known to us in which some affected members have Arnold-Chiari malformation type 1. In Okihiro syndrome, one patient with a SALL4 deletion was born with a meningomyelocele (J. Kohlhase, unpublished data). Therefore, NTDs are a rare feature associated with heterozygous SALL1 or SALL4 mutations in humans. However, it is conceivable that compound mutations in SALL1, SALL2, or SALL4 may result in defective neural tube closure also in humans. But such a severe phenotype would implicate embryonic lethality and hence escape the clinical diagnosis of TBS or Okihiro syndrome. Mutations in SALL2 have not yet been correlated with a human disease, and this may be because of the compensatory character of the other SALL genes. This assumption is strengthened by the fact that Sall2+/− mice are phenotypically normal and that Sall2−/− embryos exhibit a reduced penetrance of the NTDs (∼10%) only in some mouse strains. It is therefore likely that the heterozygous loss of SALL2 can effectively be compensated by the related SALL proteins in human.
Acknowledgments
We thank Monika Schindler, Henning Riedesel, and Stefan Wolf for expert assistance in the generation of knockout mice; Gerd Scherer, Annette Neubüser, and Alexander Craig for critical review of the manuscript; and Pauline Soulas and Jean-Louis Pasquali for the kind gift of NZW mice.
Footnotes
Address reprint requests to Prof. Dr. Jürgen Kohlhase, Center for Human Genetics Freiburg, Heinrich-von-Stephan-Str. 5, D-79100 Freiburg, Germany. E-mail: jkohlhase@humangenetik-freiburg.de.
Supported by the Deutsche Forschungsgemeinschaft (grants Ko1850/3-2 and Ko1850/6-2 to J. K. and Graduiertenkolleg Molekulare Genetik der Entwicklung to J. K.).
J.B. and A.B. contributed equally to this study.
References
- Jürgens G. Head and tail development of the Drosophila embryo involves spalt, a novel homeotic gene. EMBO J. 1988;7:189–196. doi: 10.1002/j.1460-2075.1988.tb02799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühnlein RP, Frommer G, Friedrich M, Gonzalez-Gaitan M, Weber A, Wagner-Bernholz JF, Gehring W, Jäckle H, Schuh R. spalt encodes an evolutionary conserved zinc finger protein of novel structure which provides homeotic gene function in the head and tail region of the Drosophila embryo. EMBO J. 1994;13:168–179. doi: 10.1002/j.1460-2075.1994.tb06246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis J, Barrio R, Kafatos F. A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature. 1996;381:421–424. doi: 10.1038/381421a0. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Barrio R, Kafatos FC. Regulation of the spalt/spalt-related gene complex and its function during sensory organ development in the Drosophila thorax. Development. 1999;126:2653–2662. doi: 10.1242/dev.126.12.2653. [DOI] [PubMed] [Google Scholar]
- Kühnlein RP, Schuh R. Dual function of the region specific homeotic gene spalt during Drosophila tracheal system development. Development. 1996;122:2215–2223. doi: 10.1242/dev.122.7.2215. [DOI] [PubMed] [Google Scholar]
- Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Kohlhase J, Wischermann A, Reichenbach H, Froster U, Engel W. Mutations in the SALL1 putative transcription factor gene cause Townes-Brocks syndrome. Nat Genet. 1998;18:81–83. doi: 10.1038/ng0198-81. [DOI] [PubMed] [Google Scholar]
- Powell CM, Michaelis RC. Townes-Brocks syndrome. J Med Genet. 1999;36:89–93. [PMC free article] [PubMed] [Google Scholar]
- Surka WS, Kohlhase J, Neunert CE, Schneider DS, Proud VK. Unique family with Townes-Brocks syndrome, SALL1 mutation, and cardiac defects. Am J Med Genet. 2001;102:250–257. doi: 10.1002/1096-8628(20010815)102:3<250::aid-ajmg1479>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Li D, Dower K, Ma Y, Tian Y, Benjamin T. A tumor host range selection procedure identifies p150(sal2) as a target of polyoma virus large T antigen. Proc Natl Acad Sci USA. 2001;98:14619–14624. doi: 10.1073/pnas.251447198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Tian Y, Ma Y, Benjamin T. p150(Sal2) is a p53-independent regulator of p21(WAF1/CIP). Mol Cell Biol. 2004;24:3885–3893. doi: 10.1128/MCB.24.9.3885-3893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhase J, Hausmann S, Stojmenovic G, Dixkens C, Bink K, Schulz-Schaeffer W, Altmann M, Engel W. SALL3, a new member of the human spalt-like gene family, maps to 18q23. Genomics. 1999;62:216–222. doi: 10.1006/geno.1999.6005. [DOI] [PubMed] [Google Scholar]
- Kohlhase J, Chitayat D, Kotzot D, Ceylaner S, Froster U, Fuchs S, Montgomery T, Rösler B. SALL4 mutations in Okihiro syndrome (Duane-radial ray syndrome), acro-renal-ocular syndrome, and related disorders. Hum Mutat. 2005;26:176–183. doi: 10.1002/humu.20215. [DOI] [PubMed] [Google Scholar]
- Nishinakamura R, Matsumoto Y, Nakao K, Nakamura K, Sato A, Copeland NG, Gilbert DJ, Jenkins NA, Scully S, Lacey DL, Katsuki M, Asashima M, Yokota T. Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development. 2001;128:3105–3115. doi: 10.1242/dev.128.16.3105. [DOI] [PubMed] [Google Scholar]
- Sato A, Matsumoto Y, Koide U, Kataoka Y, Yoshida N, Yokota T, Asashima M, Nishinakamura R. Zinc finger protein sall2 is not essential for embryonic and kidney development. Mol Cell Biol. 2003;23:62–69. doi: 10.1128/MCB.23.1.62-69.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki-Yumoto M, Kobayashi C, Sato A, Fujimura S, Matsumoto Y, Takasato M, Kodama T, Aburatani H, Asashima M, Yoshida N, Nishinakamura R. The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development. 2006;133:3005–3013. doi: 10.1242/dev.02457. [DOI] [PubMed] [Google Scholar]
- Elling U, Klasen C, Eisenberger T, Anlag K, Treier M. Murine inner cell mass-derived lineages depend on Sall4 function. Proc Natl Acad Sci USA. 2006;103:16319–16324. doi: 10.1073/pnas.0607884103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, Lou Y, Yang J, Ma Y, Chai L, Ng HH, Lufkin T, Robson P, Lim B. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- Koshiba-Takeuchi K, Takeuchi JK, Arruda EP, Kathiriya IS, Mo R, Hui CC, Srivastava D, Bruneau BG. Cooperative and antagonistic interactions between Sall4 and Tbx5 pattern the mouse limb and heart. Nat Genet. 2006;38:175–183. doi: 10.1038/ng1707. [DOI] [PubMed] [Google Scholar]
- Harvey SA, Logan MP. Sall4 acts downstream of tbx5 and is required for pectoral fin outgrowth. Development. 2006;133:1165–1173. doi: 10.1242/dev.02259. [DOI] [PubMed] [Google Scholar]
- Onai T, Sasai N, Matsui M, Sasai Y. Xenopus XsalF: anterior neuroectodermal specification by attenuating cellular responsiveness to Wnt signaling. Dev Cell. 2004;7:95–106. doi: 10.1016/j.devcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Camp E, Hope R, Kortschak RD, Cox TC, Lardelli M. Expression of three spalt (sal) gene homologues in zebrafish embryos. Dev Genes Evol. 2003;213:35–43. doi: 10.1007/s00427-002-0284-6. [DOI] [PubMed] [Google Scholar]
- Farrell ER, Tosh G, Church E, Munsterberg AE. Cloning and expression of CSAL2, a new member of the spalt gene family in chick. Mech Dev. 2001;102:227–230. doi: 10.1016/s0925-4773(01)00296-9. [DOI] [PubMed] [Google Scholar]
- Onuma Y, Nishinakamura R, Takahashi S, Yokota T, Asashima M. Molecular cloning of a novel Xenopus spalt gene (Xsal-3). Biochem Biophys Res Commun. 1999;264:151–156. doi: 10.1006/bbrc.1999.1479. [DOI] [PubMed] [Google Scholar]
- Copp AJ. Neurulation in the cranial region—normal and abnormal. J Anat. 2005;207:623–635. doi: 10.1111/j.1469-7580.2005.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MJ, Juriloff DM. Mouse mutants with neural tube closure defects and their role in understanding human neural tube defects. Birth Defects Res A Clin Mol Teratol. 2007;79:187–210. doi: 10.1002/bdra.20333. [DOI] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ. Mouse models for neural tube closure defects. Hum Mol Genet. 2000;9:993–1000. doi: 10.1093/hmg/9.6.993. [DOI] [PubMed] [Google Scholar]
- Padmanabhan R. Etiology, pathogenesis and prevention of neural tube defects. Congenit Anom (Kyoto) 2006;46:55–67. doi: 10.1111/j.1741-4520.2006.00104.x. [DOI] [PubMed] [Google Scholar]
- Wiles MV, Vauti F, Otte J, Fuchtbauer EM, Ruiz P, Fuchtbauer A, Arnold HH, Lehrach H, Metz T, von Melchner H, Wurst W. Establishment of a gene-trap sequence tag library to generate mutant mice from embryonic stem cells. Nat Genet. 2000;24:13–14. doi: 10.1038/71622. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- Kohlhase J, Heinrich M, Liebers M, Froehlich Archangelo L, Reardon W, Kispert A. Cloning and expression analysis of Sall4, the murine homologue of the gene mutated in Okihiro syndrome. Cytogenet Genome Res. 2002;98:274–277. doi: 10.1159/000071048. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- Catron KM, Wang H, Hu G, Shen MM, Abate-Shen C. Comparison of MSX-1 and MSX-2 suggests a molecular basis for functional redundancy. Mech Dev. 1996;55:185–199. doi: 10.1016/0925-4773(96)00503-5. [DOI] [PubMed] [Google Scholar]
- Kohlhase J, Altmann M, Archangelo L, Dixkens C, Engel W. Genomic cloning, chromosomal mapping, and expression analysis of Msal-2. Mamm Genome. 2000;11:64–68. doi: 10.1007/s003350010012. [DOI] [PubMed] [Google Scholar]
- Buck A, Archangelo L, Dixkens C, Kohlhase J. Molecular cloning, chromosomal localization, and expression of the murine SALL1 ortholog Sall1. Cytogenet Cell Genet. 2000;89:150–153. doi: 10.1159/000015598. [DOI] [PubMed] [Google Scholar]
- Böhm J, Kaiser FJ, Borozdin W, Depping R, Kohlhase J. Synergistic cooperation of Sall4 and cyclin D1 in transcriptional repression. Biochem Biophys Res Commun. 2007;356:773–779. doi: 10.1016/j.bbrc.2007.03.050. [DOI] [PubMed] [Google Scholar]
- Netzer C, Rieger L, Brero A, Zhang C-D, Hinzke M, Kohlhase J, Bohlander SK. SALL1, the gene mutated in Townes-Brocks syndrome, encodes a transcriptional repressor which interacts with TRF1/ PIN2 and localizes to pericentromeric heterochromatin. Hum Mol Genet. 2001;10:3017–3024. doi: 10.1093/hmg/10.26.3017. [DOI] [PubMed] [Google Scholar]
- Fleming A, Copp AJ. A genetic risk factor for mouse neural tube defects: defining the embryonic basis. Hum Mol Genet. 2000;9:575–581. doi: 10.1093/hmg/9.4.575. [DOI] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, Tom C, MacDonald KB. Normal mouse strains differ in the site of initiation of closure of the cranial neural tube. Teratology. 1991;44:225–233. doi: 10.1002/tera.1420440211. [DOI] [PubMed] [Google Scholar]
- Sweetman D, Smith T, Farrell ER, Chantry A, Münsterberg A. The conserved glutamine rich region of chick csal1 and csal3 mediates protein interactions with other spalt family members. Implications for Townes-Brocks syndrome. J Biol Chem. 2003;278:6560–6566. doi: 10.1074/jbc.M209066200. [DOI] [PubMed] [Google Scholar]
- Yang J, Chai L, Gao C, Fowles TC, Alipio Z, Dang H, Xu D, Fink LM, Ward DC, Ma Y. SALL4 is a key regulator of survival and apoptosis in human leukemic cells. Blood. 2008;112:805–813. doi: 10.1182/blood-2007-11-126326. [DOI] [PMC free article] [PubMed] [Google Scholar]