Abstract
The balance between bone resorption and bone formation involves the coordinated activities of osteoblasts and osteoclasts. Communication between these two cell types is essential for maintenance of normal bone homeostasis; however, the mechanisms regulating this cross talk are not completely understood. Many factors that mediate differentiation and function of both osteoblasts and osteoclasts have been identified. The LIM protein Limd1 has been implicated in the regulation of stress osteoclastogenesis through an interaction with the p62/sequestosome protein. Here we show that Limd1 also influences osteoblast progenitor numbers, differentiation, and function. Limd1−/−calvarial osteoblasts display increased mineralization and accelerated differentiation. While no significant differences in osteoblast number or function were detected in vivo, bone marrow stromal cells isolated from Limd1−/− mice contain significantly more osteoblast progenitors compared to wild type controls when cultured ex vivo. Furthermore, we observed a significant increase in nuclear β-catenin staining in differentiating Limd1−/− calvarial osteoblasts suggesting that Limd1 is a negative regulator of canonical Wnt signaling in osteoblasts. These results demonstrate that Limd1 influences not only stress osteoclastogenesis but also osteoblast function and osteoblast progenitor commitment. Together, these data identify Limd1 as a novel regulator of both bone osetoclast and bone osteoblast development and function.
Keywords: LIM proteins, Limd1, osteoblast, osteoprogenitors, mesenchymal stem cells, Wnt signaling, β-catenin
Introduction
Bone remodeling is a normal function of healthy bone and involves two major cell types: bone forming osteoblasts and bone resorbing osteoclasts. This highly dynamic process requires large localized pools of osteoblast and osteoclast progenitor cells able to respond quickly to environmental changes or pathologic challenges. Both osteoblast and osteoclast progenitors are found in the bone marrow: osteoblasts derive from the mesenchymal bone marrow stromal cell lineage while osteoclasts are hematopoietic in origin, arising from bone marrow derived macrophages (BMDM). These two highly specialized cell populations exist in a delicate equilibrium that, when disrupted, results in pathologic consequences such as osteoporosis, osteopetrosis, and Paget’s disease. While sustaining a balance between bone resorption and bone formation is critical for maintaining normal bone homeostasis, the precise mechanisms responsible are not clearly defined.
In addition to their bone forming capacity, osteoblasts also regulate osteoclast differentiation. The earliest committed progenitors, pre-osteoblasts, produce receptor-activator of NF-κB ligand (RANKL), the major cytokine stimulating osteoclastogenesis [1]. These immature osteoblasts support osteoclast differentiation from BMDMs in vivo as well as in vitro. As the pre-osteoblast develops into a bone forming osteoblast, it no longer produces RANKL but instead produces the RANKL inhibitory factor osteoprotegrin (OPG) thereby inhibiting osteoclast differentiation and favoring bone formation. It is thought that serum ratios of RANKL to OPG may serve as a clinical measure of in vivo osteoclast activity in certain disease states [2].
LIMD1 belongs to the Ajuba/Zyxin family of cytosolic LIM domain containing proteins [3]. There are six family members: Zyxin [4], LPP [5], TRIP6 [6], Ajuba [7], LIMD1 [3], and WTIP [8]. These proteins regulate cytoskeletal dynamics and cell adhesion [9]. Not only are they components of adhesive receptor complexes in both mesenchymal and epithelial cells but they also shuttle to and from the nucleus, thereby potentially communicating environmental cell adhesive events into nuclear responses [9]. The family is divided into two subfamilies, Ajuba and Zyxin, based on a phylogenetic analysis of the LIM domains as well as distinctive N-termini and unique protein interacting partners. Ajuba, WTIP and LIMD1 belong to the Ajuba sub-family. The LIMD1 gene is located at the 3p21.3 chromosome locus in humans, a region containing quantitative trait loci (QTL) for bone and mineral density loss[10].
Recently, Limd1 was shown to interact with the p62/sequestosome protein and influence IL-1 and RANKL signaling by facilitating the assembly of a p62/TRAF6/a-PKC multi-protein complex [11, 12]. The Limd1-p62 interaction affects both NF-κB and AP-1 activity in epithelial cells and osteoclasts [11, 12]. Furthermore, Limd1−/− pre-fusion osteoclasts are defective in RANKL signaling in vitro [11]. Although basal bone density of Limd1−/− mice was not increased compared to wild type (wt), Limd1−/− mice exhibit diminished osteoclastogenesis in response to RANKL treatment, acute parathyroid hormone (PTH) stimulus, and serum induced arthritis [11]. An additional alteration in osteoblast function could contribute to the defective osteoclast stress response observed in Limd1−/−mice. Therefore, we asked whether osteoblast development and function was influenced by deletion of the Limd1 gene.
Materials and Methods
Calvarial osteoblast isolation, culture and immunofluorescence analysis
Osteoblasts were isolated from the calvaria of 4–5 day old mice as described previously [13]. Primary calvarial cells were grown in Gibco α-MEM with 10% FBS (Invitrogen, Carlsbad, CA). Immortalized cell lines were generated by infecting primary calvarial cells with retroviruses that express SV40 large T antigen (pBABE-neo-T). After three passages, only immortalized cells continued to proliferate. For mineralization assays, 5×105 primary or immortalized calvarial cells were plated in 12-well dishes and cultured in osteogenic media (α-MEM/10% FBS supplemented with 10mM β-glycerol-2-phosphate and 50μg/ml L-ascorbic acid; Sigma, St. Louis, MO). Total RNA and/or protein extracts were prepared from cells after 0, 1, 3, 6, 9 or 12 days of culture in osteogenic conditions. To detect mineralized nodules, calvarial osteoblasts cultured for 10–12 days in osteogenic media were fixed in 70% ethanol for 1 hour at 4°C, washed with water then stained with 0.4% Alizarin Red-S (Sigma, St. Louis, MO) for 10 minutes at room temperature. Cells were washed five times with water then dehydrated once with 70% Ethanol and once with 100% ethanol. The number of individual Alizarin Red positive nodules was quantified and graphed, or the percent of each well stained was calculated by threshold analysis of scanned images using ImageJ software (http://rsb.info.nih.gov/ij/).
Primary and immortalized calvarial osteoblasts were infected with pMX-myc, pMX-myc-Limd1, pMX-RFP or pMX-RFP-Limd1 retrovirus and selected in 2μg/mL blastocidin. All cells were fixed in 4% paraformaledhyde (Electron Microscopy Sciences, Hatfield, PA) stabilization buffer (127mM NaCl, 5mM KCl, 1.1mM NaH2PO4, 0.4mM KH2PO4, 2mM MgCl2, 5.5mM Glucose, 1mM EGTA, 10mM Pipes in water), permeabilized in 0.5% Triton/PBS, then treated with 6N guanidine-HCl. Blocking and immunofluorescence was performed in 3% BSA (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), 0.05% Triton X-100, 0.02% Sodium Azide (Sigma, St. Louis, MO). Primary immunofluorescence was performed using monoclonal anti-human Vinculin antibody (1:500, Sigma, St. Louis, MO), polyclonal Limd1 antiserum (1:250, described previously[11]), or β–catenin antibody (BD Biosciences, San Jose, CA, 1:500). Secondary immunofluorescence was performed using Alexa Flour 488 or 568 (Molecular Probes, Eugene, OR). Nuclei were visualized by DAPI staining (Sigma, St. Louis, MO).
Osteoblast/BMDM co-culture assays
Co-culture Assays were performed as described previously [14]. Briefly, 2×104 primary or immortalized calvarial cells were plated together with 2×104 primary bone marrow derived macrophages (BMDM) in 48 well dishes and cultured in α-MEM/10% FBS supplemented with 10nM 1α,25-Dihydroxyvitamin D3 (Biomol International, Plymouth Meeting, PA). After 10–12 days, osteoclasts were visualized by tartrate resistant acid phosphatase (TRAP) staining using a Leukocyte TRAP kit (Sigma, St. Louis, MO) per manufacturers suggestion.
Mesenchymal progenitor cell isolation and culture
Bone marrow mesenchymal progenitor colony forming assays were performed as described previously[15]. Briefly, marrow was flushed from the femora and tibiae of 6–8 week old Wt and Limd1−/− mice and plated at equal densities in 24-well dishes: 1.5 ×106 cells for fibroblast colony forming assays (CFU-F) or 2.5 ×106 cells for adipocyte colony forming assays (CFU-A) and osteoblast colony forming assays (CFU-O). Cells were maintained in α-MEM/10% FBS with or without supplements and left undisturbed for the first 7 days in culture. Afterward, media was changed every other day. After 9 days in culture without supplements, CFU-Fs were visualized using Harris Hematoxylin solution (Sigma, St. Louis, MO) per manufacturer’s suggestion. CFU-A were maintained in adipogenic cocktail (50μM indomethacin, and 5μg/mL insulin) for 9 days then fixed for 10 minutes in 10% neutral formalin (Sigma, St. Louis, MO) and stained with 0.3% Oil Red O (Sigma, St. Louis, MO) in water for 45 minutes. CFU-O were maintained in osteogenic media for 3–21 days then fixed for 10 minutes in 10% neutral formalin and stained with alkaline phosphatase staining solution for 40 minutes (1 capsule fast Blue RR salt/2mL napthol/50mL water; Sigma, St. Louis, MO). After alkaline phosphatase staining, cultures were co-stained with Alizarin Red S. For Von Kossa staining, cells were incubated with 5% AgNO3 for 1hr at room temperature in the dark, washed once with 5% sodium thiosulfate, twice with water, then exposed to ultraviolet light for 1 hour at room temperature while drying. The number of individual Von Kossa positive nodules was quantified and graphed. For alkaline phosphatase and Alizarin Red staining, the percent of each well stained was calculated by threshold analysis of scanned images using ImageJ software (http://rsb.info.nih.gov/ij/).
RT-PCR
Total cellular RNA was isolated using RNeasy kits (Qiagen Inc., Valencia, CA). First-strand cDNA was generated from 1 μg of total RNA using SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen Corporation, Carlsbad, CA) as recommended by the manufacturer. The cDNA product was diluted to a final volume of 100 μL with water. Diluted cDNA (1μL) was used as the template for PCR. Primers sequences and PCR conditions will be provided upon request.
Cell Proliferation and BrdU Staining
Cell proliferation rates were determined by plating equal numbers (100,000) of Limd1−/− and wt primary calvarial cells in osteogenic media and counting the total number of cells present after 2, 4, and 6 days in culture. For BrdU staining calvarial osteoblasts were cultured in osteogenic media for 0, 3, or 6 days then incubated with 50μM BrdU for 2hr at 37°C, washed three times with PBS, and fixed in 100% methanol for 1 minute at −20°C then 4 minutes at room temperature. Cells were washed three times with PBS then treated with 1.5M HCl for 40 minutes at 37°C, washed again then blocked for 1 hour in 15%FBS/PBS. Immunofluorescence was performed using mouse monoclonal anti-BrdU antibody (1:2000, Sigma, St. Louis, MO). Cells were counterstained with DAPI to detect nuclei. The number of BrdU positive nuclei were counted and normalized to total number of nuclei.
PTH injections and mouse bone analysis
Mice aged 16 weeks were injected subcutaneously once daily for 28 days with 400 ng human parathyroid hormone (PTH, 1–34) per g of weight (Bachem, Torrance, CA) dissolved in 0.9% saline/0.1mM acetic acid or vehicle. Animals were sacrificed at 28 days. Bone mineral density was measured each week by dual-energy x-ray absorptiometry (DEXA) using a PIXImus2 scanner (Lunar Corporation, Madison, WI). MicroCT analyses were performed on tibiae that had been dissected and embedded in 1% agarose using a SCANCO Medical microCT 40 (SCANCO USA, Inc., Southeastern, PA). For histomorphometric analyses leg bones were decalcified for two weeks in 14% EDTA, embedded in paraffin, sectioned and TRAP stained. Histomorphometry was performed using the OsteoMeasure Histomorphometry System (OsteoMetrics, Atlanta, GA).
Statistical analyses
All Statistical analyses were performed in Microsoft Excel using the paired student t-test. Error bars represent standard error of the mean.
Results
Limd1 protein expression increases during osteoblast differentiation
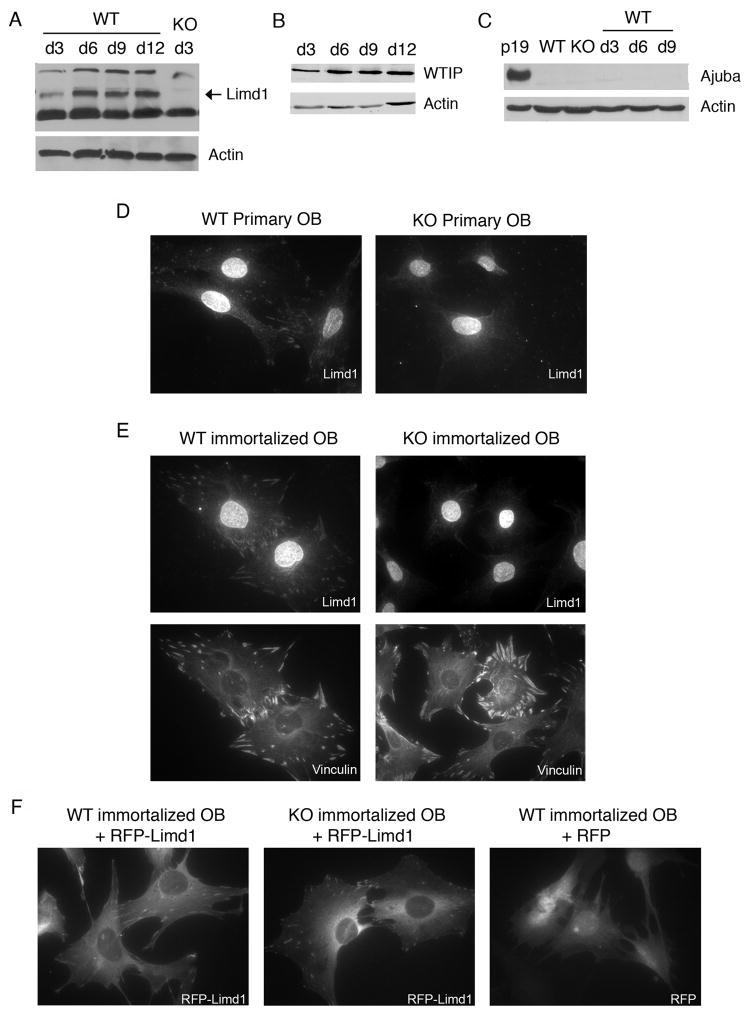
To determine the expression pattern of Ajuba family LIM proteins during osteoblast differentiation, calvarial osteoblasts isolated from newborn wt and Limd1−/− mice were cultured in osteogenic conditions. Cell lysates were prepared at various times during osteoblast differentiation and levels of the Ajuba LIM proteins determined and compared by Western Blot analysis. Limd1 protein was initially expressed at low levels, however, upon exposure to osteogenic conditions, its level of expression was dramatically increased and sustained (Fig. 1A). Analysis of other family member expression during osteoblast differentiation demonstrated that WTIP protein levels increased slightly (Fig. 1B) while the already low Ajuba levels did not appreciably change (Fig. 1C). The uniquely induced expression of Limd1 in response to osteogenic stimulation suggested that Limd1 might play a role in the regulation of osteoblast development and function.
Fig. 1.
Limd1 protein expression and localization in calvarial osteoblasts.
(A–C) Primary calvarial osteoblasts were cultured in osteogenic conditions for 3, 6, 9, or 12 days, lysed, and equal amounts of protein run on SDS-PAGE then Western blotted for the presence of the Ajuba-LIM family members Limd1 (A), WTIP (B) and Ajuba (C). Actin was used as a loading control. Primary (D) or immortalized (E) Wt or Limd1−/− calvarial osteoblasts were fixed, permeabilized and stained with Limd1 or vinculin antisera. (F) Immortalized wt or Limd1−/−calvarial osteoblasts transduced with RFP or RFP-Limd1, as indicated.
To determine the sub-cellular localization of Limd1 in osteoblast progenitor cells, primary Limd1−/− and wt calvarial osteoblasts were stained with Limd1 antiserum. Endogenous Limd1 protein was found to localize throughout the cytosol as well as to sites of cell-matrix adhesion where it co-localized with the focal adhesion protein vinculin (Fig. 1D, data not shown). The nuclear staining pattern observed following staining with Limd1 antibodies was non-specific as the nucleus of Limd1−/− cells also stained brightly (Fig. 1D). Immortalized calvarial osteoblast cell lines generated from Limd1−/− and wt primary calvarial osteoblasts were also used to determine the sub-cellular distribution of Limd1. Anti-Limd1 immunofluorescence staining of these cells revealed the same pattern as seen in primary calvarial osteoblasts, including nonspecific nuclear staining (Fig. 1E). Focal adhesions were not disrupted in Limd1−/− cells, as vinculin staining patterns were identical to those seen in wt cells (Fig. 1E). To address the question of nuclear staining, immortalized wt and Limd1−/− calvarial osteoblasts were transduced with RFP or RFP-Limd1 by retroviral infection. In both cell types, RFP-Limd1 localized throughout the cytosol and to sites of cell-matrix adhesion (Fig. 1F). There was no evidence for nuclear staining of RFP-Limd1 under these culture conditions (Fig. 1F). In sum these studies show that, under the culture conditions employed, Limd1 is present at matrix adhesion sites and in the cytosol of calvarial osteoblasts.
Limd1 influences osteoblast function in vitro
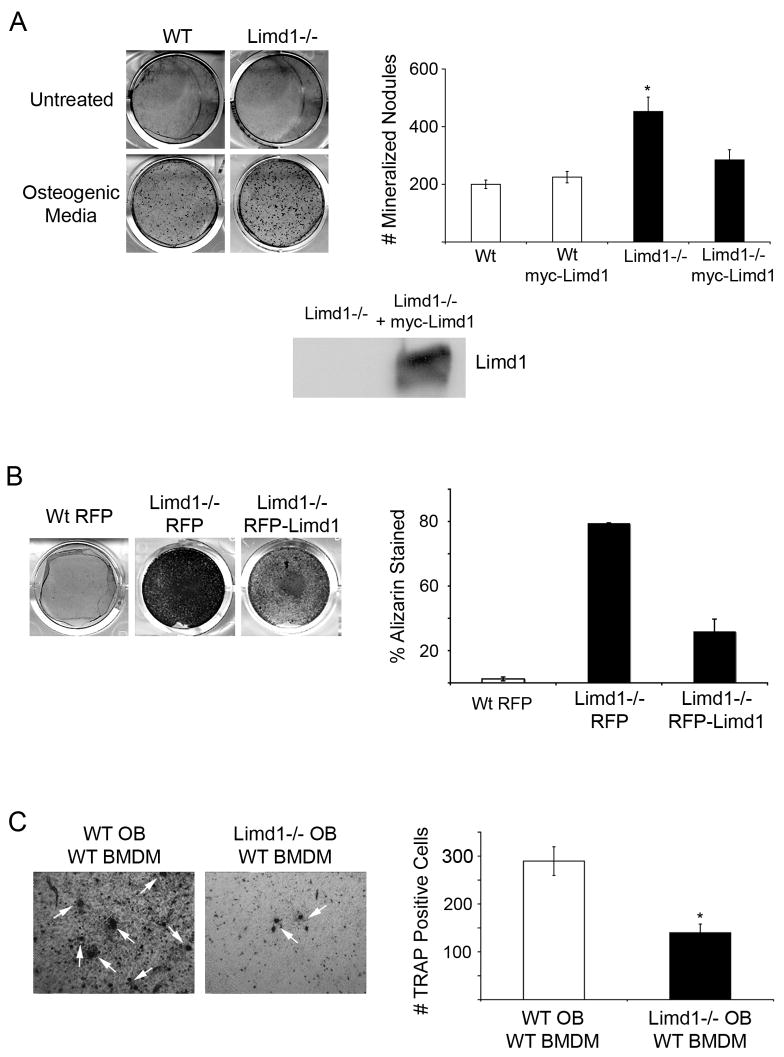
A major function of osteoblasts is to mineralize bone matrix. To determine if Limd1 affected osteoblast mineralization function, equal numbers of Limd1−/−and wt primary calvarial cells were cultured in osteogenic media for twelve days. Cultures were then fixed and stained with Alizarin Red S to visualize and quantify the extent of mineralization. Limd1−/− calvarial osteoblasts displayed significantly increased mineralization compared to wt controls (Fig. 2A). Retroviral reintroduction of the Limd1 protein into Limd1−/− cells prior to addition of osteogenic media reverted the extent of mineralization toward wt levels (Fig. 2A), indicating that this defect was due to loss of Limd1 expression. Over-expression of Limd1 in wt calvarial cells had no effect on mineralization (Fig. 2A). The capacity of immortalized calvarial cell lines to mineralize matrix was also tested. As seen with the primary cells, immortalized Limd1−/− cell lines displayed increased mineralization (Fig. 2B). Again, re-introduction of Limd1 protein expression into Limd1−/− immortalized calvarial cells prior to the addition of osteogenic media was sufficient to revert mineralization toward wt levels (Fig. 2B).
Fig. 2. Limd1−/− calvarial osteoblasts display increased mineralization and support less osteoclastogenesis than Wt controls.
(A) Primary Wt or Limd1−/− calvarial osteoblasts were cultured in normal or osteogenic media for 12 days then fixed and stained with Alizarin Red S to detect mineralization. The number of Alizarin positive nodules were counted and graphed. Data is a compilation of three different experiments, all performed in triplicate. For rescue experiments, Limd1−/− calvarial osteoblasts were infected with either myc-empty or myc-Limd1 retroviruses before exposure to osteogenic culture conditions. Aliquots of Limd1−/− cells infected with myc-empty or myc-Limd1 were lysed and equal amounts of protein run on SDS-PAGE then Western blotted for the presence of Limd1. (B) Immortalized Wt or Limd1−/− calvarial osteoblast were cultured in osteogenic conditions for 12 days then fixed, stained with Alizarin Red S, and the percentage of Alizarin Red S staining determined and graphed. (C) Equal numbers of primary Wt or Limd1−/− calvarial osteoblasts and wt BMDM were co-cultured in the presence of vitamin D for 10 days then fixed and TRAP stained to identify osteoclasts (white arrows). The number of TRAP positive osteoclasts was quantified and graphed. For rescue experiments, Limd1−/− calvarial osteoblasts were infected with either myc-empty or myc-Limd1 retroviruses before co-culturing. *Indicates a significant difference between Limd1−/− and Wt (*p<0.05, **p<0.01, ****p<0.001, student’s paired t-test).
In the presence of vitamin D, cultured calvarial osteoblasts produce sufficient quantities of cytokines to support the development and differentiation of osteoclast from BMDM [1]. To determine the capacity of Limd1−/− calvarial osteoblasts to support osteoclastogenesis, co-cultures containing equal numbers of primary wt BMDM and primary wt or Limd1−/− calvarial cells were grown in the presence of vitamin D. After ten days in culture, cells were fixed and TRAP stained for detection of osteoclasts. Significantly fewer TRAP-positive osteoclasts were generated in co-cultures containing Limd1−/− calvarial osteoblasts compared to wt calvarial osteoblasts (Fig. 2C). Similarly, immortalized Limd1−/− calvarial cells supported less osteoclast differentiation than immortalized wt control calvarial cells (data not shown). Taken together, these data indicate that Limd1 affects both osteoblast functions: the ability of immature osteoblasts to support osteoclastogenesis and the capacity of mature osteoblasts to properly mineralize matrix in vitro.
Limd1 influences osteoblast differentiation
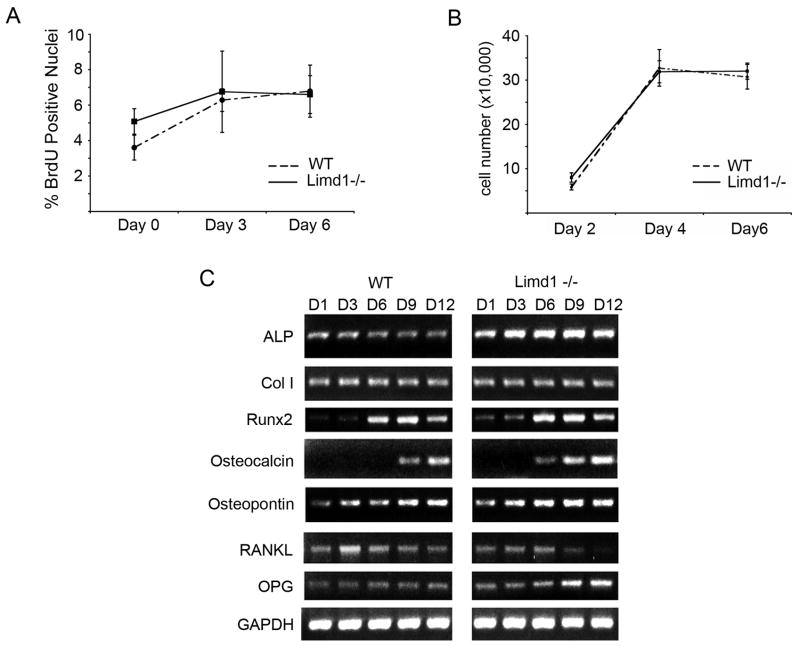
Limd1−/− calvarial osteoblasts display enhanced mineralization and support less osteoclastogenesis than wt calvarial osteoblasts. These functional deficits suggest that Limd1−/− cultures contain a more mature osteoblast population than wt: a phenomenon that could be explained by either enhanced proliferation or accelerated differentiation. Calvarial cell proliferation rates were measured by BrdU incorporation, and cell proliferation assays. Cells exposed to mineralization conditions were incubated with BrdU at three-day intervals for twelve days. BrdU uptake was determined by immunofluorescence. No difference in BrdU uptake between Limd1−/− and wt calvarial osteoblasts was observed (Fig. 3A). Similarly, when equal numbers of wt and Limd1−/− calvarial cells were cultured there was no difference in proliferation over 6 days (Fig. 3B).
Fig. 3.
Limd1 influences osteoblast differentiation but not proliferation.
(A) Primary wt or Limd1−/− calvarial osteoblasts were cultured in osteogenic media for 0, 3, or 6 days then incubated with BrdU, fixed and stained. The number of BrdU positive nuclei was normalized to the total number of nuclei. (B) Equal numbers of wt or Limd1−/− calvarial osteoblasts (100,000) were plated and cultured in osteogenic conditions for the time indicated. Total cell numbers were determined and graphed for comparison. (C) RT-PCR analysis of osteoblast-specific gene expression in RNA isolated from primary Wt or Limd1−/− calvarial osteoblasts cultured in osteogenic media for 1, 3, 6, 9, and 12 days. ALP – alkaline phosphatase; Col I – type 1 collagen; RANKL – receptor activator of NF-kB ligand; OPG – osteoprotegrin; GAPDH – glyceraldehydes-3-phosphate dehydrogenase
During differentiation, osteoblasts produce cytokines that can either positively or negatively regulate osteoclastogenesis [1]. Less differentiated pre-osteoblasts produce stimulatory RANKL, while mature, bone forming osteoblasts produce the inhibitory factor osteoprotegrin (OPG). Since Limd1−/− osteoblasts did not appropriately support osteoclastogenesis in vitro, we asked whether deletion of Limd1 affected osteoblast differentiation programs. RT-PCR analysis was performed on differentiating calvarial osteoblasts over a twelve-day time-course. Limd1−/− calvarial osteoblasts displayed enhanced expression of the early osteoblast marker alkaline phosphatase and premature expression of the mature osteoblast marker Osteocalcin suggesting that they adopted a mature osteoblast phenotype sooner than their wt counterparts (Fig. 3C). Furthermore, early repression of RANKL expression coincident with up-regulation of OPG expression was observed in Limd1−/− calvarial cells compared to wt controls (Fig. 3C). This accelerated osteoblast maturation and decreased RANKL/OPG ratio provided a potential explanation for the reduced ability of Limd1−/− calvarial cells to support osteoclast differentiation.
Limd1−/− mice have increased numbers of osteoblast progenitors
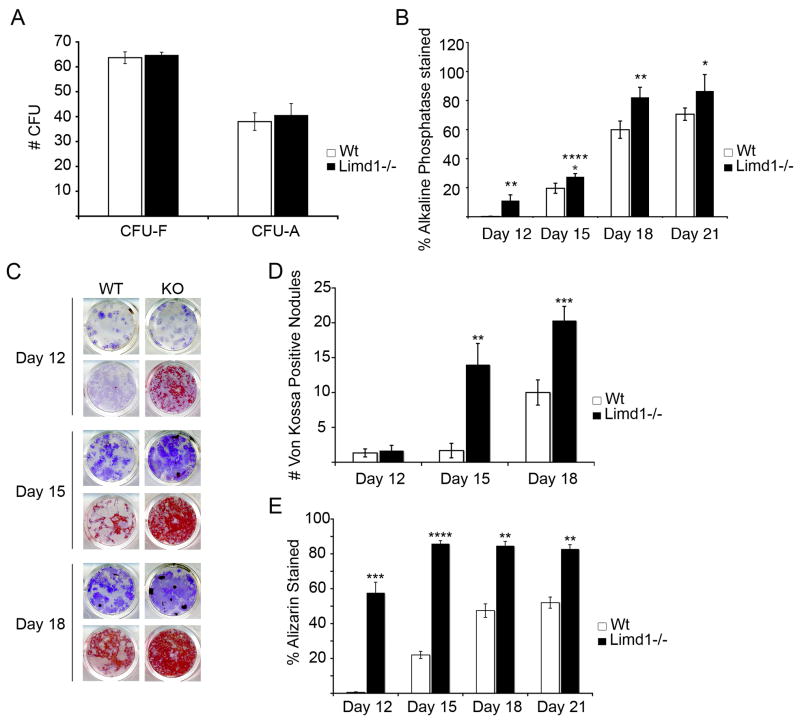
Analysis of osteoblast gene expression profiles in vitro suggested that deletion of Limd1 results in accelerated osteoblast differentiation. To address this possibility, total bone marrow was isolated from the tibiae and femora of Limd1−/− and wt mice. Cells were plated in normal, osteogenic, or adipogenic culture conditions for nine days and the number of fibroblast (CFU-F), osteoblast (CFU-O) and adipocyte (CFU-A) colony forming units scored. While the number of CFU-F and CFU-A formed was not affected by the absence of Limd1, an increase in the number of CFU-O was observed (Fig. 4A,B). Alkaline phosphatase staining of CFU-O revealed significant increases in the number of individual Limd1−/− osteoblast progenitor colonies compared to wt controls at early time points (days 3 and 6, data not shown). At later times colonies begin to fuse precluding a valid enumeration of single colonies so total plate threshold analyses was performed. As seen at early times the increase in alkaline phosphatase staining in Limd1−/− cultures persisted out to 21 days (Fig. 4B).
Fig. 4.
Limd1−/− mice have increased numbers of osteoblast progenitors.
(A) A comparison of the number of hematoxylin positive fibroblast colony forming units (CFU-F) or Oil Red O positive adipocyte colony forming unit (CFU-A) present after culturing Limd1−/− and Wt mice bone marrow stromal cells in normal or adipogenic media for 9 days. (B) A comparison of the number of alkaline phosphatase positive CFU-O present after culturing Limd1−/− and wt bone marrow stromal cells in osteogenic media for the number of days indicated. (C–E) Limd1−/− and Wt marrow stromal cells were cultured in osteogenic media for the number of days indicated days then fixed and stained for the presence of alkaline phosphatase (purple, all wells). Mineralization was visualized by Von Kossa (black, top row for each time point) or Alizarin Red S (red, bottom row for each time point) and the number of Von Kossa positive CFU-O (D) or Alizarin positive CFU-O (E) enumerated. *Indicates a significant difference between Limd1−/− and Wt (*p<0.05, **p<0.01, ***p<0.005, ****p<0.001, as determined by the student’s paired t-test).
Although a basal increase in osteoblast progenitor number in Limd1−/− mice could account for the in vitro enhanced mineralization, this analysis did not measure osteoblast progenitor differentiation rates. To test differentiation, bone marrow stromal cells grown in osteogenic media were fixed at three-day intervals for twenty-one days and stained for the presence of Alkaline Phosphatase to detect the number of CFU-O, while either Alizarin Red S or Von Kossa staining was used to determine the extent of mineralization. This analysis not only verified that Limd1−/− cultures contained more CFU-O but also showed that they exhibited significantly enhanced mineralization compared to wt controls at all time points (Fig. 4B–E). As early as twelve days, an increase in staining by Alizarin Red was clearly evident in Limd1−/− cultures compared to wt controls (Fig. 4C and E). At 15 days both Alizarin Red and Von Kossa staining was significantly increased in cultures of Limd1−/− cells (Fig. 4C–E). Even after twenty-one days, wt cultures did not reach the level of mineralization observed in Limd1−/− cultures (Fig. 4D,E). These results demonstrated that loss of Limd1 leads to increased in vivo osteoblast progenitor numbers and confirmed that Limd1−/− osteoblast progenitors exhibited accelerated differentiation in ex vivo cultures compared to wt controls.
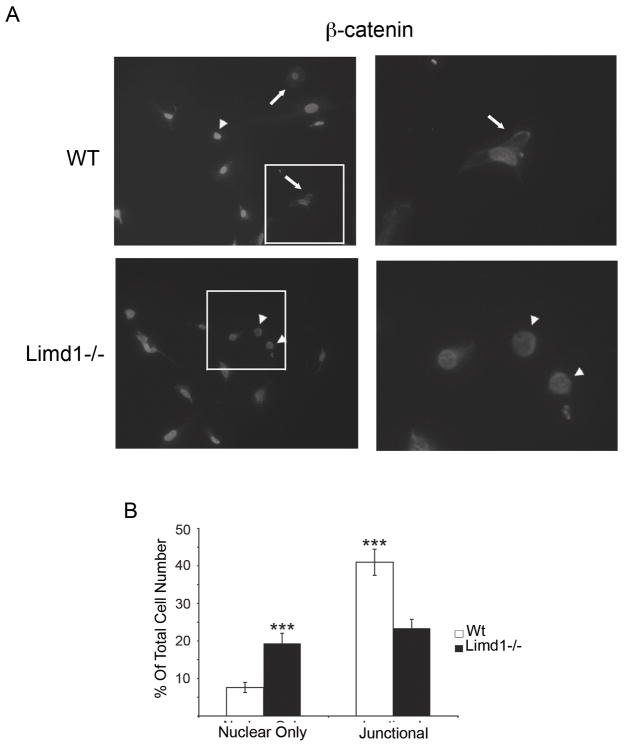
Limd1−/− mice have increased nuclear β-catenin staining compared to wt controls
Recently, the Limd1-related LIM protein, Ajuba, was shown to negatively regulate Wnt signaling by interacting with GSK-3β, promoting β-catenin phosphorylation, and thus β-catenin destruction[16]. Canonical Wnt signaling is an essential pathway for osteoblast development and differentiation: activation of Wnt signaling results in cellular accumulation of β-catenin followed by nuclear translocation and activation of tissue specific target genes [16–18]. To determine if Limd1, like Ajuba, affects Wnt signaling, the sub-cellular localization of β-catenin in primary wt and Limd1−/− calvarial osteoblasts differentiated in mineralization conditions for 5 days was determined. Increased nuclear accumulation of β-catenin is indicative of enhanced Wnt signaling. The number of cells containing nuclear only, nuclear and cytosolic, cytosolic only, and junctional β-catenin was quantified and compared (Fig. 5A and B). While no difference in the number of cells containing cytosolic only β-catenin was observed, Limd1−/− calvarial osteoblasts displayed significantly more nuclear only β-catenin compared to wt controls (data not shown and Fig. 5B). Furthermore, significantly less junctional β-catenin was found in Limd1−/− calvarial osteoblasts compared to wt controls (Fig. 5B). Together, these data indicate that Limd1 is a negative regulator of Wnt signaling in osteoblasts.
Fig. 5.
Limd1−/− calvarial cells have more nuclear β-catenin than Wt controls.
(A) Primary Wt or Limd1−/− calvarial osteoblasts were fixed, permeabilized and stained for β-catenin and DAPI. Cells with nuclear only β-catenin (arrowheads) and junctional β-catenin (arrows) are highlighted. (B) The percentage of cells with nuclear only β-catenin and the percentage of cells containing junctional β-catenin was determined and graphed. (***p<0.005, as determined by the student’s paired t-test).
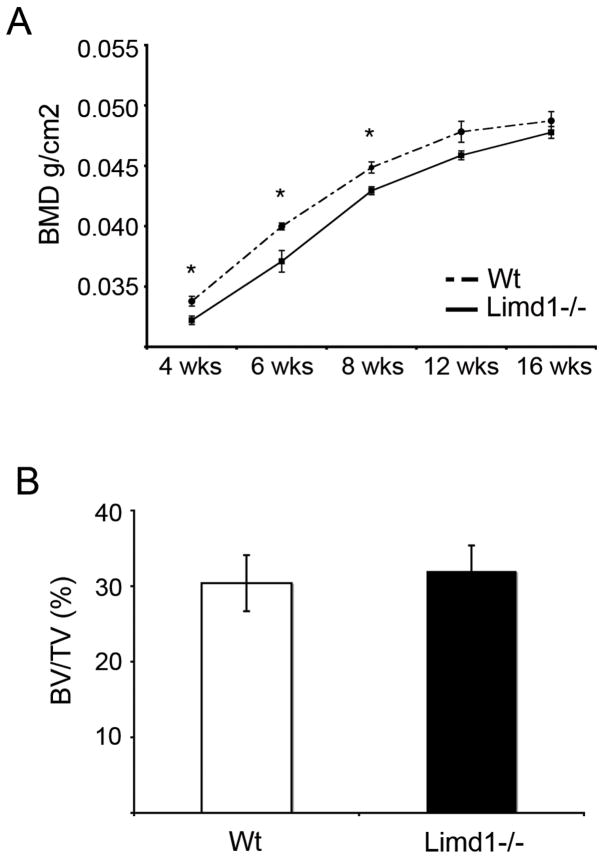
Limd1−/− mice do not display increased bone density at steady state or in response to PTH stress stimulus
While our in vitro data suggests that Limd1 is important for normal osteoblast differentiation and function, no increase in whole animal bone density was observed between Limd1−/− and wt mice by Dual Energy X-Ray Absorptiometry (DEXA) (Fig. 6). Interestingly, Limd1−/− mice had a significantly lower whole animal BMD compared to wt controls up to 8 weeks of age (Fig. 6A). This difference was reconciled by 12 weeks of age (Fig. 6A). Moreover, micro-CT analysis of tibiae isolated from Limd1−/− and wt mice at 6 weeks of age demonstrated no difference in bone mineral density (data not shown), suggesting the bones of Limd1−/− are likely not abnormal.
Fig. 6.
Basal bone density is not increased in Limd1−/− mice.
(A) Dual energy x-ray absorptiometry (DEXA) analysis was performed on a cohort of Limd1−/− (n=9) and Wt (n=8) mice at 4, 6, 8 12 and 16 weeks of age. (B) Micro-CT analysis of tibiae from Limd1−/− (n=3) and Wt (n=3) mice at 6 weeks of age. (*p<0.05, as determined by the student’s paired t-test).
Recent studies demonstrated that loss of Limd1 contributes to osteoclast differentiation in vivo but only in response to stress stimuli [11]. Therefore to determine if Limd1 influenced osteoblast stress response in vivo, sustained PTH treatment was used to directly stimulate osteoblasts to proliferate and subsequently differentiate. Hormone was administered to 16 week old mice daily for four weeks and bone density was measured weekly by DEXA. Analysis by both micro-CT and histomorphometry following four weeks of PTH stress stimulus treatment showed no significant differences in bone density (Table 1) or bone growth (data not shown) between PTH treated groups. Interestingly, however, Limd1−/− mice exhibited a trend toward increased osteoblast number per total area and increased bone surface covered by osteoblasts after four weeks of PTH treatment compared to wt, while no differences were observed between the corresponding control vehicle treated groups (Table 1).
Table 1.
Histomorphometric analysis of animals treated for four weeks with PTH (Limd1−/− n=7; Wt n=6) or vehicle (Limd1−/− n=4; Wt n=4).
| Parameters | Abbreviation | Wt Vehicle
(n=4) |
Wt PTH
(n=4) |
Limd1−/− vehicle
(n=4) |
Limd1−/− PTH
(n=4) |
|---|---|---|---|---|---|
| Bone volume/total volume | BV/TV (%) | 16.1+/−0.7 | 38.5+/−3.4 | 17+/−2 | 35.3+/−2.1 |
| Bone surface/bone volume | BS/BV | 10.3+/−0.2 | 20.8+/−1.2 | 9.3+/−0.7 | 20.7+/−0.5 |
| Trabecular thickness | Tb.Th. (um) | 31.3+/−1.7 | 36.7+/−2.5 | 36.9+/−4.2 | 34.1+/−1.8 |
| Trabecular spacing | Tb.Sp.(um) | 162.4+/−3.3 | 60.4+/−5.9 | 183+/−18.4 | 62.6+/−2.9 |
| Trabecular number | Tb.N. (/mm) | 5.2+/−0.1 | 10.4+/−0.6 | 4.7+/−0.4 | 10.4+/−0.2 |
| Osteoclast number/total area | N.Oc./T.Ar. | 475+/−88 | 655+/−97 | 481+/−139 | 533+/−112 |
| Osteoclast number/bone perimeter | N.Oc./B.Pm. | 45.4+/−7.5 | 31.5+/−4.4 | 50.1+/−12.1 | 25.6+/−5.0 |
| Osteoblast number/total area | N.Ob./T.Ar. | 61+/−17.5 | 277.3+/−73 | 70.5+/−33.6 | 380.6+/−80 |
| Osteoblast number/bone perimiter | N.Ob./B.Pm. | 6.0+/−1.9 | 13.2+/−3.3 | 7.6+/−3.6 | 18.2+/−3.4 |
Discussion
Here we demonstrate that the Ajuba-related LIM protein Limd1 regulates osteoblast differentiation and function. In the absence of Limd1, primary calvarial osteoblasts cultured ex vivo display increased mineralization and accelerated differentiation with no change in proliferation rates. Furthermore, marrow osteoblast progenitor numbers determined in ex vivo cultures were increased in Limd1−/− mice compared to wt controls. Despite these clear and significant effects of Limd1 deletion on osteoblast development in ex vivo cultures, basal bone homeostasis and PTH-induced bone osteoblastogenesis was unchanged compared wt mice.
Compensation by other members of the Ajuba LIM protein family is one possible explanation for the lack of an in vivo phenotype. Although protein expression levels of the family members Ajuba and WTIP are either low or do not substantially change during osteoblast progenitor differentiation or increase to compensate for the absence of Limd1 (data not shown), both are present in developing osteoblasts and bone marrow stroma. If they indeed compensate for the absence Limd1 during development, then these compensatory effects are likely not cell-autonomous effects since Limd1−/− osteoblasts cultured ex vivo exhibit phenotypic change.
Primary cells cultured, ex vivo, or outside their physiologically relevant environmental niche(s) often behave differently. This likely represents the effects of environmental regulators, both positive and negative. For instance, blood progenitor cells in the bone marrow require specific growth factors to survive, proliferate, and differentiate in ex vivo culture systems [19]. Many of these factors are secreted by or expressed on the surface of cells of the bone marrow stroma. In the mice used herein, the Limd1 gene is deleted in all cells in vivo, thus, it is possible that in the absence of Limd1 expression factors regulating osteoblast development, secreted by other, non-osteoblast stromal cells or bone marrow hematopoietic cells, are altered. Loss of Limd1 in non-osteoblast cells could result in a microenvironment that limits the enhanced development of Limd1−/− osteoblast progenitors observed in ex vivo cultures. Tissue specific deletion of a conditional Limd1 allele or reciprocal bone marrow transplantation experiments would address such possibilities.
It is intriguing that Limd1 levels increase during osteoblast differentiation yet loss of Limd1 results in accelerated differentiation. Its upregulation could be required to inhibit a parallel positive regulatory pathway that is likewise upregulated during osteoblast differentiation. Limd1 may act not only to repress osteoblast lineage commitment in undifferentiated cells, but also in the induction of mature osteoblast genes in committed osteoblast cells. These multiple functions could be dictated by distinct subcellular pools of the Limd1 protein, as has been described for β-catenin [16–18], or through interaction with specific factors during different stages of osteoblast commitment and differentiation. Many LIM domain containing proteins, have different roles depending on their subcellular localization, cell type, and interacting partners [9]. The data presented here provides a potential mechanism by which Limd1 acts as a negative regulator of osetoblast differentiation. Enhanced Wnt signaling as manifest by increased nuclear localization of β-catenin in differentiating Limd1−/− calvarial osteoblasts could explain both the increased in vivo osteoblast progenitor numbers as well as the accelerated differentiation of Limd1−/− stromal cells cultured ex vivo. Wnt/β-catenin signaling is an essential component of osteoblast lineage commitment, differentiation, and function [18, 20]. Augmentation of this signaling pathway in mice results in increased osteoblast progenitor numbers as well as enhanced bone formation [20]. Therefore, up-regulation of Wnt/β-catenin signaling, in the absence of Limd1, could explain not only the increase in osetoprogenitor cell number but also the accelerated osteoblast differentiation observed in Limd1−/− cultured cells.
Ajuba and Limd1 interact with the p62/sequestosome protein, a cytosolic protein important in bone osteoclast regulation [21]. Through this interaction, Limd1 influences assembly of an intracellular p62/TRAF6/a-PKC complex [11, 12] which contributes to RANKL-mediated osteoclastogenesis [22, 23]. In the absence of Limd1, pre-fusion osteoclasts were defective in RANKL signaling in vitro [11]. Furthermore, decreased osteoclastogenesis in response to RANKL, PTH, and inflammatory stimuli was observed in vivo [11]. RANKL is produced, in part, by immature osteoblasts in vivo. Limd1−/− osteoblasts did not support osteoclastogenesis of wt BMDM and RT-PCR analysis of differentiating Limd1−/−osteoblast progenitors demonstrated that the RANKL/OPG ratio was decreased thereby favoring inhibition of osteoclastogenesis. Therefore, in addition to its direct effects on RANKL signaling in osteoclasts, Limd1 also affects the ability of osteoblasts to support osteoclastogenesis. In conclusion, defective stress osteoclastogenesis of Limd1−/− mice is likely due to not only to Limd1’s cell intrinsic role in osteoclast cell differentiation but also its role in osteoblast development and function.
Activating mutations in the p62 gene are present in both familial and sporadic Paget’s disease [24], a disease in which both osteoclast and osteoblast functions are altered. A recent report demonstrated that osteoblasts isolated from Paget’s patients display decreased mineralization and inhibited differentiation through repression of Wnt signaling [25]. All reported p62 Pagetic mutations are predicted to not interfere with its interaction with Limd1, however [11]. This raises the possibility that the interaction between Limd1 and p62 contributes to normal osteoblast function, perhaps through negative regulation of Wnt/β-catenin signaling. Pagetic p62-Limd1 complexes may enhance Limd1’s apparent inhibitory role upon osteoblast progenitor commitment, differentiation and function. Understanding how Limd1 regulates these events will provide insight into the complex mechanisms controlling osteoblast differentiation in vivo.
Acknowledgments
We would like to thank Deborah Novack, Roberto Civitelli, and Marie Demay for thoughtful discussion, Crystal Idleburg for excellent histology work, Marina Kisseleva, Ellen Langer and Shannon Macauley for helpful suggestions. This work was supported by grants from the NIH (R01 CA85839) and Pfizer/Washington University Biomedical Research program to GDL. DEXA analysis was supported by CNRU grant DK56341. HFL was supported by the Siteman Cancer Center Cancer Biology Pathway, the Kauffman Fellowship in Life Sciences Entrepreneurship, and a grant from the NIH/NHLBI (T32 HL07088).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roodman GD. Regulation of osteoclast differentiation. Ann N Y Acad Sci. 2006;1068:100–9. doi: 10.1196/annals.1346.013. [DOI] [PubMed] [Google Scholar]
- 2.Rogers A, Eastell R. Circulating osteoprotegerin and receptor activator for nuclear factor kappaB ligand: clinical utility in metabolic bone disease assessment. J Clin Endocrinol Metab. 2005;90:6323–31. doi: 10.1210/jc.2005-0794. [DOI] [PubMed] [Google Scholar]
- 3.Kiss H, Kedra D, Yang Y, Kost-Alimova M, Kiss C, O’Brien KP, Fransson I, Klein G, Imreh S, Dumanski JP. A novel gene containing LIM domains (LIMD1) is located within the common eliminated region 1 (C3CER1) in 3p21.3. Hum Genet. 1999;105:552–9. doi: 10.1007/s004399900188. [DOI] [PubMed] [Google Scholar]
- 4.Crawford AW, Beckerle MC. Purification and characterization of zyxin, an 82,000-dalton component of adherens junctions. J Biol Chem. 1991;266:5847–53. [PubMed] [Google Scholar]
- 5.Petit MM, Mols R, Schoenmakers EF, Mandahl N, Van de Ven WJ. LPP, the preferred fusion partner gene of HMGIC in lipomas, is a novel member of the LIM protein gene family. Genomics. 1996;36:118–29. doi: 10.1006/geno.1996.0432. [DOI] [PubMed] [Google Scholar]
- 6.Yi J, Beckerle MC. The human TRIP6 gene encodes a LIM domain protein and maps to chromosome 7q22, a region associated with tumorigenesis. Genomics. 1998;49:314–6. doi: 10.1006/geno.1998.5248. [DOI] [PubMed] [Google Scholar]
- 7.Goyal RK, Lin P, Kanungo J, Payne AS, Muslin AJ, Longmore GD. Ajuba, a novel LIM protein, interacts with Grb2, augments mitogen-activated protein kinase activity in fibroblasts, and promotes meiotic maturation of Xenopus oocytes in a Grb2- and Ras-dependent manner. Mol Cell Biol. 1999;19:4379–89. doi: 10.1128/mcb.19.6.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srichai MB, Konieczkowski M, Padiyar A, Konieczkowski DJ, Mukherjee A, Hayden PS, Kamat S, El-Meanawy MA, Khan S, Mundel P, Lee SB, Bruggeman LA, Schelling JR, Sedor JR. A WT1 co-regulator controls podocyte phenotype by shuttling between adhesion structures and nucleus. J Biol Chem. 2004;279:14398–408. doi: 10.1074/jbc.M314155200. [DOI] [PubMed] [Google Scholar]
- 9.Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–31. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 10.Wilson SG, Reed PW, Bansal A, Chiano M, Lindersson M, Langdown M, Prince RL, Thompson D, Thompson E, Bailey M, Kleyn PW, Sambrook P, Shi MM, Spector TD. Comparison of genome screens for two independent cohorts provides replication of suggestive linkage of bone mineral density to 3p21 and 1p36. Am J Hum Genet. 2003;72:144–55. doi: 10.1086/345819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y, Zhao H, Luderer HF, Epple H, Ross FP, Teitelbaum SL, Longmore GD. The LIM protein, LIMD1, regulates AP-1 activation through an interaction with TRAF6 to influence osteoclast development. J Biol Chem. 2006 doi: 10.1074/jbc.M607399200. [DOI] [PubMed] [Google Scholar]
- 12.Feng Y, Longmore GD. The LIM protein Ajuba Influences Interleukin-1-Induced NF-kB Activation by Affecting the Assembly and Activity of the Protein Kinase Cz/p62/TRAF6 Signaling Complex. MCB. 2005:21. doi: 10.1128/MCB.25.10.4010-4022.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semerad CL, Christopher MJ, Liu F, Short B, Simmons PJ, Winkler I, Levesque JP, Chappel J, Ross FP, Link DC. G-CSF potently inhibits osteoblast activity and CXCL12 mRNA expression in the bone marrow. Blood. 2005;106:3020–7. doi: 10.1182/blood-2004-01-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi N, Akatsu T, Udagawa N, Sasaki T, Yamaguchi A, Moseley JM, Martin TJ, Suda T. Osteoblastic cells are involved in osteoclast formation. Endocrinology. 1988;123:2600–2. doi: 10.1210/endo-123-5-2600. [DOI] [PubMed] [Google Scholar]
- 15.Castro CH, Shin CS, Stains JP, Cheng SL, Sheikh S, Mbalaviele G, Szejnfeld VL, Civitelli R. Targeted expression of a dominant-negative N-cadherin in vivo delays peak bone mass and increases adipogenesis. J Cell Sci. 2004;117:2853–64. doi: 10.1242/jcs.01133. [DOI] [PubMed] [Google Scholar]
- 16.Haraguchi K, Ohsugi M, Abe Y, Semba K, Akiyama T, Yamamoto T. Ajuba negatively regulates the Wnt signaling pathway by promoting GSK-3beta-mediated phosphorylation of beta-catenin. Oncogene. 2007 doi: 10.1038/sj.onc.1210644. [DOI] [PubMed] [Google Scholar]
- 17.Church VL, Francis-West P. Wnt signalling during limb development. Int J Dev Biol. 2002;46:927–36. [PubMed] [Google Scholar]
- 18.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 19.Zubler RH. Ex vivo expansion of hematopoietic stem cells and gene therapy development. Swiss Med Wkly. 2006;136:795–9. doi: 10.4414/smw.2006.11424. [DOI] [PubMed] [Google Scholar]
- 20.Liu F, Kohlmeier S, Wang CY. Wnt signaling and skeletal development. Cell Signal. 2007 doi: 10.1016/j.cellsig.2007.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duran A, Serrano M, Leitges M, Flores JM, Picard S, Brown JP, Moscat J, Diaz-Meco MT. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Dev Cell. 2004;6:303–9. doi: 10.1016/s1534-5807(03)00403-9. [DOI] [PubMed] [Google Scholar]
- 22.Sanz L, Sanchez P, Lallena MJ, Diaz-Meco MT, Moscat J. The interaction of p62 with RIP links the atypical PKCs to NF-kappaB activation. Embo J. 1999;18:3044–53. doi: 10.1093/emboj/18.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanz L, Diaz-Meco MT, Nakano H, Moscat J. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. Embo J. 2000;19:1576–86. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morissette J, Laurin N, Brown JP. Sequestosome 1: mutation frequencies, haplotypes, and phenotypes in familial Paget’s disease of bone. J Bone Miner Res. 2006;21 Suppl 2:P38–44. doi: 10.1359/jbmr.06s207. [DOI] [PubMed] [Google Scholar]
- 25.Naot D, Bava U, Matthews B, Callon KE, Gamble GD, Black M, Song S, Pitto RP, Cundy T, Cornish J, Reid IR. Differential gene expression in cultured osteoblasts and bone marrow stromal cells from patients with Paget’s disease of bone. J Bone Miner Res. 2007;22:298–309. doi: 10.1359/jbmr.061108. [DOI] [PubMed] [Google Scholar]