Abstract
Retinoid X receptors (RXRs) function as ligand-activated transcription factors and are obligatory components of a large number of nuclear receptor heterodimers. RXRs help regulate diverse physiological responses including the cancer prevention responses of cell proliferation, inflammation, cell differentiation, and apoptosis. Since RXRs represent important targets for cancer chemoprevention, an ultrafiltration mass spectrometry-based assay was developed to facilitate the discovery of potential chemoprevention agents that bind to human RXRα. Natural and synthetic ligands for RXRα including 9-cis-retinoic acid, docosahexaenoic acid and LG100268 could be detected and identified in DMSO or even complex matrices such as extracts of marine bacteria. Specific binding of ligands to RXRα was demonstrated through competitive binding using ultrafiltration LC-MS-MS, and ligands could be ranked in order of affinity for RXRα. Therefore, ultrafiltration LC-MS-MS is suitable for the screening of complex mixtures such as natural product extracts for the discovery of new ligands to RXRα.
Keywords: retinoid X receptor, cancer chemoprevention, high throughput screening, LC-MS-MS
Introduction
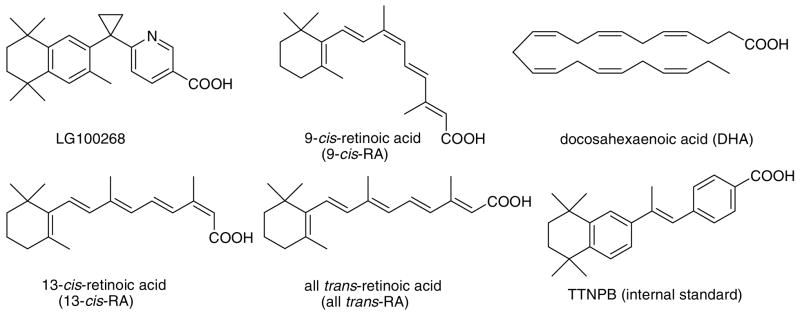
The retinoic acid receptors (RARs) and the retinoid X receptors (RXRs) are two families of nuclear receptors, which are the receptors for retinoic acids. RARs bind all trans-retinoic acid (all trans-RA) and 9-cis-retinoic acid (9-cis-RA), whereas RXRs bind only the 9-cis-RA stereoisomer (See structures in Figure 1).1 RXRs include the forms RXRα, RXRβ, and RXRγ and are obligatory components of a large number of nuclear receptor heterodimers that include the RARs, the vitamin D receptor, thyroid hormone receptors, peroxisome proliferator-activated receptors, and many orphan receptors.2–5 RXRs also form heterodimers with NGFIB and NURR1, which are two members of the small nerve growth factor-induced clone B subfamily that can interact with DNA as monomers and homodimers.1 The interaction of receptors with RXR increases their DNA-binding efficiency, and then these heterodimers act as ligand-dependent transcriptional regulators by binding to specific DNA-response elements found in the promoter region of target genes.1 It has been reported that the RXR partner does not exhibit a significant preference for any of the three subtypes in most of the cases.1
Figure 1.
Structures of the RXRα ligands and other test compounds used during this investigation.
Because ligand activation of RXRs have potentially pleiotropic effects on numerous biological pathways, RXR selective ligands (rexinoids) have potential use as therapeutic agents for the treatment or chemoprevention of cancer and metabolic diseases.6,7 Rexinoids might elicit the beneficial pharmacological activities of retinoic acids without toxic side effects such as teratogenicity and hypervitaminosis.1 The synthetic rexinoid LGD1069 (bexarotene, Targretin) was recently approved for treating refractory advanced-stage cutaneous T-cell lymphoma. However, adverse side effects such as induction of hyperglyceridemia, hypercholesterolemia, central hypothyroidism, and headache were observed.8 As an important new class of therapeutic agents, an efficient screening assay is needed to identify ligands of RXR. Therefore, we developed a new screening assay based on ultrafiltration mass spectrometry.
Electrospray mass spectrometry has been used for the direct characterization of RXR-ligand complexes.9–11 These studies demonstrated the principle that ligands with high affinity for RXR may be distinguished from compounds that bind non-specifically and with low affinity. However, this approach has not been extended to include the screening of mixtures of compounds such as natural product extracts or combinatorial libraries for the discovery of RXR ligands or for ranking ligands according to their affinity for RXR.
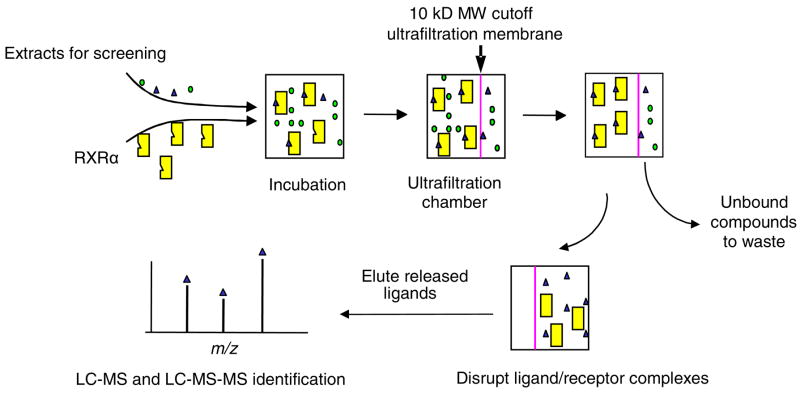
Ultrafiltration mass spectrometry was invented and developed for the screening of combinatorial library mixtures and natural product extracts in order to identify ligands to macromolecular targets, such as adenosine deaminase,12,13 dihydrofolate reductase,14 cyclooxygenase-2,15 serum albumin,16 and estrogen receptors.17 During ultrafiltration mass spectrometric screening, ligands in a mixture are allowed to bind to a receptor such as a protein therapeutic target, and then ultrafiltration is used to separate the receptor-ligand complexes from unbound low mass compounds. Finally, the ligands are released by disrupting the ligand-receptor complexes using organic solvent, a pH change, or other destabilizing conditions, and then analyzed using mass spectrometry or liquid chromatography-mass spectrometry. Control assays are carried out using inactive receptor or no receptor to test for non-specific binding to the macromolecule or the ultrafiltration membrane. This process is summarized in Figure 2.
Figure 2.
Experimental design of ultrafiltration LC-MS-MS screening of solutions for ligands to human RXRα.
In this study, an ultrafiltration mass spectrometry assay was developed for the screening of natural product extracts and combinatorial libraries for the discovery of ligands to human RXRα. Using this assay, drugs or drug leads may be discovered that might be useful for chemoprevention or cancer therapy. In addition, the utility of this assay for ranking RXR ligands according to their binding affinity was investigated.
Experimental section
Chemical and reagents
All organic solvents were HPLC grade or better and were purchased from Fisher (Hanover Park, IL). The RXR agonist LG100268 was purchased from CVChem (Cary, NC), and 9-cis-RA, 13-cis-retinoic acid (13-cis-RA), all trans-RA, docosahexaenoic acid (DHA), and 4-[(E)-2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid (TTNPB) (see structures in Figure 1) were purchased from Sigma-Aldrich (St. Louis, MO). The human RXRα ligand binding domain was expressed and purified as described previously and confirmed to be a homodimer by size-exclusion chromatography and mass spectrometry.18 A dichloromethane extract of a culture of a Streptomyces species from marine sediment was used as a natural product matrix for screening.19
Binding to RXRα and ultrafiltration
In preparation for ultrafiltration screening, 85 μL binding buffer consisting of 50 mM Tris-HCl (pH 7.5), 10% glycerol, 50 mM KCl, and 1 mM EDTA, 5 μL of a sample solution in DMSO and 10 μL of RXRα (10 μM in binding buffer), were mixed and incubated for 2 h at room temperature. In some incubations, sample solutions consisted of an extract of a marine sediment bacterial culture (4 mg/mL) reconstituted in DMSO and spiked with ligands for RXRα or binding buffer spiked with ligands for RXRα. For the competitive binding assay, the samples consisted of equimolor mixtures (1.0 or 0.2 μM) of LG100268, 9-cis-RA and DHA in DMSO. After incubation, each mixture was filtered thro ugh a Microcon (Millipore, Bedford, MA) YM-10 centrifugal filter containing a regenerated cellulose ultrafiltration membrane with a 10,000 MW cutoff by centrifugation at 13,000 g for 10 min at 4°C. The RXRα-ligand complexes were washed three times with 200 μL aliquots of 30 mM ammonium acetate (pH 7.5) followed by centrifugation again at 13,000 g for 15 min at 4 °C to remove the unbound compounds. For the two-filter method, the washed RXRα-ligand solution (~10 μL) was transferred to a second filter where it was treated with 200 μL of 90% methanol in deionized water to disrupt the receptor-ligand complexes. For the one-filter method, the washed RXRα-ligand solution was not transferred. The released ligands were then isolated from the denatured protein by centrifugation at 13,000 g for 15 min. The solvent in the ultrafiltrate was evaporated under vacuum, and the ligands were reconstituted in 50 μL of methanol/water (80:20, v/v) containing 200 nM TTNPB as an internal standard for analysis using liquid chromatography-tandem mass spectrometry (LC-MS-MS) as described below. For comparison, control analyses were carried out that were identical except for the use of denatured RXRα.
LC-MS-MS
Aliquots (10 μL each) of each reconstituted ultrafiltrate were analyzed using an Applied Biosystems (Foster City, CA) API 4000 triple quadrupole mass spectrometer equipped with Shimadzu (Columbia, MD) LC-10A pumps and a Leap (Carrboro, NC) HTS PAL autosampler. HPLC separations were carried out using a Thermo Quest (San Jose, CA ) column (3 μm, 2.1 × 30 mm) with a mobile phase consisting of isocratic 80:20 (v/v) methanol/10 mM aqueous ammonium acetate at a flow rate of 400 μL/min. Negative ion electrospray at an ion source temperature of 350 °C was used with tandem mass spectrometry for the detection of each compound. During MS-MS, collision-induced dissociation was carried out followed by multiple reaction monitoring (MRM) with a dwell time of 200 ms/ion as summarized in Table 1. Data were acquired and analyzed using Analyst software version 1.2 (Applied Biosystems).
Table 1.
Precursor/product ion pairs and parameters for multiple reaction monitoring LC -MS-MS of compounds used in this investigation.
| Compounds | Precursor ion→ Product ion (m/z) | Declustering Potential | Entrance Potential | Collision Energy | Collision Cell Exit Potential |
|---|---|---|---|---|---|
| LG100268 | 362.2→317.9 | −85 | −10 | −32 | −17 |
| 9-cis-RA | 299.2→255.0 | −85 | −10 | −22 | −13 |
| DHA | 327.3→283.2 | −68 | −10 | −15 | −17 |
| 13-cis-RA | 299.2→255.0 | −85 | −10 | −22 | −13 |
| all trans-RA | 299.2→255.0 | −85 | −10 | −22 | −13 |
| TTNPB | 347.3→303.2 | −95 | −10 | −26 | −19 |
Results and discussion
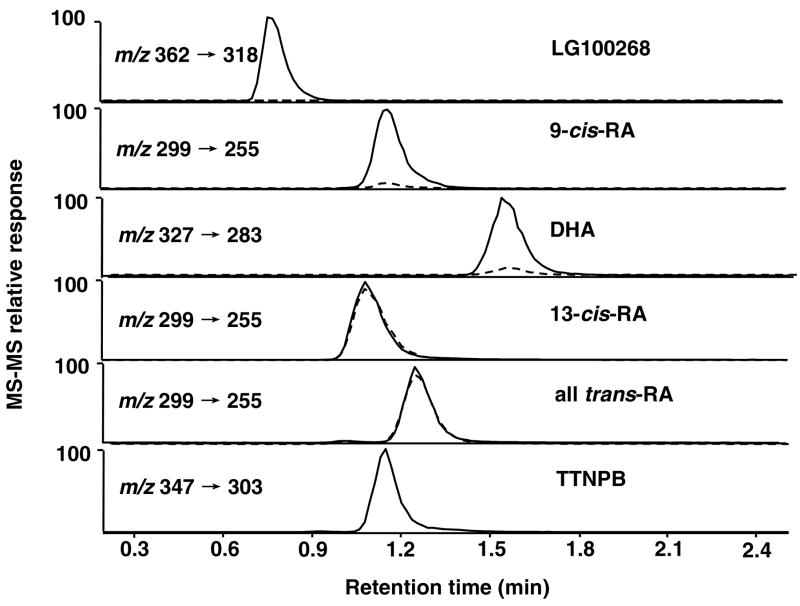
The RXRα ligands, LG100268, 9-cis-RA and DHA,1 and the structurally similar non-ligands, 13-cis-RA and all trans-RA, were used for the development of the ultrafiltration screening assay. Figure 3 shows the LC-MS-MS analysis of an ultrafiltrate obtained following the incubation of LG100268, 9-cis-RA, DHA, 13-cis-RA, and all trans-RA with 1 μM recombinant human RXRα. As a control for non-specific binding, the LC-MS-MS analysis of another ultrafiltrate is shown in Figure 3 that was prepared in an identical manner except for the use of denatured RXRα. In both LC-MS-MS analyses, TTNPB was used as an internal standard.
Figure 3.
Ultrafiltration LC-MS-MS screening of LG100268, 9-cis-RA, DHA, 13-cis-RA, and all trans-RA for binding to RXRα. The concentration of RXRα and each compound was 1 μM, and the incubations were carried out for 2 h. TTNPB was added to the samples as an internal standard immediately before analysis using LC-MS-MS with negative ion electrospray. The control incubations (dashed lines) containing denatured RXRα were used to test for non-specific binding and adsorption of sample to the ultrafiltration apparatus. Enhancement of HPLC peak areas in the experimental incubations (solid lines) indicates specific binding of ligands to RXRα. Note that the known ligands, LG100268, 9-cis-RA and DHA, but not 13 -cis-RA and all trans-RA, which are not known to bind to RXRα, was shown to bind.
When specific binding to RXRα occurred, the peak area of the ligand was greater in the chromatogram corresponding to incubation with active RXRα than in the chromatogram of sample incubated with denatured RXRα. As shown in Figure 3, LG100268, 9-cis-RA and DHA showed enhanced peak areas relative to the control chromatograms. The peaks for 9-cis-RA and DHA were enhanced approximately 10-fold compared to the corresponding control peaks representing non-specific binding, and the signal for LG100268 was enhanced to an even greater extent. Since the peaks corresponding to 13-cis-RA and all trans-RA could not be distinguished from the control incubations using denatured RXRα, the ultrafiltration screening assay confirmed that these compounds were not ligands. These results demonstrate the specificity of this ultrafiltration screening method for the detection of RXRα ligands such as 9-cis-RA but not its non-ligand isomers 13-cis-RA and all trans-RA.
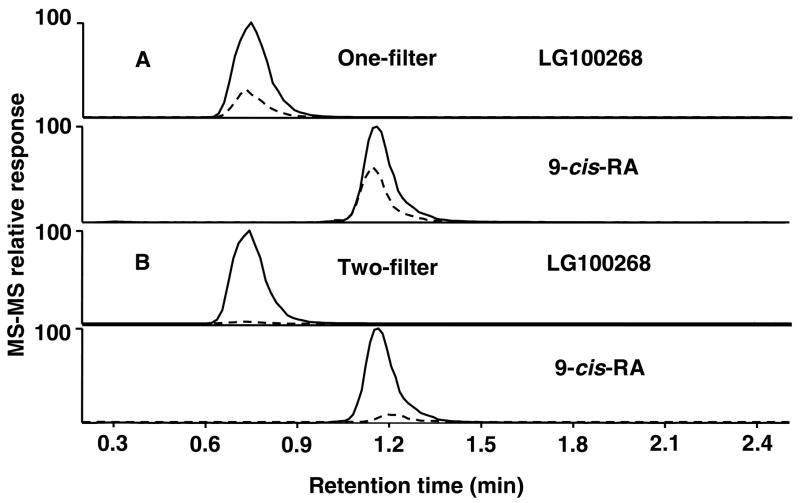
Since RXRα ligands such as LG100268 and 9-cis-RA tend to be very hydrophobic, the use of ultrafiltration for screening can result in high non-specific binding that might interfere with the detection of specifically bound ligands. By replacing the ultrafiltration filter before the ligand dissociation and elution step (see Figure 2) with a clean filter uncontaminated by ligands, the level of non-specific binding was decreased more than 5-fold as shown in Figure 4, which compares ultrafiltration RXRα screening results for LG100268 and 9-cis-RA obtained using one or two filters. By adding a second filter to the screening process, the specific binding (expressed as a ratio of the ligand peak area for the experiment to that of the control) increased from 3.8 ± 0.4 to 26.9 ± 5.2 (N = 3) for LG100268 and from 2.1 ± 0.4 to 10.6 ± 1.1 (N = 3) for 9-cis-RA.
Figure 4.
Comparison of specific and non-specific binding of LG100268 and 9-cis-RA to recombinant human RXRα during ultrafiltration LC-MS-MS screening using the one-filter and two-filter procedures. In the MRM chromatograms, the solid lines represent incubations with active RXRα, and the dashed lines represent the use of denatured RXRα as a control for non-specific binding. The concentrations of ligand and protein were each 1 μM, and each incubation was carried out for 2 h. A) When the RXRα-ligand complexes were washed with ammonium acetate to remove unbound compounds and then dissociated using methanol/water (90:10; v/v) using the same ultrafiltration filter, non-specific binding to the membrane produced a strong signal for LG100268 and 9-cis-RA. B) Transferring the RXRα-ligand complexes after the washing step to a clean ultrafiltration filter for dissociation resulted in much less non-specific binding.
Ultrafiltration LC-MS-MS screening data may be expressed in terms of the percentage of ligand specifically bound to a receptor relative to the total amount of the ligand in the incubation. Defined by Nikolic et al.15 as an “enrichment factor”, this value is an indicator of the relative affinity of ligands for a receptor. Sun et al.17 showed that enrichment factors may be used to rank ligands in order of their affinities for a receptor. In order to calculate enrichment factors for RXRα, a quantitative LC-MS-MS assay was developed for the measurement of ligands LG100268, 9-cis-RA and DHA. The calibration curves for all three compounds were linear (r2 >0.99) from the limit of quantitation to 1 μM. The limits of detection of LG100268, 9-cis-RA and DHA were 0.036 pg (10 pM, 10 μL injection), 0.090 pg (30 pM, 10 μL injection) and 0.082 pg (25 pM, 10 μL injection), respectively, based on a signal-to-noise ratio of 3:1. The limits of quantitation for these compounds were 0.108 pg (30 pM, 10 μL injection), 0.300 pg (100 pM, 10 μL injection) and 0.262 pg (80 pM, 10 μL injection), respectively, for a signal-to-noise ratio of 10:1.
The enrichment factors for the ultrafiltration LC-MS-MS screening assay are shown in Figure 3 and others are shown in Table 2. At test concentrations of 1 μM for RXRα and each ligand (assayed individually), the highest and lowest enrichment factors were 14.9 ± 2.6% and 0.9 ±0.2% for LG100268 and DHA, respectively (N = 3). The enrichment factor for 9-cis-RA was intermediate at 11.7 ± 1.8%. These factors are consistent with the reported affinities of these compounds, which are 3 ± 1 nM20 for LG100268 and 11.7 nM21 for 9-cis-RA. DHA is a much lower affinity compound compared to 9-cis-RA with an EC50 of 50–100 μM.22 Compounds with no measurable affinity for RXRα, such as 13-cis-RA and all trans-RA (Figure 3) have enrichment factors of 0%. At a test concentration of 0.2 μM for each ligand and 1 μM for RXRα, the enrichment factors for LG100268, 9 -cis-RA and DHA were 41.9 ± 4.6%, 37.4 ± 7.4% and 1.6 ± 0.7%, which are also consistent with the reported affinities for these ligands. These results also show that enrichment factors are dependent upon the relative concentrations of ligand and receptor.
Table 2.
Ultrafiltration LC-MS-MS signal enrichment factorsa for ligands to RXRα (1 μM).
| RXRα ligand | Ligand (1 μM) | Ligand (0.2 μM) | Mixture (3 ligands, 1 μM each) | Mixture (3 ligands, 0.2 μM each) | Kd |
|---|---|---|---|---|---|
| LG100268 | 14.9 ± 2.6b | 41.9 ± 4.6 | 7.3 ± 2.5 | 27.4 ± 5.4 | 3 ± 1 nM20 |
| 9-cis-RA | 11.7 ± 1.8 | 37.4 ± 7.4 | 6.2 ± 1.0 | 26.1 ± 2.0 | 11.7 nM21 |
| DHA | 0.9 ± 0.2 | 1.6 ± 0.7 | LODc | LOD | Lower affinity compared to 9-cis-RA22 |
Enrichment factor = (amount of compound specifically bound)/(total amount of compound in incubation) × 100 %.
Mean ± std dev (N = 3).
The percent DHA bound to RXRα could not be calculated for the mixture because the signal was reduced to near the limit of detection by competition with the higher affinity ligands LG100268 and 9-cis-RA.
When LG100268, 9-cis-RA and DHA were assayed with 1 μM RXRα as equimolar mixtures of 1 μM or 0.2 μM, only LG100268 and 9-cis-RA were identified as ligands. The enrichment factors for LG100268 and 9-cis-RA were 7.3 ± 2.5% and 5.5 ± 1.0% in 1 μM mixture, 27.4 ± 5.4 and 26.1 ± 2.0 in 0.2 μM mixture, respectively. Because the amount of RXRα available for binding was the same as assayed individually, the higher affinity ligands LG100268 and 9-cis-RA excluded DHA through competition for the binding site. This experiment demonstrated that binding of these ligands to RXRα was specific to the active site.
To test for possible interference from common solvents used in high throughput screening such as DMSO or complex natural product matrices such as a bacterial culture extract, LG100268 and 9-cis-RA were screened using ultrafiltration LC-MS-MS either in DMSO or in a matrix consisting of an extract of a marine bacterial culture (4 mg/mL). The enrichment factors for LG100268 in binding buffer, DMSO and the bacterial culture extract, were 13.1%, 14.6% and 16.6%, respectively; and for 9-cis-RA, the enrichment factors were 10.9%, 11.8% and 9.0%, respectively. These results indicate that both LG100268 and 9-cis-RA could be detected as ligands in the presence of DMSO or an extract of a bacterial culture.
Conclusions
An ultrafiltration LC-MS-MS screening assay has been developed for the discovery of ligands to human RXRα. The method was optimized to minimize potential interference due to non-specific binding of hydrophobic compounds to the ultrafiltration membrane. The use of DMSO to dissolve samples, which is common practice for combinatorial libraries used for high throughput screening, did not interfere with the assay. Because no interference was observed from an extract of a bacterial cell culture, this ultrafiltration method should be suitable for screening samples in complex matrices such as natural product extracts. Finally, this assay shows high sensitivity and only requires 100 pmol of RXRα and ligand per assay. Overall, ultrafiltration LC-MS-MS is a powerful new method for the discovery and characterization of RXRα ligands.
Acknowledgments
The work was supported by grant 5 P01 CA48112 from the National Cancer Institute. We thank Drs. Michael B. Sporn, Guowen Liu and Dejan Nikolic for helpful suggestions and discussions.
References
- 1.Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H. Pharmacol Rev. 2006;58:760–772. doi: 10.1124/pr.58.4.7. [DOI] [PubMed] [Google Scholar]
- 2.Yu VC, Delsert C, Andersen B, Holloway JM, Devary OV, Naar AM, Kim SY, Boutin JM, Glass CK, Rosenfeld MG. Cell. 1991;67:1251–1266. doi: 10.1016/0092-8674(91)90301-e. [DOI] [PubMed] [Google Scholar]
- 3.Leid M, Kastner P, Lyons R, Nakshatri H, Saunders M, Zacharewski T, Chen JY, Staub A, Garnier JM, Mader S. Cell. 1992;68:377–395. doi: 10.1016/0092-8674(92)90478-u. [DOI] [PubMed] [Google Scholar]
- 4.Zhang XK, Hoffmann B, Tran PB, Graupner G, Pfahl M. Nature. 1992;355:444–446. doi: 10.1038/355441a0. [DOI] [PubMed] [Google Scholar]
- 5.Sporn MB, Suh N. Nat Rev Cancer. 2002;2:5374–543. doi: 10.1038/nrc844. [DOI] [PubMed] [Google Scholar]
- 6.Dragnev KH, Petty WJ, Shah S, Biddle A, Desai NB, Memoli V, Rigas JR, Dmitrovsky E. J Clin Oncol. 2005;23:87574–8764. doi: 10.1200/JCO.2005.01.9521. [DOI] [PubMed] [Google Scholar]
- 7.Shulman AI, Mangelsdorf DJ. N Engl J Med. 2005;353:6044–615. doi: 10.1056/NEJMra043590. [DOI] [PubMed] [Google Scholar]
- 8.Lowe MN, Plosker GL. Am J Clin Dermatol. 2000;1:2454–50. 2514–2. doi: 10.2165/00128071-200001040-00006. [DOI] [PubMed] [Google Scholar]
- 9.Lengqvist J, Mata De Urquiza A, Bergman AC, Willson TM, Sjovall J, Perlmann T, Griffiths WJ. Mol Cell Proteomics. 2004;3:6924–703. doi: 10.1074/mcp.M400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Lengqvist J, Alvelius G, Jornvall H, Sjovall J, Perlmann T, Griffiths WJ. J Am Soc Mass Spectrom. 2005;16:16314–1640. doi: 10.1016/j.jasms.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Lengqvist J, Mata de Urquiza A, Perlmann T, Sjovall J, Griffiths WJ. J Mass Spectrom. 2005;40:14484–1461. doi: 10.1002/jms.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Breemen RB, Huang CR, Nikolic D, Woodbury CP, Zhao YZ, Venton DL. Anal Chem. 1997;69:21594–2164. doi: 10.1021/ac970132j. [DOI] [PubMed] [Google Scholar]
- 13.Zhao YZ, van Breemen RB, Nikolic D, Huang CR, Woodbury CP, Schilling A, Venton DL. J Med Chem. 1997;40:40064–4012. doi: 10.1021/jm960729b. [DOI] [PubMed] [Google Scholar]
- 14.Nikolic D, van Breemen RB. Comb Chem High Throughput Screen. 1998:474–55. [PubMed] [Google Scholar]
- 15.Nikolic D, Habibi 4-Goudarzi S, Corley DG, Gafner S, Pezzuto JM, van Breemen RB. Anal Chem. 2000;72:3853–3859. doi: 10.1021/ac0000980. [DOI] [PubMed] [Google Scholar]
- 16.Gu C, Nikolic D, Lai J, Xu X, van Breemen RB. Comb Chem High Throughput Screen. 1999;2:353–359. [PubMed] [Google Scholar]
- 17.Sun Y, Gu C, Liu X, Liang W, Yao P, Bolton JL, van Breemen RB. J Am Soc Mass Spectrom. 2005;16:271–279. doi: 10.1016/j.jasms.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan X, Broderick D, Leid ME, Schimerlik MI, Deinzer ML. Biochemistry. 2004;43:909–917. doi: 10.1021/bi030183c. [DOI] [PubMed] [Google Scholar]
- 19.Soria-Mercado IE, Prieto-Davo A, Jensen PR, Fenical W. J Nat Prod. 2005;68:904–910. doi: 10.1021/np058011z. [DOI] [PubMed] [Google Scholar]
- 20.Boehm MF, Zhang L, Zhi L, McClurg MR, Berger E, Wagoner M, Mais DE, Suto CM, Davies JA, Heyman RA. J Med Chem. 1995;38:3146–3155. doi: 10.1021/jm00016a018. [DOI] [PubMed] [Google Scholar]
- 21.Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, Evans RM, Thaller C. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 22.de Urquiza AM, Liu S, Sjoberg M, Zetterstrom RH, Griffiths W, Sjovall J, Perlmann T. Science. 2000;290:2140–2144. doi: 10.1126/science.290.5499.2140. [DOI] [PubMed] [Google Scholar]