Summary
Phytopathogenic bacteria suppress plant innate immunity and promote pathogenesis by injecting proteins called type III effectors into plant cells using a type III protein secretion system. These type III effectors use at least three major strategies to alter host responses. One strategy is to alter host protein turnover, either by direct cleavage or by modulating ubiquitination and targeting to the 26S proteasome. Another strategy involves alteration of RNA metabolism by transcriptional activation or ADP-ribosylation of RNA-binding proteins. A third major strategy is to inhibit the kinases involved in plant defence signalling, either by removing phosphates or by direct inhibition. The wide array of strategies bacterial pathogens employ to suppress innate immunity suggest that circumvention of innate immunity is critical for bacterial pathogenicity of plants.
Introduction
Gram-negative bacterial plant pathogens such as Pseudomonas syringae, Ralstonia solanacearum and Xanthomonas campestris infect their hosts by entering through wounds or natural openings. Once inside they produce virulence factors that alter the host providing an environment beneficial for the bacteria and suppressing host defences. Among the most important of these virulence factors are proteins called type III effectors (T3Es) that are injected directly into host cells using a type III protein secretion system (T3SS) known as the hypersensitive reaction and pathogenicity (Hrp) T3SS system [1]. Bacteria that are compromised in the Hrp T3SS are extremely reduced in virulence, demonstrating that T3Es are critical for successful pathogenesis [2]. The virulence activity of several T3Es is thought to be due to their ability to suppress plant innate immunity.
Plant innate immunity controls the activation of innate immune responses using mitogen-activated protein kinase (MAPK) cascades and hormones such as salicylic acid (SA). These defences include the deposition of lignin and callose in the cell wall, transcription of pathogenesis related (PR) genes, and the production of antimicrobial compounds and reactive oxygen species [3]. Plant innate immunity can be divided into two branches based on the types of microbial molecules recognized.
One branch recognizes conserved molecules essential to many microbes called pathogen-associated molecular patterns (PAMPs) or microbe-associated molecular patterns (MAMPs). PAMP recognition occurs by pattern recognition receptors (PRRs) predominately localized in the extracellular matrix. In plants these PRRs are PAMP receptor-like kinases and this recognition, which historically has been known as basal defense, is now referred to as PAMP-triggered immunity (PTI) [4,5,6].
The other branch recognizes specific pathogen effectors (historically referred to as avirulence (Avr) proteins) using nucleotide binding site-leucine rich repeat (NB-LRR) resistance (R) proteins. This recognition response is associated with the long-standing gene-for-gene hypothesis and is now known as effector-triggered immunity (ETI) [4]. For bacterial pathogens the only proteins known to induce ETI are T3Es. ETI has overlapping signalling pathways with PTI but generally results in a faster and more intense defence response, one outcome of which is a rapid and localized programmed cell death known as the hypersensitive response (HR).
ETI, in some cases, may be caused by the direct recognition of an effector by an R protein but a current hypothesis, known as the guard hypothesis [7], is that many R proteins detect the modifications of host targets made by specific pathogen effectors. Thus, R proteins detect effector activities rather than the proteins themselves and act to guard or monitor important host proteins for signs of pathogen ingress. Bacteria must suppress or evade both PTI and ETI responses in order to be successful pathogens and T3Es appear to play a major role in this suppression. Thus, the combination of host PRRs and R proteins, and the pathogen’s T3Es largely decides whether the pathogen or the plant will be successful in this molecular tête-à-tête. An important area of research in plant-pathogen interactions is the determination of how these T3Es function. These studies provide insight into how pathogen virulence and plant innate immunity work, as well as uncovering new enzyme activities.
Widespread screens to determine the effect of T3Es on plant immunity outputs demonstrate that certain T3Es can suppress the plant innate immunity [8,9,10] or up-regulate hormones that are antagonistic to SA and other defence responses [11,12,13]. This review will focus on recent progress in elucidating T3Es activities, plant targets and mechanisms of action. The impact of this new information on the guard hypothesis will also be addressed. The review is divided into sections based on the following T3E activities: protein turnover; RNA expression or stability; protein phosphorylation and dephosphorylation; and clues about T3E activities based on interactions with host proteins. Their sites of action and enzymatic activities are summarized in Fig. 1 and Table 1.
Fig. 1. Plant targets and activities of type III effectors from phytobacterial pathogens.
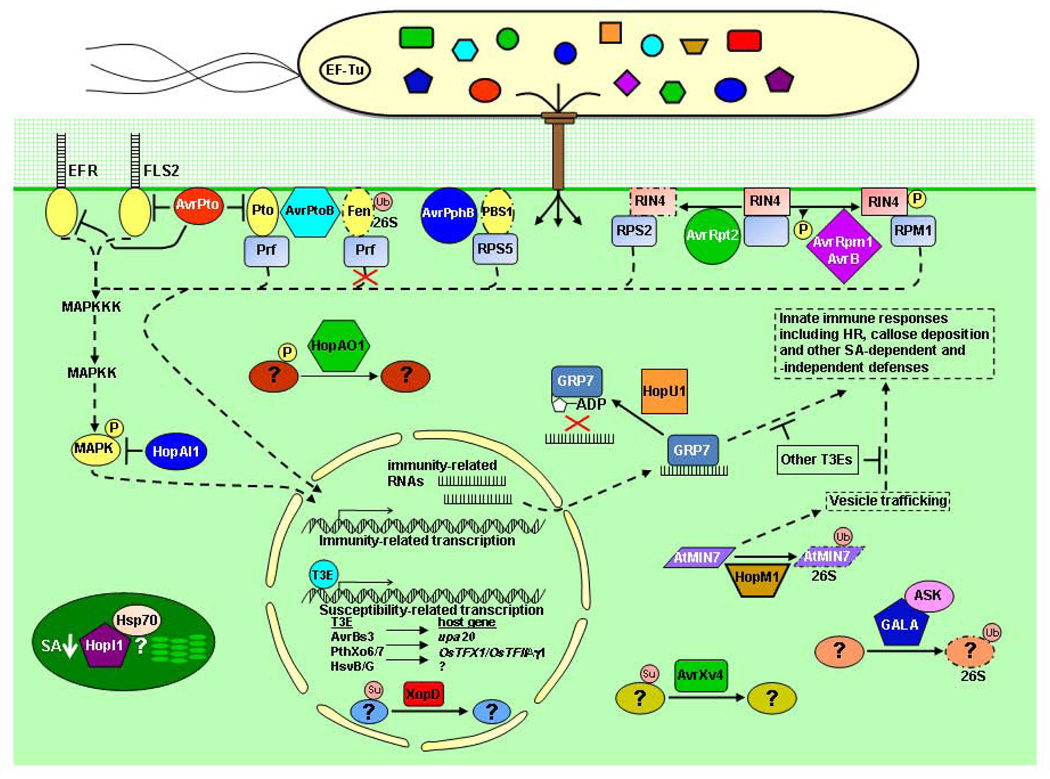
Bacterial plant pathogens inject many different type III effectors (T3Es) into plant cells via the type III secretion system. The activities of T3Es can be recognized by plant resistance (R) proteins inducing effector-triggered immunity (ETI). The R protein RPM1 causes ETI by recognizing the phosphorylation (P) of RIN4 by the T3Es AvrRpm1 and AvrB, while RPS2 causes ETI upon the cleavage of RIN4 by the T3E AvrRpt2. The T3E AvrPphB degrades the PBS1 kinase inducing RPS5-dependent ETI. The R protein Prf recognizes the interaction of the Pto kinase with AvrPto or AvrPtoB eliciting ETI, however, AvrPtoB ubiquinates (Ub) the Fen kinase targeting it for degradation and preventing recognition by Prf. Plants can also use receptor kinases such as EFR or FLS2 to detect pathogen-associated molecular patterns (PAMPs). This leads to PAMP-triggered immunity (PTI), which like ETI, induces innate immune responses. AvrPto inhibits the kinase activity of Pto, FLS2, and EFR. The HopAI1 T3E is a phosphothreonine lyase that suppresses MAPKs. The HopAO1 T3E is a protein tyrosine phosphatase whose target is unknown. The HopU1 T3E is a mono-ADP-ribosyltransferase that modifies GRP7 glycine-rich RNA-binding protein and likely prevents it from binding RNA. The HopM1 T3E causes the ubiquination and degradation via the 26S proteasome of AtMIN7, which may be involved in vesicle trafficking. The GALA T3Es contain F-box domains and can interact with plant ASK proteins (part of an SCF-type E3 ubiquitin ligase complex). GALAs are predicted to change the ubiquitination status of host proteins. The T3Es XopD and AvrXv4, which function in different locations in the plant cell, are isopeptidases that remove SUMO (Su) from host proteins. The chloroplast localized, J domain-containing T3E HopI1, suppresses salicylic acid (SA) production and may associate with Hsp70. T3Es AvrBs3, PthXo6/7 and HsvG/B bind to specific promoters in the nucleus inducing the transcription of genes favoring pathogenesis. Broken lines indicate plant responses and solid lines T3E activities.
Table 1.
T3E activities and plant targets
| T3E | Species | Activity | Target | Reference |
|---|---|---|---|---|
| AvrB | P. syringae pv. glycinea race 0 | Induces phosphorylation | RIN4/RAR1 | [50,52] |
| AvrBs3 | X. campestris pv. vesicatoria race 1 | Transcription activator-like | upa20/Bs3 | [35,36] |
| AvrPphB | P. syringae pv. phaseolicola race 3 | Cysteine protease | PBS1 | [14] |
| AvrPto | P. syringae pv. tomato JL1065 | Kinase inhibitor | Pto/EFR/FLS2 | [47,49] |
| AvrPtoB | P. syringae pv. tomato DC3000 | E3 ubiquitin ligase | Fen | [29,31] |
| AvrRpm1 | P. syringae pv. glycinea race 0 | Induces phosphorylation | RIN4 | [48] |
| AvrRpt2 | P. syringae pv. tomato T1 | Cysteine protease | RIN4 | [18,19] |
| AvrXa27 | X. oryzae pv. oryzae PXO99A | Transcription activator-like | Xa27 | [38] |
| AvrXv4 | X. campestris pv. vesicatoria T3 | DeSUMOylating cysteine protease | unknown | [24] |
| GALA | R. solanacearum GMI1000 | F-box and LRR domains | ASK(s) | [28] |
| HopAI1 | P. syringae pv. tomato DC3000 | Phosphothreonine lyase | MPK3/MPK6 | [42] |
| HopAO1 | P. syringae pv. tomato DC3000 | Protein tyrosine phosphatase | unknown | [44,45] |
| HopI1 | P. syringae pv. maculicola ES4326 | J-domain protein (possible Hsp70 co-chaperone) | unknown | [55] |
| HopM1 | P. syringae pv. tomato DC3000 | unknown | AtMIN7 and others | [34] |
| HopU1 | P. syringae pv. tomato DC3000 | Mono-ADP-ribosyltransferase | GRP7 and other RNA-binding proteins | [41] |
| HsvB | Pantoea agglomerans pv. betae 4188 | Transcriptional activator-like | unknown | [40] |
| HsvG | Pantoea agglomerans pv. gypsophilae 824-1 | Transcriptional activator-like | unknown | [40] |
| PthXo1 | X. oryzae pv. oryzae PXO99A | Transcriptional activator-like | Os8N3 | [39] |
| PthXo6/7 | X. oryzae pv. oryzae PXO99A | Transcriptional activator-like | OsTFX1 OsTFIIA γ1 | [37] |
| XopD | X. campestris pv. vesicatoria 85-10 | DeSUMOylating cysteine protease | unknown | [22] |
T3Es that impact host protein turnover
One way for T3Es to suppress plant innate immunity is to remove or inactivate its components. Certain T3Es are proteases that can remove these components by degradation. For example, AvrPphB from P. syringae is a papain-like cysteine protease that cleaves the Arabidopsis protein kinase PBS1 [14]. PBS1 exists in a complex with the R protein RPS5, which recognizes its cleavage and initiates ETI [15]. This recognition makes AvrPphB an obvious liability for P. syringae on RPS5-containing plants. These results support the guard hypothesis since they suggest that RPS5 is guarding PBS1 and perceives its modification by AvrPphB. Arabidopsis pbs1 mutants are not more susceptible to P. syringae. Thus, it is unclear whether P. syringae benefits from cleaving PBS1 in plants lacking RPS5. Therefore, PBS1 may be either a redundant virulence target or a plant protein that resembles a virulence target (i.e., a decoy) used by the plant to detect AvrPphB activity.
Another P. syringae T3E, AvrRpt2, is a staphopain-like cysteine protease that cleaves RIN4 [16], a negative regulator of PTI [17]. It seems counter intuitive that RIN4 is a virulence target since its loss de-represses PTI, which would apparently have a negative impact on pathogenic success. However, RIN4 interacts with at least two other T3Es and two R proteins suggesting that inactivating it may destabilize this complex and benefit the pathogen. One of these R proteins, RPS2, recognizes the cleavage of RIN4 by AvrRpt2 and induces ETI [18,19]. This finding was one of the first to provide molecular evidence in support of the guard hypothesis. AvrRpt2 likely targets additional host proteins since it can cleave other plant proteins and P. syringae virulence is increased by its presence in plants that lack RIN4 [20,21]. Additionally, AvrRpt2 was recently shown to be capable of altering auxin physiology [12], whether this is a direct or indirect effect remains to be determined.
Two families of T3Es (XopD and YopJ) are cysteine proteases/isopeptidases that specialize in the cleavage of ubiquitin or SUMO (small ubiquitin-like modifiers) from proteins. As the addition of a single ubiquitin or SUMO to proteins can alter their activity, it is likely that the de-ubiquitinating/de-SUMOylating activity of these T3Es alters the activity or stability of host proteins. XopD is a X. campestris pv. vesicatoria (Xcv) T3E that is translocated to the nucleus where it is believed to mimic endogenous plant SUMO isopeptidases [22] as its activity is specific to plant SUMOs [23].
YopJ is a well-studied human pathogen T3E from Yersinia spp. whose family members in Xcv include AvrXv4, AvrBsT, AvrRxv and XopJ. These T3Es appear to be SUMO isopeptidases as the expression of AvrXv4 in planta leads to a reduced level of SUMOylated proteins [24]. AvrXv4 is cytoplasmic [24], whereas XopJ is targeted to the plasma membrane [25] suggesting they have different targets in vivo. Yersinia YopJ has de-ubiquitinating, de-SUMOylating and acetylating activities and targets MAPK signalling pathways [26,27], therefore these YopJ family members may also possess acetyltransferase activity and their plant targets may also be involved in MAPK signalling.
Rather than possessing protease activity some T3Es use the host ubiquitination machinery and 26S proteasome to alter protein levels. For instance, Ralstonia solanacearum GALA T3Es have an F-Box domain and a leucine-rich repeat domain and interact with Arabidopsis ASK proteins [28•]. F-box and ASK proteins are part of ubiquitin E3 ligase complexes that attach ubiquitin to specific proteins and polyubiquitinated proteins are subsequently degraded by the 26S proteasome. Therefore, GALAs probably alter host protein regulation or turnover. They also increase the virulence of R. solanacearum, as a mutant lacking all 7 GALAs has severely reduced growth on Arabidopsis [28•]
A T3E from P. syringae, AvrPtoB (also known as HopAB2), contains a C-terminal E3 ligase domain [29] that ubiquitinates the tomato protein kinase Fen (originally identified as a cause of sensitivity to the pesticide fenthion [30]) leading to the degradation of Fen by the 26S proteasome [31••]. However, ETI occurs when the R protein Prf recognizes the interaction of Fen with AvrPtoB truncations that lack the E3 ligase domain. Thus, AvrPtoB has apparently evolved to evade ETI by acquiring an E3 ligase domain that marks Fen for degradation [31••]. Interestingly, some plants seem to have countered AvrPtoB’s E3 ligase activity by evolving the Pto kinase, which cannot be ubiquinated by AvrPtoB and induces Prf-mediated ETI. AvrPtoB appears to have multiple plant targets since it suppresses PTI-induced MAPK pathways in Arabidopsis [32]. It will be important to determine whether Fen and Pto are virulence targets or virulence decoys – if they are decoys evolved to ‘trick’ T3Es it would be tempting to speculate that, in a manner similar to AvrPto described below, PAMP receptor kinases may be the ‘real’ virulence targets of AvrPtoB.
A final example of a T3E that alters protein turnover is HopM1 (formerly HopPtoM) from P. syringae [33]. It is present in the endomembrane fraction of plant cells where it enhances AtMIN7 degradation by targeting it to the 26S proteasome [34••]. AtMIN7 is an adenosine diphosphate ribosylation factor guanine nucleotide exchange factor (ARF-GEF). HopM1 may target AtMIN7 for degradation in order to disrupt vesicle transport as ARF-GEFs have been implicated in vesicle trafficking. Vesicle transport is necessary for the export of defence compounds to the cell wall and apoplast, so its disruption would impair cell wall-based defences [34••]. HopM1 has no obvious similarity to known components of the host ubiquitination machinery, so exactly how it targets AtMIN7 to the proteasome remains to be established.
Targeting host transcription or RNA stability
T3Es can also modify protein levels by altering host transcription, which can lead to increased host susceptibility. T3Es that target host transcription include AvrBs3, PthXo6, and PthXo7 from Xanthomonas spp., and HsvG and HsvB from Pantoea agglomerans. AvrBs3 alters transcription by binding to the promoter region of the pepper transcription factor upa20 and increasing its expression. This up-regulation of upa20 is partly responsible for the hypertrophy of mesophyll cells seen when AvrBs3 is overexpressed in plants [35••]. In resistant plants AvrBs3 also binds to the promoter and activates the transcription of its cognate R gene, Bs3, leading to ETI [36••]. The encoded product of Bs3 is a flavin-dependent monooxygenase rather than an NB-LRR protein and it causes ETI by an unknown mechanism [36••]. The case of AvrBs3 recognition could be considered as a variation of the guard hypothesis, in that instead of guarding a protein the R gene is guarding a DNA sequence. This could work by the promoter of Bs3 acting as a decoy target for AvrBs3 by mimicking the promoter of upa20, which is apparently a ‘real’ virulence target.
AvrBs3 belongs to a family of transcription activator-like T3Es. Two members of this family, PthXo6 and PthXo7 increase the expression of the rice genes OsTFX1 (a bZIP transcription factor) and OsTFIIAγ1 (a small subunit of the transcription factor IIA), respectively [37•]. Loss of virulence in PthXo6 mutants can be rescued by over-expression of OsTFX1 but it is not yet known if PthXo6 directly binds to the promoter of OsTFX1 [36•]. Two other host genes Xa27 [38] and Os8N3 [39] are specifically up-regulated by the T3Es AvrXa27 and PthXo1, respectively. Rice plants silenced for Os8N3 RNA result in plants that are less susceptible to X. oryzae pv. oryzae supporting the idea that Os8N3 is a susceptibility gene [39]. Although less established than with the Xanthomonas T3Es similar activities are emerging for the P. agglomerans T3Es HsvG and HsvB [40]. Collectively, these examples suggest that the use of T3Es to up-regulate expression of genes such as transcription factors that co-regulate many genes is a common virulence strategy employed by bacterial pathogens. These T3E-induced proteins likely represent susceptibility factors that make the plant more favourable to bacterial colonization. Whether they cause the suppression of PTI or modify the cellular environment to benefit the pathogen remains to be determined.
The P. syringae pv. tomato HopU1 (formerly HopPtoS2) T3E targets RNA metabolism rather than mRNA production. HopU1 is a mono-ADP-ribosyltransferase (ADP-RT) that ADP ribosylates several Arabidopsis RNA-binding proteins in vitro, including the glycine-rich RNA-binding protein GRP7 [41••]. Furthermore, this ADP-RT activity is necessary for HopU1’s ability to suppress PTI and ETI and Arabidopsis plants lacking GRP7 are more susceptible to P. syringae [41••]. ADP-RTs are well-characterized toxins of animal pathogens but not previously shown to be associated with RNA-binding proteins. The ADP-ribosylation of RNA-binding proteins by HopU1 possibly decreases their ability to bind, stabilize or process RNA [41••]. It is not known how this alteration of RNA-binding leads to the suppression of innate immunity or if it alters specific or general RNA metabolism. Nevertheless, the post-transcriptional control of RNA by T3Es represents an exciting new pathogenic strategy to suppress innate immunity.
Phosphorylation or dephosphorylation of host proteins
PTI and ETI use kinase based signalling pathways, perhaps most prominently MAPK pathways. These kinases represent ideal T3E targets as multiple signal transduction pathways often converge at them and disabling them results in the suppression of many downstream responses. As these pathways work by sequentially phosphorylating downstream components, modifying their phosphorylation states inhibits them preventing activation of host defences.
Two T3Es that seem to use this strategy are HopAI1 and HopAO1 (formerly HopPtoD2). HopAI1 from P. syringae has phosphothreonine lyase activity and irreversibly inactivates MAPKs [42••]. HopAI1 binds to animal MAPKs [42••] and MPK3 and MPK6 of Arabidopsis that are involved in defence responses to P. syringae [43]. HopAI1’s broad specificity leaves open the possibility that its in vivo targets may include other MAPKs.
HopAO1 is a protein tyrosine phosphatase that contributes to virulence of P. syringae by suppressing PTI and ETI [44,45,46]. Because MAPKs are phosphorylated on tyrosine residues these are attractive putative targets for HopAO1. However, HopAO1 does not appear to act on MPK3 and MPK6 [46]. Therefore, its direct target(s) remain to be identified.
A slightly different strategy for modifying phosphorylation is used by the P. syringae T3E AvrPto, which interacts with the PAMP receptor kinases FLS2 and EFR, inhibiting their ability to autophosphorylate and activate MAPK signalling cascades [47••]. Similarly to AvrPtoB discussed above, AvrPto induces Pto-Prf-mediated ETI [48]. AvrPto inhibits the kinase activity of Pto [49•]. However, its induction of Prf-mediated ETI appears to be independent of this inhibition [49•]. Currently, the virulence target of AvrPto appears to be PAMP receptor kinases even though it also inhibits the kinase activity of Pto. This suggests that Pto may be an R protein-guarded decoy and that other T3Es that interact with it, such as AvrPtoB, may also target PAMP receptor kinases.
Clues about T3E activities based on interactions with host proteins
Host proteins that interact with T3Es have been identified through a variety of methods. Sometimes these interactors have provided clues to the T3E function. In other cases the protein-protein interaction itself is vital to the activity of the T3E by either stabilizing or destabilizing host protein complexes. This class of T3Es includes AvrB, AvrRpm1 and HopI1 from P. syringae. In pioneering research, AvrB and AvrRpm1 was found to interact with RIN4 leading to its hyperphosphorylation, which apparently allows it to be recognised by the R protein RPM1 inducing ETI [50,51]. AvrB has also been found to interact in vivo with RAR1 [52]. Moreover, RAR1 was identified in a genetic screen for Arabidopsis mutants insensitive to AvrB [52]. RAR1 is an attractive candidate target for an AvrB target because, together with SGT1 and HSP90, it stabilizes R proteins and, therefore, is a primary component of ETI.
Protein-protein interactions also appear to be important for HopI1 (formerly HopPmaI) a choloroplast-targeted T3E with a potential J domain [53]. J domains are often found in the co-chaperones of Hsp70, which direct proteins to Hsp70 and activate the ATPase activity and protein folding ability of Hsp70 [54]. The J domain of HopI1 is necessary for its virulence activity and overexpression of HopI1 in Arabidopsis causes thylakoid remodelling and the suppression of SA accumulation [55]. However, how the possible Hsp70 co-chaperone activity of HopI1 leads to these outcomes remains to be established. The targets and activities of the T3Es discussed here are summarized in Fig. 1 and Table 1.
Concluding remarks
It is clear that although much remains to be understood about how T3Es interfere with plant innate immunity several underlying themes are becoming apparent. The first is that the guard hypothesis probably accurately describes the perception of T3E activities inside plant cells. The open question now seems to be whether the guarded T3E targets identified such as Pto or RIN4 represent real virulence targets or decoys. If they indeed represent decoys this information could be exploited to identify their actual targets using similar logic employed for AvrPto’s targets (i.e., PAMP receptor kinases). In the fascinating co-evolution that has occurred between bacterial pathogens and plants it seems clear that PTI evolved prior to ETI. Did the evolution of PTI suppression provide the selection pressure necessary for ETI evolution? If so, it seems plausible that other R proteins may be guarding decoys of PAMP receptor kinase complexes.
A second emerging theme is that T3Es often have multiple targets many of which alter different aspects of innate immunity. These targets seem to function at every possible level of PTI and ETI pathways including the receptors themselves, downstream signal transduction pathways, and transcriptional and post-transcriptional responses. In this manner T3Es have apparently evolved to have the highest chance of suppressing sufficient innate immune responses to allow for the success of the bacterial pathogen. T3Es likely co-evolved to target a particularly important node of regulation or component of innate immunity. Because T3Es are targeting the innate immune response at seemingly every level, studying T3E targets promises to identify important unknown components of innate immunity and represent exciting tools for exploring plant biology.
Acknowledgements
We apologize to our colleagues whose papers were not cited here due to space limitations. Research in our laboratory is supported in part by grants from the National Science Foundation (Award No. 0544447), United States Department of Agriculture (Award No. 2007-35319-18336), and the National Institutes of Health (Award No. 1R01AI069146-01A2).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Alfano JR, Collmer A. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 2004;42:385–414. doi: 10.1146/annurev.phyto.42.040103.110731. [DOI] [PubMed] [Google Scholar]
- 2.Jin Q, Thilmony R, Zwiesler-Vollick J, He SY. Type III protein secretion in Pseudomonas syringae. Mibrobes Infect. 2003;5:301–310. doi: 10.1016/s1286-4579(03)00032-7. [DOI] [PubMed] [Google Scholar]
- 3.Loake G, Grant M. Salicylic acid in plant defence-the players and protagonists. Cur. Opin. Plant Biol. 2007;10:466–472. doi: 10.1016/j.pbi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 5.Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U S A 2007. 2007;104:19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van der Biezen EA, Jones JD. Plant disease resistance proteins and the gene-for–gene concept. Trends Biochem. Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- 8.Espinosa A, Alfano JR. Disabling surveillance: bacterial type III secretion system effectors that suppress innate immunity. Cell Microbiol. 2004;6:1027–1040. doi: 10.1111/j.1462-5822.2004.00452.x. [DOI] [PubMed] [Google Scholar]
- 9.Nomura K, Melotto M, He SY. Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr. Opin. Plant Biol. 2005;8:1–8. doi: 10.1016/j.pbi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Abramovitch RB, Anderson JC, Martin GB. Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell. Biol. 2006;7:601–611. doi: 10.1038/nrm1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Egea PR, Bogre L, Grant M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007;26:1467–1473. doi: 10.1038/sj.emboj.7601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl. Acad. Sci. USA. 2007;104:20131–20136. doi: 10.1073/pnas.0704901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thilmony R, Underwood W, He SY. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 2006;46:34–53. doi: 10.1111/j.1365-313X.2006.02725.x. [DOI] [PubMed] [Google Scholar]
- 14.Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:101–112. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- 15.Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 2003;49:1537–1546. doi: 10.1046/j.1365-2958.2003.03666.x. [DOI] [PubMed] [Google Scholar]
- 17.Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S, Dangl JL, Mackey D. Two Pseudomonas type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 18.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 19.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 20.Lim MT, Kunkel BN. The Pseudomonas syringae type III effector AvrRpt2 promotes virulence independently of RIN4, a predicted virulence target in Arabidopsis thaliana. Plant J. 2004;40:790–798. doi: 10.1111/j.1365-313X.2004.02251.x. [DOI] [PubMed] [Google Scholar]
- 21.Chisholm ST, Dahlbeck D, Krishnamurthy N, Day B, Sjolander K, Staskawicz BJ. Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc Natl Acad Sci U. S. A. 2005;102:2087–2092. doi: 10.1073/pnas.0409468102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hotson A, Chosed R, Shu H, Orth K, Mudgett MB. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 2003;50:377–389. doi: 10.1046/j.1365-2958.2003.03730.x. [DOI] [PubMed] [Google Scholar]
- 23.Chosed R, Tomchick DR, Brautigam CA, Mukherjee S, Negi VS, Machius M, Orth K. Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases. J. Biol. Chem. 2007;282:6773–6782. doi: 10.1074/jbc.M608730200. [DOI] [PubMed] [Google Scholar]
- 24.Roden J, Eardley L, Hotson A, Cao Y, Mudgett MB. Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol. Plant Microbe Interact. 2004;17:633–643. doi: 10.1094/MPMI.2004.17.6.633. [DOI] [PubMed] [Google Scholar]
- 25.Thieme F, Szczesny R, Urban A, Kirchner O, Hause G, Bonas U. New type III effectors from Xanthomonas campestris pv. vesicatoria trigger plant reactions dependent on a conserved N-myristoylation motif. Mol. Plant Microbe Interact. 2007;20:1250–1261. doi: 10.1094/MPMI-20-10-1250. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 27.Sweet CR, Conlon J, Golenbock DT, Goguen J, Silverman N. YopJ targets TRAF proteins to inhibit TLR-mediated NF-κB, MAPK and IRF3 signal transduction. Cellular Microbiol. 2007;9:2700–2715. doi: 10.1111/j.1462-5822.2007.00990.x. [DOI] [PubMed] [Google Scholar]
- •28. Angot A, Peeters N, Lechner E, Vailleau F, Baud C, Gentzbittel L, Sartorel E, Genschik P, Boucher C, Genin S. Ralstonia solanacearum requires F-box-like domain-containg type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14620–14625. doi: 10.1073/pnas.0509393103. This study showed that T3Es from Ralstonia solanacearum called GALA proteins contain F-box domains and interact with Arabidopsis Skp1-like proteins that are part of E3 ligases involved in protein ubiquitination and turnover. A R. solanacearum mutant deficient in all seven GALAs lost its ability to cause disease on Arabidopsis.
- 29.Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB. Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2851–2856. doi: 10.1073/pnas.0507892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin GB, Frary A, Wu T, Brommonschenkel S, Chunwongse J, Earle ED, Tanksley SD. A member of the tomato Pto gene family confers sensitivity to fenthion resulting in rapid cell death. Plant Cell. 1994;6:1543–1652. doi: 10.1105/tpc.6.11.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••31. Rosebrock TR, Zeng L, Brady JJ, Abramovitch RB, Xiao F, Martin GB. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature. 2007;448:370–374. doi: 10.1038/nature05966. The authors showed the C-terminal E3 ligase activity of AvrPtoB promotes ubiquitination and destruction of Fen kinase via the 26S proteasome. Co-expression of Fen with N-terminal AvrPtoB, but not with full length AvrPtoB activates Prf-dependent programmed cell death.
- 32.He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nurnbereger T, Sheen J. Specific bacterial suppressors of MAMP signalling upstream of MAPKKK in Arabidopsis innate immunity. Cell. 2006;125:563–575. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 33.DebRoy S, Thilmony R, Kwack YB, Nomura K, He S-Y. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9927–9932. doi: 10.1073/pnas.0401601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••34. Nomura K, DebRoy S, Lee YH, Pumplin N, Jones J, He S-Y. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. The authors discovered that the Pseudomonas syringae T3E HopM1 causes the ubiquitination and degradation of AtMIN7 via 26S proteasome. AtMIN7 encodes a member of the adenosine diphosphate (ADP) ribosylation factor (ARF) guanine nucleotide exchange factor (GEF) protein family that function in vesicle trafficking. This suggests HopM1 promotes the virulence of P. syringae by interfering with the host’s vesicle trafficking.
- ••35. Kay S, Hahn S, Marios E, Hause G, Bonas U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science. 2007;318:648–651. doi: 10.1126/science.1144956. This paper reported that the T3E AvrBs3 from Xanthomonas campestris binds to the promoter and induces the expression of upa20, a bHLH transcription factor and master regulator of cell enlargement, to cause hypertrophy in mesophyll cells.
- ••36. Romer P, Hahn S, Jordan T, Strass T, Bonas U, Lahaye T. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science. 2007;318:645–648. doi: 10.1126/science.1144958. The authors showed that AvrBs3 binds to the Bs3 promoter to cause transcriptional activation of this resistance gene. This is a novel way to induce the expression of a resistance gene by a T3E.
- •37. Sugio A, Yang B, Zhu T, White FF. Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAγ1 and OsTFX1 during bacterial blight of rice. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10720–10725. doi: 10.1073/pnas.0701742104. This paper showed that the T3Es PthoXo6 and PthoXo7 from Xanthomonas oryzae transcriptionally activate OsTFX1 and OsTFllAγ1, respectively. OsTFX1, when ectopically expressed in the host, substitutes for the role of pthoXo6 in virulence, acting as a host susceptible gene.
- 38.Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, Yin Z. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature. 2005;435:1122–1125. doi: 10.1038/nature03630. [DOI] [PubMed] [Google Scholar]
- 39.Yang B, Sugio A, White FF. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nissan G, Manulis-Sasson S, Weinthal D, Mor H, Sessa G, Barash I. The type III effectors HsvG and HsvB of gall-forming Pantoea agglomerans determine host specificity and function as transcriptional activators. Mol. Microbiol. 2006;61:1118–1131. doi: 10.1111/j.1365-2958.2006.05301.x. [DOI] [PubMed] [Google Scholar]
- ••41. Fu ZQ, Guo M, Jeong B-J, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–289. doi: 10.1038/nature05737. This study identified a T3E from Pseudomonas syringae, HopU1, as a mono ADP-ribosyltransferase (ADP-RT). This class of enzyme has never been reported in plant pathogens or plants. HopU1 suppresses the outputs of plant innate immunity dependent on its ADP-RT catalytic site. Using a proteomic approach, the authors identified HopU1 substrates to be glycine-rich RNA binding proteins, which are new substrates of ADP-RT. More importantly, this paper suggests a novel strategy employed by a bacterial pathogen where the ADP-ribosylation of plant RNA binding proteins results in posttranscriptional inhibition of host innate immunity.
- ••42. Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou JM, Shao F. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 2007;315:1000–1003. doi: 10.1126/science.1138960. This paper demonstrated that the T3E HopAI from Pseudomonas syringae is a phosphothreonine lyase that irreversibly inactivates kinases.
- 43.Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, Chen S, Tang X, Zhou JM. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host and Microbe. 2007;1:175–185. doi: 10.1016/j.chom.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Bretz JR, Mock NM, Charity JC, Zeyad S, Baker CJ, Hutcheson SW. A translocated protein tyrosine phosphatase of Pseudomonas syringae pv. tomato DC3000 modulates plant defence response to infection. Mol. Microbiol. 2003;49:389–400. doi: 10.1046/j.1365-2958.2003.03616.x. [DOI] [PubMed] [Google Scholar]
- 45.Espinosa A, Guo M, Tam VC, Fu ZQ, Alfano JR. The Pseudomonas syringae type III-secreted protein HopPtoD2 possesses protein tyrosine phosphatase activity and suppresses programmed cell death in plants. Mol. Microbiol. 2003;49:377–387. doi: 10.1046/j.1365-2958.2003.03588.x. [DOI] [PubMed] [Google Scholar]
- 46.Underwood W, Zhang S, He SY. The Pseudomonas syringae type III effector tyrosine phosphatase HopAO1 suppresses innate immunity in Arabidopsis thaliana. Plant J. 2007;52:658–672. doi: 10.1111/j.1365-313X.2007.03262.x. [DOI] [PubMed] [Google Scholar]
- ••47. Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, Zhou JM. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Current Biology. 2007;18:74–80. doi: 10.1016/j.cub.2007.12.020. The Pseudomonas syringae T3E AvrPto was found to interact with the PRRs FLS2 and EFR and inhibit their kinase activities to block plant innate immunity. In the resistant host, Pto competes with FLS2 for AvrPto binding, and the binding of Pto with AvrPto triggers a Prf-dependent defence.
- 48.Wu AJ, Andriotis VM, Durrant MC, Rathjen JP. A patch of surface-exposed residues mediates negative regulation of immune signalling by tomato Pto kinase. Plant Cell. 2004;16:2809–2821. doi: 10.1105/tpc.104.024141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •49. Xing W, Zou Y, Liu Q, Liu J, Luo X, Huang Q, Chen S, Zhu L, Bi R, Hao Q, Wu JW, Zhou JM, Chai J. The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature. 2007;449:243–247. doi: 10.1038/nature06109. The authors showed that AvrPto from Pseudomonas syringae has similarity to a kinase inhibitor based on the crystal structure of the AvrPto-Pto complex and that AvrPto inhibits the kinase activity of Pto in vitro. In the absence of AvrPto, the two AvrPto-interacting loops of Pto negatively regulate Prf-mediated defence. Interaction of AvrPto unlocks these two inhibition loops in Pto to activate Prf dependent host defence.
- 50.Mackey D, Holt BF, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated disease resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 51.Desveaux D, Singer AU, Wu A-J, McNulty BC, Musselwhite L, Nimchuk Z, Sondek J, Dangl JL. Type III effector activation via nucleotide binding, phosphorylation and host target interaction. PloS pathogens. 2007;3:456–469. doi: 10.1371/journal.ppat.0030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shang Y, Li X, Cui H, He P, Thilmony R, Chintamanani S, Zwiesler-Vollick J, Gopalan S, Tang X, Zhou JM. RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19200–19205. doi: 10.1073/pnas.0607279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guttman DS, Vinatzer BA, Sarkar SF, Ranall MV, Kettler G, Greenberg JT. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science. 2002;295:1722–1726. doi: 10.1126/science.295.5560.1722. [DOI] [PubMed] [Google Scholar]
- 54.Kelley WL. The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 55.Jelenska J, Yao N, Vinatzer BA, Write CM, Brodsky JL, Greenberg JT. A J domain virulence effector of Pseudomonas syringae remodels host cholorplasts and suppresses defenses. Current Biology. 2007;17:499–508. doi: 10.1016/j.cub.2007.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]


