Abstract
Kinetics of mRNA expression following a single loading event was measured using an in vivo rat tail model. Animals were instrumented and loaded in compression for 1.5 h at 1 MPa and 1 Hz. Real-time RT-PCR was used to measure mRNA levels 0, 8, 24 and 72 h after mechanical stimulation for genes associated with matrix proteins (aggrecan, collagen-I, collagen-II), proteases (MMP-2, MMP-3, MMP-13, ADAMTS-4), and their inhibitors (TIMP-1, TIMP-3) in anulus fibrosus and nucleus pulposus regions. Baseline mRNA levels were of greatest abundance for matrix proteins and lowest for proteases. The mRNA levels reached maximum levels 24 h following mechanical stimulation for the majority of genes evaluated, but some had maximum levels 8 and 72 h following loading. The mRNA levels returned to baseline levels for all genes in the nucleus 72 h following loading, but the majority of genes in the anulus remained upregulated. Results support a coordinated strategy of relative mRNA expression that varied over time beginning with inhibition of tissue breakdown, followed by synthesis of aggrecan and matrix degrading enzymes, and eventually collagen metabolism days following loading. Consequently, optimal time for tissue harvest for mRNA measurements depends on genes of interest. Results suggest attempts at anabolic remodeling must be given adequate time for metabolic processes and protein synthesis to occur, and that changes in TIMP and MMP levels may have greater potency in affecting structural protein abundance than direct changes in the structural protein messages. Results have important implications for disc remodeling and tissue engineering.
Keywords: intervertebral disc, mRNA kinetics, animal model, real-time RT-PCR, mechano-biology
INTRODUCTION
Mechanical loading on intervertebral discs is known to influence the metabolic behaviors in the nucleus pulposus and anulus fibrosus.1-3 Compression loading on the intervertebral disc in vivo is expected to result in large hydrostatic pressures for cells of the nucleus pulposus, and more complicated deformation patterns for cells of the highly oriented anulus fibrosus. Intervertebral disc cells are diverse and difficult to characterize into distinct phenotypes but include notochordal and chondrocyte-like cells in the nucleus pulposus and fibroblast-like cells in the outer anulus fibrosus.4 Intervertebral disc cells also exhibit diverse biologic responses to mechanical stimuli, depending on the loading type, magnitude, duration, and anatomic zone of cell origin.1,3,5,6 The overall goal of this study is to provide greater understanding of this diversity of responses through improved knowledge of the kinetics of the mRNA response to mechanical perturbations. This information contributes to improved understanding of healthy and damaging loads as well as improved understanding of disc mechanobiology towards optimizing cell, molecular, and tissue engineering repair techniques.
Intervertebral disc pathologies are commonly implicated in low back pain which has enormous economic consequences of more than $100 billion per year.7 Primary factors associated with intervertebral disc degeneration are aging, nutrition,8 genetics,9 and biomechanics.10 Mechanical loading of the intervertebral disc may contribute to disc degeneration by initiating tissue damage or by regulating cell-mediated remodeling events in response to mechanical stimuli. Recent animal studies point to interactions between hypermobility, innervation, and inflammation as characteristic findings in pathological and painful disc degeneration.11 In vivo animal model studies and in vitro cell culture studies further demonstrate sensitivity of disc cells to magnitude and frequency of loading with specific anabolic and catabolic responses as well as apoptosis and matrix remodeling.1,3,5,6
Degeneration favors a shift towards predominantly catabolic remodeling responses in the intervertebral disc,12-14 and molecular therapies demonstrate some promise for shifting the balance of remodeling responses from predominantly catabolic to more anabolic remodeling.12,14 This shift can occur through alterations in matrix production, enzyme production and activity, and through anti-catabolic agents such as TIMPs.14 Increases in metalloproteinases and their inhibitors have been found in human disc degeneration in autopsy and surgical specimens, including MMP-2 and -3, ADAMTS-4, and TIMP-1.15,16
Disc regeneration and tissue engineering in general require a greater understanding of the chemical and mechanical conditions needed to promote proper cell differentiation, phenotypic expression, and metabolic activity.17 Strategies for disc regeneration require knowledge of the mechanobiology that regulates disc metabolism,6 and recently presented priority research questions in the biomechanics of intervertebral disc degeneration included the determination of physiologically relevant biomechanical signals for intervertebral disc cells and those mechanisms that regulate homeostasis and deviations from this state.18
Defining the kinetics of mRNA expression in response to a mechanical loading event represents a large gap in the literature and is a research priority. The use of RT-PCR and evaluation of message levels is a key tool for monitoring cell differentiation and phenotype. In vivo loading studies used measurements of mRNA levels to separate effects of immobilization and overloading, define thresholds of loading required to maintain disc homeostasis, and distinguished loading conditions with predominantly anabolic and catabolic responses.1,10 These short-term studies were also very important in screening a wide range of independent and dependent variables to isolate those parameters that warrant long-term investigation of compositional, structural, and mechanical changes that occur under chronic loading.
These short-term measurements are typically made at only a single time point, and tissue engineering and mechanobiology studies require an improved knowledge of the kinetics of mRNA synthesis and accumulation. Furthermore, to our knowledge, no study has investigated the most appropriate time to measure gene expression levels in the disc following intervention. Transient mRNA responses are particularly relevant to short-term cell/tissue culture, and animal model studies where the measurement time may have a large effect on observed responses. In particular, it is unknown when gene expression levels peak or how long they remain altered following stimulation. This information is important to optimize mechanobiology protocols and to define appropriate recovery times after levels of loading that may maximize a regenerative (or anabolic) remodeling response, or alternately minimize a degenerative (or catabolic) remodeling response.
Therefore, the purpose of this study was to measure the transient message response following a single loading event and to determine how it varies among different genes. Specifically, this study evaluated the kinetics of mRNA expression by measuring message levels 0, 8, 24, and 72 h after a single episode of loading using real-time RT-PCR, and interpreted this response in the context of a previously proposed hypothetical model.1 A wide range in time points were chosen with consideration that mRNA kinetics will be affected by differences in basal mRNA levels, cell signaling pathways, and half-lives which all vary with the chosen gene. A single episode of loading was considered one that would be in the physiological range, have sufficient duty cycles to elicit a significant response, and not lead to chronic remodeling. The 1.5-h duration was previously shown to allow creep to reach steady state, and the magnitude and frequency were demonstrated to be in the high range of physiological loading.13 A rat tail model was used to evaluate this transient response in vivo under cyclic loading conditions, as rodent tail models are effective in defining distinct relationships between mechanical loading and biological responses in an in vivo environment.1,2
We hypothesized that a time-dependent response in mRNA levels would be observed over a 3-day period following a single loading event, the largest relative changes would be seen in the genes with low basal expression levels (proteases and their inhibitors) within 24 h, and mRNA levels would return to baseline levels within 72 h.
METHODS
Animal Model
This experiment was approved by the University of Vermont Institutional Animal Care and Use Committee. Fifty skeletally mature Wistar rats (>12 months old, n = 10 animals per group, 80% power, α = 0.05) were instrumented with an Ilizarov-type device spanning caudal disc 8–9 (“instrumented”), as described previously.19 Briefly, under general anesthesia, carbon fiber rings were attached to caudal vertebrae 8 and 9 using two orthogonal 0.8-mm diameter Kirschner wires (Fig. 1). Each animal was given subcutaneous analgesic (buprenorphine) preoperatively and 12, 24, and 36 h postoperatively. Rat tails were subjected to mechanical loading 72 h following surgery.
Figure 1.
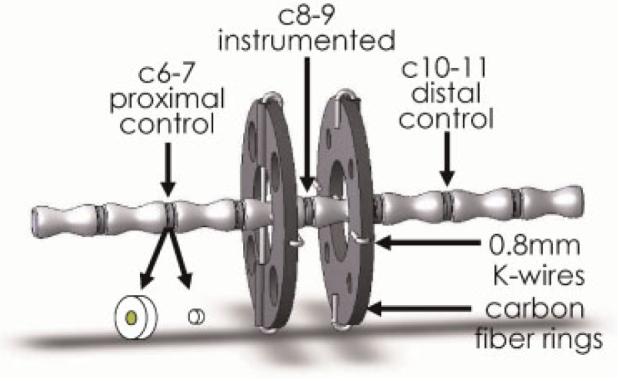
Solid model representation of instrumented rat tail including control and instrumented levels, and disc harvest involving separation of anulus fibrosus from nucleus pulposus.
Experimental Procedure
Dynamic compression using a sinusoidal waveform was applied under load control with the proximal ring held fixed and the motion of the distal ring driven by the computer-controlled actuator similar to that described previously.19 The device ensures that the rings are parallel and that loading is aligned with the axis of the tail. Throughout testing, body temperature was maintained at ∼39°C. Animals were loaded for 1.5 h under sinusoidal compression at a magnitude of 12.6 N (corresponding to 1 MPa or ∼300% body weight) and frequency of 1 Hz using methods previously described.19 The mRNA was measured for genes that represent important matrix proteins, proteases, and anti-catabolic regulators. Immediately following loading, animals were either euthanized or awakened and allowed to recover for 8, 24, or 72 h. An additional sham group was included in which rats were instrumented with rings, allowed 72 h to recover from surgery, placed under general anesthesia for 1.5 h without loading, and then immediately euthanized (Table 1). All rats were allowed unlimited food and water, and allowed to move freely at all times except while under anesthesia during both surgical and loading procedures.
Table 1.
Loading Protocols
| Load |
|||||
|---|---|---|---|---|---|
| Group | Magnitude | Frequency | Durationa | Recovery | n |
| Sham | — | — | — | 0 h | 9 |
| 0 h | 1 MPa | 1 Hz | 1.5 h | 0 h | 10 |
| 8 h | 1 MPa | 1 Hz | 1.5 h | 8 h | 10 |
| 24 h | 1 MPa | 1 Hz | 1.5 h | 24 h | 10 |
| 72 h | 1 MPa | 1 Hz | 1.5 h | 72 h | 10 |
Animals were under anesthesia during this time.
Sample Harvest and Preparation
The instrumented disc (c8–9) and two control discs (c6–7 and c10–11) were removed immediately after euthanasia and prepared for real-time RT-PCR analysis as previously described.19 Briefly, motion segments containing the loaded and control discs were removed and flash frozen in liquid nitrogen. Frozen discs were exposed with a scalpel and the nucleus and anulus regions separated using 2-mm and 5-mm diameter biopsy punches, respectively (Fig. 1). The tissue was then pulverized under liquid nitrogen and placed in TRI Reagent (Molecular Research Center, Inc., Cincinnati, OH) for RNA extraction. Samples from both proximal and distal control levels were pooled together in TRI reagent to provide a single internal control sample with an identical “effective level” as the instrumented disc. Total RNA was isolated using 1-Bromo-3-Chloro-Propane and the GenElute Mammalian Total RNA Kit (Sigma, St. Louis, MO) and reverse-transcribed to cDNA using the TaqMan reverse transcription kit (Applied Biosystems, Foster City, CA), according to the manufacturers' specifications. As described previously, to ensure there was no anulus fibrosus tissue mixed with nucleus tissue, nucleus tissue was allowed to reach room temperature (∼10 s) and any nongelatinous material was removed prior to refreezing and pulverization. This technique had no effect on 18S rRNA and the improved harvest technique decreased the variance in real-time RT-PCR results for each of the genes measured in nucleus specimens.
Real-Time PCR
The effects of mechanical stimulation were determined by measuring the mRNA levels of three anabolic genes (aggrecan, collagen-I, and collagen-II), three catabolic genes (stromelysin-MMP3, collagenase-MMP13, and aggrecanse-ADAMTS-4), two regulatory genes (TIMP-1 and TIMP-3), and an endogenous control (18S rRNA) using real-time RT-PCR (GeneAmp 7700 Sequence Detection System, Applied Biosystems). The endogenous control 18S rRNA was previously determined to be the best housekeeping gene for mechanobiology of cartilaginous tissues, and was not affected by loading in the disc.20 Data was analyzed using the comparative Ct method (see 19 for detailed description) in which mRNA levels for each gene were normalized to the endogenous control (18s rRNA corrects for differences in sample size) and values in the instrumented c8–9 level were then normalized to the average of the proximal c6–7 and distal c10–11 internal controls (to control for inter-animal variation) before being compared to the sham group. In addition, the relative abundance of each gene was estimated for the anulus and nucleus by comparing delta-Ct values. Delta-Ct values represent the mRNA normalized to the endogenous control (18S rRNA) which corrects for differences in the amount of tissue harvested. Relative abundance was determined by calculating the delta-Ct levels of each gene relative to that of the least abundant gene in each region (ADAMTS-4 in the anulus and MMP-3 in the nucleus), similar to the method used in cartilage tissue.21
Statistical Analysis
For the PCR results, two series of statistical comparisons were performed. First, a paired Student's t-test, with hypothesized mean of 1, was used to determine if mRNA levels in the instrumented c8–9 level of the sham group were significantly different from the average of the internal controls (c6–7 and c10–11) for each gene. Next, an ANOVA with Fisher's-PLSD post hoc analysis was used to determine if mRNA levels in the loaded discs changed with each of the recovery times relative to the sham group. All statistical analyses were performed with StatView software (SAS Institute, Cary, NC) and significance was taken as p < 0.05.
RESULTS
Animals tolerated the surgery well, maintained bodyweight, and had no signs of inflammation. Based on the mechanical data collected, all loadings were precisely administered at 1 MPa and 1 Hz without complication. The c8–9 specimen from one animal in the sham group was destroyed during tissue harvest and thus the results of only nine specimens are reported for that group.
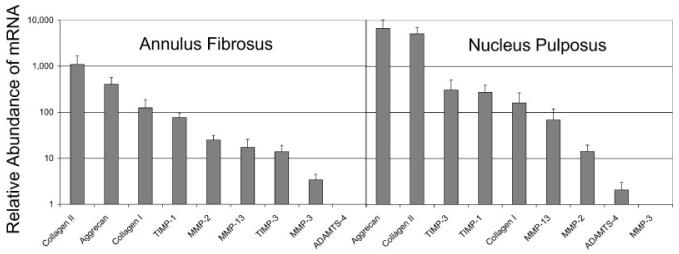
The relative abundance of mRNA (measured in noninstrumented control discs) shows that mRNA coding for matrix proteins (collagen-I, collagen-II, and aggrecan) is much more abundant in disc cells than mRNA coding for the catabolic enzymes responsible for their breakdown (Fig. 2). These results should be taken into account when considering the implications of a 10-fold change in collagen-II relative to a 40-fold change in MMP-3. The least abundant levels of mRNA were for ADAMTS-4 and MMP-3 in the anulus and nucleus, respectively.
Figure 2.
Relative abundance of mRNA measured in noninstrumented control level discs for the anulus fibrosus and nucleus pulposus. Relative mRNA is normalized by mRNA levels for ADAMTS-4 and MMP-3 which were lowest in anulus and nucleus regions, respectively.
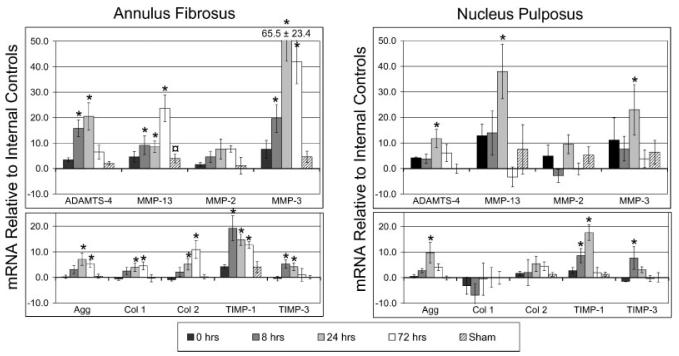
In the nucleus pulposus region, significant increases were seen in aggrecan, TIMP-1, ADAMTS-4, MMP-13, and MMP-3 (Fig. 3). Gene levels were highest for all genes 24 h after loading and were not different from sham after 72 h. No significant changes were observed in collagen-I, collagen-II, or MMP-2. In the anulus fibrosus regions, gene expression levels were greatest after 8 h for TIMP-1 and TIMP-3, greatest after 24 h for aggrecan, ADAMTS-4, and MMP-3, and after 72 h for collagen-I, collagen-II, and MMP-13. No significant changes were observed in MMP-2.
Figure 3.
Fold changes in mRNA levels relative to internal control levels (Mean ± SEM) for nine genes at 0, 8, 24, and 72 h after loading. The mRNA was isolated from pooled tissue from proximal and distal control to eliminate potential level effects. * =significantly different from sham;  = significantly different from internal controls for sham group only; p < 0.05.
= significantly different from internal controls for sham group only; p < 0.05.
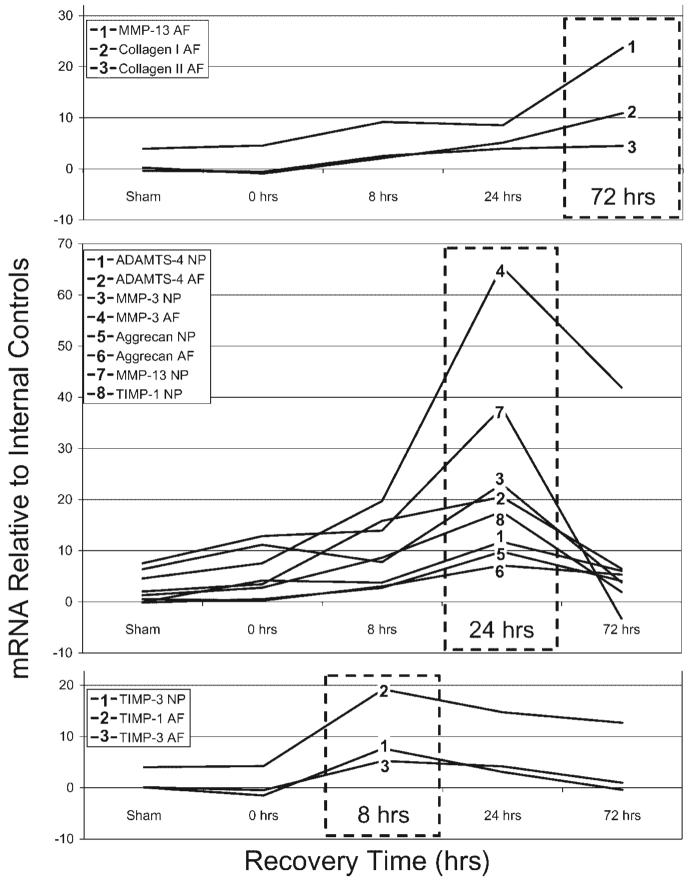
The kinetics of mRNA expression following loading varied with gene of interest and disc region, and were quantified according to time points when largest levels were recorded (Fig. 4). Regulatory genes responsible for inhibiting active enzymes (TIMP-1 and TIMP-3) responded most rapidly to loading and returned to baseline levels shortly thereafter. In general, genes in the nucleus and anulus had similar time of upregulation following stimulation, with the exception of MMP-13 which was upregulated faster in the nucleus and TIMP-1 which responded faster in the anulus. In the nucleus, all statistically significant changes in message levels returned to control levels by 72 h. However, many genes were significantly upregulated in the anulus 72 h after loading with MMP-3 and TIMP-1 levels remaining elevated at 72 h after previously reaching a maximum value, and MMP-13, collagen-I, and collagen-II levels reaching their largest measured values 72 h after stimulation. Some genes were observed to rapidly change from baseline levels such as ADAMTS-4, TIMP-3, and to some extent aggrecan, while others showed a slower response such as collagen types I and II. No genes were observed to be at maximal level immediately following the cessation of loading.
Figure 4.
Kinetics of mRNA expression relative to internal control levels for genes that had highest levels at 8, 24, and 72 h following mechanical stimulation with quantitative values for the hypothetical model given in Figure 5.
DISCUSSION
A rat tail model was used to evaluate the kinetics of mRNA expression in the intervertebral disc anulus and nucleus following a single loading event and to establish a quantitative relationship between mechanical loading and mRNA levels. To aid in interpretation of results, a hypothetical model unique to each gene and disc region (i.e., anulus vs. nucleus), represents the general shape of the transient mRNA levels anticipated (Fig. 5). This model has mRNA levels that are altered from control levels at time t1 following an event, reach a maximum amplitude (Amax) at time tmax and recover to a final amplitude (A2, i.e., the new baseline gene expression level) at time t2. A remodeling or injury response involves an upregulation following loading (Amax≥0) that never returns to baseline levels (A2>0). In the context of mechanical stimulation, the amplitudes are a specific function of both magnitude and frequency of compression joint forces,13,19 and are relative to control levels (i.e., = 0; homeostasis), >0 (i.e., upregulation), or <0 (i.e., downregulation) provided measurements are taken at times beyond the half-life of the mRNA being measured.
Figure 5.
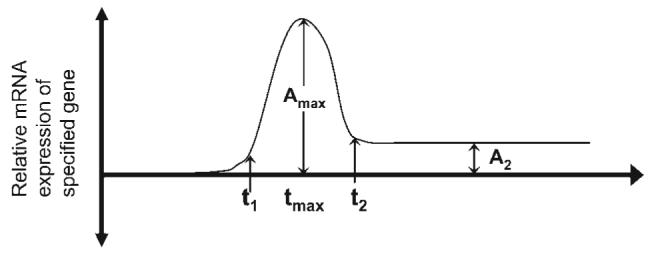
Hypothetical model for kinetic relationships between mRNA levels and mechanical stimulation specific to gene, disc region, and loading condition. Times t1, tmax, and t2 represented when relative mRNA expression level was significantly upregulated, reached a peak, and returned to baseline while Amax and A2 represented maximum and final amplitudes achieved.
Kinetic patterns of mRNA expression were observed over a 3-day period following a single loading event consistent with hypothesis 1. The transient responses were generally consistent with the hypothetical model although it is impossible to define precise timing or magnitudes based on the current methodologies and the chosen study design. The maximum measured mRNA levels were for proteases and their inhibitors which were the genes with low basal expression levels, consistent with hypothesis 2. Further, the majority of genes reached maximum observed levels 24 h following cessation of loading consistent with the hypothesis, but maximum mRNA levels were also observed 8 and 72 h following loading. Finally, mRNA levels returned to baseline levels within 72 h for some, but not all, genes in partial support of hypothesis 3.
Patterns of mRNA levels were consistent with turnover/repair in the nucleus and remodeling and/or injury in the anulus.1 In the nucleus, all mRNA levels returned to control levels (A2 = 0) representative of a turnover/repair response consistent with our hypothesis. In the anulus, however, cells exhibited mRNA levels that remained significantly upregulated for six genes (of nine measured) even 72 h after loading (A2 > 0). This continued upregulation of the anulus may represent adaptation to new steady state baseline mRNA levels consistent with a remodeling and/or injury responses where anulus cells and tissue physicochemical patterns were permanently modified. A second explanation for the continued upregulation of mRNA levels in the anulus regions is that the mRNA is not degraded before 72 h, suggestive of relatively slow metabolism. It is possible the extended changes in gene expression of the anulus could also represent a prolonged response associated with a cascade of events where initial mechanically induced changes in gene expression lead to subsequent changes (e.g., exposure to matrix breakdown products), which further influence gene expression. The mRNA levels for a few genes were not significantly altered in response to loading (MMP-2 in the anulus and nucleus and collagen-I and collagen-II in the nucleus) indicating a homeostasis response and/or lack of mechano-sensitivity of these genes under these cyclic compression loading conditions.
Based on the relative changes in gene expression, the short-term mRNA response to mechanical perturbation in vivo on important matrix proteins was suggestive of a strategy that first involved inhibiting tissue breakdown prior to accumulating structural proteins and enzymes. Due to the high baseline abundance of mRNA for collagens and aggrecan, significant upregulation of mRNA for these genes requires more mRNA and thus occurred later. The intermediate mRNA response involved increased aggrecan and matrix degrading enzymes mRNA in response to loading. The long-term mRNA response was associated with repair or anabolic remodeling modification of the matrix represented by collagen metabolism in the anulus. Results therefore suggest that attempts at anabolic remodeling must be given adequate time for metabolic processes and protein synthesis to occur and underscores the fact that MMPs and TIMPs have greater potency in altering aggrecan and collagen levels than protein synthesis alone.
The mRNA levels for aggrecan and ADAMTS-4 in both regions of the disc had maximum levels in 24-h measurements while collagens and MMP-13 (collagenase-3) in the anulus fibrosus had maximum levels in 72-h measurements. This finding complements the understanding that aggrecan turnover occurs more quickly than collagen turnover in intervertebral disc22 and cartilage.23 Differences between anulus and nucleus responses may also be attributed to the distinct cell populations and different physical signals in each region,3 and it is notable that in the nucleus collagen mRNA patterns were different with no significant alterations in collagen mRNA levels and MMP-13 reaching maximal levels at 24 h.
The mRNA levels of greatest abundance in the disc coded for genes of structural proteins, i.e., aggrecan followed closely by collagen-II in the nucleus and collagen-II followed by aggrecan and then collagen-I in the anulus fibrosus (Fig. 2). It is noteworthy that resident collagen-II and ADAMTS-4 mRNA levels were most and least abundant, respectively, in the anulus similar to that reported for articular cartilage.21 Thus, the 20-fold increase in relative mRNA expression for ADAMTS-4 in the anulus requires substantially less mRNA to be expressed than does the 10-fold increase in collagen-II mRNA observed in this study.
The kinetic mRNA response for a gene depends on several factors. First, genes with greater basal levels of mRNA require more time for significant mRNA changes to accumulate in order to achieve similar relative changes in mRNA levels. Second, cell signaling pathways and their stimuli vary for different proteins and thus kinetics are likely to vary between genes. Third, the minimum time for gene levels to return to baseline levels must be several half-lives greater than the half-life of the mRNA for that gene. Findings provided strong justification for taking measurements of mRNA levels associated with aggrecan metabolism at time points approximately 24 h past mechanical stimulation. While the 24-h time point also coincided with significantly upregulated mRNA levels for genes associated with collagen catabolism, and is therefore a reasonable time point for making such measurements, the maximum levels of collagen-I and II mRNA occurred at substantially later times and had not returned to baseline levels 72 h after mechanical stimulation.
The mRNA kinetics may also be affected by alterations in cell pressurization and strain which vary in vivo with load duration, magnitude, and frequency. It is likely that gene expression profiles change immediately after the initiation of loading, and, in a prior study on load duration effects, 30 min of cyclic loading was sufficient to induce measurable changes in mRNA expression levels in vivo.24 The single loading event used in this study was of 1.5-h duration which was chosen since it corresponded to a time when disc creep reached steady state24 so that alterations in cell strain due to viscoelasticity would not confound interpretation of the mRNA kinetic results. The magnitude and frequency were chosen since measurable changes in mRNA levels for several anulus and nucleus genes were previously observed,13 and these levels were predicted to be on the high end of physiological loading.24 The disc height change and water loss in rat caudal discs under 1-Hz cyclic compression was found to depend most strongly on peak load magnitudes and not average load values.25 The distinct anulus and nucleus responses may be associated with different biomechanical signals within each disc region, and it was reported that water redistribution from nucleus to anulus occurs with an initial disc bulge followed by a reduction in disc volume over time under 1-Hz compression loading.25 However, cells from the anulus and nucleus also respond to mechanical stimulation distinctly.3
Due to the fact that gene expression was measured far beyond the expected half-life of the genes in question,26,27 we are able to make a distinction between homeostasis (i.e., maintenance of mRNA at baseline levels), downregulation (i.e., a decrease or complete termination of mRNA production) and upregulation (i.e., an increase in mRNA production). No significant downregulation was observed following loading for any of the genes and results indicate cyclic compression had a significant stimulatory response on 14 of 18 genes measured (9 per disc region) and support the known stimulatory effects of cyclic loading on cartilaginous tissues.28,29 We may also conclude that levels of collagen-I and collagen-II in the nucleus and MMP-2 mRNA in both nucleus and anulus were unaffected by loading under these cyclic compression conditions. The finding for MMP-2 is consistent with Hsieh and Lotz who reported no change in total MMP-2 but an increase in the percentage active enzyme with compression loading on mouse tail intervertebral discs.30 Collagen-I and collagen-II are mechanically sensitive and were upregulated in the nucleus under different loading conditions13; it is also noteworthy that collagen-I in the NP had a nonsignificant decrease in mRNA levels immediately following loading.
Levels of mRNA coding for nine genes were measured at four time points using 49 animals in order to evaluate kinetics of matrix protein metabolism in two disc regions following a single loading event. While clear kinetic patterns were established, the determination of important times when mRNA levels were upregulated, peaked, and returned to baseline is not quantitative since time cannot be resolved beyond the four points measured. For example, the prolonged anulus mRNA response suggests that longer durations than 72 h may be necessary to definitively identity a new steady state. Based on the small tissue size of rat caudal discs and limited mRNA, a trade-off was necessary between the number of time points and the number of genes being evaluated. Measurements were therefore focused on genes coding for matrix proteins, proteases, and TIMPs; future evaluation of mRNA for other matrix molecules, cytokines, and transcription factors are important for a more thorough understanding of short-term matrix remodeling. Levels of MMP-2, 3, and 13 were all slightly elevated in the sham group (and only significant for MMP-13 in the annulus) similar to what has been previously seen, which may indicate a catabolic response to surgery and/or anesthesia. However, these effects were small in comparison to those seen 24 h after loading, and given the low basal levels of these genes represent a minor effect. The sham group assessed effects of surgical intervention, and, while potential effects of anesthesia cannot be completely eliminated since only a single 0-h recovery sham group was included, it is clear that surgical interventions were very small compared to effects of loading.
In conclusion, this study evaluated kinetics of mRNA expression in the disc in response to mechanical compression. Patterns of mRNA kinetics were consistent with turnover/repair in the nucleus and remodeling and/or injury in the anulus. The magnitude and frequency of loading used in this study is predicted to be on the high end of physiological loading for the rat24 which would therefore be expected to induce these types of gene expression alterations. The kinetic results also supported a strategy of mRNA expression that first involved inhibition of tissue breakdown (through expression of TIMPs) followed by synthesis of aggrecan and matrix degrading enzymes, and then collagens. Put in the context of disc repair and regeneration, these findings might also suggest that biological strategies that prevent degeneration through anti-catabolic pathways such as upregulation of TIMPs or downregulation of MMPs may be more easily achievable and more effective than strategies that focus only on increasing structural proteins (aggrecan and collagen) alone. Results demonstrated that resident mRNA levels in control discs were most abundant for structural matrix proteins and least abundant for proteases. Results provide justification for taking measurements of mRNA levels associated with aggrecan metabolism at time points approximately 24 h after stimulation, but for collagen metabolism at later times. This improved understanding of the quantitative relationship between loading and gene expression may help elucidate the mechanisms involved with injury, may lead to more effective rehabilitation treatments, and may help optimize tissue engineering approaches for intervertebral disc repair.
ACKNOWLEDGMENTS
This research was supported by grants from the National Institutes of Health (R01 AR051146), Canadian Institutes of Health Research, and the AO Foundation, Switzerland. The authors gratefully acknowledge Dr. Keita Ito for the use of the in vivo loading device, Stephen Bell for the use of the University of Vermont Cardiology Research Lab, and the technical assistance of Justin Stinnett-Donnelly and David Korda.
REFERENCES
- 1.Iatridis JC, MacLean JJ, Roughley PJ, et al. Effects of mechanical loading on intervertebral disc metabolism in vivo. J Bone Joint Surg [Am] 2006;88(Suppl 2):41–46. doi: 10.2106/JBJS.E.01407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lotz JC. Animal models of intervertebral disc degeneration: lessons learned. Spine. 2004;29:2742–2750. doi: 10.1097/01.brs.0000146498.04628.f9. [DOI] [PubMed] [Google Scholar]
- 3.Setton LA, Chen J. Mechanobiology of the intervertebral disc and relevance to disc degeneration. J Bone Joint Surg [Am] 2006;88(Suppl 2):52–57. doi: 10.2106/JBJS.F.00001. [DOI] [PubMed] [Google Scholar]
- 4.Roberts S, Evans H, Trivedi J, et al. Histology and pathology of the human intervertebral disc. J Bone Joint Surg [Am] 2006;88(Suppl 2):10–14. doi: 10.2106/JBJS.F.00019. [DOI] [PubMed] [Google Scholar]
- 5.An HS, Masuda K. Relevance of in vitro and in vivo models for intervertebral disc degeneration. J Bone Joint Surg [Am] 2006;88(Suppl 2):88–94. doi: 10.2106/JBJS.E.01272. [DOI] [PubMed] [Google Scholar]
- 6.Lotz JC, Staples A, Walsh A, et al. Mechanobiology in intervertebral disc degeneration and regeneration; Conf Proc IEEE Eng Med Biol Soc; 2004. p. 5459. [DOI] [PubMed] [Google Scholar]
- 7.Katz JN. Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. J Bone Joint Surg [Am] 2006;88(Suppl 2):21–24. doi: 10.2106/JBJS.E.01273. [DOI] [PubMed] [Google Scholar]
- 8.Grunhagen T, Wilde G, Soukane DM, et al. Nutrient supply and intervertebral disc metabolism. J Bone Joint Surg [Am] 2006;88(Suppl 2):30–35. doi: 10.2106/JBJS.E.01290. [DOI] [PubMed] [Google Scholar]
- 9.Battie MC, Videman T, Parent E. Lumbar disc degeneration: epidemiology and genetic influences. Spine. 2004;29:2679–2690. doi: 10.1097/01.brs.0000146457.83240.eb. [DOI] [PubMed] [Google Scholar]
- 10.Stokes IA, Iatridis JC. Mechanical conditions that accelerate intervertebral disc degeneration: overload versus immobilization. Spine. 2004;29:2724–2732. doi: 10.1097/01.brs.0000146049.52152.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lotz JC, Ulrich JA. Innervation, inflammation, and hypermobility may characterize pathologic disc degeneration: review of animal model data. J Bone Joint Surg [Am] 2006;88(Suppl 2):76–82. doi: 10.2106/JBJS.E.01448. [DOI] [PubMed] [Google Scholar]
- 12.Larson JW, III, Levicoff EA, Gilbertson LG, et al. Biologic modification of animal models of intervertebral disc degeneration. J Bone Joint Surg [Am] 2006;88(Suppl 2):83–87. doi: 10.2106/JBJS.F.00043. [DOI] [PubMed] [Google Scholar]
- 13.MacLean JJ, Lee CR, Alini M, et al. Anabolic and catabolic mRNA levels of the intervertebral disc vary with the magnitude and frequency of in vivo dynamic compression. J Orthop Res. 2004;22:1193–1200. doi: 10.1016/j.orthres.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Yoon ST, Patel NM. Molecular therapy of the intervertebral disc. Eur Spine J. 2006;15(Suppl 3):379–388. doi: 10.1007/s00586-006-0155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts S, Caterson B, Menage J, et al. Matrix metalloproteinases and aggrecanase: their role in disorders of the human intervertebral disc. Spine. 2000;25:3005–3013. doi: 10.1097/00007632-200012010-00007. [DOI] [PubMed] [Google Scholar]
- 16.Roughley PJ. Biology of intervertebral disc aging and degeneration: involvement of the extracellular matrix. Spine. 2004;29:2691–2699. doi: 10.1097/01.brs.0000146101.53784.b1. [DOI] [PubMed] [Google Scholar]
- 17.Evans C. Potential biologic therapies for the intervertebral disc. J Bone Joint Surg [Am] 2006;88(Suppl 2):95–98. doi: 10.2106/JBJS.E.01328. [DOI] [PubMed] [Google Scholar]
- 18.Andersson GB, An HS, Oegema TR, Jr, et al. Intervertebral disc degeneration. Summary of an AAOS/NIH/ORS workshop, September 2005. J Bone Joint Surg [Am] 2006;88:895–899. doi: 10.2106/JBJS.F.00028. [DOI] [PubMed] [Google Scholar]
- 19.MacLean JJ, Lee CR, Grad S, et al. Effects of immobilization and dynamic compression on intervertebral disc cell gene expression in vivo. Spine. 2003;28:973–981. doi: 10.1097/01.BRS.0000061985.15849.A9. [DOI] [PubMed] [Google Scholar]
- 20.Lee CR, Grad S, MacLean JJ, et al. Effect of mechanical loading on mRNA levels of common endogenous controls in articular chondrocytes and intervertebral disk. Anal Biochem. 2005;341:372–375. doi: 10.1016/j.ab.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Lee JH, Fitzgerald JB, Dimicco MA, et al. Mechanical injury of cartilage explants causes specific time-dependent changes in chondrocyte gene expression. Arthritis Rheum. 2005;52:2386–2395. doi: 10.1002/art.21215. [DOI] [PubMed] [Google Scholar]
- 22.Sivan SS, Tsitron E, Wachtel E, et al. Age-related accumulation of pentosidine in aggrecan and collagen from normal and degenerate human intervertebral discs. Biochem J. 2006;399:29–35. doi: 10.1042/BJ20060579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Billinghurst RC, Wu W, Ionescu M, et al. Comparison of the degradation of type II collagen and proteoglycan in nasal and articular cartilages induced by interleukin-1 and the selective inhibition of type II collagen cleavage by collagenase. Arthritis Rheum. 2000;43:664–672. doi: 10.1002/1529-0131(200003)43:3<664::AID-ANR24>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 24.MacLean JJ, Lee CR, Alini M, et al. The effects of short-term load duration on anabolic and catabolic gene expression in the rat tail intervertebral disc. J Orthop Res. 2005;23:1120–1127. doi: 10.1016/j.orthres.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 25.Masuoka K, Michalek AJ, MacLean JJ, et al. Different effects of static versus cyclic compressive loading on rat intervertebral disc height and water loss in vitro. Spine. 2007;32:1974–1979. doi: 10.1097/BRS.0b013e318133d591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curtis AJ, Devenish RJ, Handley CJ. Modulation of aggrecan and link-protein synthesis in articular cartilage. Biochem J. 1992;288(Pt 3):721–726. doi: 10.1042/bj2880721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krupsky M, Kuang PP, Goldstein RH. Regulation of type I collagen mRNA by amino acid deprivation in human lung fibroblasts. J Biol Chem. 1997;272:13864–13868. doi: 10.1074/jbc.272.21.13864. [DOI] [PubMed] [Google Scholar]
- 28.Blain EJ, Gilbert SJ, Wardale RJ, et al. Upregulation of matrix metalloproteinase expression and activation following cyclical compressive loading of articular cartilage in vitro. Arch Biochem Biophys. 2001;396:49–55. doi: 10.1006/abbi.2001.2575. [DOI] [PubMed] [Google Scholar]
- 29.Fitzgerald JB, Jin M, Dean D, et al. Mechanical compression of cartilage explants induces multiple time-dependent gene expression patterns and involves intracellular calcium and cyclic AMP. J Biol Chem. 2004;279:19502–19511. doi: 10.1074/jbc.M400437200. [DOI] [PubMed] [Google Scholar]
- 30.Hsieh AH, Lotz JC. Prolonged spinal loading induces matrix metalloproteinase-2 activation in intervertebral discs. Spine. 2003;28:1781–1788. doi: 10.1097/01.BRS.0000083282.82244.F3. [DOI] [PubMed] [Google Scholar]