Abstract
Equilibrium analyses have been performed to elucidate the role of dimerization in folding and stability of dynein light chain Tctex-1. The equilibrium unfolding transition was monitored by intrinsic fluorescence intensity, fluorescence anisotropy, and circular dichroism and was modeled as a two-state mechanism where a folded dimer dissociates to two unfolded monomers without populating thermodynamically stable monomeric or dimeric intermediates. Sedimentation equilibrium and chemical cross-linking experiments performed at increasing concentrations of denaturants show no change in the association state before the unfolding transition, and are consistent with the two-state model of dissociation coupled to unfolding. A linear dependence on denaturant concentration is observed by fluorescence intensity and anisotropy before unfolding in the 0–2 M GdnCl, and 0–4 M urea concentration range. This change is not protein concentration dependent and possibly reflects relief of quenching associated with premelting conformational disorder in the vicinity of Trp 83. The data clearly show that the dissociation-coupled unfolding mechanism of Tctex-1 is different from the three-state denaturation mechanism of its structural homolog light chain LC8. The absence of a stable monomer in Tctex-1 offers insight into its functional differences from LC8.
Keywords: Dynein light chain, Tctex-1, dimerization, stability, dimer interface, equilibrium unfolding
Cytoplasmic dynein is a multisubunit, minus-end directed microtubule ATPase, which provides the motive force for intracellular processes such as the perinuclear positioning of the Golgi complex (1, 2), the localization of the mitotic spindle and centrosome separation during mitosis (3, 4) and in the transport of membraneous organelles and endocytotic vesicles. The dynein complex is composed of two heavy chain subunits joined by flexible stalk domains to a common base. The heavy chains provide the ATPase activity as well as the microtubule binding function and the base is composed of accessory subunits which couple the dynein motor to a wide variety of intracellular cargo (5). Since the heavy chains are encoded by a limited number of genes (6), it is thought that the multiple functions of dynein are regulated by the subunit complexity and interactions of the accessory subunits composed of 74 kDa intermediate chains, 50–60 kDa light intermediate chains and 8–22 kDa light chains (7–9).
Drosophila Tctex-1 is a light chain subunit of dynein with 70% sequence identity to the human protein. Molecular genetics studies in Drosophila show that null mutations of the Tctex-1 gene do not affect essential cytoplasmic dynein functions but result in complete male sterility (10). In mammals, Tctex-1 is thought to be required for dynein-mediated transport of rhodopsin to the apical surface of photoreceptor cells (11). This transport is abolished in retinitus pigmentosa, a retinal degenerative condition, as a result of mutations at the C-terminus cytoplasmic end of rhodopsin. However, Tctex-1 is not necessarily an obligate dynein subunit, and is reported to exist in cytoplasmic extracts outside the dynein complex (10, 12). Within the dynein complex, Tctex-1 along with light chain LC8 associate with the 74 kDa intermediate chain subunit (13, 14), and the three subunits form a tight sub-complex (15). The interaction of Tctex-1 with a wide array of unrelated proteins, may in part reflect interactions of Tctex-1 that are independent of the dynein motor complex (16, 17). A recently proposed dynein-independent role for Tctex-1 in actin remodeling during neuronal outgrowth is consistent with this interpretation (18).
Tctex-1 is a 25 kDa dimeric protein at physiological conditions in vitro (19, 20). Both the crystal and NMR structures show that each monomer contains a short N-terminal beta strand and two alpha helices followed by four beta strands (Figure 1), with the second beta strand of one subunit intertwined at the dimer interface with a beta strand from the other subunit, and each makes extensive intersubunit contacts (21, 22). The three-dimensional structure is very similar to that of light chain LC8 (23), which is also a dimeric protein with intertwined beta strands at the interface and shows no sequence homology to Tctex-1.
FIG 1.

Ribbon diagram of Tctex-1 showing the location of the tryptophan residues, a cysteine residue in close proximity to Trp 83, and a lysine residue at the interface that may be involved in covalent cross-linking. The image was produced with PyMol (Delano scientific LLC) (51) using PDB code 1ygt (21).
Homodimeric proteins may be divided into three classes according to their denaturation patterns: a two-state transition to fully unfolded monomers, and a three-state transition with either a monomeric or a dimeric intermediate preceding unfolding (24). Small two-state dimers require a dimeric interface for substrate binding and have a high number of intersubunit contacts. Examples of this class include DNA-RNA binding proteins such as ROP (25), Arc repressor (26), Trp aporepressor (27), and proteases such as HIV and SIV (28). The quaternary structure is essential for folding of such proteins and their primary function is binding specifically and symmetrically to other molecules. The three-state dimers that form a monomeric intermediate are in general larger, mostly enzymes that dimerize after folding, and their oligomerization may activate the monomeric subunit. Their quaternary structure is important to improve their stability and binding but not for folding of the tertiary structure. The three-state dimers with a dimeric intermediate also require the quaternary structure for folding, and in kinetic experiments, dimeric intermediates increase the rates of folding, as observed for intertwined alpha-helical homodimers (29).
LC8 is a three-state dimer which dissociates to a folded monomer before global unfolding. The folded monomer is populated at low protein concentration in physiological buffer and also at low GdnCl concentration and low pH (30). The ionization state of a single residue at the interface, His 55, controls the pH-dependent monomer dimer equilibrium (31). We have proposed that the monomer-dimer equilibrium of LC8 regulates its function and that the dimeric nature of LC8 with dual symmetric binding sites for many ligands is an essential factor in bringing its interacting partners in close proximity, and thereby promoting their dimerization or assembly (32). LC8 and Tctex-1 are both homodimers with very similar overall fold. They both bind IC74 and diverse other proteins unrelated to dynein, and may be involved in regulating dynein recognition to multiple targets. Comparison of their dimer stability and denaturation mechanisms may illuminate possible differences or similarities in their function. In addition, investigation into the mechanism of assembly of the Tctex-1 quaternary structure and the stability of the dimer interface, and comparison to dimeric proteins of the same class is a first step in understanding its roles in dynein function and various other complexes.
In the present study we show that Drosophila Tctex-1 is a two-state unfolding dimer whose unfolding and dissociation are tightly coupled. This unfolding mechanism offers insight into functional differences between Tctex-1 and its structural homolog LC8.
Materials and Methods
Stock Solutions, Buffers, and Proteins
Samples for CD and fluorescence measurements at pH 7.8 were prepared in 50 mM sodium phosphate and 10 mM TCEP was added to rule out possible contribution of intermolecular disulfide bonds to self-association. TCEP stock solutions were prepared in 50 mM phosphate buffer and adjusted with NaOH to pH 7.8. Solutions of 8 M GdnCl were prepared as described elsewhere (33), and the exact concentration was determined by refractive index. All buffers and stock solutions were freshly prepared. Tctex-1 stock solutions were dialyzed in the appropriate buffers and their concentration determined from absorbance at 280 nm using an extinction coefficient of 23470 M−1cm−1 calculated from the number of tryptophan and tyrosine residues using ProtParam (http:/us.expasy.org/tools).
Cloning, Expression and Protein Purification
The gene encoding Drosophila Tctex-1 engineered with a Factor Xa recognition site immediately 5′ to the start codon of the cDNA was subcloned into BamHI-Sal1 cleavage sites of a pET 15d expression vector (Novagen) and transformed into Escherichia coli BL21-DE3 cell lines (Novagen). Protein expression and purification followed previously published protocols (13). Purity was determined by SDS-PAGE to be > 98%. The mass obtained from electrospray ionization mass spectrometry (ESMS) was 12,479 Da (calculated mass of 12,478.2 Da).
Analytical Ultracentrifugation
Sedimentation equilibrium experiments were conducted using a Beckman XL-A analytical ultracentrifuge. Three loading concentrations were equilibrated at two speeds (28,000 and 40,000 rpm) and scanned at 280 nm using a cell path length of 3 mm. Experiments were done at 30 °C and at 4 °C. Samples were allowed to equilibrate for 16 hours at each speed and concentration, and were considered at equilibrium when sequential scans two hours apart were superimposable. Similar conditions were used for proteins in 0 M, 1 M, and 1.6 M GdnCl.
Circular Dichroism and Fluorescence Measurements
CD experiments were conducted on a JASCO 715 spectropolarimeter equipped with a built-in magnetic stirrer for fast equilibration within the cell. Unfolding experiments were recorded in a 1 mm cell for far UV CD data at 30° C using a batch-type experiment to ensure that equilibrium was achieved. Samples were equilibrated for 12 hours at room temperature and 30 minutes at 30 °C before data collection.
Intrinsic fluorescence emission spectra were determined using a Jobin Yvon/Spex spectrofluorometer. The excitation wavelength was set to 283 nm and fluorescence emission spectra were scanned from 300 nm to 380 nm, with excitation and emission bandwidths adjusted depending on the protein concentration used. To limit photobleaching at low protein concentrations, samples were stirred continuously and only one measurement per sample was recorded. The conditions (protein concentrations, buffer and temperature) of the unfolding experiments were the same as those for CD. All experiments were carried out at 30 °C using an external circulating bath. Reversibility was determined after dialysis in native buffer and comparison of the fluorescence and CD spectra of the refolded protein with those of the native protein.
Steady state intrinsic tryptophan emission anisotropy measurements were performed using Glan-Thompson quartz excitation and emission polarizers installed in the emission and excitation paths. The fluorescence intensities of the vertical and horizontal emission components on excitation and vertically polarized light were measured with a fixed emission wavelength of 360 nm. The anisotropy signals were subtracted from buffer blanks containing the same concentration of GdnCl.
Analysis of Equilibrium Denaturation Data
The data were analyzed using the two-state model for denaturation of dimeric proteins according to equation 1.
| (eq 1) |
The free energy change, ΔGH2O, was determined by a fit of the free energy ΔGi for points in the transition regions of each denaturation curve using equation 2.
| (eq 2) |
where [GdnCl] is the denaturant concentration, and m is the slope of the line given by a plot of ΔGi vs. [GdnCl] (33–36). The equilibrium constant, K, was determined using equation 3,
| (eq 3) |
where Pt is the monomeric protein concentration and fu is the fraction of unfolded monomeric protein (26).
The midpoint of the unfolding transition, Cm, was calculated using equation 4.
| (eq 4) |
In practice, free energies and other characteristic features of the denaturation curves were determined by fitting the calculated spectroscopic signal to the experimentally determined signal. Parameters yd, yu, and m were optimized for individual curves. ΔGH2O was globally fit to a data set containing all denaturation curves. The fits were performed using a χ2 procedure implemented in Microsoft Excel.
Quantitative Glutaraldehyde Crosslinking
A concentration of 8–10 μM of Tctex-1 at a given GdnCl or urea concentration was incubated for 18 hours at room temperature. Glutaraldehyde solution (25% w/v) was added to a final concentration of 0.5% and the reaction was quenched after 2 minutes with 0.4% NaBH4 solution dissolved in 0.1 M NaOH. The conditions for the cross-linking reactions were optimized so as to minimize higher order cross-linked products and this resulted in a cross-linked dimer in addition to a considerable amount of uncross-linked monomer. To facilitate Tctex-1 precipitation each sample was diluted 5-fold with water before adding sodium deoxycholate and trifluoracetic acid to a final concentration of 12 μg/ml and 3.6%, respectively. The samples were centrifuged at 14 000 rpm for 20 minutes, and the resulting pellet was washed with acetone and dried under vacuum. The pellet was then resuspended in 20 μl of SDS-PAGE gel loading buffer, boiled and loaded onto a 15% polyacrylamide gel. Bands were visualized after staining with Coomassie Blue, and their intensities were analyzed by densitometry with the Quantity one™ quantitation software (Biorad).
Results
Equilibrium Unfolding
The fluorescence emission spectra of Tctex-1 report on the environment of one or more of the three tryptophan residues per monomer at positions 40, 83, and 95. In the absence of chemical denaturants, an emission maximum wavelength of 336 nm indicates that the tryptophan residues are somewhat buried in the interior of the protein. In 1.6 M GdnCl, there is a slight red shift to a maximum wavelength of 338 nm and a significant increase in fluorescence intensity. A shift to a wavelength of 350 nm and a decrease in intensity is observed in 4 M GdnCl in which the protein is completely denatured (Figure 2).
FIG 2.
Overlay of fluorescence emission spectra of Tctex-1 in 0 M, 1.6 M, and 4 M GdnCl. The protein concentration was 1.7 μM in 50 mM phosphate buffer, pH 7.8 and 10 mM TCEP. The fluorescence spectra were obtained at 30 °C with excitation at 283 nm and emission recorded from 300–380 nm. The excitation and emission slit widths used were 4.8 and 5.0 respectively. A reference spectrum was obtained for every GdnCl concentration and subtracted from the protein spectrum.
Equilibrium unfolding profiles were obtained by monitoring changes in the far UV CD signal at 222 nm and the fluorescence intensity signal at 336 nm (Figure 3). A midpoint of unfolding of 2.52 M determined from the far UV CD data, and a similar midpoint of 2.56 M from fluorescence data suggest that unfolding is a two-state transition. Both sets of data when individually fit to a two-state model of folded dimer to an unfolded monomer give a similar free energy of unfolding of 20.5 ± 0.4 kcal/mol (CD) and 19.9 ± 0.2 kcal/mol (fluorescence), and similar m-values of −5.0 ± 0.3 kcal/mol.M−1 (CD) and −5.2 ± 0.2 kcal/mol.M−1 (fluorescence). The m-value is a measure of denaturant dependent cooperativity of native to denaturant transitions, and indicates the efficacy of the denaturant in forcing unfolding. The linear denaturant-dependent increase in tryptophan fluorescence intensity observed at increasing GdnCl concentration before unfolding reaches a maximum in 2 M GdnCl and decreases to a minimum after complete unfolding. There is no change in the CD signal in the same denaturant range. An increase in fluorescence intensity was also observed at increasing urea concentrations before complete unfolding (data not shown).
FIG 3.
Equilibrium unfolding transitions monitored by far UV CD at 222 nm (▲), and fluorescence intensity at 336 nm (●) at a protein concentration of 18.3 μM. Data were acquired at 30 °C in 50 mM sodium phosphate buffer, and 10 mM TCEP at pH 7.8. CD signal is measured as ellipticity in mdegrees. Curves were fit to a two-state model. Data were acquired using a batch-type experiment to ensure that equilibrium is achieved. A reference spectrum was obtained for every GdnCl concentration and subtracted from the protein spectrum.
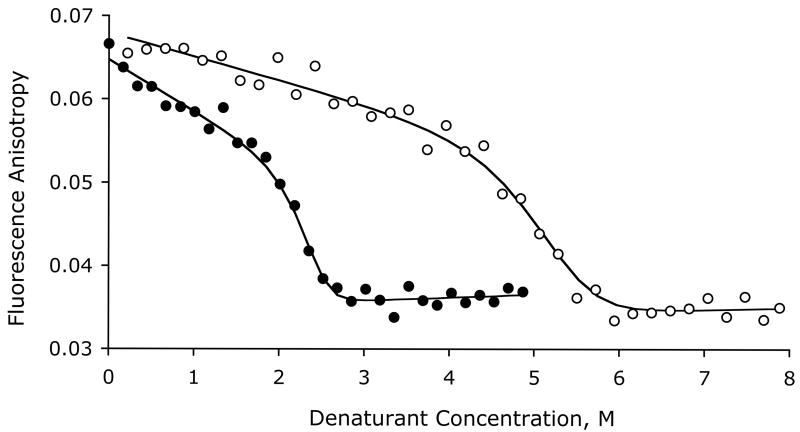
Unfolding profiles were also obtained by monitoring changes in steady-state fluorescence anisotropy at increasing urea and GdnCl concentration (Figure 4). There is a slight steady decrease in anisotropy in the 0–2 M GdnCl concentration range and 0–4 M urea concentration range, followed by a sharp decrease to a value of 0.035 representing complete unfolding. The decrease in anisotropy at higher denaturant concentration after unfolding is a consequence of increased mobility of the Trp side chains. The midpoint of unfolding is 5.11 M in urea and 2.33 M in GdnCl for the same protein concentration. Both urea and GdnCl unfolding data give a similar free energy of unfolding of 19.8 kcal/mol. The lines in Figure 4 show the results of least squares global fitting analysis using a ΔGH2O of 19.8 kcal/mol to give m-values of −2.5 kcal/mol.M−1 in urea and −5.5 kcal/mol.M−1 in GdnCl.
FIG 4.
Unfolding curves followed by changes in fluorescence anisotropy as a function of increasing urea (○), and GdnCl concentrations (●). Both experiments were performed for a protein concentration of 3.9 μM at 30 °C in 50 mM phosphate buffer, pH 7.8 with 10 mM TCEP. Excitation was set to 283 nm and emission to 360 nm.
The observed fluorescence anisotropy of native Tctex-1 is notably low (0.06 instead of 0.1 in proteins of similar size), consistent with resonance energy homo transfer between two tryptophan residues, which causes depolarization (37). Possible candidates for intertryptophanyl energy transfer are Trp 83 and 83′, that are 15 Å apart across the interface, and Trp 40 and Trp 95 that are 10 Å apart within the same subunit (Figure 1). In both cases, the two indole rings are well within the Förster distance (R0) of resonance energy transfer of one another (10–15 Å for tryptophans in a hydrophobic environment). In lysozyme, intertryptophanyl energy transfer was observed for a separation distance of 13 Å (38). The indole ring of Trp 83 at the interface is somewhat more solvent exposed than the other two, 26 Å2 of accessible surface area relative to 9 Å2 and 10 Å2 for Trp 40 and Trp 95.
Thermal denaturation of Tctex-1 is irreversible due to protein aggregation at high temperatures but the protein is stable up to 55 °C as determined by CD at 222 nm before it starts to aggregate (data not shown).
Denaturation Profiles by Chemical Crosslinking
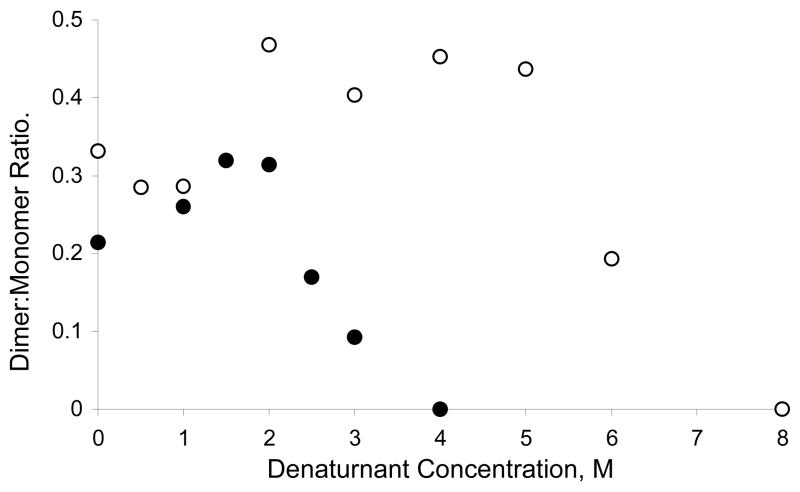
Glutaraldehyde modifies primary amines (39) and has been extensively used to investigate protein-protein interactions. Since there is a lysine residue close to the dimer interface (Figure 1), glutaraldehyde cross-linking was used to measure the stability of the dimer at increasing denaturant concentration. Figure 5 shows the apparent dimer to monomer ratios of Tctex-1 as a function of GdnCl and urea concentrations. In low denaturant concentrations before unfolding, there is no decrease in the relative population of dimer to monomer indicating the absence of monomeric intermediate. In high denaturant concentration after complete unfolding, only monomers were detected. The midpoints of the transition in both urea and GdnCl are consistent with those observed with fluorescence anisotropy indicating that there is no dissociation before unfolding, and that a higher concentration of urea relative to GdnCl is required for unfolding. While the results are somewhat noisy given the nature of the method, it is clear is that there is no dissociation preceding unfolding.
FIG 5.
Glutaraldehyde cross-linking of Tctex-1. Ratios of band intensities obtained from SDS-PAGE corresponding to the chemically cross-linked dimer relative to the monomer are plotted at increasing concentrations of GdnCl (●) for 8 μM, and urea (○) for 10 μM protein. The midpoint of dimer dissociation is consistent with dimer unfolding obtained by fluorescence spectroscopy. Both monomer and dimer bands are present at native conditions because of incomplete cross-linking.
Hydrodynamic Measurements
Sedimentation equilibrium was used to determine the association state(s) of Tctex-1 at low denaturant concentrations. Sedimentation equilibrium conducted at 4 °C in 0 M GdnCl for protein concentrations of 14.3, 28.7, and 57.5 μM were fit to a one-component ideal model with an apparent molecular weight of 24 824 ± 4 Da (actual molecular weight of 24 956 Da) indicating that Tctex-1 is a tight dimer with no detectable population of a monomeric species at these protein concentrations. Similarly, data collected on the His-tagged protein at 30 °C for a protein concentration of 22 μM in 0 M, 1 M, and 1.6 M GdnCl were fit to a one-component ideal model with an apparent molecular weight of 31 540 ± 8 Da (actual molecular weight 30 724 Da with the fusion His tag peptide) in 0 M GdnCl, 32 110 ± 10 in 1 M GdnCl, and 31 300 ± 10 in 1.6 M GdnCl. Models incorporating larger oligomeric species were also tested but did not provide acceptable variances.
Further evidence consistent with tight dimer comes from several other experiments: A sedimentation velocity coefficient of 2.6 S was obtained at three protein concentrations in the range of 4 to 36 μM at 20 °C; Tctex-1 elutes as a single peak on a size-exclusion column with a retention time that corresponds to the dimer; and dynamic light scattering gives a hydrodynamic radius of 2.30 nm which corresponds to an estimated molecular mass of 25 kDa.
Protein Concentration Dependence
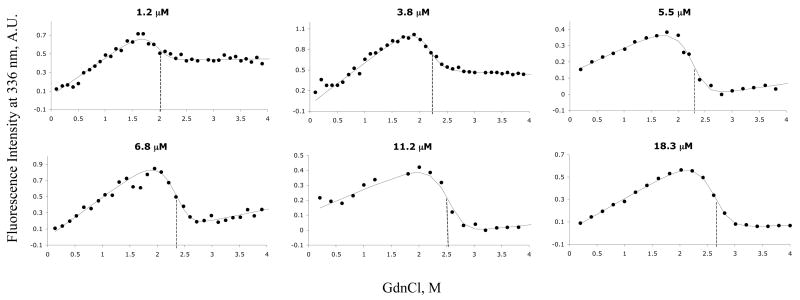
Unfolding profiles were determined from fluorescence emission intensity in the protein concentration range of 1.2 to 18.3 μM (Figure 6). Solid lines correspond to a global fit of all the data to a free energy of unfolding ΔGH2O of 19.87 ± 0.3 kcal/mol. An m-value of 5.40 ± 0.3 kcal/mol.M−1 was determined from the average m-values of all the data sets. The low uncertainty among measurements at various concentrations is a strong indication of the absence of an intermediate.
FIG 6.
GdnCl-induced equilibrium denaturation of Tctex-1 at increasing protein concentration. Unfolding profiles are shown for protein concentrations of 1.2, 3.8, 5.5, 6.8, 11.2, and 18.3 μM. The curves represent the non-linear least-squares global fit of all data to the two-state linear extrapolation model. Fluorescence intensity was recorded at 336 nm. Dotted vertical lines correspond to the transition midpoint.
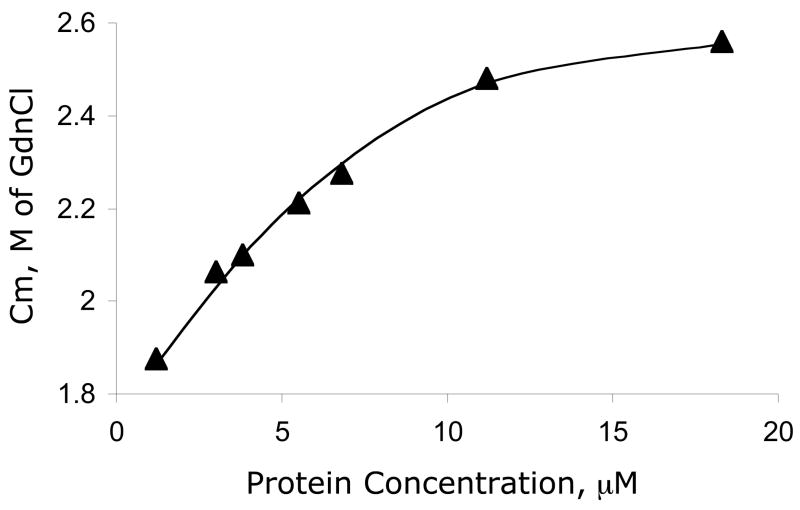
Fitting the data to a three-state model that includes either a monomeric or dimeric intermediate was tested but did not improve the fit. The linear dependence of the fluorescence intensity of the native state on the denaturant concentration was observed for all concentrations but no obvious protein concentration dependence was detected. Figure 7 shows the protein concentration dependence of the transition midpoint, Cm. Cm increases from 1.9 M GdnCl at protein concentration of 1 μM and plateaus at Cm of 2.6 M GdnCl at protein concentration of > 12 μM.
FIG 7.
The transition midpoint, Cm, for various protein concentrations. The change in Cm ranges from 1.9 M GdnCl for protein concentration of 1 μM to a Cm of 2.6 M for a protein concentration greater than 12 μM.
DISCUSSION
Dissociation and Unfolding of Tctex-1 Are Tightly Coupled
Studies on protein folding concentrate to a large extent on monomeric proteins undergoing unimolecular reactions. Studies reported here are for an intertwined tight dimeric protein that unfolds with no evidence of monomeric population at low denaturant concentrations. A combined analysis of the various spectroscopic and biochemical data show that the N2 to 2U two-state model of unfolding is sufficient to interpret the equilibrium data. The two-state model is based on the following: a similar midpoint of unfolding determined from fluorescence and far UV CD for the same protein concentration, a similar free energy of unfolding obtained from both urea and GdnCl denaturation data, a Cm that is a function of protein concentration, and a global analysis of all data fit well to a two-state unfolding mechanism with no dependence of free energy and m values on spectroscopic probes or protein concentrations (40). Sedimentation equilibrium measurements and covalent cross-linking at low denaturant concentrations also suggest that breaking the intersubunit contacts destroys the folding integrity of the monomers as there were no conditions under which structured monomers could be observed.
With a high unfolding free energy of 20 kcal/mol, Tctex-1 is a fully native dimer at physiological concentrations greater or equal to 10−14 M. The high free energy of unfolding of Tctex-1 is not unusual for a two-state unfolding dimer; E. coli Trp aporepressor, for example, which is also an intertwined dimeric protein has a free energy of unfolding of 23 kcal/mol (27). The high m-values of 5.4 kcal/mol.M−1 in GdnCl and 2.5 kcal/mol.M−1 in urea associated with dimer dissociation implies that a large surface area of 20 000 Å2 is buried upon folding, estimated according to (41) and is the same for both urea and GdnCl. This value is similar to other two-state unfolding proteins of similar size such as Trp aporepressor.
Preunfolding Fluorescence Changes
The change in fluorescence intensity and anisotropy at low denaturant concentration is not accompanied by a significant shift in the emission maximum (336 to 338 nm), nor a change in the far UV CD signal at 222 nm. Fluorescence signals are usually very sensitive to the conformational state of a macromolecule since changes in the fluorescence life times and rotational correlation times belong to the same time scale of local protein motions and fluctuations (42). However, in the absence of life time data, fluorescence changes cannot be unequivocally related to molecular details (43). The linear increase in fluorescence reported here is not likely due to solvent accessibility of a fluorophore because only a modest increase in fluorescence signal is reported for a solvent exposed tryptophan model compound, N-acetyl-L- tryptophanamide, (NATA) when GdnCl is used as denaturant (43). In our sample conditions and at excitation wavelength of 283 nm no detectable change in fluorescence of NATA was observed in the 0–2 M range of GdnCl (data not shown).
One plausible reason for the observed changes in fluorescence at low concentration of GdnCl is relief of quenching caused by small motions of amino acid side chains or increase of distance from the quencher. A likely candidate for quenching is Cys 81 which is in direct contact with the indole ring of Trp 83 (Figure 1). A cysteine side chain is a fairly strong quencher of tryptophan in solution due to excited state electron transfer (44). Since Trp 83 is on a loop that could show premelting disorder, the rise in fluorescence may be due to an increase in the average distance between Cys and Trp. This hypothesis of premelting disorder around Trp 83 can also explain the slight decrease in emission anisotropy.
Differences in Dimer Dissociation between Light Chains Tctex-1 and LC8
Although Tctex-1 and LC8 are structural homologs, their sequences are too distant to be identified as related by a Blast search. Structure-based sequence alignments between them show zero sequence identity (21). Interestingly, their mechanisms for unfolding and dissociation are also quite different. Tctex-1 is a tighter dimer than LC8 and does not form a stable monomeric intermediate in the presence of denaturants or at low pH. Comparison of their structures reveals possible explanations for these differences. In Tctex-1, subunit interactions involve a large surface area with numerous hydrophobic amino acid residues in close contact (21). In our calculations, each protomer buries 1574 Å2 of surface area, whereas in LC8 each protomer buries only 860 Å2. The major difference between structures is in the length of the intertwined β-strand at the interface. In Tctex-1 there are 10 residues per strand, while in LC8 there are 5 residues per strand. For a stable monomeric form of LC8, the shorter peptide strand, while disordered, adopts alternative turn-like conformations (45), while for Tctex-1 it appears that burial of these extended strands is essential for the folding of its tertiary structure.
Since Tctex-1 is a two-state unfolded dimer, a significant proportion of its overall stability is apparently contributed by quaternary interactions. For LC8, quaternary interactions apparently provide ~ 50% of dimer stability since the free energy of dissociation is 8.4 kcal/mol while for monomer unfolding another 7.5 kcal/mol is observed (30). While Tctex-1 and LC8 can be classified as two-state and three-state dimers, respectively, there is a fine energetic balance between two-state and three-state denaturation systems. A few mutations at the interface can result in destabilization of the dimer relative to monomer and conversion of the equilibrium denaturation from a two-state to a three-state as observed in Trp aporepressor (46). Both Tctex-1 and LC8 are highly conserved across species, and therefore, it is likely that their unfolding mechanisms are conserved as well.
For monomeric proteins, it appears that the rates and mechanisms for protein folding depend largely on the complexity of the native structure topology rather than the details of the amino acid sequence and that protein folding mechanisms are better conserved than amino acid sequences (47). For example, highly divergent dihydrofolate reductases from human, E. coli and Lactobacillus casei share complex folding mechanisms (48). Comparison of ubiquitin with the structurally related Ras binding domain shows that both proteins exhibit a similar unfolding mechanism despite little sequence homology (49). However, dimers of related proteins with high sequence and structural homology from mesophilic and thermophilic organisms have different unfolding mechanisms (50). The distinct equilibrium unfolding mechanisms reported for LC8 and Tctex-1 in the present study represent a case in which structural homologs with little sequence similarity have different unfolding pathways.
Functional Implications
Within the dynein complex, the two light chains Tctex-1 and LC8 form a tight sub-complex with intermediate chain IC74 (15). Each of the light chain homodimers binds to a contiguous segment of the disordered N-terminal domain of IC74, and thereby induces ordered structure in the intermediate chain (13). Previous results (30) show that in contrast to Tctex-1, LC8 is a moderately tight dimer that dissociates to a folded monomer which is expected to be populated under cellular conditions (31). The folded LC8 monomer does not bind IC74 (Norwood and Barbar unpublished data). The higher stability of the Tctex-1 dimer may promote its binding to two subunits of IC74 and the subsequent dimerization of IC74. Thus, Tctex-1 may serve as the primary anchor of the sub-complex of LC8, Tctex-1 and IC74.
Acknowledgments
This work was supported in part by an NSF CAREER grant MCB-0417181 to EB, GM 044757 from NIH to TSH, and by an undergraduate research fellowship to MT from the Honor’s Tutorial College at Ohio University.
We acknowledge the support of the nucleic acid and protein core and the mass spectrometry core in the OSU Environmental Health Sciences Center (NIH/NIEHS 00210), and in particular, Drs Sonia Anderson and Dean Malencik for help with analytical ultracentrifugation experiments, and Drs. Clare Woodward and Andy Karplus for critical reading of this manuscript. E.B. also wishes to thank Dr. Pat Callis of Montana State University for helpful discussion.
Abbreviations
- Tctex-1
12.5 kDa dynein light chain and also known as LC14
- LC8
10 kDa dynein light chain
- IC74
74 kDa dynein intermediate chain
- CD
circular dichroism
- HPLC
high performance liquid chromatography
- SEC
size exclusion chromatography
- DTT
dithiothreitol
- TCEP
tris(2-carboxyethyl) phosphine
- GdnCl
guanidine hydrochloride
- IPTG
isopropyl-β-D-thiogalatosidase
References
- 1.Allan VJ, Thompson HM, McNiven MA. Motoring around the Golgi. Nature Cell Biology. 2002;4:E236–E242. doi: 10.1038/ncb1002-e236. [DOI] [PubMed] [Google Scholar]
- 2.Harada A, Hirokawa N. The role of cytoplasmic dynein in the ER to Golgi transport. Molecular Biology of the Cell. 1999;10:2126. [Google Scholar]
- 3.Li M, McGrail M, Serr M, Hays TS. Drosophila cytoplasmic dynein, a microtubule motor that is asymmetrically localized in the oocyte. Journal of Cell Biology. 1994;126:1475–94. doi: 10.1083/jcb.126.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGrail M, Ludmann S, Hays TS. Analysis of Cytoplasmic Dynein Function in Drosophila Oogenesis. Molecular Biology of the Cell. 1995;6:886–886. doi: 10.1083/jcb.131.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vallee RB, Williams JC, Varma D, Barnhart LE. Dynein: An ancient motor protein involved in multiple modes of transport. Journal of Neurobiology. 2004;58:189–200. doi: 10.1002/neu.10314. [DOI] [PubMed] [Google Scholar]
- 6.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–80. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 7.Susalka SJ, Pfister KK. Cytoplasmic dynein subunit heterogeneity: implications for axonal transport. Journal of Neurocytology. 2000;29:819–829. doi: 10.1023/a:1010995408343. [DOI] [PubMed] [Google Scholar]
- 8.Tai AW, Chuang JZ, Sung CH. Cytoplasmic dynein regulation by subunit heterogeneity and its role in apical transport. Journal of Cell Biology. 2001;153:1499–1509. doi: 10.1083/jcb.153.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowman AB, Patel-King RS, Benashski SE, McCaffery JM, Goldstein LSB, King SM. Drosophila roadblock and Chlamydomonas LC7: A conserved family of dynein-associated proteins involved in axonal transport, flagellar motility, and mitosis. Journal of Cell Biology. 1999;146:165–179. [PMC free article] [PubMed] [Google Scholar]
- 10.Li MG, Serr M, Newman EA, Hays TS. The Drosophila tctex-1 light chain is dispensable for essential cytoplasmic dynein functions but is required during spermatid differentiation. Mol Biol Cell. 2004;15:3005–14. doi: 10.1091/mbc.E04-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tai AW, Chuang JZ, Bode C, Wolfrum U, Sung CH. Rhodopsin’s carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell. 1999;97:877–87. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- 12.Tai AW, Chuang JZ, Sung CH. Localization of Tctex-1, a cytoplasmic dynein light chain, to the golgi apparatus and evidence for dynein complex heterogeneity. Journal of Biological Chemistry. 1998;273:19639–19649. doi: 10.1074/jbc.273.31.19639. [DOI] [PubMed] [Google Scholar]
- 13.Makokha M, Hare M, Li M, Hays T, Barbar E. Interactions of cytoplasmic dynein light chains Tctex-1 and LC8 with the intermediate chain IC74. Biochemistry. 2002;41:4302–11. doi: 10.1021/bi011970h. [DOI] [PubMed] [Google Scholar]
- 14.Mok YK, Lo KWH, Zhang MJ. Structure of Tctex-1 and its interaction with cytoplasmic dynein intermediate chain. Journal of Biological Chemistry. 2001;276:14067–14074. doi: 10.1074/jbc.M011358200. [DOI] [PubMed] [Google Scholar]
- 15.King SM, Barbarese E, Dillman JF, Benashski SE, Do KT, Patel-King RS, Pfister KK. cytoplasmic dynein contains a family of differentially expressed light chains. Biochemistry. 1998;37:15033–15041. doi: 10.1021/bi9810813. [DOI] [PubMed] [Google Scholar]
- 16.Campbell KS, Cooper S, Dessing M, Yates S, Buder A. Interaction of p59(fyn) kinase with the dynein light chain, Tctex-1, and colocalization during cytokinesis. Journal of Immunology. 1998;161:1728–1737. [PubMed] [Google Scholar]
- 17.Yano H, Lee FS, Kong H, Chuang JZ, Arevalo JC, Perez P, Sung CH, Chao MV. Association of Trk neurotrophin receptors with components of the cytoplasmic dynein motor. Journal of Neuroscience. 2001;21:U7–U13. doi: 10.1523/JNEUROSCI.21-03-j0003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuang JZ, Yeh TY, Bollati F, Conde C, Canavosio F, Caceres A, Sung CH. The Dynein Light Chain Tctex-1 Has a Dynein-Independent Role in Actin Remodeling during Neurite Outgrowth. Dev Cell. 2005;9:75–86. doi: 10.1016/j.devcel.2005.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makokha M, Barbar E. Physical characterization of Tctex-1, a dynein light chain: Dimerization, folding and role in dynein assembly. Biophysical Journal. 2001;80:2530. [Google Scholar]
- 20.DiBella LM, Benashski SE, Tedford HW, Karrison A, Patel-King RS, King SM. The Tctex1/Tctex2 class of dynein light chains - Dimerization, differential expression, and interaction with the LC8 protein family. Journal of Biological Chemistry. 2001;276:14366–14373. doi: 10.1074/jbc.M011456200. [DOI] [PubMed] [Google Scholar]
- 21.Williams JC, Xie H, Hendrickson WA. Crystal structure of dynein light chain TCTEX-1. J Biol Chem. 2005;280:21981–6. doi: 10.1074/jbc.M414643200. [DOI] [PubMed] [Google Scholar]
- 22.Wu H, Maciejewski MW, Takebe S, King SM. Solution structure of the Tctex1 dimer reveals a mechanism for dynein-cargo interactions. Structure (Camb) 2005;13:213–23. doi: 10.1016/j.str.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 23.Liang J, Jaffrey SR, Guo W, Snyder SH, Clardy J. Structure of the PIN/LC8 dimer with a bound peptide. Nat Struct Biol. 1999;6:735–40. doi: 10.1038/11501. [DOI] [PubMed] [Google Scholar]
- 24.Mei G, Di Venere A, Rosato N, Finazzi-Agro A. The importance of being dimeric. Febs J. 2005;272:16–27. doi: 10.1111/j.1432-1033.2004.04407.x. [DOI] [PubMed] [Google Scholar]
- 25.Steif C, Weber P, Hinz HJ, Flossdorf J, Cesareni G, Kokkinidis M. Subunit interactions provide a significant contribution to the stability of the dimeric four-alpha-helical-bundle protein ROP. Biochemistry. 1993;32:3867–76. doi: 10.1021/bi00066a005. [DOI] [PubMed] [Google Scholar]
- 26.Bowie JU, Sauer RT. Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry. 1989;28:7139–43. doi: 10.1021/bi00444a001. [DOI] [PubMed] [Google Scholar]
- 27.Gittelman MS, Matthews CR. Folding and stability of trp aporepressor from Escherichia coli. Biochemistry. 1990;29:7011–20. doi: 10.1021/bi00482a009. [DOI] [PubMed] [Google Scholar]
- 28.Grant SK, Deckman IC, Culp JS, Minnich MD, Brooks IS, Hensley P, Debouck C, Meek TD. Use of protein unfolding studies to determine the conformational and dimeric stabilities of HIV-1 and SIV proteases. Biochemistry. 1992;31:9491–501. doi: 10.1021/bi00154a023. [DOI] [PubMed] [Google Scholar]
- 29.Topping TB, Hoch DA, Gloss LM. Folding mechanism of FIS, the intertwined, dimeric factor for inversion stimulation. J Mol Biol. 2004;335:1065–81. doi: 10.1016/j.jmb.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 30.Barbar E, Kleinman B, Imhoff D, Li MG, Hays TS, Hare M. Dimerization and folding of LC8, a highly conserved light chain of cytoplasmic dynein. Biochemistry. 2001;40:1596–1605. doi: 10.1021/bi002278+. [DOI] [PubMed] [Google Scholar]
- 31.Nyarko A, Cochrun L, Norwood S, Pursifull N, Voth A, Barbar E. Ionization of His 55 at the Dimer Interface of Dynein Light-Chain LC8 Is Coupled to Dimer Dissociation. Biochemistry. 2005;44:14248–55. doi: 10.1021/bi0512694. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Hare M, Hays TS, Barbar E. Dynein light chain LC8 promotes assembly of the coiled-coil domain of swallow protein. Biochemistry. 2004;43:4611–20. doi: 10.1021/bi036328x. [DOI] [PubMed] [Google Scholar]
- 33.Pace CN, Scholtz JM. In: Protein structure: a practical approach. Creighton TE, editor. Oxford University Press; Oxford: 1997. pp. 299–322. [Google Scholar]
- 34.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods in Enzymology. 1986;131:266–80. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 35.Neet KE, Timm DE. Conformational stability of dimeric proteins: Quantitative studies by equilibrium denaturation. Protein Sci. 1994;3:2167–2174. doi: 10.1002/pro.5560031202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramsay GD, Eftink MR. Analysis of multidimensional spectroscopic data to monitor unfolding of proteins. Methods Enzymol. 1994;240:615–45. doi: 10.1016/s0076-6879(94)40066-0. [DOI] [PubMed] [Google Scholar]
- 37.Runnels LW, Scarlata SF. Theory and application of fluorescence homotransfer to melittin oligomerization. Biophys J. 1995;69:1569–83. doi: 10.1016/S0006-3495(95)80030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imoto T, Forster LS, Rupley JA, Tanaka F. Fluorescence of lysozyme: emissions from tryptophan residues 62 and 108 and energy migration. Proc Natl Acad Sci U S A. 1972;69:1151–5. doi: 10.1073/pnas.69.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richards FM, Knowles JR. Glutaraldehyde as a protein cross-linkage reagent. J Mol Biol. 1968;37:231–3. doi: 10.1016/0022-2836(68)90086-7. [DOI] [PubMed] [Google Scholar]
- 40.Timm DE, Neet KE. Equilibrium Denaturation Studies of Mouse Beta-Nerve Growth- Factor. Protein Science. 1992;1:236–244. doi: 10.1002/pro.5560010205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–48. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karplus M, Petsko GA. Molecular dynamics simulations in biology. Nature. 1990;347:631–9. doi: 10.1038/347631a0. [DOI] [PubMed] [Google Scholar]
- 43.Eftink MR. The use of fluorescence methods to monitor unfolding transitions in proteins. Biophys J. 1994;66:482–501. doi: 10.1016/s0006-3495(94)80799-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, Barkley MD. Toward understanding tryptophan fluorescence in proteins. Biochemistry. 1998;37:9976–82. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- 45.Makokha M, Huang YJ, Montelione G, Edison AS, Barbar E. The solution structure of the pH-induced monomer of dynein light-chain LC8 from Drosophila. Protein Sci. 2004;13:727–34. doi: 10.1110/ps.03462204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mann CJ, Royer CA, Matthews CR. Tryptophan replacements in the trp aporepressor from Escherichia coli: probing the equilibrium and kinetic folding models. Protein Sci. 1993;2:1853–61. doi: 10.1002/pro.5560021107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alm E, Baker D. Matching theory and experiment in protein folding. Curr Opin Struct Biol. 1999;9:189–96. doi: 10.1016/S0959-440X(99)80027-X. [DOI] [PubMed] [Google Scholar]
- 48.Wallace LA, Matthews CR. Sequential vs. parallel protein-folding mechanisms: experimental tests for complex folding reactions. Biophys Chem. 2002;101–102:113–31. doi: 10.1016/s0301-4622(02)00155-2. [DOI] [PubMed] [Google Scholar]
- 49.Vallee-Belisle A, Turcotte JF, Michnick SW. raf RBD and ubiquitin proteins share similar folds, folding rates and mechanisms despite having unrelated amino acid sequences. Biochemistry. 2004;43:8447–58. doi: 10.1021/bi0359426. [DOI] [PubMed] [Google Scholar]
- 50.Bhatt AN, Prakash K, Subramanya HS, Bhakuni V. Different unfolding pathways for mesophilic and thermophilic homologues of serine hydroxymethyltransferase. Biochemistry. 2002;41:12115–23. doi: 10.1021/bi020356i. [DOI] [PubMed] [Google Scholar]
- 51.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]