Abstract
It is generally believed that proteins of the troponin complex are not expressed in smooth muscle. We have directly assayed for expression of troponin transcripts in mouse vascular smooth muscle and found that troponin sequences normally associated with fast twitch skeletal muscle (fTnT, fTnI, fTnC) were present at significant levels in the thoracic aorta. In situ hybridization experiments demonstrated that fTnT, fTnI and fTnC transcripts were expressed in the smooth muscle layer of mouse blood vessels of all sizes. Protein blot analysis using rat tissue showed that at least two members of the troponin complex, Troponin T and Troponin I, were translated in vascular smooth muscle of the aorta. Finally, immuno-fluorescence microscopy of rat aortic smooth muscle revealed that TnT and TnI are localized in a unique pattern, coincident with the distribution of tropomyosin. It seems likely therefore, that a complete troponin complex is expressed in vascular smooth muscle and is associated with the contractile machinery of the cell. These observations raise the possibility that troponins play a role in regulation of smooth muscle function.
Keywords: troponin, vascular smooth muscle, intestinal smooth muscle
INTRODUCTION
The mechanism of activation of smooth muscle contraction is fundamentally different from that of skeletal and cardiac (striated) muscle, although both are triggered by an increase in cytosolic free calcium concentration,[Ca2+]. In striated muscles, [Ca2+] binds to troponin C, leading to conformational changes in thin filament components and allowing actin-myosin interactions and cross-bridge cycling. In contrast, regulation of smooth muscle contraction is primarily through calcium-calmodulin-dependent phosphorylation of the 20 kDa regulatory light chain of myosin (MLC). Stimulation of smooth muscle cells, by either receptor binding or membrane depolarization, leads to elevated [Ca2+]. This increase activates the calmodulin dependent MLC kinase (MLCK), which catalyzes MLC phosphorylation, driving crossbridge formation. Relaxation of smooth muscle cells is due to dephosphorylation of MLC by the MLC phosphatase following the return of [Ca2+] to basal levels and subsequent release of the actin filaments (reviewed in Ito and Hartshorne, 1990; Arner and Pfitzer, 1999). Although it is clear that MLC phosphorylation is the primary regulatory pathway for the initiation of smooth muscle contraction, numerous biochemical and physiological studies over the last two decades indicate that additional regulatory mechanisms are likely to exist. For example, during the well-established latch state, high levels of force are maintained by slowly cycling dephosphorylated crossbridges as well as low levels of [Ca2+]. In fact, cross-bridge cycling rates can differ without detectable alterations in MLC phosphorylation (Siegman et al., 1984; Rembold and Murphy, 1986; Moreland et al., 1987; Hai and Murphy, 1988). These properties cannot be adequately explained by MLC phosphorylation acting as a simple switch. In addition, mutations in the heavy chain of myosin that are spatially distant from the MLC phosphorylation site, lead to activation of the MgATPase activity in the absence of MLC phosphorylation, and phorbol dibutyrate-induced contractions of smooth muscle occur in the absence of an increase in either [Ca2+] or serine-MLC phosphorylation. Once again, these studies are not consistent with the simple switch hypothesis (Fulginiti et al, 1993; Walsh et al., 1996; Rovner et al., 1995; Throckmorton et al, 1998). Thus, additional mechanisms appear to be required to account for smooth muscle contractile force generation and maintenance. Several pathways have been proposed to account for the apparent disparities between calcium levels, MLC phosphorylation and smooth muscle contractile force. In particular, a thin filament-based regulatory system for regulating smooth muscle actomyosin interactions as well as contractile force and maintenance has been suggested, principally involving the proteins caldesmon and calponin. However, no consensus has been reached on the specific roles of these proteins, that can adequately account for the contractile behavior of smooth muscle (reviewed in Ebashi and Kuwayama, 1994; Sellers, 1999. Facemire et al., 2000; Morgan and Gangopadhyay, 2001; Hai and Kim, 2005; Ogut et al., 2008).
The Troponin (Tn) complex is the sarcomeric [Ca2+] sensor for activation of striated (skeletal and cardiac) muscle contraction. The Tns are a cooperative complex of three proteins comprised of TnT (Tropomyosin interacting subunit), Troponin C ([Ca2+] interacting subunit) and Troponin I (Inhibitory subunit) that function with tropomyosin (Tm) in modulating the interaction of myosin-thick filaments and actin-thin filaments during force generation. Although the issue has not been definitively studied, the absence of a troponin complex from smooth muscle tissues is generally assumed and is widely stated in the primary literature and in reviews of smooth muscle structure and function (Johansson, 1987; Walsh, 1994; Jiang and Stephens, 1994; Owens, 1995; Endo et al., 1997; Morgan and Gangopadhyay, 2001; Stephens, 2001; Darland and D’Amore, 2001; Watanabe et al., 2003). In this report however, we provide several distinct lines of evidence showing that all Tn genes required to assemble a functional fast-twitch troponin complex are expressed in vascular tissue and immunofluorescence microscopy showed that Tn proteins are associated with tropomyosin-actin filaments. The existence of a complete troponin complex in vascular smooth muscle may suggest a role in regulation of smooth muscle function.
MATERIALS AND METHODS
RT-PCR analysis of troponin gene expression in vascular smooth muscle
For transcript analysis, the heart, thoracic aorta (dissected from the arch to diaphragm) and femoral muscle were dissected from adult mice (P6H/+ X P6H/+), taking great care to prevent contamination with adjacent tissues. Tissues were snap-frozen in liquid nitrogen, ground using a mortar and pestle, and RNA was extracted using Trizol reagent (Invitrogen Life Technologies) according to the manufacturers recommendations. cDNA was prepared using standard methods and primer pairs specific for each of the known mouse troponin transcripts were used in RT-PCR experiments, together with control sequences. Primers were chosen to avoid regions of the transcripts subject to alternative splicing. All RT-PCR products were radioactively labeled during synthesis, separated on denaturing polyacrylamide gels and detected using autoradiography.
Forward (F) and reverse (R) primers used for RT-PCR analysis of mouse transcripts: GAPDH, F- GGTGTGAACGGATTTGG, R- CATGGACTGTGGTCATG; MyoD, F- CGACTGCCTGTCCAGCATAGT, R-CCTGTTACACCTGAGACCT; SM22, F-AGCTACTCTCCTTCCAG, R- TGACACCTCAGAGCTGTC; α-tropomyosin, F- TCTCTGAACAGACGCATCC, R-CAGAGAGAACCTTGATCTCC; fTnT, F-GCAAGCCTCTGAACAT, R- CCAGCATCTCCACTCT; fTnC, F- ACCATGACGGACCAACAG, R- CTCTTCTGTCACATGCTCC; fTnI, F- GCTTGAGATCTCAGGATG, R- TCCATGCCAGACTTCTCC; sTnT, F- GTTCTGTCCAACATGG, R- ACACAGCAGGTCATGTC; sTnI, F- AGAGCTCCATGCCAAGG, R- AGAAAGATAGGTGAGTGG; cTnT, F- CCACCTGAATGAAGACCAAC, R- ACTGGCTTCTAGCTAAGC; cTnC, F- CACAGTGGACTTCGATGAG, R- CTCAGAGTCTAGAGAGC; cTnI, F- TGATGAAAGCAGCGATGC, R- TTCCATGCCACTCAGTG.
In situ hybridization of sectioned muscle tissues
Probes for all in situ hybridizations were generated from linearized plasmid templates for IMAGE mouse EST clones: SM22, (IMAGE: 5056116); fTnT, (IMAGE: 1092865); fTnI, (IMAGE: 521075); and fTnC, (IMAGE: 963956). T3 or T7 RNA polymerase transcriptional synthesis (Megascript, Ambion) incorporated digoxigenin-UTP as a label for detection (Roche). Mouse tissues were fixed overnight in 3.7% Formaldehyde in PBS, cleared in xylene and embedded in Paraplast. Ten micrometer sections were mounted on Superfrost plus charged slides (VWR), dewaxed in xylene and rehydrated from ethanol into PBS. In situ hybridization to sections was carried out using standard methods adapted from Schaeren-Wiemers and Gerfin-Moser (1993). Briefly, sections were digested in proteinase K (Ambion), refixed in 4% Paraformaldehyde in PBS, hybridized to probe at 65°C, incubated with alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche) and developed in NBT + BCIP (Roche) in the presence of levamisole (Sigma). Slides were dehydrated, mounted with Cytoseal (VWR) and photographed.
Protein blot analysis of TnT expression in vascular smooth muscle
Tissues were dissected from Sprague-Dawley rats, lysed with 2× SDS Sample Buffer (0.25M Tris-HCl pH 6.8, 10% SDS, 25% glycerol), boiled and centrifuged. Protein concentrations were measured using the BCA assay procedure (Pierce Chemical Co., Rockford, IL), with bovine serum albumin as a standard. Appropriate amounts of protein were separated by 10% SDS-PAGE and electrophoretically transferred to nitrocellulose. Antigens were detected by primary antibodies (1:5,000 and 1:1,000 dilutions of TnT and TnI respectively) followed by peroxidase-conjugated anti-mouse IgG (1:15,000 dilution). Protein bands were identified using chemiluminescent substrate (NEN, Boston, MA) exposed on X-OMAT AR film (Kodak, Rochester, NY).
Immunoprecipitation and LC-MS/MS Analysis
Snap-frozen rat aorta was ground in liquid nitrogen into a fine powder using a mortar and pestle and immunoprecipitated using a protocol modified from Fowler and Adam (1992). In brief, the aorta powder was solubilized in SDS-immunoprecipitation buffer (300 mM NaCl, 10 mM NaHPO4 pH 7.5, 5 mM EGTA, 0.2 mM EDTA, 1 mM MgCl2, 0.4% SDS, 0.02% azide plus a protease inhibitor cocktail), and boiled for 2 minutes. Next, the samples were sonicated, Triton X-100 was added (final concentration 2%), and the samples were spun in an ultracentrifuge for 30 minutes at 40,000 rpm. The supernatant was transferred to GammaBind Plus Sepharose beads (Amersham Biosciences) coated with 25 μg monoclonal anti-TnT (Sigma JLT-12) antibodies and incubated end-over-end overnight at 4°C. After subsequent washes, bound protein (and anti-troponin antibodies) was eluted from the beads in SDS sample buffer and fractionated on a 10% polyacrylamide gel. The gel was stained in silver stain and a single band, migrating at a position consistent with TnT (arrow, Figure 3E) was excised and subjected to LC-MS/MS Analysis by the Arizona Mass Spectrometry Consortium (University of Arizona).
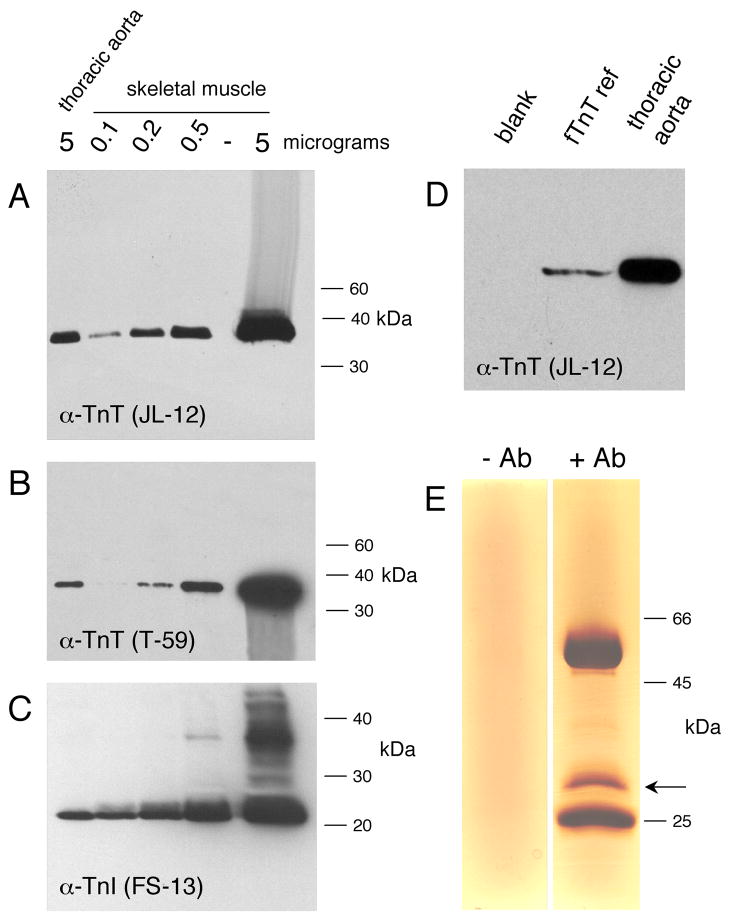
Fig. 3.
Protein blot analysis shows troponin T and I expression in aortic smooth muscle. (A). Western blot of thoracic aorta protein extract (left lane) and quadricep skeletal muscle dilution series, probed with anti-TnT (JLT-12) antibody. The amount of protein extract loaded in each lane is indicated at the top of the figure. Position of molecular weight markers is indicated at the right of the panel. (B) Identical to (A) except probed with anti-TnT (T-59) antibody. (C). Western blot of dorsal aorta protein sample (5 micrograms) showing co-migration with fTnT reference protein translated in Xenopus embryo. Lane labeled blank is embryo extract without added TnT mRNA. (D). Identical to (A) except probed with anti-TnI (FS-13) antibody. Streaks visible above and below the darkest bands were caused by movement of the film between chemiluminescent exposures.
Immunofluoresence Microscopy and Antibodies
Frozen sections of rat aorta were fixed in 3% paraformaldehyde, permeabilized in 0.2% Triton-X 100, and incubated in 2% BSA/1% normal donkey serum to minimize non-specific binding. Sections were then incubated with anti-TnT monoclonal antibodies at a 1:40 dilution (Clone JLT-12, Sigma), anti-TnT monoclonal antibodies (clone T-59; a generous gift from Dr. Stefano Schiaffino, University of Padova, Padua, Italy – Saggin et al., 1990) or anti-TnI monoclonal antibodies (10μg/ml, Spectrum, Clone FI-23) followed by AlexaFluor 594 conjugated-goat anti-mouse IgG antibodies (1:800, Molecular Probes, Eugene, OR). Sections were co-stained using rabbit polyclonal anti-chicken gizzard tropomyosin antibodies (1:40, T3651 Sigma) followed by Cy2 conjugated-goat anti-rabbit IgG antibodies (1:600, Jackson ImmunoResearch Labs). In each experiment, secondary only controls were performed: negligible background staining was observed (data not shown). Stained sections were analyzed on an Olympus IX70 microscope using a 100× (1.35 NA) objective. Micrographs were recorded as digital images (with Z-series containing 0.15 μ;m sections) using a Series 300 CCD camera (Photometrics, Tucson, AZ) and deconvolved using DeltaVision software (Applied Precision, LLC, Issaquah, WA).
RESULTS
Sequences encoding the fast twitch skeletal muscle troponin complex are expressed in the thoracic aorta of the adult mouse
Examination of mouse and human EST sequences in the GenBank database revealed Tn transcripts in numerous highly vascularized tissues, including mammary gland, kidney and lung, which do not contain striated muscle. To specifically assay for Tn transcripts in vascular smooth muscle, RNA was prepared from carefully dissected skeletal, cardiac and vascular smooth muscle tissues from adult mice and RT-PCR methods were used to assay for the presence of all eight different troponin transcripts. The results are presented in Figure 1. As initial controls, we used RT-PCR detection of skeletal muscle specific myoD, and smooth muscle specific SM22 transcripts, to demonstrate that there was no detectable cross contamination of muscle types in the tissue samples. We then proceeded to assay for expression of Tn genes in the different muscle tissues. As expected, transcripts encoding the cardiac muscle troponin genes, cTnI and cTnT were heart specific, while cTnC transcripts were present in both cardiac and skeletal muscle. Significantly, the RT-PCR results clearly show that transcripts encoding the fast twitch Tn complex, fTnI, fTnC and fTnT are expressed at significant levels in mouse thoracic aorta tissue. In addition, expression of slow-twitch TnT (sTnT) was detected in the aorta sample. The identity of each of these PCR products was confirmed by DNA sequencing (data not shown). GAPDH was used as a loading control to indicate equal amounts of total template in the different samples. In order to provide an estimate of the relative levels of transcripts encoding troponin interacting proteins in the different tissue isolates, we also detected tropomyosin sequences in the skeletal, heart and smooth muscle samples, using primers designed to recognize all tropomyosin isoforms (Fig. 1, panel labeled Tm).
Fig. 1.
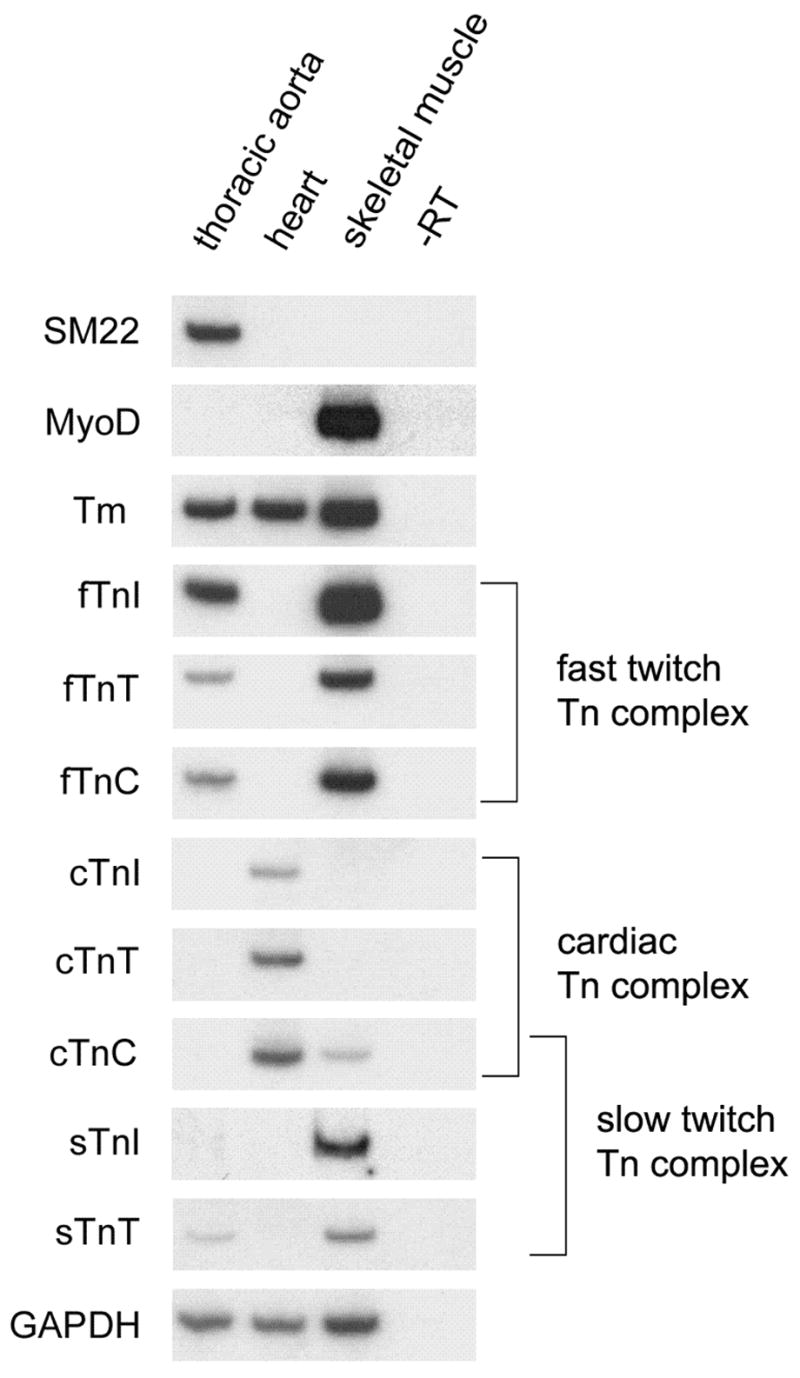
Troponin expression in mouse vascular smooth muscle. RNA preparations from thoracic aorta, heart and skeletal muscle (quadriceps) were assayed using RT-PCR for the presence of muscle markers and different troponin transcripts. Each panel is an autoradiograph from an individual polyacrylamide gel. The ubiquitously expressed GAPDH sequence acts as a loading control. Lane marked–RT is the minus reverse transcriptase control reaction.
In situ hybridization analysis of troponin expression in adult blood vessels
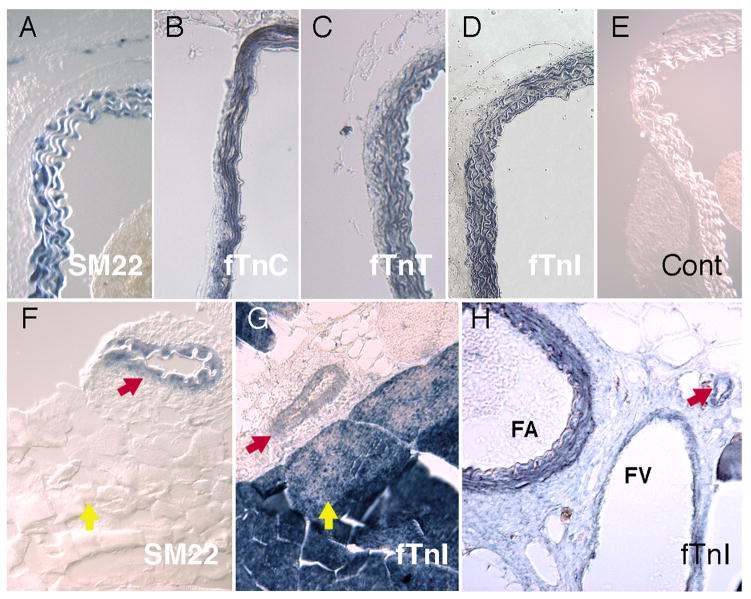
To further characterize expression of troponin transcripts in vascular tissues, we carried out in situ hybridization studies using adult mouse tissues. When assayed with a probe directed against fTnC, strong expression was detected in the smooth muscle layer of the aorta (Fig. 2B). Identical results were obtained using probes directed against the fTnT and fTnI sequences (Fig. 2C, D). In control experiments, the definitive smooth muscle marker, SM22 showed the same expression pattern (Fig. 2A), while no staining was observed in negative control experiments (Fig. 2E). In situ hybridization analysis also facilitated detection of Tn sequences within the smooth muscle layer of smaller arteries and veins that could not be easily dissected for RT-PCR analysis. For example, in situ hybridization of cross sections through the quadricep muscle of the thigh, revealed expression of fTnI mRNA in the smooth muscle layer of a small vessel located within the skeletal muscle tissue (Fig. 2G). In this panel fTnI transcripts are also detected, at significantly higher levels, within adjacent skeletal muscle tissues. This is to be expected, since skeletal muscles of the rodent predominantly express fast twitch troponin sequences (Jin et al., 1998). Note that the skeletal muscle tissues are negative for the smooth muscle marker, SM22 (Fig. 2F). In Fig. 2H, fTnI expression is visible in the thick smooth muscle of the femoral artery and at lower levels in the thinner smooth muscle layer of the adjacent femoral vein. Overall, these results indicate that troponin expression is not limited to major arteries and veins but appears to be a general feature of vascular smooth muscle tissue.
Fig. 2.
In situ hybridization analysis of troponin gene expression in vascular smooth muscle cells. Transverse sections of mouse tissues: dorsal aorta, (A-E); quadricep muscle, (F, G); and femoral artery and vein, (H). (A) In the thoracic aorta, the vascular smooth muscle cells (VMSC) of the vessel wall are specifically marked by SM22. In separate thoracic aorta sections, (B) fTnC, (C) fTnT and (D) fTnI stain VSMC however, staining is not observed in the collagen bundles within the vessel wall or the connective tissue of the adventica. (E) Control sections of thoracic aorta are not stained with either the alkaline phosophatase-conjugated antibody or endogenous sources. (F) SM22 transcripts mark VSMCs of a smaller vessel (red arrow) that penetrates the skeletal muscle of the quadricep, but does not stain the skeletal muscle fibers (yellow arrow). (G) In similar sections of quadriceps muscle, fTnI marks a small vessel (red arrow) and skeletal muscle fibers (yellow arrow). (H) fTnI is also expressed in smooth muscle of the femoral artery (FA) and femoral vein (FV) and a nearby small vessel (red arrow).
Troponin T and I proteins are expressed in thoracic aorta vascular smooth muscle
The results presented above indicate the presence of troponin mRNA in smooth muscle of the tunica media of blood vessels. However, it is possible that these troponin mRNAs are translationally regulated and that no troponin proteins are synthesized. Experiments to demonstrate the presence of the fTnI, fTnC and fTnT proteins in smooth muscle were complicated by the lack of commercial antibodies specific for each of the different troponin isoforms. However, several monoclonal antibodies are available that are specific to troponin isoforms of the same general class (i.e. that will detect all of the troponin Ts). Western blotting experiments were performed to determine whether troponin protein is indeed expressed in vascular smooth muscle (Fig. 3). The blots included a dilution series of skeletal muscle samples, so that an estimate of the relative abundance of troponin in skeletal muscle and aorta could be obtained. Rat tissues were used in these experiments to facilitate dissection of the larger amounts of thoracic aorta tissue required for protein detection. As shown in Fig, 3A, probing of protein blots with an anti-TnT monoclonal antibody (clone JLT-12) detected TnT protein in skeletal muscle, as expected, and also in thoracic aorta vascular smooth muscle. Densitometry and quantitation of the dilution series indicated that, as a proportion of total protein, the aorta contains approximately 5% of the amount of total TnT present in skeletal muscle. The migration of the protein band at 38 kDa is identical to that previously reported for the fTnT isoforms (Siedner et al., 2003). As shown in Fig. 3B, identical results were obtained when protein blots were probed using a second anti-TnT monoclonal antibody (clone T-59) (Saggin et al., 1990). The fact that the same size band is detected using two different monoclonal antibodies, each recognizing distinct epitopes, strongly suggests that the protein detected is TnT. It has been reported that anti-TnT antibodies may cross react with caldesmon (Shirinsky et al., 1989) but no bands were detected at the expected position for this protein (60.5 kDa). To establish the precise position of migration of bona fide fTnT, we prepared a marker control by translating mouse fTnT (B3e16 isoform), in the Xenopus embryo and fractionated this protein adjacent to the thoracic aorta sample (Fig. 3D). Note that the fTnT marker precisely co-migrated with the protein detected by the JLT-12 antibody in thoracic aorta, increasing confidence that the antibodies specifically detected TnT. Additional Western blot experiments were performed using an anti-TnI monoclonal antibody (clone FI-23). Once again, the antibody detects a single protein band in the aorta sample that co-migrates with the troponin I band in the skeletal muscle sample (Fig. 3C). The migration of this band relative to MW standards is identical to that previously reported for fTnI (Siedner et al., 2003). To the best of our knowledge, no commercial antibodies specific for TnC are available and so we were unable to evaluate the presence of TnC protein in aorta smooth muscle.
Previous reports have indicated that anti-TnT antibodies may also cross react with caldesmon and glyceraldehyde–3-phosphate dehydrogenase (Sanders et al., 1987; Shirinsky et al., 1989). To confirm the identity of the proteins detected by the anti-troponin T antibody we carried out immunoprecipitation isolation of protein from rat thoracic aorta followed by mass spectroscopic analysis. As shown in Fig 3E, immunoprecipitation using JLT-12 antibody resulted in isolation of a single prominent protein band. Analysis of this material by mass spectroscopy provided the sequences of three independent peptides that corresponded exactly to troponin T sequences. No non-troponin sequences were detected. We conclude that troponin T protein is expressed in smooth muscle of the thoracic aorta.
Troponin genes are expressed in mouse intestinal smooth muscle
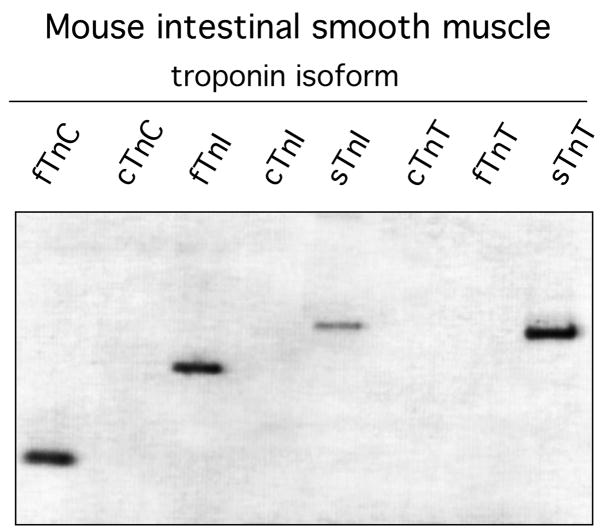
Are troponin sequences expressed in other smooth muscle tissue? Using the same PCR primers employed to analyze vascular smooth muscle, we characterized troponin expression in mouse intestine (Fig. 4). The results show that fTnC, fTnI, sTnT and sTnI are expressed in gut muscle. The first three sequences are equivalent to the troponin isoforms detected in vascular smooth muscle. However, gut muscle expresses sTnI but not fTnT detected in thoracic aorta. While confirming the presence of troponin sequences in other smooth muscle tissues, these results raise the possibility that different tissues express different complements of troponin isoforms.
Fig. 4.
Detection of troponin transcripts in mouse intestinal smooth muscle. PCR primers were the same as used for detection of troponin in vascular smooth muscle. After 40 cycles of amplification, products were fractionated on a 7% acrylamide gel.
In thoracic aorta smooth muscle, TnT and TnI are localized in a pattern consistent with association with actin filaments
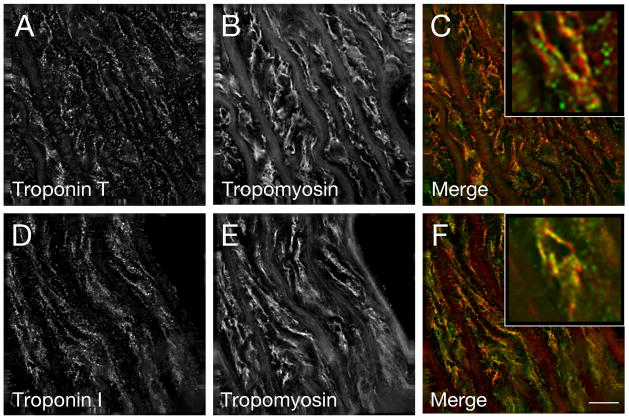
To determine the intracellular distribution of the Tn proteins in rat aorta, frozen sections were co-stained with antibodies specific for troponin I or T and smooth muscle tropomyosin. Deconvolution immunofluorescence analysis of the stained tissue revealed that all anti-Tn antibodies used (two different anti-TnT antibodies and one anti-TnI antibody) localized in a nonrandom pattern that co-localized with a portion of filaments that stain for tropomyosin (data shown for anti-TnT JLT-12 antibodies and anti-TnI FI-23 antibodies; Fig. 5A–C and D–F respectively). The observation that not all tropomyosin-stained filaments appear to contain troponin is consistent with the apparent lower levels of troponin detected in smooth muscle in comparison to skeletal muscle as predicted from Western blot analysis described above.
Fig. 5.
Immunofluorescence microscopy reveals that TnI and TnT colocalize with tropomyosin in the tunica media of the rat thoracic aorta. Frozen sections of rat aorta were immunostained for TnT (A) or TnI (D) and costained for smooth muscle tropomyosin (B, E). Merged images demonstrate co-localization of TnT (C, green) or TnI (F, green) with tropomyosin (C and F, red). Insets show a magnified view. Bar=10μm.
DISCUSSION
Troponins are expressed in adult vascular smooth muscle
Using a combination of RT-PCR and in situ hybridization methods, we examined vascular smooth muscle tissue for the presence of specific troponin transcripts. Of the eight known troponin genes, we found that mRNAs encoding four different troponin proteins, fTnI, fTnC, fTnT and sTnT, are present in vascular tissue. We note that, unlike protein detection methods, the use of oligonucleotide primers allows gene detection by PCR to be absolutely specific, since even closely related proteins are encoded by variable nucleotide sequences. The presence of isoforms of each of the TnI, C and T subclasses raises the possibility that a complete Tn complex is expressed in vascular smooth muscle. In situ hybridization confirms that Tn transcripts are expressed in the smooth muscle layer of the aorta, in a pattern indistinguishable from that of SM22, a definitive smooth muscle marker (Fig. 2). The in situ results also indicate that expression of Tn transcripts is not limited to the larger vessels but can also be observed in the smooth muscle walls of arterioles and veins (Fig. 2G, H). In protein blot experiments, two independent anti-TnT monoclonal antibodies each recognize a single band in vascular smooth muscle tissue. The predicted size of the protein is appropriate for fTnT and is indistinguishable from in vitro translated fTnT (Fig. 3C). Similarly, anti-TnI monoclonal antibody, FS-13, detects a clear band at the correct size in rat aortic smooth muscle samples. Most convincingly, immunoprecipitation followed by mass spectroscopy demonstrated the presence of TnT in the rat thoracic aorta tissue. Overall, these results strongly suggest that Tn genes are transcribed and Tn proteins are translated in vascular smooth muscle tissue of the adult rodent. To determine whether troponin sequences are expressed in other smooth muscle tissue we analyzed mouse intestinal smooth muscle (Fig. 4). Although preliminary, these results demonstrated that fTnC, fTnI, sTnI and sTnT are present in gut muscle.
Additional experiments will be required to determine the diversity of troponin expression in different smooth muscle tissues. Finally, immunofluorescent deconvolution studies of dorsal aorta tissue showed significant co-localization of TnT and TnI with tropomyosin (Fig. 5). This observation indicates that the troponin proteins are associated with actin filaments within smooth muscle tissues and places them in a position to play a functional role in regulation of smooth muscle contraction. Our experimental results are limited to tissues isolated from rat and mouse, however, we note that many different ESTs corresponding to fast twitch Tn sequences are present in human cDNA libraries prepared from tissues that do not contain striated muscle, including placenta, lung, prostate, testes and liver. Taking fTnI as an example, approximately 20% of all human EST entries for fTnI are derived from non-striated muscle tissues.
Previous studies suggested that troponin sequences are expressed in smooth muscle
Several previous studies have reported that proteins with properties similar to Tn are present in non-striated muscle tissues. For example, a protein reacting with an anti-TnT monoclonal antibody was detected in chicken gizzard tissue by Western blot analysis and immuno-fluorescence staining (Lim et al., 1983; Lim et al., 1985). In another study, monoclonal antibodies directed against cardiac troponin T detected Tn-like proteins in bovine aortic smooth muscle (Zanellato et al., 1991). This same research showed that two different antibodies against cardiac TnI also detected bands of the predicted size for TnI in protein blots of bovine aorta. In order to determine which troponins are expressed in bovine aorta, we used RT-PCR to detect troponin transcripts. This is only possible for six troponins, since the nucleotide sequences for sTnI and fTnI are not present in current databases. Our analysis showed that bovine aorta contains transcripts for cTnC, fTnC, sTnT and fTnT (Supplemental Fig. S1). Despite the fact that Zanellato et al (1991) used antibodies specific for cTnT and cTnI, we did not detect transcripts encoding these sequences in bovine thoracic aorta. At present, the reason for the discrepancy in results between the protein and nucleic acid methods is unclear. More recently, in situ hybridization experiments detected expression of cTnT transcripts in bladder smooth muscle in the mouse embryo (Wang et al., 2001) and 2D-gels showed TnT protein in renal arterioles (Pinet et al., 2004). None of the previous studies detected sequences consistent with a complete troponin complex in the respective smooth muscle tissues.
Does the troponin complex function in smooth muscle contractility?
Our results provide evidence for the presence of fast twitch troponin complex transcripts, and the corresponding proteins, in vascular smooth muscle. At present there is no evidence to suggest that such a troponin complex actually functions in regulating maintenance of smooth muscle contraction. We considered the possibility that the troponin complex might contribute to the latch mechanism that maintains contractility during conditions of low [Ca2+] and low phosphorylation of regulatory myosin light chain (Hai and Kim, 2005; Ogut et al., 2008). However, in skeletal muscle, the troponin complex blocks myosin in a weakly bound state (Jin et al., 2008; Kad et al., 2005) and it therefore seems unlikely that troponins would meet the requirements for the latch mechanism in smooth muscle .
The influence of Tns on striated muscle contractility has received increasing attention due to substantial clinical evidence correlating changes in Tn levels and Tn mutations with alternations in cardiac function and more recently in striated muscle myopathies and muscular dystrophies (Hernandez et al, 2001; Tubridy et al., 2001; Clark et al., 2002; Clarkson et al., 2004). Despite the absence of mechanism, the findings in this report suggest that the contributions of vascular smooth muscle troponins might also be taken into account when considering the etiology of skeletal myopathies. Further studies will be required to directly test whether the troponin complex does indeed play a role in regulating vascular smooth muscle contractile activity and to determine its mode of action.
Acknowledgments
The authors would like to thank Ron Lynch, David Hartshorne and James Sellers for insightful comments. The Mass Spectroscopy analysis was carried out by George Tsaprailis, Proteomics Core Director at the University of Arizona. PAK is the Allan C. Hudson and Helen Lovaas Endowed Professor of the Sarver Heart Center at the University of Arizona College of Medicine. This work was supported by the Sarver Heart Center and by the NHLBI of the NIH, grants HL63926 and CA56666 to RLH, HL57461 and HL03985 to CCG and HL74184 to PAK.
References
- Arner A, Pfitzer G. Regulation of cross-bridge cycling by Ca2+ in smooth muscle. Rev Physiol Biochem Pharmacol. 1999;134:63–146. doi: 10.1007/3-540-64753-8_3. [DOI] [PubMed] [Google Scholar]
- Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Striated muscle cytoarchitecture: an intricate web of form and function. Annu Rev Cell Dev Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- Clarkson E, Costa CF, Machesky LM. Congenital myopathies: diseases of the actin cytoskeleton. J Pathol. 2004;204:407–417. doi: 10.1002/path.1648. [DOI] [PubMed] [Google Scholar]
- Darland DC, D’Amore PA. Cell-cell interactions in vascular development. Curr Top Dev Biol. 2001;52:107–149. doi: 10.1016/s0070-2153(01)52010-4. [DOI] [PubMed] [Google Scholar]
- Ebashi S, Kuwayama H. Is phosphorylation the main physiological action of myosin light chain kinase? Can J Physiol Pharmacol. 1994;2:1377–1379. doi: 10.1139/y94-198. [DOI] [PubMed] [Google Scholar]
- Endo T, Matsumoto K, Hama T, Ohtsuka Y, Datsur G, Obinata T. Temporal and spatial expression of distinct troponin T genes in the embryonic/larval tail striated muscle and adult body wall smooth muscle of Ascidian. Cell Structure Function. 1997;22:197–203. doi: 10.1247/csf.22.197. [DOI] [PubMed] [Google Scholar]
- Facemire C, Brozovich FV, Jin JP. The maximal velocity of vascular smooth muscle shortening is independent of the expression of calponin. J Muscle Res Cell Motil. 2000;21:367–373. doi: 10.1023/a:1005680614296. [DOI] [PubMed] [Google Scholar]
- Fowler VM, Adam EJ. Spectrin redistributes to the cytosol and is phosphorylated during mitosis in cultured cells. J Cell Biol. 1992;119:1559–1572. doi: 10.1083/jcb.119.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulginiti J, Singer HA, Moreland RS. Phorbol ester-induced contractions of swine carotid artery are supported by slowly cycling crossbridges which are not dependent on calcium or myosin light chain phosphorylation. J Vasc Res. 1993;30:315–322. doi: 10.1159/000159012. [DOI] [PubMed] [Google Scholar]
- Hai CM, Murphy RA. Cross-bridge phosphorylation and regulation of latch state in smooth muscle. Am J Physiol. 1988;254:C99–C106. doi: 10.1152/ajpcell.1988.254.1.C99. [DOI] [PubMed] [Google Scholar]
- Hai CM, Kim HR. An expanded latch-bridge model of protein kinase C-mediated smooth muscle contraction. J App Physiol. 2005;98:1356–1365. doi: 10.1152/japplphysiol.00834.2004. [DOI] [PubMed] [Google Scholar]
- Hernandez OM, Housmans PR, Potter JD. Pathophysiology of cardiac muscle contraction and relaxation as a result of alterations in thin filament regulation. J Appl Physiol. 2001;90:1125–1136. doi: 10.1152/jappl.2001.90.3.1125. [DOI] [PubMed] [Google Scholar]
- Ito M, Hartshorne DJ. Phosphorylation of myosin as a regulatory mechanism in smooth muscle. Prog Clin Biol Res. 1990;327:57–72. [PubMed] [Google Scholar]
- Jiang H, Stephens NL. Calcium and smooth muscle contraction. Mol Cell Biochem. 1994;135:1–9. doi: 10.1007/BF00925956. [DOI] [PubMed] [Google Scholar]
- Jin JP, Chen A, Huan Q-Q. Three alternatively spliced mouse slow skeletal muscle troponin T isoforms: Conserved primary structure and regulated expression during postnatal development. Gene. 1998;214:121–129. doi: 10.1016/s0378-1119(98)00214-5. [DOI] [PubMed] [Google Scholar]
- Jin JP, Zhang Z, Bautista JA. Isofrom diversity, regulation and functional adaptation of troponin and calponin. Crit Rev Eukaryot Gene Expra. 2008;18:93–124. doi: 10.1615/critreveukargeneexpr.v18.i2.10. [DOI] [PubMed] [Google Scholar]
- Johansson B. Calcium and regulation of contraction: A short review. J Cardiovasc Pharmacol. 1987;10:S9–S13. doi: 10.1097/00005344-198710001-00003. [DOI] [PubMed] [Google Scholar]
- Kad NM, Kim S, Warshaw DM, VanBuren P, Baker JE. Single-myosin crossbridge interactions with actin filaments regulated by troponin-tropomyosin. Proc Natl Acad Sci USA. 2005;102:16990–16995. doi: 10.1073/pnas.0506326102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SS, Zeng-Hong T, Lemanski LF. Anti-troponin T monoclonal antibody cross-reacts with all muscle types. J Muscle Res Cell Motil. 1983;5:515–526. doi: 10.1007/BF00713258. [DOI] [PubMed] [Google Scholar]
- Lim SS, Hering GE, Borisy GG. A monoclonal anti-troponin-T cross-reacts with smooth muscle and non-muscle cells. Can J Biochem Cell Biol. 1985;63:470–478. doi: 10.1139/o85-066. [DOI] [PubMed] [Google Scholar]
- Moreland S, Little DK, Moreland RS. Calmodulin antagonists inhibit latch bridges in detergent skinned swine carotid media. Am J Physiol. 1987;252:C523–531. doi: 10.1152/ajpcell.1987.252.5.C523. [DOI] [PubMed] [Google Scholar]
- Morgan KG, Gangopadhyay SS. Signal transduction in smooth muscle. Cross-bridge regulation by thin filament-associated proteins. J Appl Physiol. 2001;91:953–962. doi: 10.1152/jappl.2001.91.2.953. [DOI] [PubMed] [Google Scholar]
- Ogut O, Yuen SL, Brozovich FV. Regulation of smooth muscle contractile phenotype by nonmuscle myosin. J Muscle Res Cell Motil. 2008 doi: 10.1007/s10974-008-9132-2. (epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Owens GK. Regulation of differentiation of vascular smooth muscle cells. Physiol Rev. 1995;75:487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- Pinet F, Poireir F, Fuchs S, Tharauz PL, Caron M, Corvol P, Michel JB, Joubert-Caron R. Troponin T as a marker of differentiation revealed by proteomic analysis of renal arterioles. FASEB J. 2004;18:585–586. doi: 10.1096/fj.03-0939fje. [DOI] [PubMed] [Google Scholar]
- Rembold CM, Murphy RA. Myoplasmic calcium, myosin phosphorylation, and regulation of the crossbridge cycle in swine arterial smooth muscle. Circ Res. 1986;58:803–815. doi: 10.1161/01.res.58.6.803. [DOI] [PubMed] [Google Scholar]
- Rovner AS, Freyzon Y, Trybus KM. Chimeric substitutions of the actin-binding loop activate dephosphorylated but not phosphorylated smooth muscle heavy meromyosin. J Biol Chem. 1995;270:30260–30263. doi: 10.1074/jbc.270.51.30260. [DOI] [PubMed] [Google Scholar]
- Saggin L, Gorza L, Ausoni S, Schiaffino S. Cardiac troponin T in developing, regenerating and denervated rat skeletal muscle. Development. 1990;110:547–554. doi: 10.1242/dev.110.2.547. [DOI] [PubMed] [Google Scholar]
- Sanders C, Stewart DH, Smillie LB. Troponin-T and glyceraldehyde-3-phophate dehydrogenase share a common antigenic determinant. J Muscle Res Cell Motil. 1987;8:118–124. doi: 10.1007/BF01753987. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- Sellers JR. Unphosphorylated crossbridges and latch: smooth muscle regulation revisited. J Muscle Res Cell Motil. 1999;20:347–349. doi: 10.1023/a:1005557724418. [DOI] [PubMed] [Google Scholar]
- Shirinsky VP, Biryukow KG, Vortnikov AV, Gusev NB. Caldesmon150, caldesmon77 and skeletal muscle troponin T share a common antigenic determinant. FEBS Lett. 1989;251:65–68. doi: 10.1016/0014-5793(89)81429-2. [DOI] [PubMed] [Google Scholar]
- Siedner S, Kruger M, Schroeter M, Metzler D, Roell W, Fleischmann BK, Hescheler J, Pfitzer G, Stehle R. Developmental changes in contractility and sarcomeric proteins from the early embryonic to the adult stage in the mouse heart. J Physiol. 2003;548:493–505. doi: 10.1113/jphysiol.2002.036509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegman MJ, Butler TM, Mooers SU, Michalek A. Ca2+ can affect Vmax without changes in myosin light chain phosphorylation in smooth muscle. Pflugers Arch. 1984;401:385–390. doi: 10.1007/BF00584340. [DOI] [PubMed] [Google Scholar]
- Stephens NL. Airway smooth muscle. Lung. 2001;179:333–373. doi: 10.1007/s004080000073. [DOI] [PubMed] [Google Scholar]
- Throckmorton DC, Packer CS, Brophy CM. Protein kinase C activation during Ca2+-independent vascular smooth muscle contraction. J Surg Res. 1998;78:48–53. doi: 10.1006/jsre.1998.5368. [DOI] [PubMed] [Google Scholar]
- Tubridy N, Fontaine B, Eymard B. Congenital myopathies and congenital muscular dystrophies. Curr Opin Neurol. 2001;14:575–582. doi: 10.1097/00019052-200110000-00005. 2001. [DOI] [PubMed] [Google Scholar]
- Walsh MP. Regulation of vascular smooth muscle tone. Can J Physiol Pharmacol. 1994;72:919–936. doi: 10.1139/y94-130. [DOI] [PubMed] [Google Scholar]
- Walsh MP, Horowitz A, Clement-Chomienne O, Andrea JE, Allen BG, Morgan KG. Protein kinase C mediation of Ca (2+)-independent contractions of vascular smooth muscle. Biochem Cell Biol. 1996;74:485–502. doi: 10.1139/o96-053. [DOI] [PubMed] [Google Scholar]
- Wang Q, Reiter RS, Huang QQ, Jin JP, Lin JJ. Comparative studies on the expression patterns of three troponin T genes during mouse development. Anat Rec. 2001;263:72–84. doi: 10.1002/ar.1078. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Yoshino Y, Morimoto S. Troponin I inhibitory peptide suppresses the force generation in smooth muscle by directly interfering with cross-bridge formation. Biochem Biophys Res Commun. 2003;307:236–240. doi: 10.1016/s0006-291x(03)01170-7. [DOI] [PubMed] [Google Scholar]
- Zanellato AM, Borrione AC, Saggin L, Giuriato L, Schiaffino S, Sartore S. Troponin T-and troponin I-like proteins in bovine vascular smooth muscle. Circ Res. 1991;68:1349–1361. doi: 10.1161/01.res.68.5.1349. [DOI] [PubMed] [Google Scholar]