Abstract
A novel chemical-enzymatic approach was developed to facilitate identification of phosphorylation sites in isolated phosphoproteins. ESI–TOF mass spectrometry was used to characterize products from the chemical-enzymatic cleavage of specific phosphorylation sites in bovine α-S1 casein and synthetic phosphopeptides containing substitutions at a single phosphorylation site. Further refinements to this approach for identification of protein phosphorylation sites and its utility for the quantification of phosphopeptides by isotope-dilution mass spectrometry are presented.
Keywords: chemical-enzymatic, phosphorylation, 2-aminoethanethiol, isotope dilution mass spectrometry, phosphoproteins, phosphopeptides, quantification
Introduction
The post-translational modification of proteins is key to understanding cellular function. Of particular interest is the reversible process of phosphorylation on serine, threonine, and tyrosine residues, which has been linked to critical cellular functions such as intracellular signaling, apoptosis, metabolism, gene repression, cell cycling, and gene transcription.1 The vast majority of these phosphorylations occur at serine and threonine residues with the formation of phosphoserine occurring approximately 10 times more often than phosphothreonine.2,3 Proteins in the cell are phosphorylated by protein kinases, which are encoded by more than 2000 genes in the human genome.4 It is estimated that at least one-third of all proteins in the proteome of a human cell are phosphorylated.5 Thus, the importance of phosphorylation has led to the development of phosphoproteomics, which simply defined, is the study of the phosphorylation state of all cellular proteins. It is estimated that there are over 100 000 potential phosphorylation sites in the human cellular proteome.3
Mass spectrometry (MS) has proven to be an important analytical technique in the characterization of complex protein mixtures from biological sources (e.g., biological fluids, cells, tissue).6,7 The increased sensitivity and high mass accuracy of MS has provided for the rapid analysis and identification of phosphorylation sites in the phosphoproteome.4,8 The challenges in characterizing phosphorylation sites has led to the development of several multi-technique approaches, with mass spectrometry often used in the final step of identification. The current trends in the identification of protein phosphorylation sites by MS have been recently reviewed.8,9
Many noteworthy examples of the application of mass spectrometry to identify phosphorylation sites have been published. These include: (1) The use of LC–MS/MS for the identification of phosphorylation by neutral ion loss of phosphoric acid (H3PO4) via gas-phase β-elimination of this ion (−98 Da) from phosphoserine and phosphothreonine.10–13 (2) Tandem MS under alkaline conditions in a negative-ion mode to scan for the loss of a phosphate groups (−79 Da), which is characteristic of phosphorylated peptides.14 (3) MALDI-TOF mass analysis of spectrum shifts resulting from phosphate loss after phosphatase treatment of phosphopeptides,15,16 and (4) MALDI-TOF post source decay experiments to identify characteristic fragments of phosphoamino acids in the resultant mass spectra.17
Recently, the high sensitivity and mass accuracy of electro-spray Fourier transform ion cyclotron resonance (FT–ICR) MS has been used as a method to analyze singly and multiply phosphorylated peptides by electron capture dissociation (ECD).18,19 ECD has been shown to be uniquely useful in phosphate mapping because the labile phosphoamino acids are stable during the fragmentation of phosphorylated peptides. Muddiman20,21 and others22 have used infrared multiphoton dissociation (IRMPD) with FT–ICR MS in order to fragment phosphopeptides for identification.
The low abundance of phosphorylated peptides and the ion suppression effects of nonphosphorylated peptides in proteolytic digests have led to the development of separation techniques, which selectively enrich and isolate phosphopeptides from the more abundant nonphosphorylated peptides. Of great importance is the technique that employs the use of Gallium (III) or Iron (III) metal ions for immobilized metal affinity chromatography (IMAC) to capture phosphopeptides from complex protein digests. The IMAC method has been successful in the enrichment of phosphopeptides that are present in low concentrations, and has thus facilitated a more reliable identification of phosphorylation sites by LC–MS/MS methods.23,24 High affinity antibody techniques have also been used to isolate phosphopeptides from nonphosphorylated peptides, and to separate phosphoproteins from complex biological fluids or cell extracts.25
A report from Barr et al.26 demonstrated the use of coupling protein cleavage with isotope dilution mass spectrometry (IDMS) for the absolute quantification of apolipoprotein A–I. IDMS has been used for over 30 years to quantify biomolecules in solution, and the routine use of this method in clinical chemistry, pharmacology, and toxicology has been reviewed.27 Barr et al., demonstrated that protein cleavage of apolipoprotein A–I could be absolutely quantified in the presence of a stable isotope labeled peptide, which was used as an internal standard (I. S.). The response of the stable isotope labeled I. S. (i.e., a synthetic peptide that corresponded to a natural peptide cleaved from apolipoprotein A–I) was then used to calculate the concentration of the unlabeled peptide by IDMS. A later study by Gerber et al.28 also reported on the use of stable isotope labeled synthetic peptides as I. S. for the absolute quantification (termed AQUA) of proteins and phosphoproteins from cell lysates by selected reaction monitoring with tandem MS. Reports from Muddiman and co-workers have also used tandem MS coupled with protein cleavage IDMS to absolutely quantify a peptide from the proteolytic cleavage of human serum albumin,29 and the biomarker prostate-specific antigen30 using synthetic peptides as stable isotope labeled I. S.
Chemical agents such as fluorophores,31 thiols,32,33 and sulfite34 have been widely used to modify dehydrated phosphoserine and phosphothreonine residues after β-elimination of the phosphate group with sodium or lithium hydroxide. These techniques either enhance the detection and identification of phosphopeptides, or they enrich the recovery of the phosphopeptides in subsequent chromatographic steps. The most successful of these reagents has been the use of ethanethiol in the final Michael addition reaction to produce an S-ethylcysteine derivative of phosphoserine or β-methyl-S-ethylcysteine derivative of phosphothreonine, which can be easily identified by Edman sequencing 32,35,36 or tandem mass spectrometry.37,38 In a recent study by Chait and co-workers,33 the thiol derivatives of phosphorylated residues were modified with biotin-linked maleimide for enrichment of phosphopeptides on avidin gels followed by sequence analysis with tandem MS.
Hathaway and co-workers39 first described a novel chemical-enzymatic method for the protease-directed cleavage of peptide bonds C-terminal to phosphoserine or phosphothreonine residues in synthetic phosphopeptides or O-linked glycosylserine residues in GalNAc-glycopeptides. This approach used β-elimination of phosphate groups with a barium catalyst in sodium hydroxide followed by Michael addition with 2-amino-ethanethiol (2-AET or cysteamine) to convert phosphorylated or glycosylated residues into lysine analogues, which could be recognized and cleaved with endoproteinase Lys-C. A later report used the same method to identify sites of protein phosphorylation in several phosphoproteins (e.g., bovine α and β-casein, β-tubulin, and GP kinase 2), and also adapted this method to solid phase supports for the one-step enrichment and modification of phosphopeptides.40
In this study, the chemical-enzymatic approach developed by Hathaway and co-workers39 was used along with LC ESI–TOF mass spectrometry to confirm sites of phosphorylation in the model phosphoprotein, bovine α-S1 casein. We have characterized some novel peptide products in addition to those expected following proteolytic cleavage of Aec-modified α-S1 casein and a synthetic α-S1 casein phosphopeptide, which have not been previously reported39,40 In addition, several synthetic phosphopeptides of the sequence α-S1 casein 106–119, which contained specific substitutions at either asparagine 114 or phosphoserine 115, were used as model peptide substrates to compare the efficiency of this approach with 2-AET and the proteases, trypsin or endoproteinase Lys-C. Some important refinements of the chemical-enzymatic approach for the identification of protein phosphorylation sites, using bovine α-S1 casein as an example, are presented, and the feasibility of this approach for the quantification of phosphorylation by IDMS is demonstrated. Although the chemical-enzymatic approach will be an invaluable tool to identify sites of protein phosphorylation, our findings herein indicate that careful attention is warranted when using this method in phosphoproteomic studies.
Materials and Methods
Materials and Synthesis of α-S1 Casein Phosphopeptides
Bovine α-S1 casein (85%), barium hydroxide, and 2-amino-ethanethiol hydrochloride (or cysteamine hydrochloride) were purchased from Sigma-Aldrich (Milwaukee, WI). Sequencing grade-modified trypsin was purchased from Promega Corporation (Madison, WI), and endoproteinase Lys-C from Roche Diagnostics (Penzberg, Germany). All other reagents and solvents were HPLC grade and purchased from Burdick & Jackson (Muskegon, MI). Phosphopeptides of α-S1 casein 106–119 containing a phosphoamino acid of serine, threonine, or tyrosine at residue 115 for this study were synthesized by Fmoc solid-phase methods on an ABI 433 Peptide Synthesizer (Applied Biosystems, Foster City, CA) in the Mayo Proteomics Research Center (Rochester, MN). Mass weight and homogeneity of each peptide product was verified by ESI–mass spectrometry (PE Sciex 165B, Applied Biosystems, Foster City, CA), and analytical reversed-phase high performance liquid chromatography (HPLC) on Jupiter C18 columns (250 × 2.0 mm, Phenomonex Inc., CA). For quantification experiments, a stable isotope-labeled derivative of the phosphopeptide α-S1 casein 106–119 was synthesized as an I. S. by substituting Val112 with its uniformly labeled stable isotope (13C5,15N, >98%) derivative. The amino acid sequences of the synthetic phosphopeptides used in this study are shown in Table 1. The total nitrogen content of synthetic phosphopeptide α-S1 casein 106–119 and its stable isotope I.S. were determined by the Scientific Research Consortium, Inc. (St. Paul, MN). From these data the net peptide content was determined (73% and 74% peptide by mass, respectively), and the subsequent concentration dependent calculations for experiments using these synthetic peptides was adjusted accordingly.
Table 1.
Amino Acid Sequences of Synthetic α-S1 Casein Phosphopeptides 106–119 and Substitutions Made
| peptide name | peptide sequence | substitutiona |
|---|---|---|
| α-S1 casein 106–119 | VPQLEIVPNpSAEER | none |
| α-S1 casein 106–119 I. S.b | VPQLEIVPNpSAEER | 15N,13C5–Val112 |
| α-S1 casein 106–119 Ala114 | VPQLEIVPApSAEER | Asn114 → Ala |
| α-S1 casein 106–119 pThr115 | VPQLEIVPNpTAEER | pSer115 → pThr |
| α-S1 casein 106–119 Lys115 | VPQLEIVPNKAEER | pSer115 → Lys |
| α-S1 casein 106–119 pTyr115 | VPQLEIVPNpYAEER | pSer115 → pTyr |
Abbreviations are as follows: pSer, phosphoserine; pThr, phosphothreonine; pTyr, phosphotyrosine
I. S., internal standard.
LC ESI–TOF Mass Spectrometry of Proteolysis Products and Data Analysis
Mass spectra of all digests and peptides were acquired on a LCT orthogonal acceleration TOF mass spectrometer (Micromass, UK) equipped with a standard electrospray ionization source operating in positiveion mode. The LCT MS was interfaced to a Shimadzu system controller (Model SCL-10Avp, Shimadzu Scientific Instruments, MD) and autosampler (SIL-10ADvp). All liquid chromatography (LC) separations were done by reverse-phase HPLC on a Jupiter Proteo C12 column (Proteo 90 Å, 4 μm, 150 × 1.0 mm, Phenomenex Inc., CA) using linear gradients of 5 to 60% acetonitrile in aqueous 0.1% formic acid (v/v). Data acquisition and instrument control were done using MassLynx v4.0 software (Micromass, UK).
Modification and Proteolysis of Synthetic Phosphopeptides of α-S1 Casein 106–119
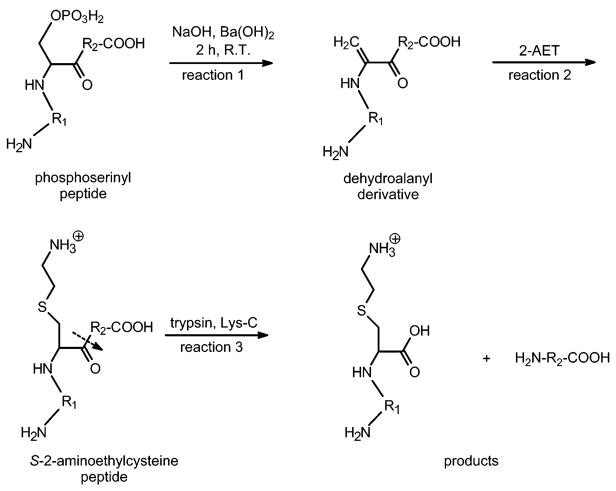
Synthetic phosphopeptides of bovine α-S1 casein comprising residues 106–119 and containing a phosphoserine, phosphothreonine, or phosphotyrosine residue at position 115 were modified by the procedure described by Knight et al.40 Briefly, 10 μg (~6.0 nmol) of each peptide was dissolved in 50 μL of a solvent of water:methylsulfoxide:ethanol (4:3:1). The β-elimination reaction at phosphoserine 115 was performed by adding 25 μL of a saturated solution of aqueous Ba(OH)2 and 5 μL 1M NaOH for 2 h, 22 °C. Michael addition to the dehydroalanine derivative of phosphoserine was performed by adding 50 μL of 1M 2-aminoethanethiol (2-AET) for 3 h, 22 °C. This method creates an S-2-aminoethylcysteine (Aec) derivative of phosphoserine, which is susceptible to proteolysis by trypsin or endoproteinase Lys-C. Conversion of phosphothreonine residues to β-methylaminoethylcysteine (Mac) in phosphopeptides was performed identically, but for 2 h at 37 °C instead of 22 °C. Reactions for the chemical-enzymatic method and protease-specific cleavage at phosphoserine are shown in Figure 1. After conversion of phosphopeptides to their Aec, or Mac, derivative, the modified peptides were desalted by RP-HPLC on an Aquapore RP-300 column (30 × 2.1 mm), and pooled fractions were dried in a Savant Speed Vac (Global Medical Instrumentation, Inc., Albertville, MN). Dried fractions were dissolved in 30 μL of 25 mM Tris buffer, pH 8.5 followed by the addition of 50 μL of sequencing grade modified trypsin (10 μg total, Promega) or endoproteinase Lys-C (5 μg total, Roche) in 50 mM Tris buffer, pH 8.5. Proteolysis was allowed to proceed for 3 h at 37 °C in a shaking water bath. Aliquots of each digest were then diluted in 0.1% formic acid (1:40), and analyzed by LC ESI–TOF mass spectrometry.
Figure 1.
Chemical modification of phosphoserine residues using β-elimination (reaction 1) and nucleophilic addition with 2-amino-ethanethiol (reaction 2) in order to produce a protease-cleavable S-2-aminoethylcysteine (Aec) lysine analogue. The Aec derivative is recognized by the proteases trypsin or Lys-C, and facilitates cleavage of the peptide bond that is C-terminal to the Aec residue (reaction 3).
Proteolysis of Synthetic Phosphopeptide α-S1 Casein 106–119 in Time Course Studies
Duplicate samples (10 μg each) of the synthetic phosphopeptide α-S1 casein 106–119 were modified with 2-AET as described for synthetic phosphopeptides above. Each Aec-modified peptide was then digested with either trypsin (10 μg total) or endoproteinase Lys-C (5 μg total) during a time course of 3 h at 37 °C in a shaking water bath. Aliquots (5 μL) of each digest were sampled at 0, 15, 30, 60, 120, and 180 min, immediately diluted in aqueous 0.1% formic acid (195 μL), and analyzed by LC ESI–TOF mass spectrometry. The ion abundances of each product in the total ion chromatogram were then quantified using the quantification application (QuanLynx) of MassLynx v4.0.
Phosphospecific Modification and Proteolysis of Bovine α-S1 Casein
Bovine α-S1 casein (23 μg, ~1 nmol) was dissolved in 30 μL 25 mM Tris buffer, pH 8.5, and porcine modified trypsin (10 μg total, Promega) in 50 μL 50 mM Tris buffer, pH 8.5 was added to the mixture. The first proteolysis was performed for 16 h at 37 °C in a shaking water bath. After digestion, 10 μL of formic acid was added to stop proteolysis, and the digest was then dried in a Savant Speed Vac. Aliquots of the α-S1 casein digest were analyzed by LC ESI–TOF mass spectrometry to confirm complete proteolysis. The digest mixture was then dissolved in 50 μL of a solvent of water: methylsulfoxide:acetonitrile (4:3:1), and the phosphopeptides were modified to their Aec derivatives as described for synthetic phosphopeptides above, except that the β-elimination reaction was performed at 37 °C for 2 h. The Aec-modified peptides were desalted by RP-HPLC on an Aquapore RP-300 column, dried in a Savant Speed Vac, and then subjected to a second trypsin proteolysis (10 μg total, Promega) for 16 h at 37 °C in a shaking water bath. Peptides from the second proteolysis were analyzed for specific products produced from cleavage of Aec-modified phosphopeptides by LC ESI–TOF mass spectrometry.
Quantification of Phosphorylation by Protein Cleavage-Isotope Dilution Mass Spectrometry
The utility of the chemical-enzymatic approach for the quantification of phosphorylation in the synthetic phosphopeptide α-S1 casein 106–119 was performed by protein cleavage-isotope dilution mass spectrometry (IDMS) as previously described.29,30 Briefly, a standard concentration curve was constructed from 10 serial dilutions (0, 0.5, 1, 2, 4, 5, 8, 10, 16, and 20 μg) of the synthetic phosphopeptide α-S1casein 106–119. After addition of the stable-isotope labeled I. S. (6.0 μg corresponding to 3.6 nmol), each dilution was subjected to β-elimination and modification with 2-AET as described for α-S1 casein phosphopeptides above. Following modification, each dilution was desalted by RP-HPLC, and then digested with trypsin (5 μg total, Promega) for 3 h at 37°C. Products from the phosphospecific cleavage of the Aec-modified peptide and I. S. were then analyzed by LC ESI–TOF mass spectrometry, and the ion abundances of each predicted product (triplicate analysis of each product in each dilution from two independent experiments) were quantified using QuanLynx application of MassLynx v4.0. The ion abundance ratio (AA/AI. S.) of each product verses the concentration ratio (CA/CI. S.) was plotted. A least-squares linear regression analysis was then applied to determine the slope (m), y-intercept (b), and correlation coefficient (R2) of each standard curve.
Results
Proteolysis of Intact α-S1 Casein Phosphoprotein
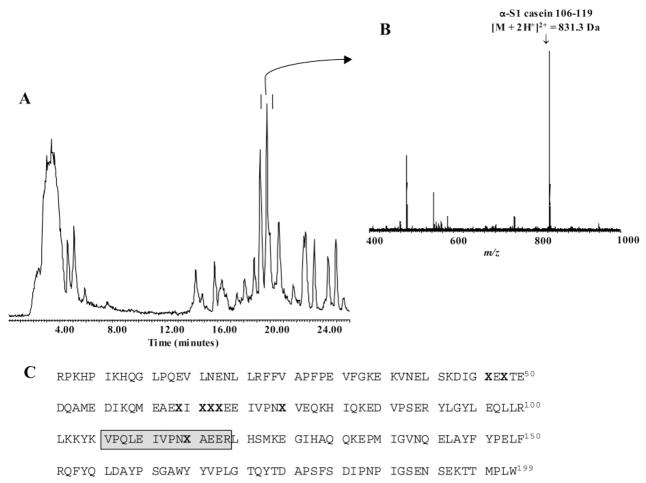
In preliminary studies, α-S1 casein was digested with trypsin and then analyzed by LC ESI–TOF MS to determine its suitability as a model phosphoprotein for more complex experiments. Ideally, the model phosphoprotein would contain an easily isolated peptide that contained only one phosphorylated residue for chemical-enzymatic analysis. This was indeed the case as we determined, by examining the total ion chromatogram (TIC) of the α-S1 casein digest (Figure 2A), the presence of a highly abundant peptide ion (M + 2H+)2+ of m/z = 831.3 (Figure 2B). Examination of the α-S1 casein amino acid sequence indicated that this corresponded to a phosphopeptide from α-S1 casein comprising residues 106–119 (Mr = 1660.6) with a single phosphorylation site at serine residue 115 (Figure 2C). With the utility of α-S1 casein as a model protein demonstrated, additional studies examining the chemical modification, proteolytic cleavage, and quantification of the abundant phosphopeptide 106–119 from digests of α-S1 casein were conducted.
Figure 2.
LC ESI–TOF MS analysis of bovine α-S1 casein digest (23 μg, 1 nmol) after trypsin proteolysis (16 h, 37 °C). (A.) Total ion chromatogram of the α-S1 casein digest depicting the elution position of the doubly charged ion of the peptide α-S1 casein 106–119 (peak in double bar). (B.) Mass spectrum of the abundant ion of phosphopeptide α-S1 casein 106–119 (M+2H+)2+ of m/z = 831.3 identified in the α-S1 casein digest. (C.) Amino acid sequence of the phosphoprotein bovine α-S1 casein, and the location of its eight phosphoserine residues (X in bold). The phosphopeptide α-S1 casein 106–119, that contains a single phosphorylation site (i.e., phosphoserine 115), is shown in the gray-boxed area of the sequence.
Chemical-Enzymatic Studies with Intact α-S1 Casein
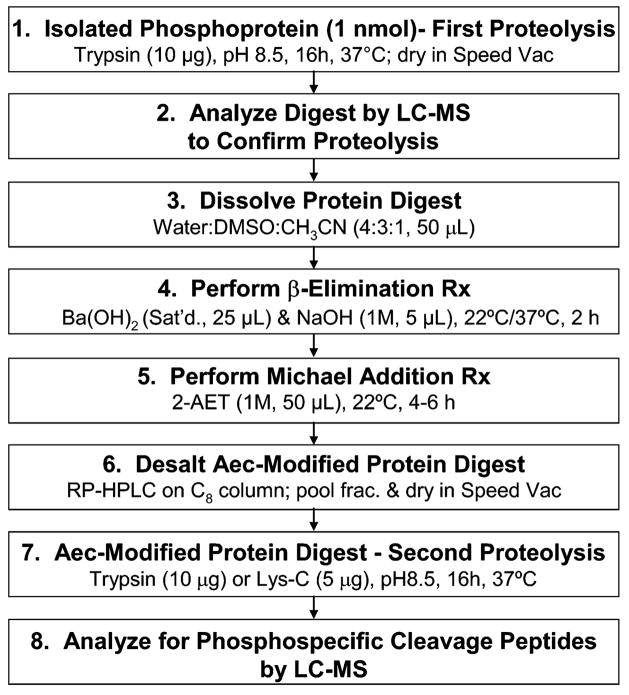
In early experiments using intact α-S1 casein as the model phosphoprotein, the chemical modification of phosphoserine residues to their S-2-aminoethylcysteine (Aec) derivatives, and the subsequent identification of Aec-modified phosphorylation sites by trypsin proteolysis were unsuccessful as previously reported.40 There were appreciable losses of the α-S1 casein phosphoprotein by precipitation in the polar-organic solvents and high hydroxide ion concentration used for the first β-elimination reaction. Therefore, we developed an alternate strategy for the conversion of phosphoserine residues to their Aec derivative in the intact α-S1 casein phosphoprotein. This strategy comprised an initial trypsin proteolysis of the α-S1 casein phosphoprotein, followed by modification of the peptides in the digest to their Aec derivatives by β-elimination and subsequent addition of 2-AET. The Aec-modified α-S1 casein digest was then subjected to a second proteolysis with trypsin, and the digest was then screened for peptides and modified peptide products by LC ESI–TOF mass spectrometry. A summary of our improved method for Aec modification and proteolysis of phosphorylation sites in an intact phosphoprotein is outlined in Figure 3.
Figure 3.
Flowchart summarizing the chemical-enzymatic method for mapping sites of protein phosphorylation by mass spectrometry in isolated phosphoproteins.
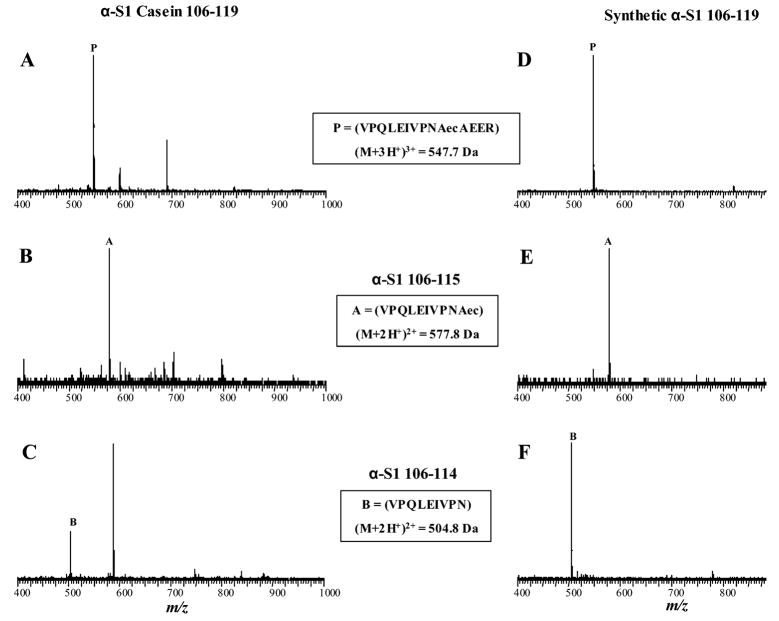
Analysis of the Aec-modified α-S1 casein digest revealed the presence of an ion (M + 3H+)3+ of m/z = 547.7 (Figure 4A, label P), which corresponded to the Aec-modified α-S1 casein peptide 106–119 (Mr = 1640.1). This peptide contained a chemical modification of phosphoserine 115 to an Aec derivative and comprised the sequence, VPQLEIVPN(Aec)AEER. In addition, proteolysis of the Aec-modified α-S1 casein digest also demonstrated the presence of two additional ions, which were presumed to originate from cleavage at residue 115 (i.e., Aec modified) in the peptide α-S1 casein 106–119. The first ion (M + 2H+)2+ of m/z = 577.8 corresponded to the peptide product α-S1 casein 106–115 (VPQLEIVPNAec) (Figure 4B, label A); a second ion (M + 2H+)2+ of m/z = 504.8 corresponded to the peptide product α-S1 casein 106–114 (VPQLEIVPN) (Figure 4C, label B). Product ions corresponding to an expected tetrapeptide α-S1 casein 116–119 (AEER) were not observed in these analyses.
Figure 4.
Mass spectra of peptide products from trypsin proteolysis of Aec-modified bovine α-S1 casein digest (1 nmol) and synthetic phosphopeptide α-S1 casein 106–119 (6 nmol). (A.) (M+3H)3+ ion of Aec-modified phosphopeptide 106–119 (VPQLEIVPNAecAEER; label P) identified in Aec-modified α-S1 casein digest. (B.) (M+2H)2+ ion of expected product α-S1 casein 106–115 (VPQLEIVPNAec; label A), and (C.) (M+2H)2+ ion of product α-S1 casein 106–114 (VPQLEIVPN; label B) following trypsin proteolysis of Aec-modified α-S1 casein digest. (D.) (M+3H)3+ ion of Aec-modified synthetic phosphopeptide 106–119 substrate (label P). (E.) (M+2H)2+ ion of expected product α-S1 casein 106–115 (VPQLEIVPNAec; label A), and (F.) (M+2H)2+ ion of product α-S1 casein 106–114 (VPQLEIVPN; label B) after trypsin proteolysis of synthetic phosphopeptide α-S1 106–119.
The detection of the doubly charged ion at m/z = 504.8, which corresponded to the peptide product α-S1 casein 106–114 (VPQLEIVPN), was novel and unexpected. The predominant presence of this peptide ion after trypsin proteolysis indicated a C-terminal cleavage of the peptide bond between asparagine 114 and the residue Aec 115 in α-S1 casein 106–119. To confirm these findings from the proteolysis of intact Aec-modified α-S1 casein digests, a detailed study of the chemical-enzymatic cleavage with a synthetic phosphopeptide of α-S1 casein 106–119 was conducted.
Chemical-Enzymatic Studies with Synthetic Phosphopep-tide α-S1 Casein 106–119
Trypsin proteolysis of an Aec-modified synthetic peptide of α-S1 casein 106–119 produced peptides with ions of identical mass to the doubly and triply charged ions detected in the proteolysis of intact Aec-modified α-S1 casein digests (Figure 4, panels D–F). The expected peptide α-S1 casein 106–115 was detected as an ion (M + 2H+)2+ of m/z = 577.8 by LC ESI–TOF MS (Figure 4E); the novel product, α-S1 casein 106–114 containing a C-terminal asparagine residue, was again detected as an ion (M + 2H+)2+ of m/z = 504.8 (Figure 4F). The amino acid sequence of the novel product, α-S1 casein 106–114, was further analyzed by tandem MS (Q-TOF API US, Micromass, UK) and confirmed to be the sequence, VPQLEIVPN (data not shown). Again, ions of the expected tetrapeptide product α-S1 casein 116–119 (AEER) were not detected in these experiments.
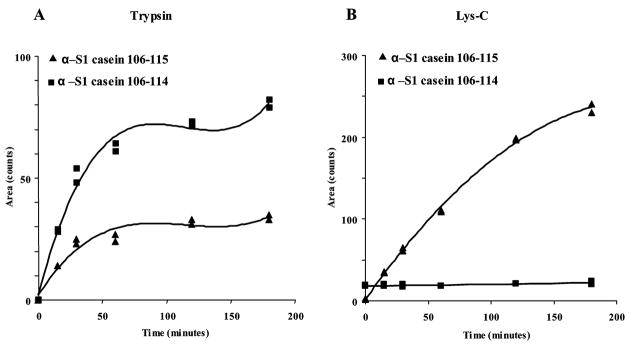
Time Course Studies with Proteolysis of Aec-Modified Synthetic α-S1 Casein 106–119
The ratios of the expected peptide (α-S1 casein 106–115) to the novel peptide (α-S1 casein 106–114) after proteolysis of synthetic α-S1 casein 106–119 substrate in a time course study were quantified by LC ESI–TOF MS. These studies were conducted to determine if the novel peptide product, α-S1 casein 106–114, resulted from a stepwise cleavage of peptide α-S1 casein 106–115 at residue Aec-115, or from an alternate cleavage of the Aec-modified peptide substrate α106–119. Time course studies with trypsin over a 3-h period of proteolysis suggested that both peptide products, α-S1 casein 106–115 (▲–▲) and α-S1 casein 106–114 (■–■), were produced from two alternate cleavages of the Aec-modified synthetic peptide substrate, α-S1 casein 106–119 (Figure 5A). Moreover, both peptide products were present in proteolytic digests of the Aec-modified peptide substrate after 24 h of proteolysis (data not shown). This longer proteolysis suggests that peptide α-S1 casein 106–115 does not convert by stepwise cleavage into the shorter peptide, α-S1 casein 106–114. However, the formation of minimal amounts of α-S1 casein 106–114 from a stepwise C-terminal clipping of α-S1 casein 106–115 cannot be completely disregarded from these studies.
Figure 5.
Time course study demonstrating the appearance of peptide products after proteolysis of synthetic Aec-modified phosphopeptide α-S1 casein 106–119. (A.) Quantification of ion abundances of peptide α-S1 casein 106–115 (▲–▲) and peptide α-S1 casein 106–114 (■–■) after trypsin proteolysis (10 μg total, 37 °C) at sampling times of 0, 15, 30, 60, 120, and 180 min (duplicate samples). (B.) Quantification of ion abundances of peptide α-S1 casein 106–115 (▲–▲) and peptide α-S1 casein 106–114 (■–■) after proteolysis with endoproteinase Lys-C (5 μg total, 37 °C) at sampling times of 0, 15, 30, 60, 120, and 180 min (duplicate samples).
The quantity of α-S1 casein 106–115 (▲–▲) produced from endoproteinase Lys-C cleavage of the modified synthetic α-S1 casein 106–119 substrate was substantially greater than the amount of the same peptide (α-S1 casein 106–115) produced by trypsin proteolysis of the substrate (Figure 5B). In addition, endoproteinase Lys-C cleavage of Aec-modified α-S1 casein 106–119 produced a minimal amount of peptide α-S1 casein 106–114 (■–■), which remained at baseline levels over a 3-hour period (Figure 5B).
Chemical-Enzymatic Studies with Alanine-Substituted Synthetic α-S1 Casein 106–119
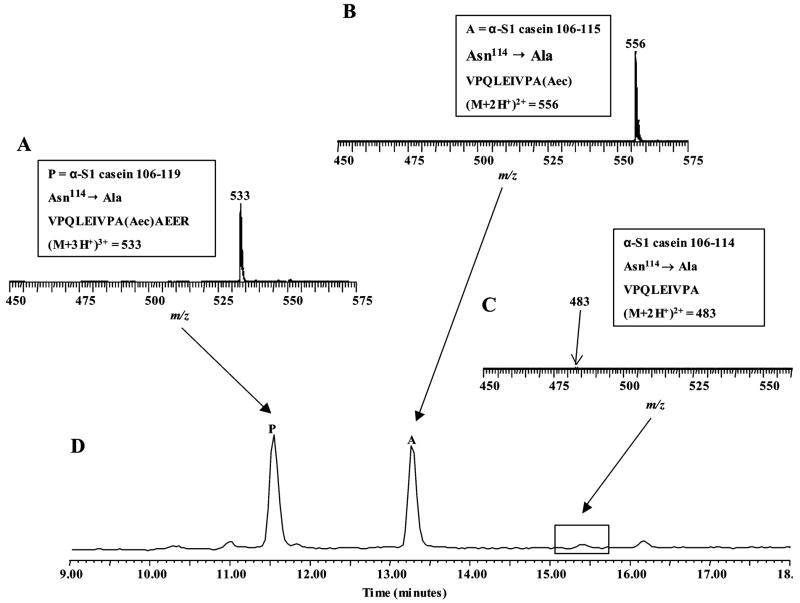
Proteolysis of Aec-modified α-S1 casein 106–119 suggested that the novel formation of α-S1 casein 106–114 is the result of a nonspecific cleavage of the peptide bond C-terminal to residue asparagine 114 (Asn-114). To verify this, a synthetic peptide of α-S1 casein 106–119 that contained the replacement of Asn-114 with alanine was made. After modification of phosphoserine 115 (pSer-115) to an Aec derivative, the alanine-substituted peptide was subjected to trypsin proteolysis and the digest was examined by LC ESI–TOF MS. Analysis of the peptide digest detected the presence of an ion (M + 3H+)3+ at m/z = 533, which corresponded to the alanine-substituted peptide that was not cleaved by trypsin (Figure 6A). A doubly charged ion, which corresponded to the peptide α-S1 casein 106–115 (VPQLEIVPAAec) and contains Aec-115 at its C-terminal end, was identified at m/z = 556 (Figure 6B). However, a doubly charged ion of m/z = 483, which corresponded to peptide α-S1 casein 106–114 (VPQLEIVPA), was almost absent in the digest of the alanine-substituted peptide (Figure 6C,D). The replacement of asparagine 114 with alanine in the wild type sequence essentially blocks a C-terminal cleavage at this position, which prevents the formation of the peptide α-S1 casein 106–114 (VPQLEIVPA). Similar to the findings with trypsin, proteolysis of the alanine-substituted peptide with endoproteinase Lys-C produced only the peptide product α-S1 casein 106–115, but not α-S1 casein 106–114 (see Table 2).
Figure 6.
LC ESI-TOF MS analysis of products from trypsin proteolysis (10 μg, 3 h, 37 °C) of alanine substituted synthetic α-S1 casein phosphopeptide 106–119 (Asn114→ Ala) after modification with 2-AET. (A.) Mass spectrum of (M+3H)3+ ion of Aec-modified alanine substituted peptide 106–119 (VPQLEIVPAAecAEER); (B.) (M+2H)2+ ion of product α-S1 casein 106–115 (VPQLEIVPAAec), and (C.) (M+2H)2+ ion of product α-S1 casein 106–114 (VPQLEIVPA). (D.) Total ion chromatogram indicating the relative elution position of the Aec-modified alanine substituted peptide (label P), peptide product α-S1 casein 106–115 (label A), and peptide product α-S1 casein 106–114 (boxed area).
Table 2.
Summary of Products from Proteolysis of Aec- or Mac-Modified Synthetic α-S1 Casein Phosphopeptides 106–119 Including Substitutions at Asn-114 or pSer-115
| modified α-S1 casein
peptides & cleavage products |
trypsin proteolysis
% peptidea |
Lys-C proteolysis
% peptidea |
|---|---|---|
| VPQLEIVPN(Aec)AEERb | 74.1 ± 0.8 | 67.5 ± 1.6 |
| α106–115/VPQLEIVPN(Aec) | 4.1 ± 2.0 | 28.9 ± 1.4 |
| α106–114/VPQLEIVPN | 21.8 ± 1.8 | 2.6 ± 0.3 |
| VPQLEIVPA(Aec)AEER | 54.9 ± 2.1 | 75.5 ± 0.6 |
| α106–115/VPQLEIVPA(Aec) | 41.2 ± 2.7 | 21.8 ± 0.5 |
| α106–114/VPQLEIVPA | 2.8 ± 0.7 | 2.7 ± 0.2 |
| VPQLEIVPN(Mac)AEER | 88.8 ± 0.4 | 97.2 ± 0.2 |
| α106–115/VPQLEIVPN(Mac) | n.d.c | n.d. |
| α106–114/VPQLEIVPN | 11.2 ± 0.4 | 2.8 ± 0.2 |
| VPQLEIVPNKAEER | 24.5 ± 1.3 | n.d. |
| α106–115/VPQLEIVPNK | 60.3 ± 6.6 | 100 |
| α 106–114/VPQLEIVPN | 15.3 ± 7.6 | n.d. |
| VPQLEIVPNpYAEER | 100 | 100 |
| α106–115/VPQLEIVPNpY | n.d. | n.d. |
| α106–114/VPQLEIVPN | n.d. | n.d. |
Amount of peptide products are expressed as a relative percentage of peptide cleavage ± the standard deviation from two independent experiments. Percentages ≤ 3.0% are background noise in the spectrum.
Abbreviations are as follows: Aec, S-2-aminoethylcysteine; Mac, β-methlyamino-ethylcysteine; pY, phosphotyrosine.
n.d.= product not detected.
Studies with Other Substituted Synthetic α-S1 Casein 106–119 Phosphopeptides
Proteolytic digests of three other substituted synthetic α-S1 casein 106–119 phosphopeptides after chemical-enzymatic treatment were analyzed by LC ESI–TOF MS. As a control for the chemical-enzymatic method, a substitution of pSer-115 with a phosphotyrosine residue was made. MS analysis demonstrated no chemical modification or cleavage of the phosphotyrosine-substituted peptide. Substitution of pSer-115 with phosphothreonine (pThr) produced a peptide with a β-methylaminoethylcysteine (Mac) derivative at position 115; this modified peptide was not efficiently cleaved by either trypsin or endoproteinase Lys-C. In addition, it was of interest to note that only peptide, α-S1 casein 106–114 (VPQLEIVPN), was detected, and not the peptide product α-S1 casein 106–115 (VPQLEIVPNMac) as expected (Table 2). Trypsin proteolysis of a peptide, which contained a substitution of pSer-115 with lysine, produced two peptides, α-S1 casein 106–115 (VPQLEIVPNK) and α-S1 casein 106–114 (VPQLEIVPN). However, only peptide α-S1 casein 106–115 was found after proteolysis of the substrate with endoproteinase Lys-C. This finding supports previous reports that trypsin, but not endoproteinase Lys-C, has a proclivity to cleave at the C-terminus of asparagine residues in proteins (Gulyás 2004). Table 2 summarizes the characterization of products from the proteolysis of synthetic α-S1 casein 106–119 phosphopeptides after chemical modification with 2-AET, including substitutions at either pSer-115 or Asn-114.
Quantification of Phosphorylation with Synthetic Phosphopeptide α-S1 Casein 106–119
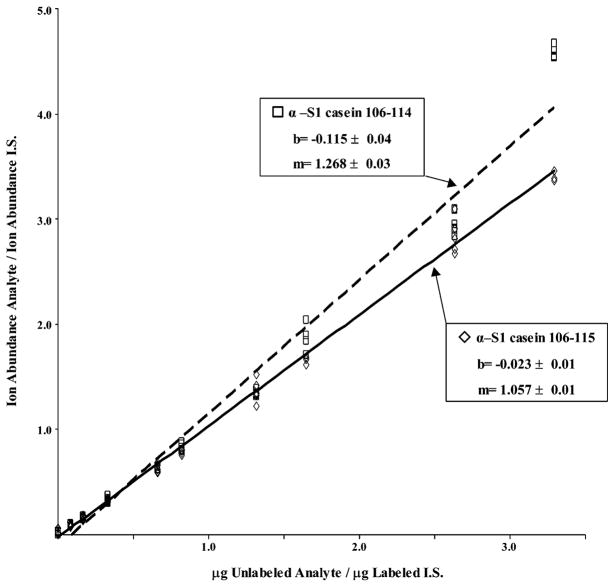
A final study focused on the quantification of phosphorylation using the chemical-enzymatic approach with Aec-modified α-S1 casein 106–119 and protein cleavage coupled with IDMS methodology previously described by this laboratory.30 A fixed amount of a stable isotope-labeled I. S. was added to 10 serial dilutions of unlabeled synthetic phosphopeptide α-S1 casein 106–119. The dilutions comprising unlabeled synthetic phosphopeptide and I. S. were then subjected to β-elimination and conversion to their Aec-modified form, followed by trypsin proteolysis. Products from the proteolysis were monitored by LC ESI–TOF MS, and the ion abundances of each product from both the unlabeled and isotope-labeled I. S. were quantified. A standard curve of products resulting from trypsin specific proteolysis of Aec-modified α-S1 casein 106–119 is shown (Figure 7). The slope (m) and y-intercept (b) for each line corresponding to product α-S1-casein 106–115 (⋄–⋄) or product α-S1-casein 106–114 (□–□) is given; a correlation coefficient of R2 = 0.99 was also determined for each line. These results illustrate the facile use of IDMS with the chemical-enzymatic approach for the quantification of phosphorylation in phosphopeptides or isolated phosphoproteins.
Figure 7.
Standard curve demonstrating the quantification of phosphorylation (AQUAP) by the chemical-enzymatic approach with synthetic α-S1 casein phosphopeptide 106–119 and its stable isotope labeled I. S. After 2-AET modification and trypsin proteolysis the standard curve for each product, α-S1 casein 106–115 (◇–◇) or α-S1 casein 106–114 (□- -□), was determined from two independent experiments as described in Materials and Methods. Least-squares linear regression statistics (y-intercept and slope) for each line is shown.
Discussion
There is no known protease that recognizes phosphoamino acids as a specificity group for phosphospecific proteolysis, and clearly the utility of such an enzyme would be invaluable as a tool for mapping phosphorylation sites in phosphoproteins. However, the use of the reagent 2-AET, described in these studies, introduces a unique site at phosphoserine and phosphothreonine residues, which can be specifically cleaved with proteolytic enzymes such as trypsin or endoproteinase Lys-C. After the well-established β-elimination of the phosphate group with hydroxide ions,32,35 the resulting dehydroalanine or de-hydrothreonine derivatives are modified by 2-AET to either an S-2-aminoethylcysteine (Aec) or β-methylaminoethylcysteine (Mac) residue, respectively. These Aec or Mac derivatives are isosteric to lysine, and are thus susceptible to proteolysis at their C-terminal side by enzymes (e.g., trypsin and endoproteinase Lys-C), which recognize lysine residues as a specificity group. Proteolysis of these lysine analogues with trypsin, or endoproteinase Lys-C, selectively hydrolyzes the amide bond adjacent to each phosphorylated residue. This digestion produces a specific cleavage pattern of peptides and phosphopeptide analogues, which can be identified by measuring their mass with several well-established techniques in mass spectrometry.8,9
Since the dehydrated phosphoamino acid is susceptible to nucleophilic addition from a variety of chemical agents, this strategy has been used to attach fluorophores,31 thiols,32,33,35,36 biotin33 and sulfites,34 in attempts to identify phosphorylation sites by either Edman N-terminal sequencing or tandem MS. As an example, Meyer et al.36 described the use of ethanethiol to introduce S-ethylcysteine residues for identification of phosphoserine residues by Edman sequencing. This reagent has also been successfully used to identify phosphorylation sites in some proteins by tandem MS.33,37,38
The specific use of an aminoalkylthiol reagent to create a protease-susceptible site at phosphorylated residues was not practically utilized until a first report by Rusnak et al.39 described the use of 2-AET to chemically target proteolysis at phosphoserine and phosphothreonine residues. Additional modifications of the chemical-enzymatic approach for mapping sites of protein phosphorylation in the proteins α- and β-casein, β-tubulin, and GRK2 were developed by Shokat and coworkers.40 Some of these modifications included the reduction of time (4 h to 1–2 h) and temperature (45 °C to 22 °C or 37 °C) for β-elimination of the phosphate group, increased barium ion concentration (3 μL to 25 μL) for phosphate catalysis, increased concentration of 2-AET (0.5 M to 1 M) for the nucleophilic addition reaction, and an improved solvent for the dissolution of most phosphopeptides from the protein digest.
However, our experience using this approach with α-S1 casein and synthetic α-S1 casein phosphopeptides, produced some further refinements to the chemical-enzymatic approach previously described.40 Most noteworthy among these refinements is an improved method for the Barium-catalyzed β-elimination of phosphate groups along with the nucleophilic addition of 2-AET. The β-elimination and nucleophilic addition reactions with intact phosphoproteins was vastly improved if the phosphoprotein (e.g., α-S1 casein in this study) was first pre-digested into smaller peptides (and phosphopeptides) by an initial trypsin proteolysis for 16–24 h (cf. Figure 3, step 1). The relatively high pH conditions and substantial concentrations of organic solvents (e.g., ethanol, methylsulfoxide) used in the initial β-elimination reaction induced substantial precipitation and loss of the intact α-S1 casein phosphoprotein (data not shown). An additional refinement from the method previously reported40 was the extension of the phosphate β-elimination reaction from 1 h to 2 h at 22 °C for peptides containing phosphoserine, and 2 h at 37 °C for peptides with phosphothreonine. This extension of time produced a complete elimination of the phosphate group in all peptide concentrations used in this study (0.5 μg to 20 μg) without apparent loss of the peptide by hydroxide-catalyzed hydrolysis (data not shown). A final refinement of was the use of a solution consisting of a mixture of 4:3:1 water/methylsulfoxide/acetonitrile for increased dissolution of the peptides from the pre-digestion of α-S1 casein (cf. Figure 3, step 3) instead of the 4:3:1 water/methylsulfoxide/ethanol mixture previously described.40
A report by Li et al.41 described the well-known problem of β-elimination at free hydroxyl groups of nonphosphorylated serine and threonine residues. However, these reactions were reported for conditions of β-elimination at high alkalinity (0.52 N NaOH) and prolonged elimination periods of 18–24 h at temperatures of 37 °C or greater. The β-elimination and modification of free hydroxyl groups of serine or threonine residues in α-S1 casein peptides was not detected in the present studies. This was presumably due to the mild conditions of the β-elimination reactions, which were performed at low alkalinity (0.06 N NaOH) for short time periods (1–2 h) and at moderate temperatures (22 °C or 37 °C).
The mass spectra contained significant amounts of the Aec-modified peptide α-S1 casein 106–119, which was not proteolyzed by either trypsin or endoproteinase Lys-C (cf. Figure 4A,D; Table 2). The inability of either protease to cleave all Aec-modified substrate is due, in part, to an epimerization reaction at the Cα atom of phosphoserine 115 during the Michael addition reaction with 2-AET. This nucleophilic addition produces a diastereomeric mixture of α-S1 casein 106–119, which contains either a R- or an S-2-aminoethylcysteine residue at position 115. A previous study demonstrated that the epimerization reaction produces an approximately equal amount of R- and S-aminoethylcysteine isomers, and that only the S-isomer is susceptible to proteolysis with trypsin.40 The persistent presence of the Aec-modified peptide α-S1 casein 106–119 after prolonged trypsin proteolysis (i.e., > 16 h) confirms these findings (data not shown). In addition, the ratio of enzyme to Aec-modified peptide substrate used in the present study for trypsin (1 nmol enzyme: 14 nmol peptide) or endoproteinase Lys-C (1 nmol enzyme: 30 nmol peptide) was fixed at the optimized enzyme substrate ratios reported previously.40 The somewhat higher concentrations of trypsin or endoproteinase Lys-C than would be recommended for ordinary protease digestion were used because Aec-modified peptides exhibit a slightly higher resistance to proteolysis, and as mentioned above, are present as diastereomeric mixtures.
The predominant appearance of the novel product α-S1 casein 106–114 (VPQLEIVPN) observed in these studies indicated that this peptide is the result of a trypsin proteolysis of the peptide bond, which is C-terminal to asparagine 114 in the Aec-modified phosphopeptide 106–119. These findings were further confirmed by the replacement of asparagine 114 with alanine, which prevented this proteolysis and the subsequent formation of this novel peptide product (cf. Figure 6; Table 2). Reports from Vestling et al.,42 and Gulyás and Medzihradszky43 have described trypsin cleavage at the C-terminus of asparagine residues, and that given enough time, trypsin proteolysis of substrates can occur indiscriminately at sites other than lysine or arginine side chains. Our studies with endoproteinase Lys-C, however, indicate that nonspecific cleavage of peptide bonds at sites other than lysine residues, rarely occur, and are usually not significant. Time course studies with endoproteinase Lys-C cleavage of Aec-modified α-S1 casein 106–119 produced insignificant (i.e., less than 3% of the total intact substrate) cleavage of the peptide bond C-terminal to asparagine114 over a hydrolysis time of 3 h (cf. Figure 5B; Table 2). A longer hydrolysis time (i.e., 16 h) with endoproteinase Lys-C was also studied, and nonspecific cleavage at asparagine 114 was not increased (data not shown).
Of particular interest was that a predominant cleavage occurred C-terminal to asparagine 114 when phosphoserine 115 was replaced with phosphothreonine, and then subsequently modified to its Mac-derivative (cf. Table 2). In this cleavage, trypsin was a more efficient protease, than endoproteinase Lys-C, but neither enzyme produced significant amounts of the expected product α-S1 casein 106–115 (VPQLEIVPNMac) from the phosphothreonine containing peptide (cf. Table 2). Our experience with the cleavage of other Aec-modified peptides (data not shown) along with those reported in this study suggest that, in general, endoproteinase Lys-C is somewhat more efficient than trypsin in the proteolysis of Aec-modified, but not Mac-modified peptides, and that Lys-C proteolysis of the Aec-modified peptides was usually complete in about 3 h at 37 °C.
An experiment using protein cleavage-isotope dilution mass spectrometry and a synthetic phosphopeptide (i.e., α-S1 casein 106–119) has demonstrated the utility of the chemical-enzymatic approach for the quantification of phosphorylation in phosphopeptides (cf. Figure 7). The extension of this approach to quantify the chemical-enzymatic cleavage of specific phosphoserine or phosphothreonine residues within an intact phosphoprotein is thus quite feasible, and will be a valuable tool for future studies in phosphoproteomics.
Finally, results from these studies indicate that careful attention to the formation of unusual products, in addition to those expected, must be considered when using the chemical-enzymatic approach to identify sites of phosphorylation in isolated phosphoproteins. The identification and characterization of novel peptide products (e.g., α-S1 casein 106–114) resulting from nonspecific proteolysis of Aec-modified phosphoserine residues, or unexpected cleavage at Mac-modified phosphothreonine residues, warrants this consideration. Nonetheless, the use of this approach will be an invaluable tool for the identification of phosphorylated sites at either serine or threonine residues since the vast majority of these modifications occur in most phosphoproteins.3
Summary
In summary, we have used a chemical-enzymatic approach coupled with LC ESI–TOF mass spectrometry to confirm sites of phosphorylation in bovine α-S1 casein, and have characterized some expected and novel peptide products from the chemical-enzymatic treatment of this model phosphoprotein, which have not been previously reported. These findings were confirmed after further study with a synthetic phosphopeptide comprising α-S1 casein residues 106–119, and with other synthetic phosphopeptides, which were used as model peptide substrates to further examine the utility of the chemical-enzymatic approach. In addition, some important refinements of the chemical-enzymatic approach for the identification of protein phosphorylation sites, using bovine α-S1 casein as an example, was presented, and the utility of this approach for the quantification of phosphorylation by isotope dilution mass spectrometry was demonstrated.
Acknowledgments
The authors wish to acknowledge the funding of these studies in part from Mayo Foundation for Medical Education and Research. The authors further acknowledge and thank Dr. H. Robert Bergen, III for his critical review of this work.
Abbreviations used
- LC
liquid chromatography
- RP-HPLC
reverse phase-high performance liquid chromatography
- MS
mass spectrometry
- LC ESI–TOF
liquid chromatography electrospray ionization-time-of-flight
- IDMS
isotope dilution mass spectrometry
- I. S.
internal standard
- 2-AET
2-aminoethanethiol
- Aec
S-2-aminoethylcysteine
- Mac
β-methylaminoethylcysteine
- TIC
total ion chromatogram
References
- 1.Cohen P. The regulation of protein function by multisite phosphorylation – a 25 year update. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 2.Hubbard MJ, Cohen P. On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem Sci. 1993;18:172–177. doi: 10.1016/0968-0004(93)90109-z. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Zha X, Tan Y, Hornbeck P, Mastrangelo A, Alessi D, Polakiewicz R, Comb M. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J Biol Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- 4.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 5.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 6.Yates JR. Mass spectrometry and the age of the proteome. J Mass Spectrom. 1998;33:1–19. doi: 10.1002/(SICI)1096-9888(199801)33:1<1::AID-JMS624>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 7.Mann M, Hendrickson RC, Pandey A. Analysis of proteins and proteomes by mass spectrometry. Annu Rev Biochem. 2001;70:437–473. doi: 10.1146/annurev.biochem.70.1.437. [DOI] [PubMed] [Google Scholar]
- 8.McLachlin DT, Chait BT. Analysis of phosphorylated proteins and peptides by mass spectrometry. Curr Opin Chem Biol. 2001;5:591–602. doi: 10.1016/s1367-5931(00)00250-7. [DOI] [PubMed] [Google Scholar]
- 9.Mann M, Ong S, Grenborg M, Steen H, Jensen O, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 10.Huddleston MJ, Annan RS, Bean MF, Carr SA. Selective detection of phosphopeptides in complex mixtures by electro-spray liquid chromatography/mass spectrometry. J Am Soc Mass Spectrom. 1993;4:710–717. doi: 10.1016/1044-0305(93)80049-5. [DOI] [PubMed] [Google Scholar]
- 11.Hunter A, Games D. Chromatographic and mass spectrometric methods for the identification of phosphorylation sites in phosphoproteins. Rapid Commun Mass Spectrom. 1994;8:559–570. doi: 10.1002/rcm.1290080713. [DOI] [PubMed] [Google Scholar]
- 12.Tholey A, Reed J, Lehmann W. Electrospray tandem mass spectrometric studies of phosphopeptides and phosphopeptide analogues. J Mass Spectrom. 1999;34:117–123. doi: 10.1002/(SICI)1096-9888(199902)34:2<117::AID-JMS769>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 13.Zhou H, Watts JD, Aebersold R. A systemic approach to the analysis of protein phosphorylation. Nat Biotechnol. 2001;19:375–378. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 14.Carr SA, Huddleston MJ, Annan RS. Selective detection and sequencing of phosphopeptides at the femtomole level by mass spectrometry. Anal Biochem. 1996;239:180–192. doi: 10.1006/abio.1996.0313. [DOI] [PubMed] [Google Scholar]
- 15.Yip TT, Hutchens TW. Mapping and sequence-specific identification of phosphopeptides in unfractioned protein digest mixtures by matrix-assisted laser desorption/ionization time-of flight mass spectrometry. FEBS Lett. 1992;308:149–153. doi: 10.1016/0014-5793(92)81264-m. [DOI] [PubMed] [Google Scholar]
- 16.Liao PC, Leykam J, Andrews PC, Gage DA, Allison J. An approach to locate phosphorylation sites in a phosphoprotein: mass mapping by combining specific enzymatic degradation with matrix-assisted laser desorption/ionization mass spectrometry. Anal Biochem. 1994;219:9–20. doi: 10.1006/abio.1994.1224. [DOI] [PubMed] [Google Scholar]
- 17.Annan RS, Carr SA. Phosphopeptide analysis by matrix assisted laser desorption time-of-flight mass spectrometry. Anal Chem. 1996;68:3413–3421. doi: 10.1021/ac960221g. [DOI] [PubMed] [Google Scholar]
- 18.Shi SD, Hemling ME, Carr SA, Horn DM, Lindh I, McLafferty F. Phosphopeptide/phosphoprotein mapping by electron capture dissociation mass spectrometry. Anal Chem. 2001;73:19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]
- 19.Stensballe A, Jensen O, Olsen K, Haselmann K, Zubarev R. Electron capture dissociation of singly and multiply phosphorylated peptides. Rapid Comm Mass Spectrom. 2000;14:1793–1800. doi: 10.1002/1097-0231(20001015)14:19<1793::AID-RCM95>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 20.Flora JW, Muddiman DC. Selective, sensitive, and rapid phosphopeptide identification in enzymatic digests using ESI–FTICR–MS with infrared multiphoton dissociation. Anal Chem. 2001;73:3305–3311. doi: 10.1021/ac010333u. [DOI] [PubMed] [Google Scholar]
- 21.Flora JW, Muddiman DC. Gas-phase ion unimolecular dissociation for rapid phosphopeptide mapping by IRMPD in a penning ion trap: An energetically favored process. J Am Chem Soc. 2002;124:6546–6547. doi: 10.1021/ja0261170. [DOI] [PubMed] [Google Scholar]
- 22.Chalmers MJ, Hakansson K, Johnson R, Smith R, Shen J, Emmett MR, Marshall AG. Protein kinase A phosphorylation characterized by tandem mass Fourier transform ion cyclotron resonance mass spectrometry. Proteomics. 2004;4:970–981. doi: 10.1002/pmic.200300650. [DOI] [PubMed] [Google Scholar]
- 23.Posewitz M, Tempst P. Immobilized gallium (III) affinity chromatography of phosphopeptides. Anal Chem. 1999;71:2883–2892. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- 24.Stensballe A, Anderson S, Jensen O. Characterization of phosphoproteins from electrophoretic gels by nanoscale Fe (III) affinity chromatography with off-line mass spectrometry analysis. Proteomics. 2001;1:207–222. doi: 10.1002/1615-9861(200102)1:2<207::AID-PROT207>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Kalo MS, Pasquale EB. Multiple in vivo tyrosine phosphorylation sites in EphB receptors. Biochemistry. 1999;38:14396–14408. doi: 10.1021/bi991628t. [DOI] [PubMed] [Google Scholar]
- 26.Barr JR, Maggio VL, Patterson DG, Jr, Cooper GR, Henderson LO, Turner WE, et al. 1996;42:1676–1682. [PubMed] [Google Scholar]
- 27.Leenheer AP, Thienpont LM. Applications of isotope dilution mass spectrometry in clinical chemistry, pharmacokinetics, and toxicology. Mass Spec Rev. 1992;11:249–307. [Google Scholar]
- 28.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci US A. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnidge DR, Hall GD, Stocker JL, Muddiman DC. Evaluation of a cleavable stable isotope labeled synthetic peptide for absolute protein quantification using LC–MS/MS. J Proteome Res. 2004;3:658–661. doi: 10.1021/pr034124x. [DOI] [PubMed] [Google Scholar]
- 30.Barnidge DR, Goodmanson MK, Klee GG, Muddiman DC. Absolute quantification of the model biomarker prostate-specific antigen in serum by LC–MS/MS using protein cleavage and isotope dilution mass spectrometry. J Proteome Res. 2004;3:644–652. doi: 10.1021/pr049963d. [DOI] [PubMed] [Google Scholar]
- 31.Fadden P, Haystead T. Quantitative and selective fluorophore labeling of phosphoserine on peptides and proteins: characterization at the attomole level by capillary electrophoresis and laser-induced fluorescence. Anal Biochem. 1995;225:81–88. doi: 10.1006/abio.1995.1111. [DOI] [PubMed] [Google Scholar]
- 32.Holmes CF. A new method for the selective isolation of phosphoserine-containing peptides. FEBS Lett. 1987;215:21–24. doi: 10.1016/0014-5793(87)80106-0. [DOI] [PubMed] [Google Scholar]
- 33.Oda Y, Nagasu T, Chait BT. Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat Biotechnol. 2001;19:379–382. doi: 10.1038/86783. [DOI] [PubMed] [Google Scholar]
- 34.Li W, Boykins R, Backlund P, Wang G, Chen H. Identification of phosphoserine and phosphothreonine as cysteic acid and methylcysteic acid residues in peptides by tandem mass spectrometric sequencing. Anal Chem. 2002;74:5701–5710. doi: 10.1021/ac020259v. [DOI] [PubMed] [Google Scholar]
- 35.Annan WD, Manson W, Nimmo JA. The identification of phosphoseryl residues during the determination of amino acid sequence in phosphoproteins. Anal Biochem. 1982;121:62–68. doi: 10.1016/0003-2697(82)90557-7. [DOI] [PubMed] [Google Scholar]
- 36.Meyer H, Hoffmann-Posorske E, Korte H, Heilmeyer L. Sequence analysis of phosphoserine-containing peptides: modification for picomolar sensitivity. FEBS Lett. 1986;204:61–66. doi: 10.1016/0014-5793(86)81388-6. [DOI] [PubMed] [Google Scholar]
- 37.Jaffe H, Veeranna H, Pant H. Characterization of serine and threonine phosphorylation sites in beta-elimination/ethanethiol addition-modified proteins by electrospray tandem mass spectrometry and database searching. Biochemistry. 1998;37:16211–16224. doi: 10.1021/bi981264p. [DOI] [PubMed] [Google Scholar]
- 38.Shen J, Smith RA, Stoll VS, Edalji R, Jakob C, Walter K, et al. Characterization of protein kinase A phosphorylation: multi-technique approach to phosphate mapping. Anal Biochem. 2004;324:204–218. doi: 10.1016/j.ab.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 39.Rusnak F, Zhou J, Hathaway GM. Identification of phosphorylated and glycosylated sites in peptides by chemically targeted proteolysis. J Biomol Technol. 2002;13:228–237. [PMC free article] [PubMed] [Google Scholar]
- 40.Knight ZA, Schilling B, Row RH, Kenski DM, Gibson BW, Shokat KM. Phosphospecific proteolysis for mapping sites of protein phosphorylation. Nat Biotechnol. 2003;21:1047–1054. doi: 10.1038/nbt863. [DOI] [PubMed] [Google Scholar]
- 41.Li W, Backlund PS, Boykins RA, Wang G, Chen HC. Susceptibility of the hydroxyl groups in serine and threonine to β-elimination/Michael addition under commonly used moderately high-temperature conditions. Anal Biochem. 2003;323:94–102. doi: 10.1016/j.ab.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 42.Vestling MM, Murphy CM, Fenselau C. Recognition of trypsin autolysis products by high-performance liquid chromatography and mass spectrometry. Anal Chem. 1990;62:2391–2394. doi: 10.1021/ac00220a025. [DOI] [PubMed] [Google Scholar]
- 43.Hunyadi-Gulyás E, Medzihradszky KF. Factors that contribute to the complexity of protein digests. DDT: Targets. 2004;3:S3–S10. [Google Scholar]