Abstract
The molecular mechanisms that control the temporal and lineage-specific accessibility, as well as the rearrangement frequency of VH genes for VH-to-DJH recombination, are not fully understood. We previously found a positive correlation between the extent of histone acetylation and the differential rearrangement frequency of individual VH genes. Here, we demonstrated that poorly rearranging VH genes are more highly associated with histone H3 dimethylated at lysine 9, a marker of repressive chromatin, than frequently rearranging VH genes. We also observed a positive relationship between the differential binding of Pax5 to individual VHS107 genes and rearrangement frequency. Furthermore, we showed that accessibility of the regions flanking the Pax5 binding site and the RSS to restriction enzyme cleavage correspond with the differential rearrangement frequency of the VHS107 family members. In addition, we found that the CpG sites located in the coding regions of VH genes are methylated in general, while the extent of DNA methylation drops dramatically near the RSS. For the VHS107 family, one CpG site located 101 bp upstream of the RSS showed variable methylation that correlates with rearrangement frequency, and the methylation status of a CpG site located 34 bp downstream of the RSS could also favor the rearrangement of V1 over V11. These findings suggest that the extent of histone modifications, chromatin accessibility, DNA methylation, as well as the differential binding of Pax5 to VH coding regions, could all influence the rearrangement frequency of individual VH genes, although some of these mechanisms are not strictly B cell specific.
Keywords: B cells, antibodies, gene rearrangement, gene regulation, repertoire development
INTRODUCTION
During B cell development, there is a precise order of immunoglobulin gene segment rearrangements, with DH to JH rearrangements occurring before VH to DJH rearrangements, and heavy chains rearranging before light chains. The observation that germline transcription of unrearranged gene segments immediately precedes rearrangements gave rise to the accessibility hypothesis (Yancopoulos and Alt, 1985). According to this model, each step of the V(D)J rearrangement process must be preceded by acquisition of an accessible chromatin structure that allows RAG proteins to recognize and cleave recombination signal sequences (RSSs) (Yancopoulos and Alt, 1985). To test this hypothesis, different indicators of active chromatin structure have been analyzed, including nuclease sensitivity, DNA hypomethylation, and covalent modifications of histone tails.
DNase I analysis has shown that initiation of V(D)J recombination at the Ig loci is preceded by increased nuclease sensitivity and changes in chromatin structure (Maës et al., 2001). Furthermore, it has been demonstrated that demethylation of L chain genes occurs during B cell development, and is associated with onset of V(D)J recombination (Goodhardt et al., 1993; Mostoslavsky et al., 1998). More recently, several studies have demonstrated that loci active in V(D)J recombination contain nucleosomes with acetylated histone proteins (McMurry and Krangel, 2000; Chowdhury and Sen, 2001; Johnson et al., 2003), and that changes in histone acetylation are dynamic during B cell differentiation. Furthermore, acetylation of histones is not uniform throughout the VH locus, but is narrowly confined to promoters, coding regions and RSSs, with lower levels of histone acetylation in intergenic regions (Johnson et al., 2003; Espinoza and Feeney, 2005).
It has been shown by several groups, including our own, that individual VH genes rearrange at different frequencies (Love et al., 2000). For instance, the three functional members of the VHS107 family have very different rearrangement frequencies in vivo, with V1 rearranging five times more often than V11, and forty times more often than V13 (Love et al., 2000). Furthermore, all twenty VH7183 family members rearrange at very different frequencies in vivo (Williams et al., 2001), with VH81X rearranging considerably more often. By using freshly isolated pro-B cells, we provided the first in vivo demonstration that the extent of acetylation of histone proteins is one of the mechanisms by which rearrangement frequency of individual VH genes may be regulated (Espinoza and Feeney, 2005). In that work, a positive correlation was found between the extent of the association with acetylated histones and relative rearrangement frequency in the VHS107 and VH7183 families (Espinoza and Feeney, 2005).
The V(D)J recombination process is also tightly controlled by transcription factors. DJH-distal VH gene segments are blocked from undergoing VH-to-DJH rearrangement in pro-B cells from Pax5-deficient mice (Urbanek et al., 1994; Nutt et al., 1997; Hesslein et al., 2003). Until recently, there was evidence for two roles of Pax5 in controlling VH-to-DJH rearrangements: (a) facilitating rearrangement of distal VH genes by inducing IgH locus contraction (Fuxa et al., 2004), and (b) removing histone H3 methylated at lysine 9, a marker of repressed chromatin, from the VH locus, making VH genes accessible for VH-to DJH recombination (Johnson et al., 2004). In addition, we recently reported that Pax5 binds to VH coding regions and to the RAG1/RAG2 complex, which may facilitate RAG mediated recombination of individual VH genes (Zhang et al., 2006).
Acetylation of histones is not sufficient to permit V(D)J recombination, suggesting that other epigenetic modifications could also be important for fully inducing accessibility resulting in V(D)J recombination (Guo et al., 2002; Sikes et al., 2002; Hesslein et al., 2003; Jackson et al., 2005). In this study, we examine acetylation on histone H4 and dimethylation of lysine 9 on histone H3 (H3/K9me2) by chromatin immunoprecipitation (ChIP), using freshly isolated B220+ pro-B cells and thymocytes from μMT mice, and NIH-3T3 fibroblasts. We find that only in pro-B cells is there a positive correlation between the association level of individual VH genes with acetylated H4 and the relative rearrangement frequency in the VHS107 and VH7183 families. Furthermore, we demonstrate that association with H3/K9me2 inversely correlates with the differential rearrangement frequency of individual VHS107 and VH7183 genes in pro-B cells. Our DNA methylation study in the VHS107 family shows that the CpG site located 18 bps downstream of the Pax5 binding site (+113 relative to the start of the coding region) is highly methylated in all three VH genes, whereas a CpG site closer to the RSS (100 bps upstream of the RSS) shows differential methylation that correlates with rearrangement frequency. Strikingly, however, the CpG sites located in the coding regions are more highly methylated than the ones located near the RSS, and this CpG methylation pattern is similar in pro-B cells and thymocytes. Lastly, we find for two sites shared among all VHS107 family members, one located near the Pax5 binding site and the other downstream the RSS, that there is a positive relationship between rearrangement frequency and differential chromatin accessibility as determined by restriction enzyme cleavage of intact nuclei.
MATERIALS AND METHODS
Mice
The μMT mice (Kitamura et al., 1991), which have a targeted disruption of a membrane exon in the gene encoding the IgM constant region resulting in a complete block of B cell development at the pro-B cell step, were maintained in specific pathogenic-free animal facilities at The Scripps Research Institute. Experiments were approved and performed according to the regulatory standards of the IACUC committee.
Cell isolation and Cell lines
Bone marrow (BM) cells were isolated from 6 week old μMT mice by crushing femoral bones with a mortar and pestle. B220+ cells were pre-enriched by first staining BM cells with PE-conjugated B220 antibody, followed by purification using anti-PE microbeads (Miltenyi Biotech, Auburn, CA). In other experiments, CD19+ cells were pre-enriched by using anti-CD19 microbeads (Miltenyi Biotech, Auburn, CA) followed by staining with FITC-conjugated CD19 antibody. In both cases, viable pro-B cells (B220+ or CD19+) were sorted by flow cytometry. Thymocytes were obtained from 4-6 week old μMT mice. NIH-3T3 fibroblasts were maintained in DMEM (Gibco/Invitrogen, Carlsbad, CA) supplemented with 10% FBS. The cell line 18.8 (Alt et al., 1981), a pre-B cell line, was maintained in RPMI 1640 (Gibco/Invitrogen, Carlsbad, CA) supplemented with 10% FBS and 50 μmol/L β-mercaptoethanol.
In vivo Chromatin Immunoprecipitation (ChIP) and Real-time PCR analysis
Chromatin immunoprecipitations were performed as previously described (Espinoza and Feeney, 2005). Antibodies used included anti-acetylated H4 at lysines K5, K8, K12 and K16 (Cat. 06-866), and anti-dimethylated H3 at lysine 9 (Cat. 07-441), all obtained from Upstate (Lake Placid, NY). Real-time PCR was performed using the Quantitec SYBR PCR Kit (Qiagen, Valencia, CA), and the 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). The input DNA and the immunoprecipitated (bound) DNA were first quantified using PicoGreen® dye (Invitrogen, Carlsbad, CA). The input DNA was diluted in 1X Tris-EDTA to match the concentration of bound DNA. The real-time PCR was carried out using 0.2 ng DNA at 95 C for 15 min, followed by 45 cycles at 94 C for 15 sec, 60 C for 30 sec, and 72 C for 30 sec. Data were collected at 72 C. A melting curve analysis step was built at the end of the cycling program to verify quality of the PCR products. The sequences of the primers used and their location were previously described (Espinoza and Feeney, 2005). The specificity of each PCR reaction was first confirmed by sequencing the amplified product. Enrichment of active genes in our ChIP preparations was assessed by real-time PCR using primers for active genes (actin and β2M), and for an inactive gene (neuregulin). Data is presented as relative to actin for each amplified DNA sequence, which was determinated as: 2^ [(Ct input sample − Ct bound sample) − (Ct input actin − Ct bound actin)], where Ct is the cycle threshold.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts from the 18.8 cell line was prepared according to Dignam et al. (1983). Complementary oligonucleotides (Table 1) were annealed by heating equal molar mixture of the upper and lower strands to 95 C for 5 minutes in annealing buffer (10 mM Tris-HCl, pH 8.0, 50 mM NaCl and 1 mM EDTA). The oligonucleotide mixture was removed from the heating block and allowed to cool slowly down to RT on the bench (∼ 3 h). Double stranded oligonucleotides were labeled with [γ-32P] ATP using T4 polynucleotide kinase. The binding reaction consisted of 1.5 pmol labeled double-stranded oligonucleotide and 1 to 4 μg of nuclear extracts in a total volume of 20 μl. Before adding the labeled double-stranded oligonucleotide, the nuclear extract was preincubated in binding buffer [10 mM Hepes, pH 7.6, 50 mM KCl, 2.5 mM MgCl2, 10% glycerol, 1mM DTT and 150 ng/μl poly (dI−dC)−(dI−dC)] for 15 minutes at RT. For supershift assays, 0.3 μg of a monoclonal anti-Pax5 antibody (sc 13146, Santa Cruz Biotechnology, Santa Cruz, CA) were added to the binding reaction at the 15 minutes preincubation step. Labeled double-stranded oligonucleotides were added to the binding reaction and incubated for additional 25 minutes at RT. The reaction mixture was then loaded onto a 6% polyacrylamide gel (29:1, acrylamide/bisacrylamide ratio) and run at 126 V for 1 h at 4 C. After electrophoresis, the gels were dried and autoradiographed.
TABLE 1.
Oligonucleotide sequences
| Sequence (5'- 3') | Tm °C | |
|---|---|---|
| DNA methylation | ||
| V1 | AGTGTGAGGTGAAGCTGGTGGA ATTATATCTCCAAAATTTCCCTCTAACCAAAA GGTTTATTTTTAGTGATTTTTATATGGAGTGG (nested) ATTATATCTCCAAAATTTCCCTCTAACCAAAA (nested) |
55 55 |
| V11 | AGTGTGAGGTGAAGCTGGTGGA TACAATTCTATACAATTACCACTATCC TGTGTAATTTTTGGGTTTATTTTTATTGA (nested) TACAATTCTATACAATTACCACTATCC (nested) |
55 55 |
| V13 | GAGTCAAAATATTGAAGTTTTA AACAATTCTATTCAATTATCAC GTATTATATGAAGTGAGTTTTGA (nested) AACAATTCTATTCAATTATCAC (nested) |
49 50 |
| 81Xa | GAGAAGAGGCTGGAGTTGGT CTCCATTTTTATCCCTACATTTC |
55 |
| Chromatin accessibility | ||
| Nla III close to Pax5 site | ||
| V107-For V1ac-5' V11ac-5' V13ac-5' V107ac-5' BW4 |
CACTTTTAMATGGTAATTTATGG GTATGTACCAGCTTTCTCTAC TGAACAAGGGATGCTGAGTATC GGAACAAGGGATGCTGAGTACG GTATCCAGTGTGAGGTGAAG CCGGGAGATCTGAATTCTAG |
52 55 55 55 55 55 |
| Nla III close to RSS | ||
| V107-5' V1-5' V11-5' V13-5' V107co-5' BW3 |
TGGGTCCGCCAGCCTCCAG AGACTGGAGTGGATTGCTGCAAG AGGCACTTGAGTGGTTGGGTTTT CAGCCTCCAGGGAAGTCACC AACAGAGTAYAGTGCATC CCGGGAGATCTGAATTCTG |
60 55 55 55 47 55 |
| Germline transcription | ||
| J558 Vβ5b Actin |
CTAACCATGGGATGGAGCTGGATC TGACGAATTCAGGATGTGGTTACAAC CTCCTGGGAACAAGTTCAGCAA GATTAAGTTACAGAAAGCCAGTAGC TCTGGCTCCTAGCACCATGAAGA GGGACTCATCGTACTCCTGCTTG |
60 60 60 |
| Oligos for EMSAs | ||
| V1 V11 V13 CD19 |
GTGCAACTTCTGGGTTCACCTTCAGTGATTTCTACATG CATGTAGAAATCACTGAAGGTGAACCCAGAAGTTGCAC GTGCAACTTCTGGGTTCACCTTCACTGATTACTACATG CATGTAGTAATCAGTGAAGGTGAACCCAGAAGTTGCAC GTGAAGCTTCTGGATTCACCTTCACTGATTACTACATG CATGTAGTAATCAGTGAAGGTGAATCCAGAAGCTTCAC CTAGACAGACACCCATGGTTGAGTGCCCTCCAGT ACTGGAGGGCACTCAACCATGGGTGTCTGTCTAG |
|
Primers were taken from Johnson et al. (2004).
Primers were taken from Fuxa et al. (2004).
DNA methylation analysis
For methylation analysis, 1 μg of genomic DNA was treated using the CpGenome™ Fast DNA Modification Kit (Chemicon, Temecula, CA) according to the manufacturer's specification. Bisulphite-treated DNA was subjected to nested PCRs. Individual PCR products were ligated into the T/A cloning vector pCR® 2.1-TOPO (Invitrogen, Carlsbad, CA). Two independent bisulphite convertions were performed, and at least 10 bisulphite-converted clones were sequenced per individual bisulphite conversion. Primers used in the PCR reactions are listed in Table 1.
Chromatin accessibility assay
Cell nuclei were prepared according to Hempel and Ferrier (2004) with some modifications (Espinoza and Feeney, 2006). For restriction enzyme digestion, 106 nuclei were treated with Nla III for 10 minutes at 37 C, followed by overnight digestion with proteinase K. DNA was then treated with RNase/DNase free (Roche, Palo Alto, CA), and the purified DNA was collected by isopropanol precipitation. The detection and quantification of chromatin cleavage was performed as recently described (Espinoza and Feeney, 2006). First-strand synthesis reactions were performed with Pfu DNA polymerase (Stratagene, San Diego, CA) using 200 ng of restriction enzyme digested DNA and family-specific forward primers V107-For or V107-5' (Table 1). After overnight ligation of the unidirectional BW linker at 17°C, linker-ligated DNA samples were used in a first round of PCR (18 cycles) using the V107-For or V107-5' primers paired with the BW4 or BW3 primers, respectively. Fifty picograms of each amplified product were utilized for SYBR Green real-time PCR, using gene specific forward primers, each paired with the respective linker primer (Table 1). The differential amplification efficiency of each pair of primers was determined as recently described (Espinoza and Feeney, 2006). Briefly, purified PCR products for V1, V11 and V13 were diluted to the same number of amplicon copies, and a standard curve was created. For normalization purposes, the whole product signal was measured by using a family-specific forward primer (V107ac-5' or V107co-5') and the BW4 or BW3 primers, respectively (Table 1). After determination of the Ct values for each primer combination (Ctcom), the Ct value from the whole product signal (Ctp) was subtracted from Ctcom (Ctcom − Ctp). The difference was named Ct control. Chromatin accessibility of individual gene family members were calculated as 2−(Ct sample − Ct control), where Ct sample is the Ct value for the gene under study.
RNA preparation and RT-PCR
Total RNA was extracted from pro-B cells and thymocytes of μMT mice with RNeasy Plus Mini Kit (Qiagen, Valencia, CA). Between 0.1 and 0.2 μg of total RNA was reverse-transcribed for 30 minutes using the QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA). After reverse transcription, PCR was carried out (primers, Table 1) using AmpliTaq Gold® DNA polymerase (Applied Biosystems, Foster City, CA) under the following conditions: denaturation at 95°C for 10 min followed by 40 cycles at 95 C for 30 sec, 60 C for 30 sec, and 72 C for 1 min. The amplification was ended with 10-min incubation at 72 C.
RESULTS
Only in pro-B cells does the level of association of individual VH gene members with acetylated histone H4 correlate with their rearrangement frequency
We previously showed a positive correlation between the extent of histone H3 and H4 acetylation and the relative rearrangement frequency for the VHS107, VH7183 and VHJ558 families in pro-B cells from μMT mice (Espinoza and Feeney, 2005). Furthermore, in that study, we found that induction of H3 acetylation is specific to the B lineage (Espinoza and Feeney, 2005). To demonstrate if there is lineage specificity in the differential level of acetylated histone H4 associated with VH genes, we performed ChIP on freshly isolated μMT thymocytes, which are predominantly CD4+CD8+ pre-T cells, and NIH-3T3 fibroblasts. We found that NIH-3T3 fibroblasts displayed little acetylation of H4 histones associated with any of the VH genes analyzed (Fig. 1C). Furthermore, although thymocytes showed an increased level of H4 acetylation as compared to NIH-3T3 fibroblasts (Fig. 1B), it is only in pro-B cells where the level of association of individual VH members with acetylated H4 histones varies, and correlates with their rearrangement frequency (Fig. 1A). Thus, we conclude that there is not differential association of acetylated H4 with individual VH genes in thymocytes (Fig. 1B), as was clearly observed in pro-B cells (Fig. 1A).
Figure 1.
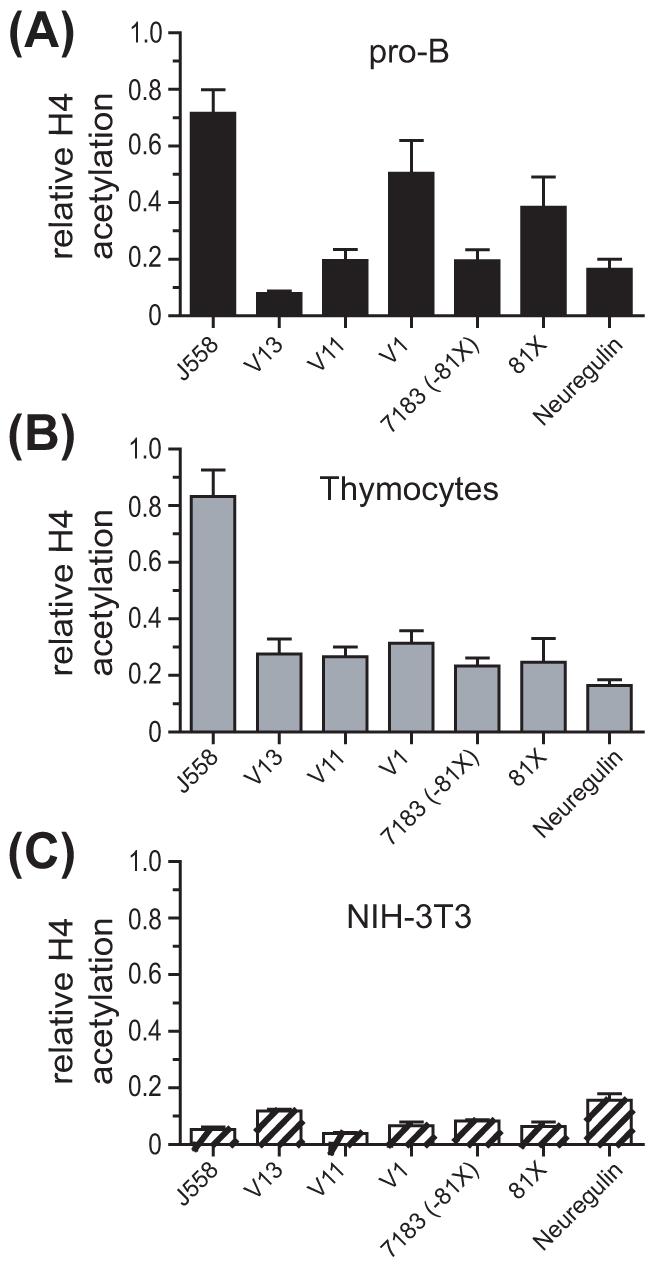
H4 acetylation is associated with frequently rearranging VH genes in pro-B cells. ChIP assays were performed on (A) adult BM pro-B cells (B220+), (B) thymocytes, and (C) NIH-3T3 fibroblasts. Analysis was performed by real-time PCR. Data are expressed relative to the actin gene (actin = 1). Results represent the mean ± SEM of 3-4 independent ChIP preparations. Data from panel A were previously published (Espinoza and Feeney, 2005) and are being used with permission.
Higher association of dimethylated H3/K9 with VH genes in non-B lineage cells
It has been shown that VH genes are associated with dimethylated histone H3 at lysine 9 (H3/K9me2), a marker of repressed chromatin, before the onset of VH-to-DJH rearrangements (Johnson et al., 2004). Since removal of that modification seems to be required to promote VH rearrangements (Johnson et al., 2004), we wanted to test the hypothesis that H3/K9me2 is more frequently removed from highly rearranging VH genes than from poorly rearranging VH genes. To test our hypothesis, we performed ChIP assays on freshly isolated pro-B cells and thymocytes from μMT mice and on NIH-3T3 fibroblasts. We found that the extent of association of H3/K9me2 was inversely correlated with the differential rearrangement frequency of VHS107 and VH7183 family members in pro-B cells (Fig. 2A). Thus, the poorly rearranging V13 gene had the highest association with H3/K9me2 in the VHS107 family, followed by V11, and then by V1, which had the least association with this repressive modification (Fig. 2A). Furthermore, the highly rearranging VH81X gene was more depleted of H3/K9me2 in pro-B cells, as compared to the less frequently rearranging VH7183 family members (Fig. 2A). Thymocytes showed a slightly higher association of most VH genes with H3/K9me2, although the VH81X gene had low association with this histone modification in both pro-B cells and thymocytes (Fig. 2B). All VH genes studied were highly associated with H3/K9me2 in NIH-3T3 fibroblasts (Fig. 2C).
Figure 2.
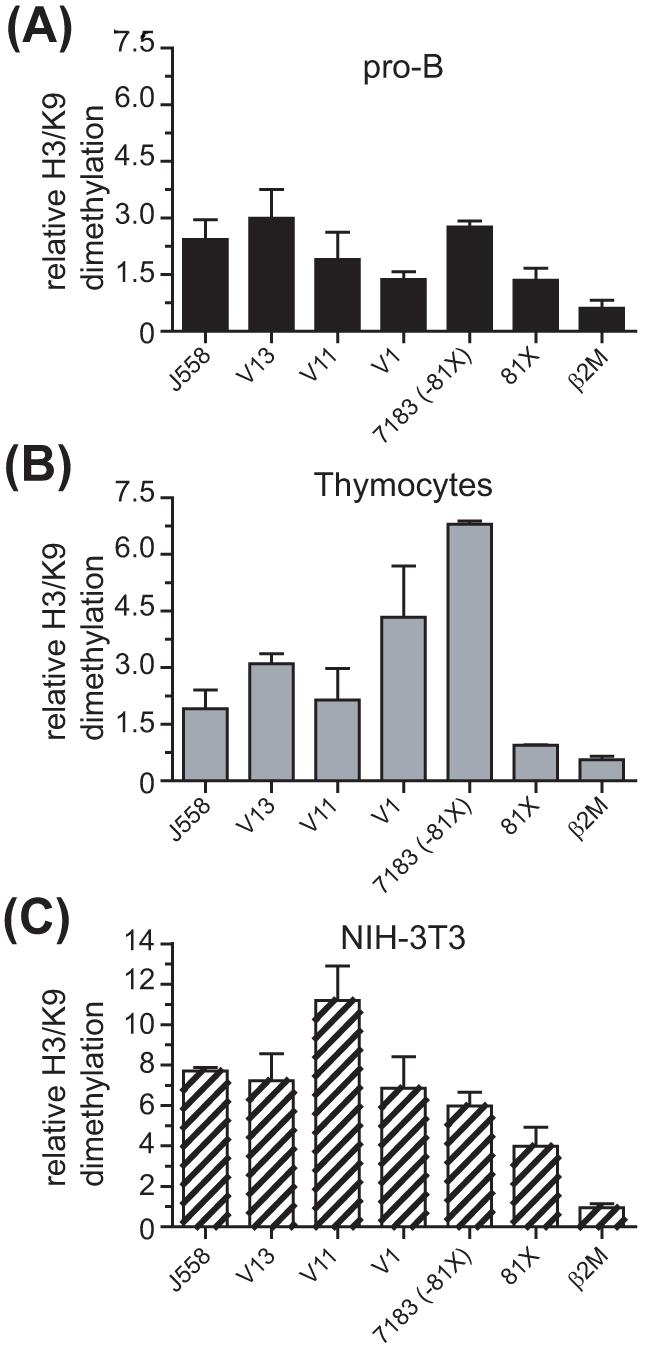
Poorly rearranging VH genes are associated with dimethylated H3/K9 in pro-B cells. ChIP assays were performed on (A) adult BM pro-B cells (B220+), (B) thymocytes, and (C) NIH-3T3 fibroblasts. Analysis was performed by real-time PCR. Data are expressed relative to the actin gene (actin = 1). The neuregulin values for pro-B cells, thymocytes and NIH-3T3 fibroblasts were 15.79±2.83, 15.01±3.51, and 5.59±1.6, respectively Results represent the mean ± SEM of 2 independent ChIP preparations.
Taken together, our data show that association with H3/K9me2 inversely correlates with differential rearrangement frequency of VHS107 and VH7183 genes in pro-B cells. Furthermore, the comparable levels of H3/K9me2 associated with some VH genes in pro-B cells and thymocytes suggest that this histone modification is unlikely to be sufficient to block rearrangement of those VH genes in thymocytes.
Binding of Pax5 to individual VHS107 family members correlates with their rearrangement frequency
We have shown that the transcription factor Pax5 can bind to VH coding regions and interact with the RAG1/RAG2 complex to directly facilitate VH-to-DJH rearrangements (Zhang et al., 2006). By electrophoretic mobility shift assays (EMSAs), using oligo probes representing the region between +63 bps to +102 bps relative to the start of the V1, V11 and V13 coding regions (Fig. 3A), and an oligo containing a Pax5 site derived from the mouse CD19 promoter, we demonstrated that Pax5 binds to all three VHS107 family members, but the binding of Pax5 was much stronger to V1 and V11 than to V13 (Fig. 3B). The formation of Pax5-DNA complexes were verified by supershift assays using a monoclonal antibody to Pax5 (Fig. 3C). Furthermore, cold competition experiments showed that Pax5 bound 18.5-times less efficiently to V13, as compared to the high affinity Pax5 binding to V1 and V11 (data not shown). We conclude that for the VHS107 family, there is a positive correlation between the high-affinity binding sites for Pax5 in V1 and V11 coding regions and their preferential recombination. Conversely, our results suggest that the weak binding of Pax5 to V13 may be one of the reasons why V13 can not rearrange efficiently.
Figure 3.
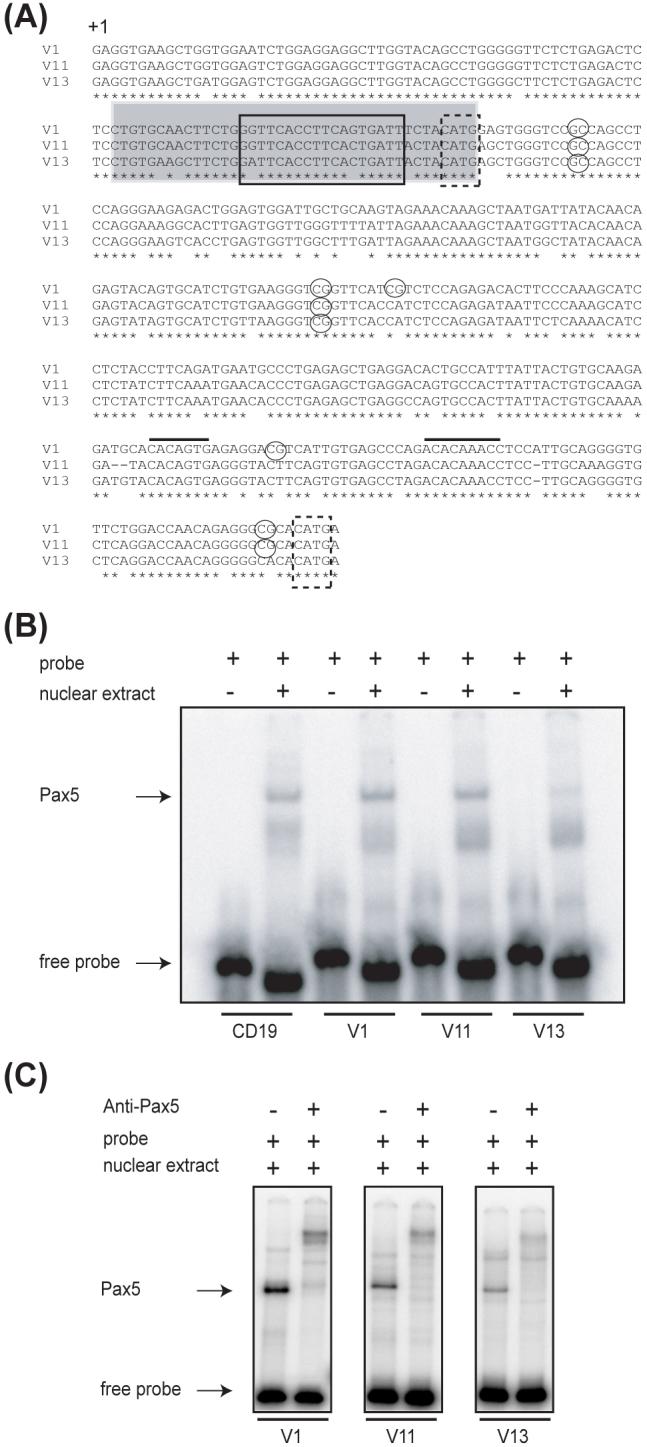
Differential binding of Pax5 to individual members of the VHS107 family. (A) Alignment of the VHS107 family members. Solid line box, Pax5 binding site; gray shaded box, oligos used in EMSAs; dashed boxes, Nla III sites; circles, CpG sites; black lines, RSS elements. The start of the coding region is indicated with +1. (B) Binding of Pax5 to individual Pax5 binding sites using double-stranded labeled oligos from the CD19, V1, V11 and V13 genes and nuclear extracts from 18.8 cells. (C) Supershift assays performed with anti-Pax5 antibody.
In pro-B cells and thymocytes, DNA methylation is more prevalent in the coding regions than near the RSS
It is known that methylation of DNA at CpG sites can suppress gene expression through effects on DNA binding proteins and chromatin structure (Fuks, 2005). Because several studies have shown that DNA methylation can inhibit recombination (Cherry and Baltimore, 1999; Whitehurst et al., 2000; Nakase et al., 2003), we attempted to determine in the VHS107 family if the extent of CpG methylation varied for individual VH genes. We analyzed all three VHS107 family members as well as the VH81X gene by bisulphite conversion of genomic DNA from freshly isolated pro-B cells and thymocytes of μMT mice.
We found that the CpG site located 18 bps downstream of the Pax5 site in framework 1 of the coding region (+ 113 relative to the start of the coding region) was highly methylated in all VHS107 family members in pro-B cell and also in thymocytes (Fig. 4A, B, and C). However, the CpG site located 111 bps downstream of the Pax5 site (+205 relative to the start of the coding region) was differentially methylated in pro-B cells (Fig. 4A, B, and C). Thus, 50%, 75% and 100% of all the clones had that CpG site methylated in V1, V11 and V13, respectively, correlating very well with the relative rearrangement frequency of these three genes. Furthermore, this CpG site was more methylated in thymocytes than in pro-B cells for V1 (Fig. 4A), whereas there were no significant differences in methylation between pro-B cells and thymocytes for the same CpG position in the V11 and V13 gene segments (Fig. 4B and C).
Figure 4.
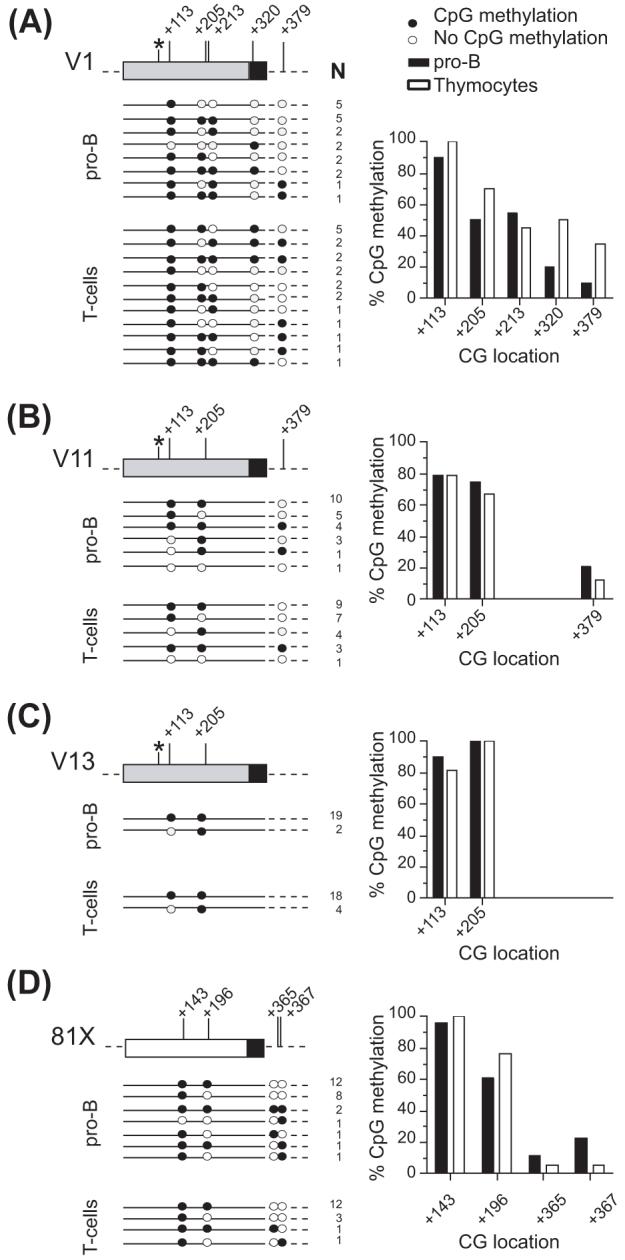
DNA methylation patterns of individual VH gene segments in pro-B cells (CD19+) and thymocytes. The schematic representation and the percentage of CpG methylation for the (A) V1, (B) V11, (C) V13, and (D) 81X genes are depicted. Asterisks show the location of the Pax5 binding site (position +94). The numbering is relative to the start of the coding region, and indicates the position of CG sites. N is the number of sequences with the same pattern of DNA methylation.
In contrast to the coding regions, the CpG sites near the RSS were predominantly demethylated. We analyzed the methylation status of a CpG site located 34 bps downstream of the V1 and V11 RSS elements (+379 relative to the start of the coding region). Unfortunately, the V13 gene does not have this CpG site. We found that in pro-B cells, only 10% and 21% of all clones were methylated in V1 and V11, respectively, at that site (Fig. 4A and B). Furthermore, in thymocytes the same CpG site was methylated in 35% of all V1 clones. It is possible that the differential methylation at the CpG site located 3' of the V1 and V11 RSS elements could favor the rearrangement of V1 over V11. In addition, the CpG site located in the spacer of the V1 RSS element (+320 relative to the start of the coding region) was methylated in only 20% of the clones in pro-B cells, but 50% in thymocytes (Fig. 4A). Thus, it is likely that demethylation of this site is critical for V1 rearrangement to occur.
For the VH81X gene, as previously reported by Johnson et al. (2004), we found that the CpG site at position +143 (relative to the start of the coding region) was methylated in all clones analyzed, whereas the second site (+196) was partially methylated (62% of all clones, Fig. 4D). The two CpG sites located downstream of the RSS in the VH81X gene were 10% and 23% methylated, respectively. The same CpG methylation pattern of the VH81X gene was observed not only in pro-B cells but also in thymocytes, suggesting that the low association of H3/K9me2 with VH81X in pro-B cells and thymocytes (Fig. 2A and B) correlates with the low DNA methylation at CpG sites surrounding the RSS.
Thus, our DNA methylation study indicates that (1) DNA methylation is much higher in VH coding regions than near the RSS, (2) the extent of DNA methylation does vary between individual VH genes, and (3) the extent of demethylation is similar in both pro-B cells and thymocytes, except for the frequently rearranging V1 gene.
Chromatin accessibility of individual VHS107 family members correlates with their rearrangement frequency
We used a restriction enzyme accessibility assay to determine whether association of acetylated histones with rearranging VH genes could be reflected in differential chromatin accessibility (Fig. 5A). We searched for restriction enzyme sites which could be shared among all VHS107 family members, and found two Nla III restriction sites for analysis: one site located 8 bps downstream of the Pax5 consensus binding site (+102 relative to the start of the coding region) (Zhang et al., 2006), and a second site located 41 bps downstream of the RSS (+386 relative to the start of the coding region) (Fig. 5A).
Figure 5.
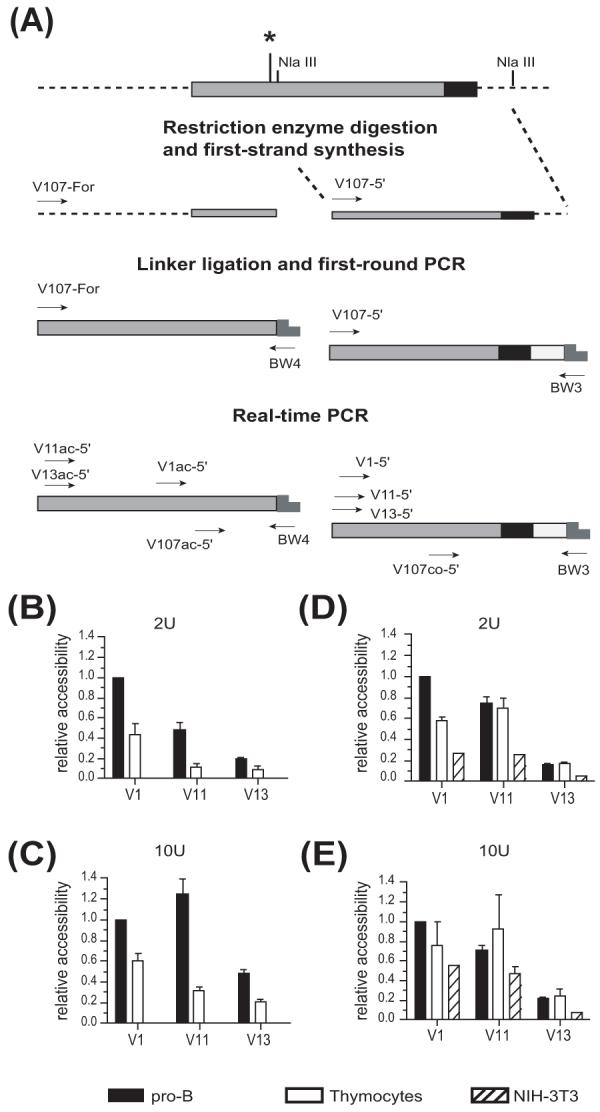
Chromatin accessibility correlates with rearrangement frequency. (A) Schematic representation of the VHS107 coding region (dark gray) showing the location of the RSS (black), the Nla III restriction enzyme sites, and the step by step strategy used to measure chromatin accessibility. Asterisk shows the location of the Pax5 binding site. Chromatin accessibility was assayed in the region flanking the Pax5 site at (B) 2 U, and (C) 10 U of enzyme in pro-B cells (CD19+), thymocytes and NIH-3T3 fibroblasts. Analysis of the chromatin accessibility surrounding the RSS was also performed at (D) 2 U and (E) 10 U of enzyme in pro-B cells (CD19+), thymocytes and NIH-3T3 fibroblasts. Data were obtained by real-time PCR, and are expressed relative to the accessibility of the V1 gene in pro-B cells (= 1). Results represent the mean ± SEM of 4 real-time PCRs from 2 independent preparations.
When analyzing the region adjacent to the Pax5 binding site, we found that at 2 U of enzyme, V1 was more accessible than V11, and V11 was more accessible than V13 in pro-B cells (Fig. 5B). At 10 U of enzyme, V1 and V11 had similar chromatin accessibility, and both were still more accessible than V13 (Fig. 5C). Chromatin accessibility of the VHS107 gene family members was lower in thymocytes than in pro-B cells, and there was no measurable accessibility in NIH-3T3 fibroblasts (Fig. 5B and C).
Analysis of the chromatin accessibility surrounding the region located downstream of the RSS also indicated a correlation between chromatin accessibility and rearrangement frequency for the individual VHS107 family members in pro-B cells at both 2 U and 10 U of enzyme (Fig. 5D and E). Surprisingly, we found that thymocytes and even NIH-3T3 fibroblasts showed very similar relative pattern of accessibility, although lower accessibility was observed at 2 U of enzyme where V1 was about 2-fold and 4-fold more accessible in pro-B cells than in thymocytes and NIH-3T3 fibroblasts, respectively (Fig. 5D). Furthermore, V11 and V13 genes displayed similar accessibility in pro-B cells and thymocytes, while the accessibility of those genes in NIH-3T3 fibroblasts was lower (Fig. 5D). Overall, this data suggest that accessibility at this site is not B-lineage specific. We conclude that increased accessibility of V1 and V11 at the region located 8 bps downstream of the Pax5 consensus binding site paralleled the strong binding of Pax5 to both V1 and V11 genes but not to V13. Furthermore, the relative rearrangement frequency of V1, V11 and V13 correlates very well with the restriction endonuclease accessibility of those VH genes, but this accessibility is only partially pro-B cell specific.
Low levels of VH germline transcription in thymocytes
Because our chromatin accessibility assays showed that thymocytes had some level of accessibility on both VHS107 gene segments analyzed, we wanted to test whether we could detect any VHJ558 germline transcripts in thymocytes. We performed reverse transcription on total RNA from μMT pro-B cells and thymocytes using random hexamer primers, followed by PCR or real-time PCR with family forward and reverse primers located in the leader and the RSS spacer region, respectively. We found very low levels of germline VHJ558 cDNA products in thymocytes (Fig. 6). Quantification by real-time PCR showed that they represented only 0.4% of the level observed in pro-B cells (data not shown). Thus, we conclude that VH gene germline transcripts are far more abundant in pro-B cells than in thymocytes, which agrees with the fact that VH-to-DJH rearrangements only occur in pro-B cells.
Figure 6.
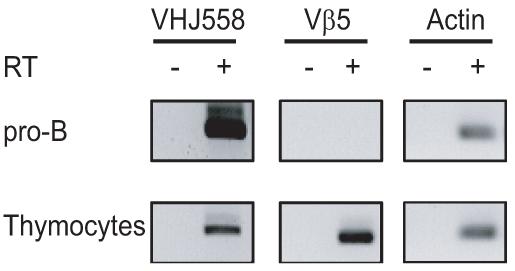
VH germline transcription in thymocytes. Transcripts of the indicated genes were analyzed by RT-PCR using pro-B cell (CD19+) and thymocyte cDNA samples. Above lanes, with (+) or without (−) reverse transcriptase (RT).
DISCUSSION
To further elucidate the epigenetic mechanisms regulating accessibility of individual VH genes to V(D)J recombination, we investigated the modification status of histone proteins, changes in DNA methylation and chromatin accessibility associated with VHS107 and VH7183 family members. We have previously shown a positive correlation between the extent of histone H3 and H4 acetylation and the differential rearrangement frequency of individual VHS107 and VH7183 family members in freshly isolated pro-B cells (Espinoza and Feeney, 2005). Furthermore, by assessing the acetylation status of histone H3 associated with VH genes in non-B cells, we found that induction of H3 acetylation was specific to the B-cell lineage (Espinoza and Feeney, 2005).
In the present report, we have performed a comparative study in pro-B cells and thymocytes from μMT mice, and in NIH-3T3 fibroblasts, analyzing the association of individual VH genes with acetylated H4 and H3/K9me2. We show that the positive correlation between rearrangement frequency among the VHS107 and VH7183 family members and their level of enrichment in acetylated H4 fractions is B-cell specific. Furthermore, we find that the previously observed preferential association of H3/K4me2 with the VHJ558 genes and to a lesser extent with the V1 gene (Espinoza and Feeney, 2005) is only observed in pro-B cells (data not shown).
Because it has been proposed that removal of H3/K9me2, a maker of repressive chromatin, seems to be required to promote VH-to-DJH rearrangements (Johnson et al., 2004), we tested the hypothesis that H3/K9me2 was preferentially removed from highly rearranging VH genes but not from poorly rearranging VH genes. Indeed, we find that in pro-B cells, poorly rearranging VH genes show higher association with H3/K9me2. Interestingly, the VH81X gene shows the same low level of association with this repressive modification not only in pro-B cells, but also in thymocytes. This observation indicates that association with H3/K9me2 is not the predominant reason for the lack of VH gene rearrangements in thymocytes. Our results differ from that of a previous report in which VHJ558 genes and the VH81X gene were highly associated with H3/K9me2 in thymocytes, but not in Rag−/− pro-B cells (Johnson et al., 2004). However, as in that study, we also show low association of the V11 RSS gene region with H3/K9me2 in thymocytes.
A likely interpretation of our ChIP data is that only a small proportion of pro-B cells contain acetylated histones H3 and H4 associated with frequently rearranging genes (e.g. V1 and 81X). Conversely, a higher proportion of pro-B cells may still have their VH genes associated with H3/K9me2 and not be accessible for rearrangement. However, we cannot rule out that these modifications occur on the same chromatin fragment. In that case, an alternative interpretation will be that increasing levels of both H3 and H4 acetylation could surpass the repressive effect of H3/K9me2 when these histone modifications are associated with frequently rearranging VH genes in pro-B cells.
We also show that Pax5 binds to the coding regions of all three members of the VHS107 family, but the binding of Pax5 is almost 20-fold stronger to V1 and V11 than to V13. Since we have demonstrated that the binding of Pax5 to VH genes can facilitate recombination (Zhang et al., 2006), we propose that the presence of a strong Pax5 binding site in V1 and V11, but not in the infrequently rearranging V13, may be another mechanism by which non-random VH gene usage is promoted. Analysis of the methylation status of a CpG site located just 18 bp downstream of the Pax5 site (+113 relative to the start of the coding region) suggests that methylation at that position does not interfere with the binding of Pax5 to individual VHS107 family members in vivo. With the exception of the CpG site located 111 bp downstream of the V1 Pax5 site (+205 relative to the start of the coding region), which is more methylated in thymocytes than in pro-B cells, we find in general the same DNA methylation pattern in pro-B cells and in thymocytes: higher CpG methylation in VH coding regions, and hypomethylation of the CpG sites located near of the RSS. Our data together with a previous report suggest that recombination can take place at VH genes when DNA methylation occurs in the coding region (Johnson et al., 2004). It has been proposed that DNA methylation in the RSS directly inhibits V(D)J cleavage and induces an inaccessible chromatin configuration (Cherry and Baltimore, 1999; Whitehurst et al., 2000; Nakase et al., 2003). In agreement with these observations, we show that a CpG site within the spacer of the V1 RSS element is 80% demethylated.
It has been shown that DNA demethylation of the kappa locus occurs at a late stage in the process of acquiring accessibility for recombination, and it takes place monoallelically (Mostoslavsky et al., 1998; Goldmit et al., 2005). Our data infer that DNA demethylation in VH genes may be limited to the region in the vicinity of the RSS. Also, since CpG sites located near the RSS are > 80% demethylated in pro-B cell and thymocytes, we suggest that DNA demethylation may neither be B-cell specific nor be restricted to one allele at the VH locus as it is in the kappa locus.
Gene rearrangement in the immune system is preceded by increased chromatin accessibility (Yancopoulos and Alt, 1985). Accordingly, we report here the first chromatin accessibility analysis on individual members of the VHS107 family. We find a positive correlation between rearrangement frequency and chromatin accessibility for the three VHS107 family members in pro-B cells at two different sites: one located near the Pax5 binding site, and the second located 3' of the RSS. Interestingly, the region located 8 bps downstream of the Pax5 site (+102 relative to the start of the coding region) shows differential chromatin accessibility for the three genes analyzed, although all members share a methylated CpG site 18 bps downstream of the Pax5 site (+113 relative to the start of the coding region). This observation indicates that chromatin accessibility and DNA methylation in that region are independent events.
Analysis of the region surrounding the RSS also shows a positive correlation between chromatin accessibility and rearrangement frequency in pro-B cells. However, thymocytes and NIH-3T3 fibroblasts show similar accessibility in that region, suggesting that accessibility at this site is not B-lineage specific. One interpretation of our data is that the Nla III site located downstream of the RSS may be situated in the DNA linker between two positioned nucleosomes. In that situation, the restriction enzyme site will be accessible in all cell types.
The data presented in this paper demonstrate that the infrequently rearranging VHS107 gene, V13, has the weakest Pax5 binding site, the lowest accessibility to restriction enzyme cleavage, the lowest degree of histone acetylation, the highest extent of H3/K9me2, and the highest CpG methylation values, while the most frequently rearranging V1 gene displays the opposite pattern. We propose that these genetic and epigenetic differences are likely to affect recombination frequency in vivo.
Subnuclear location is a mechanism by which transcription and recombination of the IgH locus is regulated during lymphocyte development (Kosak et al., 2002). In pro-B nuclei, the IgH locus is centrally located and undergoes large-scale compaction. This compacted state facilitates long-range V-to-DJ rearrangements, particularly of the distal VHJ558 genes (Kosak et al., 2002), and it is mediated by the transcription factor Pax5 (Fuxa et al., 2004). In thymocytes, however, the inactive IgH locus is mainly localized at the nuclear periphery through an interaction with the nuclear lamina or its associated proteins, but not with centromeric heterochromatin (Kosak et al., 2002). Increasing evidence supports the role of the nuclear periphery as a repressive compartment for transcriptional silencing in higher eukaryotes (Kosak and Groudine, 2004). Therefore, although our chromatin accessibility study indicates that VHS107 genes are accessible in their coding and RSS region in thymocytes, the pericentric localization of the IgH locus may be sufficient to inhibit any chance of VH-to-DJH recombination in those cells.
Taken together, our data show that several factors seem to differentiate frequently from infrequently rearranging VH genes. These include (1) high association of VH genes with both acetylated H3 and H4, (2) decreased association of VH genes with the repressive modification H3/K9me2, (3) the extent of DNA hypomethylation surrounding RSSs, (4) the presence of strong versus weak Pax5 binding sites in the coding region, and (5) increasing chromatin accessibility at the Pax5 site in the VH coding region and near the RSS. In the permissive environment of pro-B cells, these factors are likely to contribute to the unequal rearrangement potential of individual VH genes.
ACKNOWLEDGEMENTS
We thank R. Rouse and J. Menendez for critical reading of the manuscript. This work was supported by the grant AI 52313 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alt F, Rosenberg N, Lewis S, Thomas E, Baltimore D. Organization and reorganization of immunoglobulin genes in A-MULV-transformed cells: rearrangement of heavy but not light chain genes. Cell. 1981;27:381–390. doi: 10.1016/0092-8674(81)90421-9. [DOI] [PubMed] [Google Scholar]
- Cherry SR, Baltimore D. Chromatin remodeling directly activates V(D)J recombination. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10788–10793. doi: 10.1073/pnas.96.19.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D, Sen R. Stepwise activation of the immunoglobulin μ heavy chain gene locus. EMBO J. 2001;20:6394–6403. doi: 10.1093/emboj/20.22.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza CR, Feeney AJ. The extent of histone acetylation correlates with the differential rearrangement frequency of individual VH genes in pro-B cells. J. Immunol. 2005;175:6668–6675. doi: 10.4049/jimmunol.175.10.6668. [DOI] [PubMed] [Google Scholar]
- Espinoza CR, Feeney AJ. Quantifying chromatin accessibility of individual gene family members by combining ligation-mediated PCR with real-time PCR. Biotechniques. 2006;41:404–408. doi: 10.2144/000112263. [DOI] [PubMed] [Google Scholar]
- Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmit M, Ji Y, Skok J, Roldan E, Jung S, Cedar H, Bergman Y. Epigenetic ontogeny of the Igκ locus during B cell development. Nat. Immunol. 2005;6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- Goodhardt M, Cavelier P, Doyen N, Kallenbach S, Babinet C, Rougeon F. Methylation status of immunoglobulin kappa gene segments correlates with their recombination potential. Eur. J. Immunol. 1993;23:1789–1795. doi: 10.1002/eji.1830230809. [DOI] [PubMed] [Google Scholar]
- Guo J, Hawwari A, Li H, Sun Z, Mahanta SK, Littman DR, Krangel MS, He YW. Regulation of the TCRα repertoire by the survival window of CD4+CD8+ thymocytes. Nat. Immunol. 2002;3:469–476. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- Hempel WM, Ferrier P. Restriction endonuclease accessibility as a determinant of altered chromatin structure. Methods Mol. Biol. 2004;287:53–63. doi: 10.1385/1-59259-828-5:053. [DOI] [PubMed] [Google Scholar]
- Hesslein DG, Pflugh DL, Chowdhury D, Bothwell AL, Sen R, Schatz DG. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 2003;17:37–42. doi: 10.1101/gad.1031403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A, Kondilis HD, Khor B, Sleckman BP, Krangel MS. Regulation of T cell receptor β allelic exclusion at a level beyond accessibility. Nat. Immunol. 2005;6:189–197. doi: 10.1038/ni1157. [DOI] [PubMed] [Google Scholar]
- Johnson K, Angelin-Duclos C, Park S, Calame KL. Changes in histone acetylation are associated with differences in accessibility of VH gene segments to V-DJ recombination during B-cell ontogeny and development. Mol. Cell. Biol. 2003;23:2438–2450. doi: 10.1128/MCB.23.7.2438-2450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Pflugh DL, Yu D, Hesslein DG, Lin KI, Bothwell AL, Thomas-Tikhonenko A, Schatz DG, Calame K. B cell-specific loss of histone 3 lysine 9 methylation in the VH locus depends on Pax5. Nat. Immunol. 2004;5:853–861. doi: 10.1038/ni1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura D, Roes J, Kühn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin μ chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- Kosak ST, Groudine M. Form follows function: The genomic organization of cellular differentiation. Genes Dev. 2004;18:1371–1384. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Love VA, Lugo G, Merz D, Feeney AJ. Individual VH promoters vary in strength, but the frequency of rearrangement of those VH genes does not correlate with promoter strength nor enhancer-independence. Mol. Immunol. 2000;37:29–39. doi: 10.1016/s0161-5890(00)00023-7. [DOI] [PubMed] [Google Scholar]
- Maës J, O'Neill LP, Cavelier P, Turner BM, Rougeon F, Goodhardt M. Chromatin remodeling at the Ig loci prior to V(D)J recombination. J. Immunol. 2001;167:866–874. doi: 10.4049/jimmunol.167.2.866. [DOI] [PubMed] [Google Scholar]
- McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of V(D)J recombination. Science. 2000;287:495–498. [PubMed] [Google Scholar]
- Mostoslavsky R, Singh N, Kirillov A, Pelanda R, Cedar H, Chess A, Bergman Y. κ chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev. 1998;12:1801–1811. doi: 10.1101/gad.12.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase H, Takahama Y, Akamatsu Y. Effect of CpG methylation on RAG1/RAG2 reactivity: implications of direct and indirect mechanisms for controlling V(D)J cleavage. EMBO Rep. 2003;4:774–780. doi: 10.1038/sj.embor.embor904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Urbanek P, Rolink A, Busslinger M. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 1997;11:476–491. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- Sikes ML, Meade A, Tripathi R, Krangel MS, Oltz EM. Regulation of V(D)J recombination: a dominant role for promoter positioning in gene segment accessibility. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12309–12314. doi: 10.1073/pnas.182166699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- Whitehurst CE, Schlissel MS, Chen J. Deletion of germline promoter PDβ1 from the TCRβ locus causes hypermethylation that impairs Dβ1 recombination by multiple mechanisms. Immunity. 2000;13:703–714. doi: 10.1016/s1074-7613(00)00069-8. [DOI] [PubMed] [Google Scholar]
- Williams GS, Martinez A, Montalbano A, Tang A, Mauhar A, Ogwaro KM, Merz D, Chevillard C, Riblet R, Feeney AJ. Unequal VH gene rearrangement frequency within the large VH7183 gene family is not due to recombination signal sequence variation, and mapping of the genes shows a bias of rearrangement based on chromosomal location. J. Immunol. 2001;167:257–263. doi: 10.4049/jimmunol.167.1.257. [DOI] [PubMed] [Google Scholar]
- Yancopoulos GD, Alt FW. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. [PubMed] [Google Scholar]
- Zhang Z, Espinoza CR, Yu Z, Stephan R, He T, Williams GS, Burrows PD, Hagman J, Feeney AJ, Cooper MD. Transcription factor Pax5 (BSAP) transactivates the RAG-mediated VH-to-DJH rearrangement of immunoglobulin genes. Nat. Immunol. 2006;7:616–624. doi: 10.1038/ni1339. [DOI] [PubMed] [Google Scholar]


