Abstract
Differences in immune control of HIV-1 infection are often attributable to the highly variable HLA class I molecules that present viral epitopes to cytotoxic T-lymphocytes. In our immunogenetic analyses of 429 HIV-1 discordant Zambian couples (infected index partners paired with cohabiting seronegative partners), several HLA class I variants in index partners were associated with contrasting rates and incidence of HIV-1 transmission within a 12-year study period. In particular, A*3601 on the A*36-Cw*04-B*53 haplotype was the most unfavorable marker of HIV-1 transmission by index partners, while Cw*1801 (primarily on the A*30-Cw*18-B*57 haplotype) was the most favorable, irrespective of the direction of transmission (male to female or female to male) and other commonly recognized co-factors of infection, including age and genital ulcer/inflammation. The same HLA markers were further associated with contrasting viral load levels in index partners, but they had no clear impact on HIV-1 acquisition by the seronegative partners. Thus, HLA class I gene products not only mediate HIV-1 pathogenesis and evolution but also influence heterosexual HIV-1 transmission. {This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publishers of The JI, holds the copyright to this manuscript. This manuscript has not been copyedited or subjected to editorial proofreading by The JI; hence it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the United States National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org}.
Keywords: AIDS, Comparative Immunology, Human, MHC
As molecules responsible primarily for antigen presentation and immune surveillance, human leukocyte antigen (HLA)4 products are best known for their extensive allelic diversity (1–4), which is considered essential to the constant combat with a wide range of human pathogens (5). Epidemiologic and experimental analyses of patients with chronic or terminal HIV-1 infection have revealed multiple HLA class I alleles that can differentially regulate virologic, immunologic and clinical outcomes, often through their preferential targeting of conserved or variable HIV epitopes for cytotoxic T-lymphocyte (CTL) responses (6–8). Such CTL responses also lead to predictable HIV-1 mutations and immune escape, as documented in patients from Australia (9, 10), Sub-Saharan Africa (10, 11), and North America (8, 12).
The profound impact of diverse HLA alleles (designated by 4 digits) and allele groups (designated by 2 digits or serologic specificities) on HIV-1 evolution and pathogenesis is best illustrated by B57, which is by far the most favorable HLA factor in the context of HIV-1 infection (reviewed in refs. 13, 14). Regardless of HIV-1 clade (e.g., subtypes A, B, and C), B57 (mostly B5701 and B5703) predominantly and persistently directs CTL recognition of highly conserved HIV-1 Gag epitopes (e.g., IW9 and KF11) (8, 15). As the infection progresses, viruses with mutations within and around these epitopes accumulate. Despite diminished immune protection, viremia often remains low, as viral replication fitness is apparently compromised. Upon transmission to individuals without B57 and closely related alleles like B5801, viruses with the mutated Gag epitopes usually regain the wild-type sequence (11, 16). In contrast, unfavorable alleles like B35 and B53 preferentially respond to Nef epitopes (8); subsequent Nef mutations are typically associated with disease progression (12). Thus, heterogeneity in immune control and rate of disease progression following HIV-1 infection is to some degree related to pathways mediated by HLA products.
Favorable HLA factors with a clear impact on HIV-1 viral load directly benefit the infected individual by delaying disease progression (13, 14), in a manner and degree similar to that observed with HIV-specific monotherapy (reviewed in ref. 17). These HLA factors may further benefit seronegative individuals who are at high risk of acquiring infection because viral load in plasma or genital secretions of a seropositive partner is highly predictive of HIV-1 transmission potential (18, 19). Our study of HIV-1 discordant couples (infected index partners paired with cohabiting seronegative partners) from Lusaka, Zambia has yielded clear evidence that viral transmission from index to seronegative partners can vary according to specific HLA class I alleles or haplotypes, often as a result of their influences on plasma viral load of the index partners.
Materials and Methods
Study population and HIV-1 transmission as the primary outcome
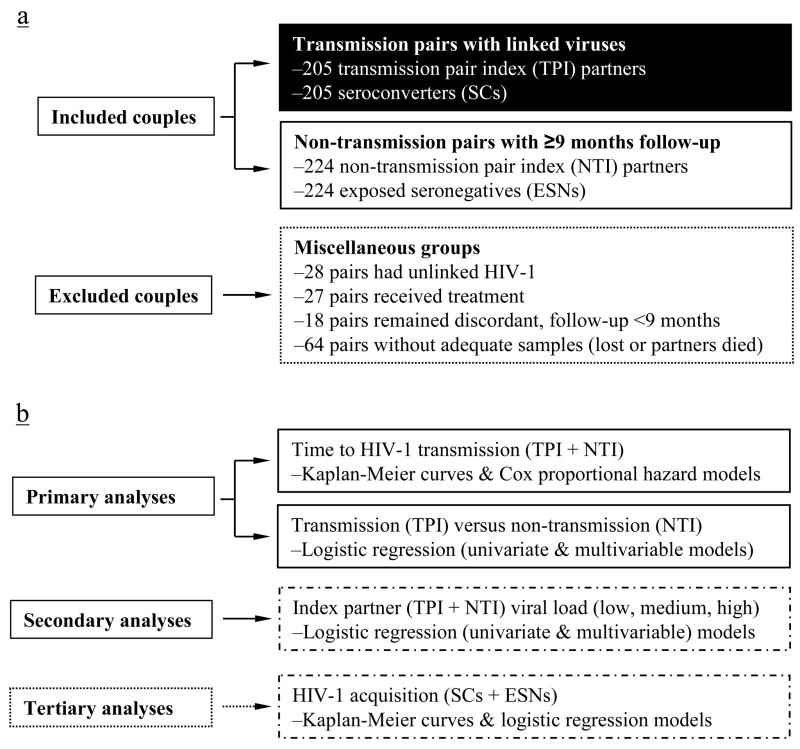
From 1995 to 2006, HIV-1 discordant Zambian couples were identified and enrolled continuously by investigators who are now part of the Rwanda/Zambia HIV-1 Research Group in Lusaka, Zambia. Procedures for initial screening, voluntary counseling and testing, as well as prospective (quarterly) medical examination have been described elsewhere (19, 20). As the primary outcome measure, heterosexual HIV-1 transmission within couples was determined every three months by rapid serological tests (the Dipstick HIV-1/HIV-2 antibody screening assay and the Capillus latex aggregation confirmatory test), coupled with the Uni-Gold Recombigen HIV test (Trinity Biotech USA, St Louis, MO), and followed by viral sequencing and phylogenetic analyses to identify viral subtype (clade) and to confirm viral linkage within couples (19, 21). This work was approved by local institutional review boards and informed consent was obtained from all study participants according to the human experimentation guidelines set forth by the US Department of Health and Human Services. With longitudinal data censored on 31 December 2006, all couples with ≥9 months (274 days) of follow-up without any antiretroviral treatment within that interval were eligible for primary analyses of HIV-1 transmission by the index partners. Of 566 HIV-1 discordant couples enrolled, 429 remained after exclusions due to a) unlinked/ambiguous viruses in newly infected (seroconverted) partners, b) antiretroviral treatment of index partners during follow-up, c) insufficient follow-up, or d) lack of usable biologic specimens (for HLA typing or viral sequencing) (Fig. 1a).
FIGURE 1.
Classification of 566 Zambian couples (1132 individuals) enrolled for longitudinal study from 1995 to 2006 (Panel a), along with the statistical approach to analyzing correlates of HIV-1 transmission and acquisition (Panel b). Characteristics of 429 (205 + 224) couples included in the final analyses are detailed in Table I. For all association analyses, HLA class I factors (common haplotypes or their component alleles) serve as the independent variables, while non-genetic factors (age, direction of HIV-1 transmission, genital ulcer/inflammation) are treated as covariates. Of note, tertiary analyses of HIV-1 acquisition are restricted to HLA factors identified by primary analyses of HIV-1 transmission.
HIV-1 viral load (RNA copies per mL of plasma) in index partners as secondary outcome
HIV-1 RNA copies in patient plasma were measured universally by the Roche Amplicor 1.0 assay (Roche Diagnostics Systems Inc., Branchburg, NJ) in laboratories certified by the Virology Quality Assurance Program of the AIDS Clinical Trials Group. The assay was highly comparable with other commercial viral assays (Chiron Quantiplex HIV-1 branched DNA, Roche Amplicor version 1.5, and nucleic acid sequence-based amplification – NASBA HIV-1 RNA QT) (22), with a lower limit of detection at 400 RNA copies per ml of plasma. In earlier analyses (19), index partners with medium (104–105 copies/ml) and high (>105) concentrations of HIV-1 RNA transmitted viruses more readily than those with low-level viremia (<104 copies/mL). Thus, these three categories of viral load were treated as a separate outcome for secondary analyses (Fig. 1b) and they were further assessed as predictive covariates in analyses of primary outcome.
PCR-based genotyping of HLA class I variants
Genomic DNA was extracted from whole blood or buffy coats using QIAamp blood kits (QIAGEN Inc., Chatsworth, CA). HLA class I genotyping was performed using a combination of PCR-based techniques, including PCR with sequence-specific primers (SSP) (Dynal/Invitrogen, Brown Deer, WI), automated reference-strand conformation analysis (RSCA) (Dynal/Invitrogen), automated sequence-specific oligonucleotide (SSO) probe hybridization (Innogenetics, Alpharetta, GA), and automated sequencing-based typing (SBT) (Abbott Molecular, Inc., Des Plaines, IL). These techniques achieved medium- to high-resolution typing of HLA class I alleles in the study population (23–26). Novel alleles and ambiguities were also resolved by confirmatory SBT using capillary electrophoresis and the ABI 3130xl DNA Analyzer (Applied Biosystems, Foster City, CA). Occasionally, alleles could not be distinguished by their exon 2 to exon 3 sequences that encode the peptide-binding groove. In these circumstances, a letter “g” (for group) was added to the most probable allele in assignment and reported as such (e.g., Cw*1701g for alleles Cw*1701, Cw*1702, and Cw*1703) (27).
Manual and computational assignment of local and extended HLA haplotypes
Common HLA-B (B) and HLA-C (C) haplotypes were initially assigned manually according to known patterns of strong linkage disequilibrium (LD) for well-documented and fully resolved alleles observed in African-Americans (28) and for medium-resolution typing results from a native African (Rwandan) population (29). Observation of common B–C haplotypes in homozygous state provided additional assurance of accuracy in manual haplotype assignment. Infrequent or rare haplotypes were inferred after the common haplotypes were assigned. Further evaluation of haplotypes involving alleles from two loci (pairwise) or all three class I loci used the expectation-maximization (EM) algorithm in SAS Genetics (SAS Institute, Cary, NC), with relative LD (D′) and correlation coefficients (r) as key measures. Haplotypes with a statistical probability ≥75% in the EM algorithm were considered reliable on an individual basis. For those with lower probabilities, the predicted probability values were incorporated in the overall estimate of probable haplotype frequencies.
Analyses of HLA genotypic heterogeneity and associations
Software packages in SAS (version 9.1.3) and SAS Genetics (SAS Institute, Cary, NC) were used to test: 1) Hardy-Weinberg equilibrium (HEW) of HLA class I alleles at each locus, before and after stratification by HIV-1 transmission or acquisition status (global log likelihood χ2 tests), 2) genetic heterogeneity between patient groups (global log likelihood χ2 tests), especially for transmission pair index (TPI) partners versus non-transmission pair index (NTI) partners, 3) primary analyses to determine associations of local and extended HLA haplotypes or their component alleles with time to or status of heterosexual HIV-1 transmission (several procedures, see below), 4) secondary (confirmatory) analyses to test the relationships of HLA variants to low, medium and high HIV-1 viral load in chronically infected index partners (TPI + NTI) (logistic regression models), 5) tertiary analyses of HIV-1 acquisition by the initially seronegative partners. With an overall focus on HIV-1 transmission by index partners, several analytic strategies (Fig. 1b) were applied to test the main hypothesis that HLA class I alleles (2- or 4-digit specificities) or haplotypes mediate heterosexual HIV-1 transmission, probably through regulation of HIV-1 viral load. The magnitude and strengths of HLA-associated effects were gauged by: 1) time-dependent hazard ratios and 95% confidence intervals (CIs) defined by Cox proportional hazard models; 2) relative odds of becoming HIV-1 transmitters during the 12-year study period, as calculated from 2 by 2 contingency tables of population frequencies, with 95% CIs obtained from the SAS FREQ procedure; 3) deviation from proportional distribution among index partners with low, medium, and high viral load. Reduced multivariable models were used to determine the relative impact of HLA and other recognized factors relevant to HIV-1 transmission and viral load. In particular, non-genetic factors, including age, direction of HIV-1 transmission (MTF for male-to-female and FTM for female-to-male), and genital ulcer/inflammation (GUI) in index and initially seronegative partners (seroconverters = SCs; exposed seronegatives = ESNs) were treated as covariates. As homozygosity with any individual HLA variant is rare, all these tests assumed dominant/codominant HLA function, with provisional statistical significance set at the nominal p value of ≤0.050 in univariate analyses. To reduce the number of statistical tests (and the chance for spurious associations), analyses targeted only major HLA variants (alleles and haplotypes) found at frequency ≥0.01 (out of 2N, i.e., number of chromosomes) or in ≥10 individuals. The selective tertiary analyses of HIV-1 acquisition were confined to HLA factors identified by primary analyses of HIV-1 transmission.
Results
Classification and characteristics of Zambian couples at the end of follow-up
Using data from 10,278 person-visits for the initially seronegative (non-index) partners, 429 couples eligible for primary analysis here were classified into 205 virologically linked transmission pairs and 224 non-transmission pairs (involving 108 female and 116 male index partners) (Table I). For the 205 transmission pair index (TPI) partners and 224 non-transmission pair index (NTI) partners, enrollment dates were highly comparable, as were within-couple age differences (ageΔ, 6.8 ± 4.2 years in the TPI group and 7.5 ± 5.2 years in the NTI group, p >0.10). On the other hand, mean and median follow-up time (FUT) of NTI was nearly double that of TPI (p <0.001).
Table I.
Classification and characterization of 429 Zambian couples available to this study (as of 31 December 2006)
| Characteristics a | By individuals a | By couples a | ||||||
|---|---|---|---|---|---|---|---|---|
| Paired index partners | Non-index partners | Transmission pairs | Non-transmission pairs | |||||
| TPI | NTI | SC | ESN | FTM | MTF | F+M− | M+F− | |
| Sample size | 205 | 224 | 205 | 224 | 81 | 124 | 108 | 116 |
| Enrollment dates | ||||||||
| Earliest | 27-Mar-95 | 27-Mar-95 | 27-Mar-95 | 27-Mar-95 | 27-Mar-95 | 30-Mar-95 | 27-Mar-95 | 15-May-95 |
| Latest | 30-Aug-05 | 17-Jan-06 | 30-Aug-05 | 17-Jan-06 | 30-Aug-05 | 6-Jan-05 | 2-Nov-05 | 17-Jan-06 |
| Age (years) | ||||||||
| Men’s (mean ± SD) | 32.2 ± 7.2 | 35.5 ± 8.3 | 32.2 ± 7.2 | 35.5± 8.3 | 31.1 ± 6.4 | 33.0 ± 7.5 | 35.3 ± 8.6 | 35.6 ± 8.0 |
| Women’s (mean ± SD) | 25.7 ± 5.9 | 28.0 ± 6.5 | 25.7 ± 5.9 | 28.0 ± 6.5 | 25.4 ± 5.7 | 25.9 ± 6.1 | 27.9 ± 5.8 | 28.1 ± 7.0 |
| AgeΔ (mean ± SD) | 6.8 ± 4.2 | 7.5 ± 5.2 | 6.8 ± 4.2 | 7.5 ± 5.2 | 6.1 ± 4.0 | 7.3 ± 4.3 | 7.4 ± 5.5 | 7.7 ± 4.7 |
| FUT: mean/median | 779/545 | 1464/1097 | 779/545 | 1464/1097 | 771/540 | 784/546 | 1618/1358 | 1320/861 |
| Genital ulcer/inflammation | ||||||||
| Index partners | 48.3%b | 21.9%b | NA | NA | 55.6%d | 43.6%f | 25.0%d | 19.0%f |
| Non-index partners | NA | NA | 45.9%c | 10.7%c | 42.0%e | 48.4%g | 12.0%e | 9.5%g |
F+M−, female index partner with a male non-index partner; M−F+, male index partner with a female non-index partner. FUT in days. Note GUI is taken in the 6 months prior to transmission for the transmission pairs or at end of follow-up for nontransmission pairs.
p <0.001 in these pairwise comparisons (by logistic regression).
Consistent with earlier observations in the Zambian study population (19), male-to-female transmission (124 pairs) was more common than female-to-male transmission (81 pairs) (Table I). Among index partners of the transmission couples, 48.3% had genital ulcer/inflammation (GUI) within the 6 months prior to seroconversion of the non-index (recipient) partners. In contrast, GUI was less common among the non-transmitting index partners (21.9%, in the 6 months before end of follow-up) (p <0.001). The proportion of GUI was also statistically different between recipient partners who became infected (seroconverted = SC group, 45.9%) and those who remained uninfected (exposed seronegative = ESN group, 10.7%) (p <0.001). These differences persisted in stratified analyses of female-to-male and male-to-female transmission (Table I). Accordingly, GUI in all partners was retained as a covariate in subsequent analyses of HLA genotypes.
Global tests of HLA class I variants at the 2-digit resolution (allele group)
At 2-digit resolution, which is largely equivalent to serologic specificity, 12 HLA-A alleles, 18 HLA-B alleles, 11 HLA-C alleles, 21 local (HLA-B–HLA-C = B–C) haplotypes, and 25 extended (3-locus = A–B–C) haplotypes were found at frequencies ≥0.10 in the 429 Zambian couples (1716 chromosomes). Within the 429 paired index partners (858 chromosomes), the respective numbers (12, 17, 11, 23, and 22) were similar. The major B–C haplotypes assigned manually correlated extremely well with haplotypes inferred using the EM algorithm, with correlation coefficients (Pearson and Spearman) all exceeding 0.99 in analyses of overall data from the 429 Zambian couples and in analyses of either index (TPI + NTI) or initially seronegative (ESN + SC) Zambians.
Three global tests generated evidence for a high level of heterogeneity in the frequencies of HLA class I genotypes between TPI and NTI subjects as a result of HIV-1 transmission. First, overall distribution of extended haplotypes differed between TPI and NTI subjects (p = 7.1 × 10−12 by log likelihood χ2 tests), mostly due to the major haplotypes with predicted frequencies ≥0.010 (p = 4.4 × 10−9, Table II). Second, in tests of 22 extended (A–B–C) haplotypes with individual frequencies ≥0.010, four differed in their distribution between the two patient groups (p <0.010) (Table II). Third, distribution of HLA-A diplotypes in the TPI (rather than NTI) patients deviated from HWE (p <0.001), whereas the distribution of HLA-B and HLA-C diplotypes all conformed to HWE in the two patient groups (data not shown – available upon request).
Table II.
Major HLA class I haplotypes in 429 paired index partners (PIPs) with or without HIV-1 transmission events
| Major local (B–C) Haplotypes and Observed Frequencies (Freq) a | Major Extended (A–B–C) Haplotypes and Estimated Frequencies (Freq) b | ||||||
|---|---|---|---|---|---|---|---|
| 23 most frequent c | Freq_PIP | Freq_NTI | Freq_TPI | 22 most frequent d | Freq_PIP | Freq_NTI | Freq_TPI |
| B*42-Cw*17 | 0.129 | 0.147 | 0.110 | A*30-B*42-Cw*17 | 0.069 | 0.074 | 0.065 |
| B*53-Cw*04 | 0.077 | 0.065 | 0.090 | A*30-B*57-Cw*18 | 0.041 | 0.048 | 0.033 |
| B*14-Cw*08 | 0.072 | 0.080 | 0.063 | A*30-B*14-Cw*08 | 0.033 | 0.035 | 0.032 |
| B*15-Cw*02 | 0.070 | 0.071 | 0.068 | A*36-B*53-Cw*04e | 0.031 | 0.015 | 0.048 |
| B*07-Cw*07 | 0.059 | 0.047 | 0.073 | A*68-B*15-Cw*03 | 0.030 | 0.038 | 0.022 |
| B*15-Cw*03 | 0.057 | 0.063 | 0.051 | A*68-B*14-Cw*08 | 0.028 | 0.035 | 0.020 |
| B*57-Cw*18 | 0.055 | 0.065 | 0.044 | A*68-B*07-Cw*07 | 0.022 | 0.013 | 0.031 |
| B*58-Cw*06 | 0.040 | 0.031 | 0.049 | A*74-B*15-Cw*02 | 0.022 | 0.022 | 0.022 |
| B*45-Cw*16 | 0.036 | 0.027 | 0.046 | A*01-B*81-Cw*18 | 0.017 | 0.022 | 0.012 |
| B*58-Cw*07 | 0.034 | 0.038 | 0.029 | A*23-B*15-Cw*02 | 0.016 | 0.017 | 0.015 |
| B*81-Cw*18 | 0.031 | 0.038 | 0.024 | A*30-B*15-Cw*02 | 0.016 | 0.010 | 0.022 |
| B*35-Cw*04 | 0.029 | 0.033 | 0.024 | A*30-B*08-Cw*07 | 0.014 | 0.012 | 0.016 |
| B*44-Cw*04 | 0.028 | 0.031 | 0.024 | A*29-B*42-Cw*17e | 0.013 | <0.001 | 0.028 |
| B*08-Cw*07 | 0.027 | 0.027 | 0.027 | A*23-B*07-Cw*07e | 0.013 | <0.001 | 0.028 |
| B*45-Cw*06 | 0.024 | 0.027 | 0.022 | A*30-B*15-Cw*03 | 0.013 | 0.013 | 0.012 |
| B*13-Cw*06 | 0.016 | 0.022 | 0.010 | A*66-B*58-Cw*06 | 0.013 | 0.013 | 0.012 |
| B*39-Cw*12 | 0.014 | 0.016 | 0.012 | A*02-B*42-Cw*17f | 0.012 | 0.024 | <0.001 |
| B*44-Cw*07 | 0.014 | 0.016 | 0.012 | A*30-B*45-Cw*16 | 0.012 | 0.009 | 0.015 |
| B*51-Cw*16 | 0.013 | 0.011 | 0.015 | A*02-B*45-Cw*16 | 0.011 | 0.008 | 0.015 |
| B*18-Cw*07 | 0.012 | 0.011 | 0.012 | A*23-B*53-Cw*04 | 0.011 | 0.009 | 0.014 |
| B*15-Cw*04 | 0.010 | 0.004 | 0.017 | A*02-B*53-Cw*04 | 0.011 | 0.012 | 0.009 |
| B*49-Cw*07 | 0.010 | 0.007 | 0.015 | A*23-B*45-Cw*06 | 0.010 | 0.020 | 0.000 |
| B*53-Cw*06 | 0.010 | 0.004 | 0.017 | 22 combined | 0.459 | 0.448 | 0.472 |
| 23 combined | 0.869 | 0.882 | 0.856 | ||||
Local haplotypes are assigned manually and confirmed by the EM algorithm.
Tabulation of extended haplotypes based on the EM algorithm.
Restricted to those with predicted frequencies ≥0.010 and sorted in order of descending frequencies (overall p = 0.197 between TPI and NTI, by log likelihood χ2 test).
Also restricted to those with frequencies ≥0.010 and sorted in order of descending frequencies (overall p = 4.4 × 10−9 between TPI and NTI, by log likelihood χ2 test).
All enriched in the TPI group (p ≤0.010).
Enriched in the NTI group (p ≤0.010).
Global tests based on all members of 429 couples also indicated that LD between HLA-B and HLA-C variants (p <1.0 × 10−140) was much stronger than LD between HLA-A and HLA-B variants (p = 1.1 × 10−69) or HLA-A and HLA-C variants (p = 1.1 × 10−53), as reflected by pairwise tests of individual 2-digit allele groups from each locus (Table III). For the index partners alone, LD patterns were the same, with p values ranging from 3.4 × 10−116 (B–C LD) to 0.005 (A–C LD). A total of 30 pairs of HLA variants had p <0.0001 in tests of correlation coefficient (r) and relative LD (D′) (Table III); the strongest LD was between B*42 and Cw*17 and between B*14 and Cw*08 in the entire cohort (r = 0.94, D′ = 1.00 and r = 0.85, D′ = 0.95, respectively), as well as in the subset of index partners (r = 0.93, D′ = 1.00 and r = 0.85, D′ = 0.91, respectively). The strength of LD provided strong rationale for analysis of major haplotypes (local and extended) as separate entities in association analyses.
Table III.
Pairwise linkage disequilibrium (LD) analyses of major HLA class I variants
| HLA pair a | Entire cohort (N = 858) b | Paired index partners only (N = 429) b | ||||||
|---|---|---|---|---|---|---|---|---|
| Frequency | r | D′ | p | Frequency | r | D′ | p | |
| A*01-B*81 | 0.014 | 0.42 | 0.53 | <1.0 × 10−20 | 0.007 | 0.39 | 0.73 | 1.0 × 10−15 |
| A*01-Cw*18 | 0.013 | 0.27 | 0.494 | 2.1 × 10−15 | 0.007 | 0.27 | 0.72 | 1.5 × 10−8 |
| A*02-B*45 | 0.020 | 0.14 | 0.198 | 2.8 × 10−5 | 0.024 | 0.17 | 0.20 | 0.0006 |
| A*02-Cw*16 | 0.028 | 0.23 | 0.316 | 6.9 × 10−12 | 0.030 | 0.26 | 0.34 | 5.3 × 10−8 |
| A*29-B*13 | 0.011 | 0.27 | 0.54 | 2.3 × 10−15 | 0.010 | 0.33 | 0.77 | 7.9 × 10−12 |
| A*30-B*42 | 0.062 | 0.28 | 0.48 | 2.2 × 10−16 | 0.063 | 0.31 | 0.55 | 2.7 × 10−10 |
| A*30-Cw*17 | 0.065 | 0.27 | 0.44 | 2.3 × 10−15 | 0.061 | 0.25 | 0.42 | 2.9 × 10−7 |
| A*34-B*44 | 0.013 | 0.36 | 0.53 | <1.0 × 10−20 | 0.017 | 0.51 | 0.61 | <1.0 × 10−20 |
| A*36-B*53 | 0.039 | 0.44 | 0.58 | <1.0 × 10−20 | 0.049 | 0.45 | 0.60 | <1.0 × 10−20 |
| A*36-Cw*04 | 0.038 | 0.31 | 0.53 | <1.0 × 10−20 | 0.049 | 0.32 | 0.56 | 1.8 × 10−11 |
| A*66-B*58 | 0.022 | 0.32 | 0.53 | <1.0 × 10−20 | 0.023 | 0.30 | 0.46 | 6.4 × 10−10 |
| A*66-Cw*06 | 0.018 | 0.24 | 0.42 | 1.1 × 10−12 | 0.019 | 0.21 | 0.36 | 1.1 × 10−5 |
| A*68-Cw*03 | 0.024 | 0.18 | 0.22 | 2.3 × 10−7 | 0.021 | 0.17 | 0.25 | 0.0003 |
| B*07-Cw*07 | 0.054 | 0.50 | 0.87 | <1.0 × 10−20 | 0.048 | 0.48 | 0.83 | <1.0 × 10−20 |
| B*08-Cw*07 | 0.027 | 0.33 | 0.80 | <1.0 × 10−20 | 0.029 | 0.41 | 1.00 | <1.0 × 10−20 |
| B*13-Cw*06 | 0.014 | 0.29 | 0.73 | <1.0 × 10−20 | 0.009 | 0.27 | 0.74 | 2.7 × 10−6 |
| B*14-Cw*08 | 0.070 | 0.85 | 0.95 | <1.0 × 10−20 | 0.080 | 0.85 | 0.91 | <1.0 × 10−20 |
| B*15-Cw*02 | 0.081 | 0.60 | 0.84 | <1.0 × 10−20 | 0.074 | 0.58 | 0.72 | <1.0 × 10−20 |
| B*15-Cw*03 | 0.054 | 0.42 | 0.66 | <1.0 × 10−20 | 0.045 | 0.43 | 0.69 | <1.0 × 10−20 |
| B*18-Cw*07 | 0.019 | 0.21 | 0.51 | 3.7 × 10−10 | 0.026 | 0.24 | 0.43 | 8.7 × 10−7 |
| B*35-Cw*04 | 0.029 | 0.33 | 0.79 | <1.0 × 10−20 | 0.027 | 0.25 | 0.62 | 2.6 × 10−7 |
| B*42-Cw*17 | 0.101 | 0.94 | 1.00 | <1.0 × 10−20 | 0.095 | 0.93 | 1.00 | <1.0 × 10−20 |
| B*44-Cw*04 | 0.026 | 0.21 | 0.41 | 5.8 × 10−10 | 0.023 | 0.20 | 0.50 | 2.6 × 10−5 |
| B*45-Cw*16 | 0.042 | 0.58 | 0.60 | <1.0 × 10−20 | 0.056 | 0.70 | 0.74 | <1.0 × 10−20 |
| B*49-Cw*07 | 0.011 | 0.23 | 0.93 | 1.4 × 10−11 | 0.009 | 0.19 | 0.72 | 7.6 × 10−5 |
| B*51-Cw*16 | 0.015 | 0.41 | 0.85 | <1.0 × 10−20 | 0.007 | 0.25 | 0.71 0 | 2.1 × 10−7 |
| B*53-Cw*04 | 0.092 | 0.65 | 0.87 | <1.0 × 10−20 | 0.119 | 0.72 | 0.95 | <1.0 × 10−20 |
| B*57-Cw*18 | 0.047 | 0.70 | 0.85 | <1.0 × 10−20 | 0.036 | 0.70 | 0.87 | <1.0 × 10−20 |
| B*58-Cw*06 | 0.050 | 0.44 | 0.47 | <1.0 × 10−20 | 0.064 | 0.54 | 0.59 | <1.0 × 10−20 |
| B*81-Cw*18 | 0.029 | 0.49 | 0.71 | <1.0 × 10−20 | 0.021 | 0.43 | 0.60 | <1.0 × 10−20 |
Those with p <0.001 and frequency ≥0.010; pairs with the strongest LD (r >0.80) are underlined; pairs involving Cw*18 are in bold.
Frequencies are estimates according to the EM algorithm; r and D′ are based on the PROC ALLELE procedure in SAS Genetics (version 9.1.3).
HLA class I variants in index partners in relation to heterosexual HIV-1 transmission
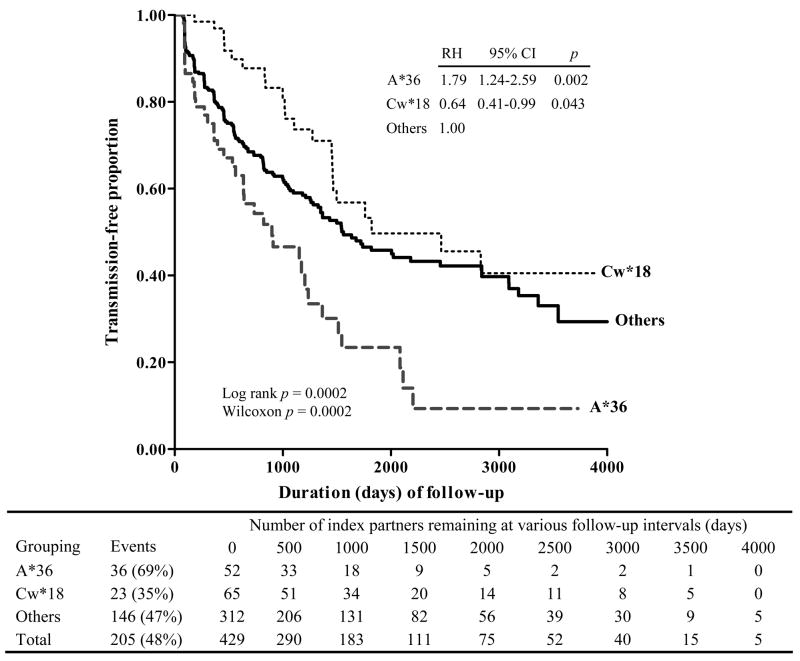
In univariate analyses of all major HLA variants, only A*36 showed a positive association with HIV transmission at p <0.01 (for proportional hazard analysis). At p <0.05, haplotype A*36-B*53-Cw*04 also showed positive association. Two other variants, B*57 and Cw*18, and their haplotype, B*57-Cw*18, were negatively associated with HIV-1 transmission. Multivariable models demonstrated that A*36 and Cw*18 were the two major factors independently associated with HIV-1 transmission (adjusted RH = 1.79 and 0.64, p = 0.002 and p = 0.043, respectively) (Fig. 2). Within the study period, 69% of index partners with A*36 transmitted HIV-1 to their seronegative partners (median transmission-free time = 899 days, 95% CI = 454 to 1,235 days), while 35% of index partners with Cw*18 and without A*36 did so (median transmission-free time = 1,823 days, 95% CI = 1,460 to >4,000 days). These estimates of infection-free time differed between the two patient groups in both log-rank (p <0.001) and Wilcoxon (p <0.001) tests; they further differed from those (median = 1,560 days, 95% CI = 1,327 to 2,458 days) for the remainder of index partners without A*36 or Cw*18 (p ≤0.041 and ≤0.019 by log-rank and Wilcoxon tests, respectively). After further statistical adjustment for age difference within couples, direction of viral transmission (MTF and FTM), and GUI, these contrasting HLA relationships remained intact (Table IV). Replacement of Cw*18 by B*57 led to very similar findings (Table IV and Fig. 3b). In addition, all but one individual who had both A*36 and Cw*18 (n = 4) or A*36 and B*57 (n = 5) behaved just like those with A*36 alone (data not shown).
FIGURE 2.
HLA class I factors (A*36 and Cw*18) with the most contrasting relationships to heterosexual HIV-1 transmission among 429 discordant Zambian couples. Both log-rank and Wilcoxon tests of significance are shown. For index partners grouped according to HLA profile, multivariable relative hazards (RH) of HIV-1 transmission are based on Cox proportional hazards models. Four individuals with both A*36 and Cw*18 are treated as part of the A*36 group as they behave like A*36+ instead of Cw*18+ patients. Estimates of RH and OR for A*36 and Cw*18 remain statistically significant (adjusted p <0.05) when the four patients with both A*36 and Cw*18 are either moved to the A36- and Cw*18- reference group or excluded from multivariable analyses.
TABLE IV.
Multifactorial influences on HIV-1 transmission, as defined by multivariable models without considering index partner’s viral load
| Factors tested in models | Cox proportional hazard model a | Logistic regression model | ||||
|---|---|---|---|---|---|---|
| RH | 95% CI | Adjusted p | OR | 95% CI | Adjusted p | |
| Best reduced model a | ||||||
| A*36 (n = 52) | 1.77 | 1.21–2.59 | 0.002 | 2.62 | 1.30–5.27 | 0.007 |
| Cw*18 (n = 65) | 0.59 | 0.37–0.92 | 0.020 | 0.57 | 0.31–1.03 | 0.064 |
| AgeΔ (per year) | 0.96 | 0.93–0.99 | 0.007 | 0.95 | 0.91–0.99 | 0.034 |
| Male to female transmission | 1.47 | 1.10–1.96 | 0.009 | 1.69 | 1.10–2.59 | 0.017 |
| Genital ulcer/inflammation | 2.45 | 1.84–3.25 | <0.0001 | 3.43 | 2.21–5.33 | <0.0001 |
| Alternative model b | ||||||
| A*36 (n = 52) | 1.82 | 1.25–2.66 | 0.002 | 2.73 | 1.37–5.49 | 0.005 |
| B*57 (n = 45) | 0.62 | 0.37–1.04 | 0.067 | 0.66 | 0.33–1.32 | 0.240 |
| AgeΔ (per year) b | 0.96 | 0.93–0.99 | 0.007 | 0.96 | 0.91–0.99 | 0.035 |
| Male to female transmission | 1.45 | 1.08–1.93 | 0.012 | 1.67 | 1.09–2.56 | 0.019 |
| Genital ulcer/inflammation | 2.41 | 1.82–3.20 | <0.0001 | 3.38 | 2.18–5.24 | <0.0001 |
Best model has factors with adjusted (multivariable) p ≤0.05; four patients with both A*36 and Cw*18 are treated as part of the A*36 group.
B*57 replacing Cw*18 (the two variants are in strong linkage disequilibrium); five patients with both A*36 and B*57 are treated as part of the A*36 group.
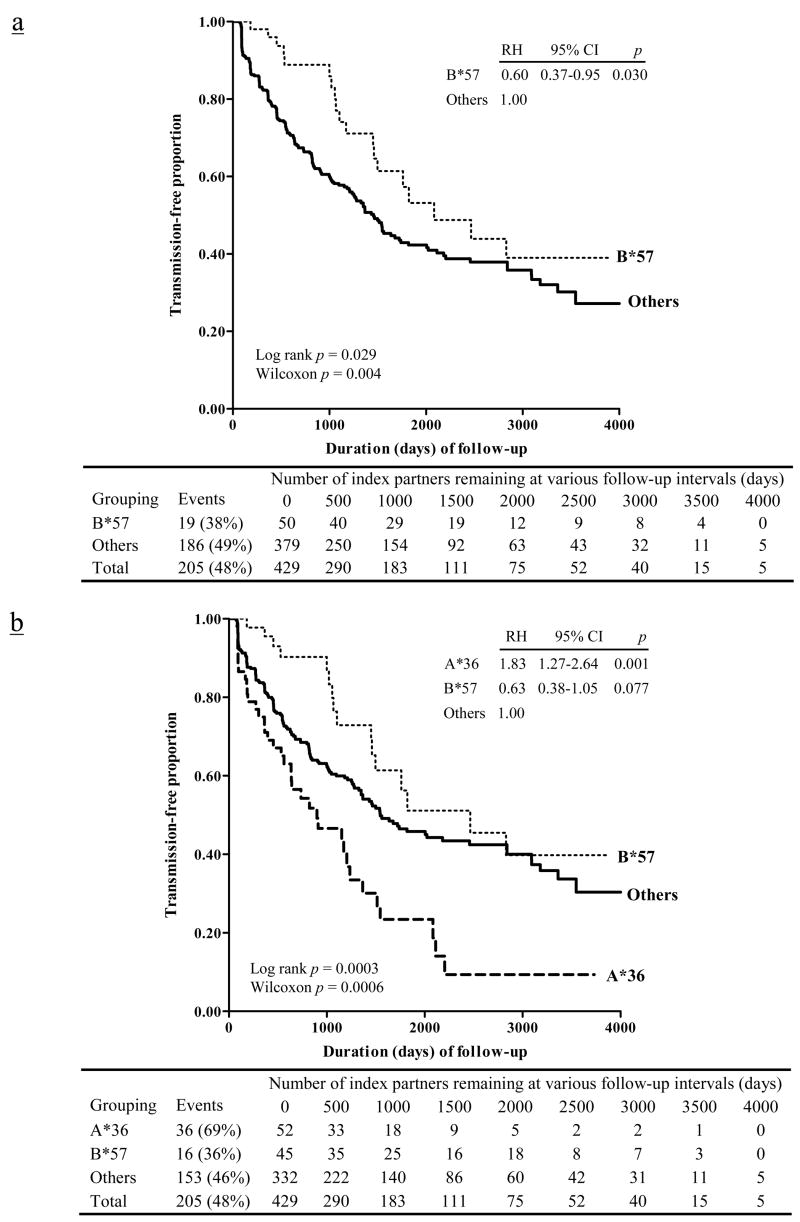
FIGURE 3.
Kaplan-Meier curves showing the relationship of HLA-B*57 to heterosexual HIV-1 transmission among 429 HIV-1-infected Zambians with cohabiting seronegative partners. Relative hazards (RH) of HIV-1 transmission are based on Cox proportional hazards models, before (panel a) and after (panel b) considering the opposing effect of HLA-A*36. Five individuals who have both A*36 and B*57 behave like A*36-positive patients (with three transmission events). For clarity, they are treated as part of the A*36 group (panel b).
A reduced logistic regression model confirmed the relationships of A*36 and Cw*18 to HIV-1 transmission (Table IV). For A*36, the adjusted odds ratio (OR) for HIV-1 transmission status was 2.62 (95% confidence interval or CI = 1.30–5.27) and for Cw*18, the OR was 0.57 (95% CI = 0.31–1.03). Age, direction of transmission, and GUI also showed independent associations based on multivariable analyses (adjusted p ≤0.034 for all). In an alternative logistic regression model, the relationship of B*57 to HIV-1 transmission status (Fig. 3a) was somewhat weaker (adjusted OR = 0.66, 95% CI = 0.33–1.32) in the presence of other more prominent factors (adjusted p ≤0.035 for all).
HLA class I variants in relation to HIV-1 viral load in index partners
In 202 TPIs and 218 NTIs, HIV-1 viral load was measured at a median interval of 546 days after enrollment. Consistent with earlier observations (19), TPIs more frequently had high (>105) or medium (104–105 copies/ml) than low (<104 copies/mL) concentrations of viral RNA in plasma, while the opposite was seen in NTIs. For example, 106 (52.3%) of TPIs and 65 (29.8%) of NTIs had high viral load (p <0.0001). Patients with very high viral load (>5.0 × 105 copies/mL), which might be indicative of acute-phase or late-stage infection, was also more common in TPIs (13.9%) than NTIs (4.6%) (p = 0.0001).
Index partners with A*36, which was associated with accelerated HIV-1 transmission, were over-represented in the subgroups defined by high and medium viral load when compared with patients defined by low viral load (univariate p = 0.037 in test for trend) (Table V). The association of A*36 with viral load diminished after statistical adjustments for age, sex, and membership of patient groups (TPI and NTI) (p = 0.072). Association with the A*36-B*53-Cw*04 haplotype did not better account for these findings. Likewise, patients with HLA class I alleles and haplotypes showing association or a tendency toward association with delayed HIV-1 transmission were always enriched among index partners with low viral load, as reflected by analyses of Cw*18 alone (adjusted p <0.0001), Cw*18 without B*57 (p = 0.002), Cw*18 without B*81 (p = 0.007), and B*81 alone (p = 0.011) (Table V).
TABLE V.
HIV-1 viral load in index partners with HLA alleles and haplotypes showing putative associations with subsequent viral transmission to paired seronegative partners
| HLA factors a | Distribution across VL categories: n (%)b | Test for trend | ||||
|---|---|---|---|---|---|---|
| Low (<104) N = 77 | Medium (104–105) N = 172 | High (>105) N = 171 | p | Adjusted pc | ||
| A*36 and related | ||||||
| A*36+ | 5 (6.5%) | 20 (11.6%) | 27 (15.8%) | 0.037 | 0.072 | |
| A*36+, B*53+, Cw*04+ | 2 (2.6%) | 12 (7.0%) | 14 (8.2%) | 0.064 | 0.233 | |
| A*36+, B*53− | 3 (3.9%) | 8 (4.7%) | 12 (7.0%) | 0.261 | 0.318 | |
| Cw*18 and related | ||||||
| Cw*18+ | 24 (31.2%) | 28 (16.3%) | 15 (8.8%) | <0.0001 | <0.0001 | |
| Cw*18+, B*57+ | 14 (18.2%) | 17 (9.9%) | 12 (7.0%) | 0.011 | 0.020 | |
| Cw*18+, B*57− | 10 (13.0%) | 11 (6.4%) | 3 (1.8%) | 0.0004 | 0.0002 | |
| Cw*18+, B*81+ | 9 (11.7%) | 11 (6.4%) | 4 (2.3%) | 0.003 | 0.001 | |
| Cw*18+, B*81− | 15 (19.5%) | 17 (9.9%) | 11 (6.4%) | 0.003 | 0.007 | |
| B*57+ | 15 (19.5%) | 19 (11.1%) | 16 (9.4%) | 0.037 | 0.089 | |
| B*81+ | 11 (14.3%) | 14 (8.1%) | 8 (4.7%) | 0.010 | 0.011 | |
| Cw*18−, (B*57 or B*81)+ | 3 (3.9%) | 5 (2.9%) | 8 (4.7%) | 0.623 | 0.256 | |
| Simplest modeld | ||||||
| A*36+ | 5 (6.5%) | 20 (11.6%) | 27 (15.8%) | 0.153 | 0.220 | |
| Cw*18+, A*36− | 24 (31.2%) | 25 (14.5%) | 14 (8.2%) | <0.0001 | <0.0001 | |
| Others (A*36−, Cw*18−) | 48 (62.3%) | 127 (73.8%) | 130 (76.0%) | Reference | NA | |
Listed here are those relevant to A*36 and Cw*18, for their contrasting influences on HIV-1 transmission (e.g., Table IV and Fig. 2); +, presence; −, absence.
Classification of index partners is the same as in Table I and Fig. 1; nine patients (two with Cw*18) are excluded from tabulations here due to lack of viral load data.
After statistical adjustment for any differences attributable to age (>40 versus ≤40, as reported earlier (23)), sex, and group membership (TPI or NTI); NA, not applicable.
This model follows the same analytic approach as in Fig. 2.
The association of Cw*18 with low viral load could not be fully captured by B*57 and B*81 despite their tight LD with Cw*18 (r = 0.88, D′ = 0.97 when the two HLA-B alleles were treated as one entity). For example, 16 patients with the two HLA-B variants but without Cw*18 (i.e., Cw*18−, B*57+, or B*81+) were in viral load categories comparable with the categories of patients without those alleles (adjusted p = 0.256) (Table V). No informative test could be done for the two patients who had Cw*18 in the absence of B*57 or B*81 (data not shown), but data from the 24 patients with Cw*18 and without B*57, along with 43 others with Cw*18 and without B*81, all seemed to suggest that the effect of Cw*18 on viral load was almost exclusively due to the two haplotypes, i.e., B*57-Cw*18 and B*81-Cw*18 (adjusted p = 0.020 and 0.001, respectively) (Table V). In the simplest multivariable model derived from analyses of HIV-1 transmission status (Fig. 2 and Table IV), Cw*18 remained as a major predictor of low viral load (p <0.0001).
Joint analyses of host and viral factors in index partners in relation to HIV-1 transmission
A close relationship between viral load in index partners and the likelihood of HIV-1 transmission status during follow-up was recognized earlier in the Zambian cohort (19). When high and medium index partner viral load (relative to the reference low level) were included in regression models as co-factors for HIV-1 transmission, only age, GUI, and HLA-A*36 were retained as independent contributors (adjusted RH for A*36 = 1.71, 95% CI = 1.17–2.49; OR = 2.42, 95% CI = 1.21–4.85) (Table VI, reduced model). In contrast, neither B*57 nor Cw*18 or its haplotype remained as independent contributor (protective factor) when viral load was included in the multivariable model (e.g., adjusted RH for Cw*18 = 0.75, p = 0.212) (Table VI, full model).
TABLE VI.
Multivariable models that account for both host and viral factors provisionally associated with differential HIV-1 transmission from 429 index Zambians to their seronegative partners
| Models accounting for host and/or viral factorsa | Relative hazard (95% CI)b | Adjusted p | Odds ratio (95% CI) | Adjusted p |
|---|---|---|---|---|
| Reduced model | ||||
| A*36 | 1.71 (1.17–2.49) | 0.005 | 2.42 (1.21–4.85) | 0.013 |
| AgeΔ (per year) | 0.96 (0.94–0.99) | 0.008 | 0.95 (0.91–1.00) | 0.039 |
| GUIc | 2.23 (1.67–2.98) | <0.001 | 3.24 (2.10–5.00) | <0.001 |
| Viral load: highd | 2.41 (1.47–3.95) | <0.001 | 4.18 (2.19–7.98) | <0.001 |
| Viral load: mediumd | 1.78 (1.07–2.95) | 0.027 | 2.16 (1.13–4.11) | 0.020 |
| Full model | ||||
| A*36 | 1.74 (1.18–2.57) | 0.005 | 2.48 (1.19–5.17) | 0.016 |
| Cw*18 e | 0.75 (0.47–1.18) | 0.212 | 0.81 (0.43–1.53) | 0.521 |
| AgeΔ (per year) | 0.96 (0.94–0.99) | 0.007 | 0.96 (0.91–0.999) | 0.045 |
| GUIc | 2.24 (1.68–2.99) | <0.001 | 3.08 (1.97–4.83) | <0.001 |
| Viral load: highd | 2.24 (1.36–3.72) | 0.002 | 3.98 (2.05–7.71) | <0.001 |
| Viral load: mediumd | 1.73 (1.04–2.89) | 0.036 | 2.11 (1.10–4.07) | 0.025 |
Other putative factors tested earlier (Tables I–II and IV–V) drop out the final models (adjusted p >0.05).
Based on the Wilcoxon proportional hazards models.
In either index (initially seropositive) or nonindex (initially seronegative) partners.
Three categories of HIV-1 viral load in the index partners are defined as high (>105 copies/ml), medium (104–105), and low (<104, which serves as the reference group).
In tight LD with both B*57 (primarily B*5703) and B*81 (Table III).
Other confirmatory analyses
Several alternative analytic strategies confirmed the major HLA associations presented above. For example, the relationships of A*36 and Cw*18 to HIV-1 viral load in index partners were confirmed in linear regression models in which log10-transformed viral load was treated as a continuous outcome measure (no deviation from normal distribution). The quantifiable viral load differences (i.e., adjusted beta estimates) independently attributable to A*36 and Cw*18 were always within 0.50 log10 (data not shown).
Tertiary analyses of HIV-1 acquisition among the initially seronegative Zambians
In selective analyses of the 429 initially seronegative Zambians (ESNs + SCs), GUI in either partner contributed to HIV-1 acquisition (p <0.0001), with adjusted RH ≥4.37 and OR = 7.14 in two multivariable models (data available from J.T.). In contrast, neither A*36 nor Cw*18 nor B*57 carried by the initially seronegative Zambians was associated with acquisition of infection, although again A*36 tended to be unfavorable, with adjusted RH ≤1.45 (p ≥0.071) in proportional hazard models and OR ≤1.85 (p ≥0.066) in logistic regression models (extra Table available from J.T.).
Discussion
Our immunogenetic evaluation of epidemiologic and clinical data collected continuously on HIV-1-discordant Zambian couples produced the first evidence that HLA class I genotypes in the index partners might differentially influence the occurrence and rate of heterosexual HIV-1 transmission during a 12-year study period. In particular, primary analyses clearly pointed to A*36 and Cw*18 in index partners as the major predictors of viral transmission, while secondary analyses revealed contrasting levels of viral load attributable to these HLA genotypes. Overall, these findings extend the well-established role of HLA class I molecules in HIV-1 pathogenesis (13, 14, 17, 30) and viral evolution (31) to include their further impact on viral transmission from index to seronegative partners. Our work also implies that current understanding of “resistance” and “susceptibility” to HIV-1 infection, as inferred typically from analyses comparing exposed and seronegative individuals with exposed SCs (32), is incomplete insofar as partner characteristics are missing in such studies.
As documented earlier for the Zambian cohort (23–26), A*3601 is the lone allele in the A*36 group. The allele is rare in Asian and European populations, with frequencies close to zero (27, 28, 33); it is also rare in African-Americans (28) and South Africans (34). Thus, little is known about its functional attributes. In terms of linkage disequilibrium, A*3601 does not seem to tag (i.e., r2 >0.80) any particular non-HLA variants (single nucleotide polymorphisms or SNPs) in the HLA class I region (4), but detection of A*3601 on the extended haplotype A*36-Cw*04-B*53 in Zambians implies that B*53 may be a potential contributing factor, since B*53 has been considered unfavorable in at least one African (Rwandan) cohort, as well as in Caucasians (13, 26). However, B*53 itself (exclusively B*5301 in Zambians) did not show appreciable impact on HIV-1 transmission or index partner viral load. Thus, the mechanisms underlying the strong relationship of A*3601 in index partners to heterosexual HIV-1 transmission deserve further investigation.
Notably, the association of A*3601 with increased rate as well as incidence of heterosexual HIV-1 transmission remained strong even after statistical adjustment for viral load and other independent co-factors of infection. The weak association between A*3601 and viral load was also seen in our earlier analyses of 168 index partners and 91 SCs (23). While index partner viral load categories can fluctuate from time to time and for various reasons during the study interval, our incorporation of viral load as a categorical variable should be insensitive to modest (<5-fold) changes in viral load. Of course, a more critical evaluation of A*3601 or its related haplotype on index partner viral load may require serial and frequent sampling, especially for the few patients who appeared to have low or medium level viral load despite having A*3601 (Table V). Collection of longitudinal viral load data from A*3601-positive seroconverters should yield that valuable information. In addition, multiple studies (35–43) have demonstrated that viral shedding in the genital tract, rectal mucosa, and semen can be independent of plasma viral load. Those observations raise the alternative possibility that the dynamics of local viral shedding may differ in some of the Zambians with A*3601. For example, local flora, mucosal innate immunity, and co-infection with other pathogens like herpes simplex virus type 2 (44) are key factors in viral shedding that may be influenced by HLA genotypes.
For Cw*18, which is equivalent to Cw*1801 in Zambians (23), the association with reduced rate and incidence of HIV-1 transmission was almost entirely attributable to two haplotypes involving B*57 and B*81, respectively, in the index partners and to the collective impact of these haplotypes on index partner viral load. B*57, especially B*5701 and B*5703, have been widely recognized as favorable factors in HIV-1-infected individuals (reviewed in refs. 13, 14, 17), whereas B*81 (exclusively B*8101 in Zambians) has been considered favorable in more recent studies of native Africans (25, 34, 45). In Caucasian populations, B*5701 on the B*5701-Cw*0602 haplotype (no Cw*18) is tagged by a SNP (rs2395029) at the HCP5 locus (4, 46), but experimental studies have repeatedly demonstrated that B*5701 is responsible for effective and often long-lasting immune control of HIV-1 infection (8, 47, 48). The protein products of B*5703 and B*8101 (B5703 and B8101, respectively) are capable of presenting HIV-1 epitopes (Gag epitopes in particular) known to be restricted by B5701 (15, 49, 50). Therefore, in the same Zambian population where both B*5703 and B*8101 are in tight LD with Cw*1801, Cw*1801 actually tracks the effect of two favorable HLA-B alleles that can target conserved HIV-1 epitopes for protective cytotoxic T-lymphocyte responses (15, 34, 45, 50) and related function (51). As a cautionary note, Zambians with Cw*18 and without B*57 or B*8101 were too infrequent to allow a clear separation of HLA-B alleles from the two B–C haplotypes (e.g., Table V). Moreover, two independent research teams have already identified four Cw1801-restricted epitopes: VI9 and FF9 in p24 Gag, VL9 in integrase, and YI9 in gp160 (6, 34, 52), suggesting that the Cw1801 allele itself can also play an important role in mediating CTL responses. Thus, a combination of favorable HLA-B and HLA-C alleles likely underlie the apparent relationships of Cw*1801 to HIV-1 transmission and viral load in more than 60 index partners (Tables IV to VI).
By conventional univariate analyses, HLA-B*57 and the B*57-Cw*18 haplotype were also negatively associated with HIV-1 transmission in Zambians. These relationships were first consistent with the well-recognized role of the B57 product in HIV-1-specific CTL responses and then supported by the association of B*57 with index partners’ HIV-1 viral load (Table V). Although multivariable models dismissed HLA-B*57 and the B*57-Cw*18 as major contributors when the stronger effect (risk) of A*36 and index partner viral load were treated as co-factors, HIV-1 transmission events within the first six to seven years (i.e., 2,192–2,557 days) of follow-up did indicate a clear advantage of B*57 and the B*57-Cw*18 haplotype, as can be inferred from Fig. 3. The relative contribution of B*57 and other favorable HLA class I alleles (like B*8101 and Cw*1801) to immune control of HIV-1 infection is expected to diminish with time, due to viral immune escape and accumulation of compensatory mutations during chronic infection (11, 12, 53–55). Accordingly, it may be particularly important to concentrate on HIV-1-related outcomes (i.e., viral load and transmission) during early infection in the effort to identify favorable HLA factors and other correlates of protective immunity.
Viral load during untreated, chronic HIV-1 infection reflects the equilibrium between viral replication and the effect of host adaptive immunity. Population-based studies have clearly established the predictive value of plasma (cell-free) viral load for heterosexual HIV-1 transmission (18, 19) as well as time to AIDS, especially in Caucasian males (56–58). Assuming that viral load in most of the chronically infected Zambians (i.e., index partners) can serve as a proxy for set-point viral load, patients with low viral load because of Cw*18 or its linked HLA-B alleles may experience a more benign course of disease. However, quantifying the dual impact of viral load on heterosexual transmission and HIV-1 pathogenesis will be difficult in populations where antiretroviral therapy has become increasingly available in the past few years. Confirmatory research will likely depend on analysis of other well-established cohorts of HIV-1 discordant couples with no or limited access to treatment.
Very high levels of viral load (often millions of RNA copies per ml of plasma) during the brief period (usually within the first nine weeks) of acute-phase infection (59) have been associated with highest rates of HIV-1 transmission per coital act (60). Since index partners in the Zambian cohort were identified by serology rather than viral load or p24 antigen tests, our work here might have missed acutely infected patients, although some index partners (4.6% NTIs and 13.9% TPIs) indeed had viral load greater than 500,000 copies per ml of plasma. However, unlike set-point viral loads that differ greatly from one chronically infected patient to another, acute-phase viral loads are so uniformly high that they would likely overwhelm any differential effect of genetic variation. Moreover, our use of viral load as a categorical variable is supported by the recent notion that viral transmission potential in chronically infected Zambians and Ugandans (61) does not increase substantially once set-point viral loads reach 100,000 copies/mL.
In other studies of paired HIV-1 donors and recipients, mother-to-child transmission (MTCT) has provided some unique models for evaluating the varying roles of HLA class I alleles, haplotypes, and diversity (heterozygosity) in HIV-1 infection and/or pathogenesis (62–68). Although consensus findings remain elusive, several maternal HLA class I (mostly HLA-B) alleles, maternal HLA homozygosity, as well as the degree of allele sharing between mothers and infants, seem to influence MTCT in one way or another (62, 63, 66, 68). Neither A*36 nor Cw*18 has been reported as critical to vertical HIV-1 transmission, which differs from viral transmission among discordant couples (adults) in two ways: 1) MTCT recipients (infants) and donors always share not only 50% or more of their HLA class I alleles but many other genetic traits throughout the nuclear genome as well and 2) the immature immune system in the infants probably expedites HIV-1 transmission, especially when HLA-adapted viral mutants are being transmitted (67, 69). In any case, maternal HLA alleles previously associated with vertical HIV-1 transmission have invariably differed from infant HLA alleles associated with acquisition of infection. Likewise, the two HLA class I alleles (A*3601 and Cw*1801) strongly implicated in our analyses of viral transmission by the index partners clearly lack association with HIV-1 acquisition by seronegative Zambians. Therefore, within paired donors and recipients, there is more to be learned about the mechanisms of adaptive and innate immunity that control the process of viral transmission as distinct from those that mediate viral acquisition.
Acknowledgments
We thank staff and study participants in the Zambia-Emory HIV-1 Research Project for their valuable contributions to various aspects of this study. We are also indebted to P. Farmer for sample management, W. Song for technical assistance, and J. Michael Kilby for critical reading of an earlier version of this manuscript.
Footnotes
This work was supported in part by National Institute of Allergy and Infectious Diseases (NIAID), through grants AI40951 (to S.A.), AI41530 (to J. Michael Kilby), AI41951 (to R.A.K.), AI51173 (to J.T.), and AI64060 (to E.H). J.T. is the recipient of an Independent Scientist Award (AI76123) from NIAID.
Abbreviations used in this paper: ageΔ, age difference; CI: confidence interval; D′, relative linkage disequilibrium; EM, expectation-maximization; ESN, exposed seronegative; Freq, frequency; FTM; female-to-male; FUT, follow-up time; GUI, genital ulcer/inflammation; HIV-1, human immunodeficiency virus type 1; HIV+, HIV-1 seropositive (infected); HIV−, HIV-1 seronegative (uninfected); HLA, human leukocyte antigen; HWE, Hardy-Weinberg equilibrium; LD, linkage disequilibrium; MTCT, mother-to-child transmission; MTF, male-to-female; NA, not applicable; OR, odds ratio; PIP, paired index partner; RH, relative hazard; SC, seroconverter; TPI: transmission pair index; NTI; non-TPI.
Disclosures
The authors have no financial conflict of interest associated with this work.
References
- 1.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC, Jr, Wright MW, Wain HM, Trowsdale J, Ziegler A, Beck S. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 2.Schreuder GM, Hurley CK, Marsh SG, Lau M, Fernandez-Vina MA, Noreen HJ, Setterholm M, Maiers M. HLA dictionary 2004: summary of HLA-A, -B, -C, -DRB1/3/4/5, -DQB1 alleles and their association with serologically defined HLA-A, -B, -C, -DR, and -DQ antigens. Hum Immunol. 2005;66:170–210. doi: 10.1016/j.humimm.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Geraghty DE, Hansen JA, Hurley CK, Mach B, Mayr WR, Parham P, Petersdorf EW, Sasazuki T, Schreuder GM, Strominger JL, Svejgaard A, Terasaki PI, Trowsdale J. Nomenclature for factors of the HLA system, 2004. Hum Immunol. 2005;66:571–636. doi: 10.1016/j.humimm.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 4.de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, Ke X, Monsuur AJ, Whittaker P, Delgado M, Morrison J, Richardson A, Walsh EC, Gao X, Galver L, Hart J, Hafler DA, Pericak-Vance M, Todd JA, Daly MJ, Trowsdale J, Wijmenga C, Vyse TJ, Beck S, Murray SS, Carrington M, Gregory S, Deloukas P, Rioux JD. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes AL. Natural selection and the diversification of vertebrate immune effectors. Immunol Rev. 2002;190:161–168. doi: 10.1034/j.1600-065x.2002.19012.x. [DOI] [PubMed] [Google Scholar]
- 6.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 7.Honeyborne I, Prendergast A, Pereyra F, Leslie A, Crawford H, Payne R, Reddy S, Bishop K, Moodley E, Nair K, van der Stok M, McCarthy N, Rousseau CM, Addo M, Mullins JI, Brander C, Kiepiela P, Walker BD, Goulder PJ. Control of human immunodeficiency virus type 1 is associated with HLA-B*13 and targeting of multiple gag-specific CD8+ T-cell epitopes. J Virol. 2007;81:3667–3672. doi: 10.1128/JVI.02689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bansal A, Yue L, Conway J, Yusim K, Tang J, Kappes J, Kaslow RA, Wilson CM, Goepfert PA. Immunological control of chronic HIV-1 infection: HLA-mediated immune function and viral evolution in adolescents. AIDS. 2007;21:2387–2397. doi: 10.1097/QAD.0b013e3282f13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharya T, Daniels M, Heckerman D, Foley B, Frahm N, Kadie C, Carlson J, Yusim K, McMahon B, Gaschen B, Mallal S, Mullins JI, Nickle DC, Herbeck J, Rousseau C, Learn GH, Miura T, Brander C, Walker B, Korber B. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- 11.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas SA, St John A, Roach TA, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker BD, Goulder PJ. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 12.Brumme ZL, Brumme CJ, Heckerman D, Korber BT, Daniels M, Carlson J, Kadie C, Bhattacharya T, Chui C, Szinger J, Mo T, Hogg RS, Montaner JS, Frahm N, Brander C, Walker BD, Harrigan PR. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 2007;3:e94. doi: 10.1371/journal.ppat.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang J, Kaslow RA. The impact of host genetics on HIV infection and disease progression in the era of highly active antiretroviral therapy. AIDS. 2003;17:S51–S60. doi: 10.1097/00002030-200317004-00006. [DOI] [PubMed] [Google Scholar]
- 14.O’Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36:565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- 15.Gillespie GM, Pinheiro S, Sayeid-Al-Jamee M, Alabi A, Kaye S, Sabally S, Sarge-Njie R, Njai H, Joof K, Jaye A, Whittle H, Rowland-Jones S, Dorrell L. CD8+ T cell responses to human immunodeficiency viruses type 2 (HIV-2) and type 1 (HIV-1) gag proteins are distinguishable by magnitude and breadth but not cellular phenotype. Eur J Immunol. 2005;35:1445–1453. doi: 10.1002/eji.200526007. [DOI] [PubMed] [Google Scholar]
- 16.Leslie A, Kavanagh D, Honeyborne I, Pfafferott K, Edwards C, Pillay T, Hilton L, Thobakgale C, Ramduth D, Draenert R, Le Gall S, Luzzi G, Edwards A, Brander C, Sewell AK, Moore S, Mullins J, Moore C, Mallal S, Bhardwaj N, Yusim K, Phillips R, Klenerman P, Korber B, Kiepiela P, Walker B, Goulder P. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J Exp Med. 2005;201:891–902. doi: 10.1084/jem.20041455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nolan D, Gaudieri S, John M, Mallal S. Impact of host genetics on HIV disease progression and treatment: new conflicts on an ancient battleground. AIDS. 2004;18:1231–1240. doi: 10.1097/00002030-200406180-00001. [DOI] [PubMed] [Google Scholar]
- 18.Quinn TC, Wawer MJ, Sewankambo N, Serwadda D, Li C, Wabwire-Mangen F, Meehan MO, Lutalo T, Gray RH. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N Engl J Med. 2000;342:921–929. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- 19.Fideli US, Allen S, Musunda R, Trask S, Hahn B, Mulenga J, Kasolo FC, Vermund SH, Aldrovandi G. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 (HIV-1) in Africa. AIDS Res Hum Retroviruses. 2001;17:901–910. doi: 10.1089/088922201750290023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen S, Meinzen-Derr J, Kautzman M, Zulu I, Trask S, Fideli U, Musonda R, Kasolo F, Gao F, Haworth A. Sexual behavior of HIV discordant couples after HIV counseling and testing. AIDS. 2003;17:733–740. doi: 10.1097/00002030-200303280-00012. [DOI] [PubMed] [Google Scholar]
- 21.Trask SA, Derdeyn CA, Fideli U, Chen Y, Meleth S, Kasolo F, Musonda R, Hunter E, Gao F, Allen S, Hahn BH. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. J Virol. 2002;76:397–405. doi: 10.1128/JVI.76.1.397-405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoesley CJ, Allen S, Raper JL, Musonda R, Niu YF, Gao F, Squires KE, Aldrovandi GM. Comparative analysis of commercial assays for the detection and quantification of plasma HIV-1 RNA in patients infected with HIV-1 subtype C. Clin Infect Dis. 2002;35:323–325. doi: 10.1086/341490. [DOI] [PubMed] [Google Scholar]
- 23.Tang J, Tang S, Lobashevsky E, Myracle AD, Fideli U, Aldrovandi G, Allen S, Musonda R, Kaslow RA. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J Virol. 2002;76:8276–8284. doi: 10.1128/JVI.76.16.8276-8284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao W, Tang J, Dorak MT, Song W, Lobashevsky E, Cobbs CS, Wrensch MR, Kaslow RA. Molecular typing of human leukocyte antigen and related polymorphisms following whole genome amplification. Tissue Antigens. 2004;64:286–292. doi: 10.1111/j.0001-2815.2004.00295.x. [DOI] [PubMed] [Google Scholar]
- 25.Lazaryan A, Lobashevsky E, Mulenga J, Karita E, Allen S, Tang J, Kaslow RA. Human leukocyte antigen B58 supertype and human immunodeficiency virus type 1 infection in native Africans. J Virol. 2006;80:6056–6060. doi: 10.1128/JVI.02119-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang J, Tang S, Rivers CA, Dorak MT, Penman-Aguilar A, Aldrovandi G, Allen S, Karita E, Musonda R, Kaslow RA. Distinct and evolving HLA class I profiles in native Africans with high prevalence of HIV/AIDS. In: Hansen J, editor. Immunobiology of the Human MHC. IHWG Press; Seattle: 2006. pp. 402–407. [Google Scholar]
- 27.Cano P, Klitz W, Mack SJ, Maiers M, Marsh SG, Noreen H, Reed EF, Senitzer D, Setterholm M, Smith A, Fernandez-Vina M. Common and well-documented HLA alleles: Report of the ad-hoc committee of the American Society for Histocompatiblity and Immunogenetics. Hum Immunol. 2007;68:392–417. doi: 10.1016/j.humimm.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Cao K, Hollenbach J, Shi X, Shi W, Chopek M, Fernandez-Vina MA. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol. 2001;62:1009–1030. doi: 10.1016/s0198-8859(01)00298-1. [DOI] [PubMed] [Google Scholar]
- 29.Tang J, Naik E, Costello C, Karita E, Rivers C, Allen S, Kaslow RA. Characteristics of HLA class I and class II polymorphisms in Rwandan women. Exp Clin Immunogenet. 2000;17:185–198. doi: 10.1159/000019138. [DOI] [PubMed] [Google Scholar]
- 30.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Klenerman P, McMichael A. AIDS/HIV. Finding footprints among the trees. Science. 2007;315:1505–1507. doi: 10.1126/science.1140768. [DOI] [PubMed] [Google Scholar]
- 32.Kaslow RA, Dorak T, Tang JJ. Influence of host genetic variation on susceptibility to HIV type 1 infection. J Infect Dis. 2005;191:S68–77. doi: 10.1086/425269. [DOI] [PubMed] [Google Scholar]
- 33.Hansen JA. Immunobiology of the Human MHC. IHWG Press; Seattle, Washington: 2006. [Google Scholar]
- 34.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJ. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 35.Rasheed S. Infectivity and dynamics of HIV type 1 replication in the blood and reproductive tract of HIV type 1-infected women. AIDS Res Hum Retroviruses. 1998;14(Suppl 1):S105–118. [PubMed] [Google Scholar]
- 36.Tachet A, Dulioust E, Salmon D, De Almeida M, Rivalland S, Finkielsztejn L, Heard I, Jouannet P, Sicard D, Rouzioux C. Detection and quantification of HIV-1 in semen: identification of a subpopulation of men at high potential risk of viral sexual transmission. AIDS. 1999;13:823–831. doi: 10.1097/00002030-199905070-00012. [DOI] [PubMed] [Google Scholar]
- 37.Gupta P, Leroux C, Patterson BK, Kingsley L, Rinaldo C, Ding M, Chen Y, Kulka K, Buchanan W, McKeon B, Montelaro R. Human immunodeficiency virus type 1 shedding pattern in semen correlates with the compartmentalization of viral Quasi species between blood and semen. J Infect Dis. 2000;182:79–87. doi: 10.1086/315644. [DOI] [PubMed] [Google Scholar]
- 38.Kovacs A, Wasserman SS, Burns D, Wright DJ, Cohn J, Landay A, Weber K, Cohen M, Levine A, Minkoff H, Miotti P, Palefsky J, Young M, Reichelderfer P. Determinants of HIV-1 shedding in the genital tract of women. Lancet. 2001;358:1593–1601. doi: 10.1016/S0140-6736(01)06653-3. [DOI] [PubMed] [Google Scholar]
- 39.Fiore JR, Suligoi B, Saracino A, Di Stefano M, Bugarini R, Lepera A, Favia A, Monno L, Angarano G, Pastore G. Correlates of HIV-1 shedding in cervicovaginal secretions and effects of antiretroviral therapies. AIDS. 2003;17:2169–2176. doi: 10.1097/00002030-200310170-00004. [DOI] [PubMed] [Google Scholar]
- 40.Zuckerman RA, Whittington WL, Celum CL, Collis TK, Lucchetti AJ, Sanchez JL, Hughes JP, Sanchez JL, Coombs RW. Higher concentration of HIV RNA in rectal mucosa secretions than in blood and seminal plasma, among men who have sex with men, independent of antiretroviral therapy. J Infect Dis. 2004;190:156–161. doi: 10.1086/421246. [DOI] [PubMed] [Google Scholar]
- 41.Coombs RW, Lockhart D, Ross SO, Deutsch L, Dragavon J, Diem K, Hooton TM, Collier AC, Corey L, Krieger JN. Lower genitourinary tract sources of seminal HIV. J Acquir Immune Defic Syndr. 2006;41:430–438. doi: 10.1097/01.qai.0000209895.82255.08. [DOI] [PubMed] [Google Scholar]
- 42.Cummins JE, Christensen L, Lennox JL, Bush TJ, Wu Z, Malamud D, Evans-Strickfaden T, Siddig A, Caliendo AM, Hart CE, Dezzutti CS. Mucosal innate immune factors in the female genital tract are associated with vaginal HIV-1 shedding independent of plasma viral load. AIDS Res Hum Retroviruses. 2006;22:788–795. doi: 10.1089/aid.2006.22.788. [DOI] [PubMed] [Google Scholar]
- 43.Pilcher CD, Joaki G, Hoffman IF, Martinson FE, Mapanje C, Stewart PW, Powers KA, Galvin S, Chilongozi D, Gama S, Price MA, Fiscus SA, Cohen MS. Amplified transmission of HIV-1: comparison of HIV-1 concentrations in semen and blood during acute and chronic infection. AIDS. 2007;21:1723–1730. doi: 10.1097/QAD.0b013e3281532c82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corey L, Wald A, Celum CL, Quinn TC. The effects of herpes simplex virus-2 on HIV-1 acquisition and transmission: a review of two overlapping epidemics. J Acquir Immune Defic Syndr. 2004;35:435–445. doi: 10.1097/00126334-200404150-00001. [DOI] [PubMed] [Google Scholar]
- 45.Geldmacher C, Currier JR, Herrmann E, Haule A, Kuta E, McCutchan F, Njovu L, Geis S, Hoffmann O, Maboko L, Williamson C, Birx D, Meyerhans A, Cox J, Hoelscher M. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. J Virol. 2007;81:2440–2448. doi: 10.1128/JVI.01847-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, De Luca A, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, Connors M. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Migueles SA, Laborico AC, Imamichi H, Shupert WL, Royce C, McLaughlin M, Ehler L, Metcalf J, Liu S, Hallahan CW, Connors M. The differential ability of HLA B*5701+ long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral gag sequences. J Virol. 2003;77:6889–6898. doi: 10.1128/JVI.77.12.6889-6898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferrari G, Currier JR, Harris ME, Finkelstein S, de Oliveira A, Barkhan D, Cox JH, Zeira M, Weinhold KJ, Reinsmoen N, McCutchan F, Birx DL, Osmanov S, Maayan S. HLA-A and -B allele expression and ability to develop anti-Gag cross-clade responses in subtype C HIV-1-infected Ethiopians. Hum Immunol. 2004;65:648–659. doi: 10.1016/j.humimm.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 50.Stewart-Jones GB, Gillespie G, Overton IM, Kaul R, Roche P, McMichael AJ, Rowland-Jones S, Jones EY. Structures of three HIV-1 HLA-B*5703-peptide complexes and identification of related HLAs potentially associated with long-term nonprogression. J Immunol. 2005;175:2459–2468. doi: 10.4049/jimmunol.175.4.2459. [DOI] [PubMed] [Google Scholar]
- 51.Dong T, Stewart-Jones G, Chen N, Easterbrook P, Xu X, Papagno L, Appay V, Weekes M, Conlon C, Spina C, Little S, Screaton G, van der Merwe A, Richman DD, McMichael AJ, Jones EY, Rowland-Jones SL. HIV-specific cytotoxic T cells from long-term survivors select a unique T cell receptor. J Exp Med. 2004;200:1547–1557. doi: 10.1084/jem.20032044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boutwell CL, Essex M. Identification of HLA class I-associated amino acid polymorphisms in the HIV-1C proteome. AIDS Res Hum Retroviruses. 2007;23:165–174. doi: 10.1089/aid.2006.0131. [DOI] [PubMed] [Google Scholar]
- 53.Yu XG, Lichterfeld M, Chetty S, Williams KL, Mui SK, Miura T, Frahm N, Feeney ME, Tang Y, Pereyra F, Labute MX, Pfafferott K, Leslie A, Crawford H, Allgaier R, Hildebrand W, Kaslow R, Brander C, Allen TM, Rosenberg ES, Kiepiela P, Vajpayee M, Goepfert PA, Altfeld M, Goulder PJ, Walker BD. Mutually exclusive T-cell receptor induction and differential susceptibility to human immunodeficiency virus type 1 mutational escape associated with a two-amino-acid difference between HLA class I subtypes. J Virol. 2007;81:1619–1631. doi: 10.1128/JVI.01580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karlsson AC, Iversen AK, Chapman JM, de Oliviera T, Spotts G, McMichael AJ, Davenport MP, Hecht FM, Nixon DF. Sequential broadening of CTL responses in early HIV-1 infection is associated with viral escape. PLoS ONE. 2007;2:e225. doi: 10.1371/journal.pone.0000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crawford H, Prado JG, Leslie A, Hue S, Honeyborne I, Reddy S, van der Stok M, Mncube Z, Brander C, Rousseau C, Mullins JI, Kaslow R, Goepfert P, Allen S, Hunter E, Mulenga J, Kiepiela P, Walker BD, Goulder PJ. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J Virol. 2007;81:8346–8351. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mellors JW, Rinaldo CR, Jr, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 57.Lyles RH, Munoz A, Yamashita TE, Bazmi H, Detels R, Rinaldo CR, Margolick JB, Phair JP, Mellors JW. Natural history of human immunodeficiency virus type 1 viremia after seroconversion and proximal to AIDS in a large cohort of homosexual men. Multicenter AIDS Cohort Study. J Infect Dis. 2000;181:872–880. doi: 10.1086/315339. [DOI] [PubMed] [Google Scholar]
- 58.Mellors JW, Margolick JB, Phair JP, Rinaldo CR, Detels R, Jacobson LP, Munoz A. Prognostic value of HIV-1 RNA, CD4 cell count, and CD4 Cell count slope for progression to AIDS and death in untreated HIV-1 infection. JAMA. 2007;297:2349–2350. doi: 10.1001/jama.297.21.2349. [DOI] [PubMed] [Google Scholar]
- 59.Grossman Z, Meier-Schellersheim M, Paul WE, Picker LJ. Pathogenesis of HIV infection: what the virus spares is as important as what it destroys. Nat Med. 2006;12:289–295. doi: 10.1038/nm1380. [DOI] [PubMed] [Google Scholar]
- 60.Wawer MJ, Gray RH, Sewankambo NK, Serwadda D, Li X, Laeyendecker O, Kiwanuka N, Kigozi G, Kiddugavu M, Lutalo T, Nalugoda F, Wabwire-Mangen F, Meehan MP, Quinn TC. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J Infect Dis. 2005;191:1403–1409. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- 61.Fraser C, Hollingsworth TD, Chapman R, de Wolf F, Hanage WP. Variation in HIV-1 set-point viral load: epidemiological analysis and an evolutionary hypothesis. Proc Natl Acad Sci USA. 2007;104:17441–17446. doi: 10.1073/pnas.0708559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacDonald KS, Embree J, Njenga S, Nagelkerke NJ, Ngatia I, Mohammed Z, Barber BH, Ndinya-Achola J, Bwayo J, Plummer FA. Mother-child class I HLA concordance increases perinatal human immunodeficiency virus type 1 transmission. J Infect Dis. 1998;177:551–556. doi: 10.1086/514243. [DOI] [PubMed] [Google Scholar]
- 63.Polycarpou A, Ntais C, Korber BT, Elrich HA, Winchester R, Krogstad P, Wolinsky S, Rostron T, Rowland-Jones SL, Ammann AJ, Ioannidis JP. Association between maternal and infant class I and II HLA alleles and of their concordance with the risk of perinatal HIV type 1 transmission. AIDS Res Hum Retroviruses. 2002;18:741–746. doi: 10.1089/08892220260139477. [DOI] [PubMed] [Google Scholar]
- 64.Kuhn L, Abrams EJ, Palumbo P, Bulterys M, Aga R, Louie L, Hodge T. Maternal versus paternal inheritance of HLA class I alleles among HIV-infected children: consequences for clinical disease progression. AIDS. 2004;18:1281–1289. doi: 10.1097/00002030-200406180-00006. [DOI] [PubMed] [Google Scholar]
- 65.Farquhar C, Rowland-Jones S, Mbori-Ngacha D, Redman M, Lohman B, Slyker J, Otieno P, Obimbo E, Rostron T, Ochieng J, Oyugi J, Bosire R, John-Stewart G. Human leukocyte antigen (HLA) B*18 and protection against mother-to-child HIV type 1 transmission. AIDS Res Hum Retroviruses. 2004;20:692–697. doi: 10.1089/0889222041524616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winchester R, Pitt J, Charurat M, Magder LS, Goring HH, Landay A, Read JS, Shearer W, Handelsman E, Luzuriaga K, Hillyer GV, Blattner W. Mother-to-child transmission of HIV-1: strong association with certain maternal HLA-B alleles independent of viral load implicates innate immune mechanisms. J Acquir Immune Defic Syndr. 2004;36:659–670. doi: 10.1097/00126334-200406010-00002. [DOI] [PubMed] [Google Scholar]
- 67.Sanchez-Merino V, Nie S, Luzuriaga K. HIV-1-specific CD8+ T cell responses and viral evolution in women and infants. J Immunol. 2005;175:6976–6986. doi: 10.4049/jimmunol.175.10.6976. [DOI] [PubMed] [Google Scholar]
- 68.Mackelprang RD, John-Stewart G, Carrington M, Richardson BA, Rowland-Jones S, Gao X, Mbori-Ngacha D, Mabuka J, Lohman-Payne B, Farquhar C. Maternal HLA homozygosity and mother-child HLA concordance increase the risk of vertical transmission of HIV-1. J Infect Dis. 2008;197:1156–1161. doi: 10.1086/529528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goulder PJ, Brander C, Tang Y, Tremblay C, Colbert RA, Addo MM, Rosenberg ES, Nguyen T, Allen R, Trocha A, Altfeld M, He S, Bunce M, Funkhouser R, Pelton SI, Burchett SK, McIntosh K, Korber BT, Walker BD. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001;412:334–338. doi: 10.1038/35085576. [DOI] [PubMed] [Google Scholar]