Abstract
The phylogenetic group termed OP5 was originally discovered in the Yellowstone National Park hot spring and proposed as an uncultured phylum; the group was afterwards analyzed by applying culture-independent approaches. Recently, a novel thermophilic chemoheterotrophic filamentous bacterium was obtained from a hot spring in Japan that was enriched through various isolation procedures. Phylogenetic analyses of the isolate have revealed that it is closely related to the OP5 phylum that has mainly been constructed with the environmental clones retrieved from thermophilic and mesophilic anaerobic environments. It appears that the lineage is independent at the phylum level in the domain Bacteria. Therefore, we designed a primer set for the 16S rRNA gene to specifically target the OP5 phylum and performed quantitative field analysis by using the real-time PCR method. Thus, the 16S rRNA gene of the OP5 phylum was detected in some hot-spring samples with the relative abundance ranging from 0.2% to 1.4% of the prokaryotic organisms detected. The physiology of the above-mentioned isolate and the related environmental clones indicated that they are scavengers contributing to the sulfur cycle in nature.
Since Woese culminated in his review from 1987 (36) the phylogenetic relationships among various prokaryotic organisms based on their 16S rRNA gene sequences, the method has been widely used in prokaryotic taxonomy and the 16S rRNA gene sequences of almost all cultured microorganisms have been determined. In addition, culture-independent 16S rRNA-based molecular analyses have been performed, leading to the discovery of many uncultured phyla (so-called “candidate phyla”), and it has been recognized that most microorganisms in nature have not yet been successfully cultivated (2, 27, 28). Although unexplored environments, such as the deep subterranean sphere, still remain, microbial phyla have been extensively analyzed and established by molecular approaches (10, 11, 29). There are 24 bacterial phyla that include one or more cultivated representatives in the taxonomic outline of Bergey's Manual of Systematic Bacteriology (http://141.150.157.80/bergeysoutline/main.htm). Moreover, the phylum Lentisphaerae, which was originally recognized as the candidate phylum VadinBE97, was newly described to include marine isolates (5). In addition to these phyla of cultured bacteria, Rappé and Giobannoni concluded that the number of recognized candidate phyla is 25 in the domain Bacteria (29).
Although molecular surveys and phylogenetic analyses of 16S rRNA genes have detected many candidate phyla in nature, pure culture isolates from candidate phyla have been obtained only in a few cases, for example, from the phyla Gemmatimonadetes (37) and Lentisphaerae (5). Thermodesulfobium narugense (22) and Caldithrix abyssi (20) are likely to belong to new phylum-level lineages, but their higher taxa have not been established yet (in the taxonomic outline of Bergey's Manual of Systematic Bacteriology, T. narugense and C. abyssi are included in the phyla Firmicutes and Deferribacteres, respectively). These novel representatives will be very important not only for the establishment of their phylogenetic positions but also for the presence of possible novel phenotypic features and their applications as well as new gene resources.
The candidate phylum OP5 has been assigned as one of the uncultured bacterial phyla and was originally represented by the environmental clone sequences retrieved from the Obsidian Pool (OP) of Yellowstone National Park (12). Recently, we isolated a novel thermophilic chemoheterotroph from a hot spring in Otari, Nagano Prefecture, Japan, which appeared to be placed into the OP5 phylum. In the present study, a phylogenetic analysis and field survey of bacteria belonging to the OP5 phylum, including a cultivated representative, have been conducted, and the possible roles of the OP5 phylum will be discussed.
MATERIALS AND METHODS
Microorganisms and growth conditions.
Strain AZM16c01 was isolated as follows in this study. Escherichia coli NBRC 3301 (K-12) and Methanothermobacter thermautotrophicus NBRC 100330T (ΔHT) were used as standards for a quantitative PCR analysis. The cultivating media for these strains were NBRC media 802 and 398 (26), respectively, and the cultivating temperatures were 37 and 65°C, respectively.
Study areas, sample collection, and measurements.
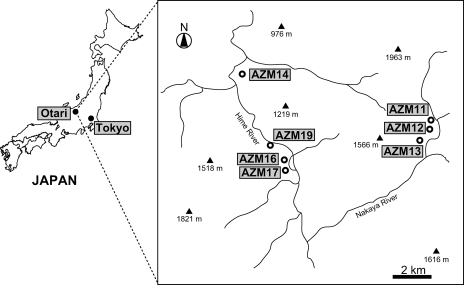
Hot-water samples were collected from Otari in Nagano Prefecture, Japan, on 10 to 13 April 2007 (Fig. 1). Some hot waters sprang out at various sites in Otari, and seven sites were selected for the study (Table 1). We collected water samples AZM11, AZM12, and AZM19 from the flowing sites. The hot-water samples AZM13 and AZM14 were obtained from a light-shielding storage tank that is deployed a few hundred meters apart from the flowing site (AZM13) or a well drilled to a 5-m depth below the ground (AZM14). The hot-water sample AZM16 was obtained from the bottom of a drilling well at a depth of 400 m. The hot-water sample AZM17 was obtained from a well that was drilled adjacent to that of the AZM16 sample but to a shallower depth (200 m). In addition to the hot-water sample, white-colored microbial mats, which were confirmed with the overflows of hot water on the surface of the tank at the AZM14 sampling site, were also collected. For the cultivations, 50-ml volumes of hot-water samples were collected and anaerobically stored in vials with butyl-rubber stoppers and aluminum caps at 4°C before inoculation. For the molecular analysis, microbial cells in 2 liters of hot-water samples were immediately collected onto a 0.2-μm-pore-sized polyvinylidene difluoride membrane filter (Millipore) by filtration using a vacuum pressure and stored at −80°C until extraction of the microbial DNA. The filtrates were stored in a polypropylene bottle and provided for the chemical analysis described below. For the enumeration of total cell densities, one-tenth of the volume of a neutralized 38% formaldehyde solution was added to 100 ml of hot-water samples and kept in the refrigerator at 4°C for 12 h. Microbial cells in the samples were filtered and collected on a 0.2-μm-pore-sized polycarbonate membrane (Nuclepore) under a vacuum pressure below 0.02 MPa.
FIG. 1.
Location of the sampling sites in Otari, Nagano Prefecture, Japan. Seven hot-spring samples were collected at the sites indicated by the open circles.
TABLE 1.
Characteristics of the sampling sites in Otari, Japan
| Sampling site | Temp (°C)
|
pH | Conductivity (ms cm−1) | Concn (mM)
|
|||
|---|---|---|---|---|---|---|---|
| Hot-spring water | For cultivation | Cl− | SO42− | HCO3− | |||
| AZM11 (flowing) | 45 | 45 | 6.8 | 2.75 | 1.49 | 0.005 | 31.7 |
| AZM12 (flowing) | 48 | 45 | 6.8 | 3.29 | 1.89 | 0.005 | 37.9 |
| AZM13 (flowing) | 88 | 80 | 8.7 | 3.42 | 5.89 | 0.15 | 36.1 |
| AZM14 (5-m depth) | 58 | 55 | 6.2 | 2.24 | 11.30 | 0.72 | 8.2 |
| AZM16 (400-m depth) | 70 | 70 | 7.3 | 3.52 | 9.47 | 0.81 | 28.9 |
| AZM17 (200-m depth) | 38 | 6.8 | 1.21 | 2.47 | 0.60 | 10.7 | |
| AZM19 (flowing) | 38 | 6.4 | 1.80 | 4.95 | 0.075 | 15.9 | |
Temperature, pH, and electron conductivity of the hot-water samples were measured on site by a temperature probe (resistance temperature probe), a pH meter (D-13; Horiba), and a conductivity meter (ES-14; Horiba), respectively. The concentrations of Cl− and SO42− in the filtered water samples were analyzed using an ion chromatograph (DX-100; Dionex). The concentration of HCO3− was determined by alkalinity titration using HCl according to the potentiometric titration technique with Gran-function evaluation (7). The total microbial cell densities in the hot-water samples were enumerated under a fluorescent microscope (Olympus) by 4′6-deamidino-2-phenylindole (DAPI) staining as described previously (33).
Enrichment and isolation of microorganisms.
In order to enrich and isolate anaerobic chemoheterotrophs, AP13SRL medium under N2/CO2 (80:20 [vol/vol]; 150 kPa) was used. The AP13SRL medium was composed of the following salts and solutions (liter−1): 0.05 g K2HPO4, 0.09 g KH2PO4, 0.25 g MgSO4·7H2O, 0.15 g CaCl2·2H2O, 0.25 g NH4Cl, 1 g Bacto yeast extract (Difco), 1.1 g sodium lactate, 1.4 g Na2SO4, 2.5 g Na2S2O3·5H2O, 1 ml trace-element solution (23), 2 ml vitamin solution (23), 0.25 g Na2CO3, 0.3 g cystein-HCl, and 0.3 g Na2S·9H2O. The medium was prepared in vials with butyl-rubber stoppers and aluminum caps under N2/CO2. For the enrichments and routine cultivations, 50-ml vials containing 20 ml of the liquid medium were used. For the isolation, a colony formation was performed on the medium solidified with 0.6% (wt/vol) gellan gum and 1 g liter−1 MgCl2·6H2O.
DNA extraction, PCR, sequencing, and quantitative PCR.
The extraction and purification of the genomic DNA of microorganisms were performed as previously described (24). The 16S rRNA gene was amplified by using the following primer sets: 27f and 1492r (16) for the domain Bacteria and Ar0023mLF (5′-TcY gGt TKA TCC TG-3′, the lowercase letters indicating locked nucleic acids [17]) and Ar1530R (5′-GGA GGT GAT CCA GCC G-3′) for the domain Archaea. Approximately 100 ng of genomic DNA was used as a template under the following cycling conditions: initial AmpliTaq Gold activation at 95°C for 9 min, followed by 35 cycles of denaturation at 95°C for 20 s, annealing at 50°C for 20 s, extension at 72°C for 60 s, and a final extension step at 72°C for 10 min. The reaction mixture (30 μl) contained 1× PCR buffer, 3.5 mM MgCl2, 10 mM deoxynucleoside triphosphate mixture, 1.25 U AmpliTaq Gold (each from Applied Biosystems), and 0.4 μM of each forward and reverse primer. The PCR product was purified using the PCR-M clean-up system (Viogene) and sequenced using the BigDye terminator v3.1 cycle sequencing kit with a 3130xl genetic analyzer (both from Applied Biosystems). The following six primers were used for sequencing of the PCR products of the bacterial 16S rRNA gene: 515F (5′-GTG CCA GCA GCC GCG GT-3′), 785F (5′-GGA TTA GAT ACC CTG GTA GTC-3′), 536R (5′-GTA TTA CCG CGG CTG CTG-3′), 802R (5′-TAC CAG GGT ATC TAA TCC-3′), 27f, and 1492r.
The genomic DNA of the microbial cells on the filters retrieved from the hot-water samples was extracted using the Ultraclean soil DNA kit (MoBio). An analysis of a quantitative PCR was performed by using the Power Sybr green PCR master mix with a 7500 real-time PCR system (both from Applied Biosystems). A reaction mixture for the PCR was prepared according to the manufacturer's instructions, and the PCR conditions were as follows: AmpErase uracil N-glycosylase activation at 50°C for 2 min, AmpliTaq Gold activation and the uracil N-glycosylase deactivation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and extension at 60°C for 1 min. The following 16S rRNA gene-targeted PCR primer sets were used: 341f (25) and 530r (16) for the domain Bacteria, A344F (4) and 530r (16) for the domain Archaea, and OP5_230F (5′-GGC TCA TCT CCT ATC AGC-3′) and 341r (16) for cluster 1 of the OP5 phylum (see Fig. 3). The OP5_230F primer was designed using the probe design tool of the ARB software package (19). For the construction of the template standards for each primer set, the dilution series of the 16S rRNA gene PCR products of E. coli, M. thermautotrophicus, and strain AZM16c01 were used. These PCR products were used in each real-time PCR analysis to calculate the copy number of the 16S rRNA genes in the sample of hot waters. Two tests were performed to confirm the specificity of the real-time PCR assay. First, a melting-curve analysis was performed after the amplification. Second, the sizes of the PCR products were confirmed by gel electrophoresis.
FIG. 3.
An NJ tree based on the 16S rRNA gene sequences of strain AZM16c01, environmental clones of the OP5 phylum, and recognized bacterial lineages (a slightly updated bacterial data set described by Hugenholtz [10] was used as the main reference sequences). After alignment was performed with the ARB program, 1,315 positions were used for analyses. The shaded wedges indicate phyla with cultivated representatives, and the unshaded wedges indicate phyla currently represented only by environmental sequences. The numbers in parentheses indicate phyla that are divided into separated clusters. Bootstrap values are indicated at the corresponding nodes (NJ tree/ML tree). The probability scores at branching points in the OP5 phylum obtained with four analysis methods (NJ, ML, MP, and Bayesian) using three phyla (OP5, Proteobacteria, and Chloroflexi) are indicated by solid circles (above 95% by all the phylogenetic analysis methods) and open circles (>85% bootstrap probability supported by two analyses). Bar, 0.02 changes per sequence position.
Phylogenetic analyses.
For the phylogenetic analyses, a bacterial domain reference data set described by Hugenholtz (10) was used and slightly modified according to the recent reports of novel lineages (5, 20, 22). The 16S rRNA gene sequences related to the OP5 phylum were obtained from public databases (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/; greengenes, http://greengenes.lbl.gov/cgi-bin/nph-index.cgi; SILVA, http://www.arb-silva.de/) and added to the ARB database (19). Only near-complete 16S rRNA gene sequences (more than 1,300 nucleotides) were used for the analysis. Putative chimeric sequences were identified with the Mallard program (3) after the sequences were aligned with that of E. coli (U00096) using ClustalX (35). The 16S rRNA gene sequences were aligned against an ARB data set (ssu_jan04.arb version) using the ARB program, and the resulting alignment was manually refined based on primary and secondary structural considerations. The alignment data set was extracted using the ARB default filter (bacteria_rr5_dec04), and 1,315 positions were used for constructing phylogenetic trees. Phylogenetic trees were inferred by neighbor-joining (NJ), maximum-likelihood (ML), and maximum-parsimony (MP) with the software packages of ClustalX (31, 35), MOLPHY (1, 8), and PAUP version 4 (D. L. Swofford, Sinauer Associates, Sunderland, MA), respectively, using previously described methods and parameters (22). In addition, the posterior probabilities of branching points were estimated by Bayesian inference using MrBayes 3.1 (9, 30) with the default settings and the GTR model with gamma-distributed rate variation across sites and a proportion of invariable sites (nst, 6; rates, invgamma). A total of 300,000 generations were calculated for the data sets, and a cladogram with the posterior probabilities with each split and a phylogram with mean branch lengths were obtained with a parameter burn-in of 7,500.
Nucleotide sequence accession number.
The 16S rRNA gene sequence of strain AZM16c01 has been deposited in GenBank/EMBL/DDBJ under the accession number AB428365.
RESULTS
Site description.
The sampling sites along with the characteristics of hot springs are summarized in Fig. 1 and Table 1, respectively. The sampling sites are geographically distant and separated by mountains and rivers. The temperatures of the hot springs visited ranged from 38 to 88°C, and the pHs were almost neutral (pH 6.4 to 7.3) except for AZM13 (pH 8.7). Except for the AZM14 sampling site, microbial mats were not observed around the vents of the sampling sites. Microbial mats developed along the overflows of hot water outside but not inside the storage tank at the AZM14 sampling site. The total cell densities of the hot-water samples were determined by DAPI staining and were 104 to 105 cells ml−1, except for the AZM12 sample (Table 2). Microbial cells in the hot-water sample from AZM12 were rarely detected, unlike in other samples.
TABLE 2.
Direct cell counts and relative abundances of Bacteria, Archaea, and cluster 1 of the OP5 phylum calculated by real-time PCR analysis
| Sample | DAPI counts (cells ml−1) | Abundance of the 16S rRNA gene no. (%)
|
||
|---|---|---|---|---|
| Bacteria | Archaea | Cluster 1 of OP5 | ||
| AZM11 | 6.08 × 104 | 99.3 | 0.7 | NDa |
| AZM12 | <104 | 70.5 | 29.5 | ND |
| AZM13 | 3.46 × 104 | Detectedb | Failedb | ND |
| AZM14 | 1.46 × 105 | 31.3 | 68.7 | 0.7 |
| AZM14 (mat) | 96.5 | 3.5 | 0.9 | |
| AZM16 | 4.96 × 104 | 87.7 | 12.3 | 1.4 |
| AZM17 | 1.08 × 105 | 75.2 | 24.8 | 0.2 |
| AZM19 | 6.60 × 105 | 93.8 | 6.2 | 1.2 |
ND indicates that the value is below the detection limit (<102 copies).
The real-time PCR for Archaea did not yield positive results for AZM13 (see Results for details).
Enrichment and isolation.
For the primary enrichment of anaerobic chemoheterotrophs, 2 ml of each hot-water sample (AZM11, AZM12, AZM13, AZM14, and AZM16) was inoculated into AP13SRL medium and incubated at temperatures nearly corresponding to the respective habitats (Table 1). After a 10-day incubation, microbial growth was observed microscopically in the cultures of AZM11, AZM14, and AZM16, and then the enriched cultures were transferred to fresh AP13SRL medium. The procedure was repeated several times. Because the cells in each culture were almost uniformly distributed after several transfers, their genomic DNA was extracted and the bacterial 16S rRNA gene was amplified by PCR and sequenced. Determination of the partial sequences revealed that bacteria belonging to the OP5 phylum were enriched in the culture of AZM16. Therefore, we focused on the enrichment of AZM16 and tried to obtain a pure isolate. After 1 week of incubation at 70°C on the solid medium, uniform white colonies (approximately 0.3 mm in diameter) were observed. After a second purification step with the same solid medium, a strain named AZM16c01 was successfully isolated from the enriched AZM16 sample.
Physiology of the isolate.
Morphologically, the cells of strain AZM16c01 were gram-negative, filamentous, and 0.3 μm in diameter, with the length ranging from 2 to 5 μm (Fig. 2). Many multicellular filaments were interwoven and formed flock-like aggregates. Strain AZM16c01 was able to grow in AP13SRL medium without sulfate, lactate, and vitamins but not in the medium without thiosulfate or yeast extract. Because strain AZM16c01 did not show growth in AP13SRL medium without thiosulfate, we concluded that the bacterium was able to grow by anaerobic respiration but not by fermentation. The ability of strain AZM16c01 to utilize various electron acceptors and substrates was then investigated (Table 3). Growth was not observed under the atmosphere of H2/CO2 (80:20 [vol/vol] at 150 kPa), nor under N2/CO2/O2 (78/20/2, 75/20/5 and 60/20/20 [vol/vol/vol] at 150 kPa). Instead of thiosulfate, strain AZM16c01 was able to use elemental sulfur and sulfite as electron acceptors in the presence of yeast extract but not sulfate, fumarate, nitrate, or nitrite. Growth was not observed in the medium containing various kinds of sugars, alcohols, or short-chain fatty acids to substitute for yeast extract and was not improved by supplementation with 0.1 g liter−1 yeast extract.
FIG. 2.

Phase-contrast micrograph of strain AZM16c01. Scale bar, 4 μm.
TABLE 3.
Growth conditions of strain AZM16c01a
| Substrate | Electron acceptor | Growthb |
|---|---|---|
| Yeast extract | None | − |
| O2 (2%) | − | |
| O2 (5%) | − | |
| O2 (20%) | − | |
| SO42− (10 mM) | − | |
| S2O32− (10 mM) | + | |
| SO32− (2 mM) | + | |
| S0 (1% [wt/vol]) | + | |
| Fumarate (10 mM) | − | |
| NO3− (10 mM) | − | |
| NO2− (2 mM) | − | |
| NO2− (5 mM) | − | |
| H2 | S2O32− (10 mM) | − |
| H2 + acetate (10 mM) | S2O32− (10 mM) | − |
| Various organic componentsc | S2O32− (10 mM) | − |
Gas phases were N2/CO2, except for the conditions of 2% O2, 5% O2, and 20% O2 as electron acceptors and H2 as an electron donor, for which the gas phases were N2/CO2/O2 (78:20:2, 75:20:5, and 60:20:20 [vol/vol]; 150 kPa) and H2/CO2 (80:20 [vol/vol]; 150 kPa), respectively.
−, negative; +, positive.
The following substrates were tested: acetate (10 mM and 30 mM), butyrate (10 mM), formate (10 mM and 30 mM), fumarate (10 mM), glutamate (10 mM), lactate (10 mM), malate (10 mM), propionate (10 mM), pyruvate (10 mM), succinate (10 mM), l-arginine (10 mM), l-cysteine (10 mM), arabinose (5 mM), fructose (5 mM), galactose (5 mM), glucose (5 mM), inositol (5 mM), mannose (5 mM), raffinose (5 mM), sucrose (5 mM), xylose (5 mM), 5% (vol/vol) CH4, methanol (5 mM), ethanol (2 mM), 2-propanol (2 mM), starch (1 g liter−1), Bacto Casamino Acids (1 g liter−1; Difco), and polypeptone (1 g liter−1; Nihon Seiyaku).
Phylogenetic analysis.
Almost the full-length 16S rRNA gene sequence of strain AZM16c01 was determined (1,481 bp; accession number AB428365). Comparative sequence analysis indicated that strain AZM16c01 was not closely related to any known cultured microorganisms in recognized phyla (less than 85% sequence similarity). Instead, it was closely affiliated with the following four environmental clones related to the candidate phylum OP5: clone SRI-280 from a hot spring mat in Iceland (sequence similarity, 99.4%; accession number AF255600) (32), clone OPS107 from the Yellowstone OP hot spring (sequence similarity, 98.8%; accession number AF027049) (12), clone pCOF_65.7_B9 from the Yellowstone Coffee Pots hot spring (sequence similarity, 98.8%; accession number EU156145), and clone SR_FBR_E18 from the sulfate-reducing community in a fluidized-bed reactor (sequence similarity, 98.6%; accession number AY340822) (15).
By knowing these results, we looked more carefully into the phylogenetic status of the OP5 phylum. We searched for sequences in the public databases and eliminated inferred chimeric sequences. Eighteen sequences were obtained as authentic sequences of the OP5 phylum. The phylogenetic topology of the OP5 phylum including strain AZM16c01 was constructed by the NJ method with related environmental clones and representative sequences of recognized bacterial phyla (Fig. 3). Although the analysis using 366 sequences caused the separation of some phyla (Firmicutes, Proteobacteria, and Chloroflexi), it was confirmed that strain AZM16c01 was associated with the sequences belonging to the OP5 lineage, and it represented an independent phylum-level lineage in the domain Bacteria with a high bootstrap value (93%). The result obtained with the ML method was similar to that obtained with the NJ method (date not shown), but results could not be obtained with the MP and Bayesian methods, because only a limited number of comparative positions was available for the analysis.
To make the phylogenetic relationship clearer, the sequences belonging to the phyla OP5, Proteobacteria, and Chloroflexi were extracted from the data set, and trees by using various methods were also constructed. The OP5 phylum was clearly and completely separated from the phyla Proteobacteria and Chloroflexi, and its independence was strongly supported by the probability scores calculated with all methods (data not shown). The probability scores at individual nodes in the OP5 phylum obtained with four phylogenetic analysis methods (NJ, ML, MP, and Bayesian) are shown in Fig. 3. In the phylum OP5, strain AZM16c01 formed a cluster with environmental clone sequences retrieved from thermophilic environments except for clone SR_FBR_E18 that was retrieved from a reactor at 35°C (cluster 1 in Fig. 3), and the maximum 16S rRNA gene sequence divergence for the cluster was 16.2%. The clone sequences except for cluster 1 of the OP5 phylum were obtained from anaerobic mesophilic environments. The sequence divergence for the OP5 phylum as a whole was 27.2%.
Environmental quantification.
For the field estimation of bacteria belonging to the OP5 phylum in hot springs, quantitative real-time PCR assays were performed (Table 2). The relative abundance of cluster 1 of the OP5 phylum was calculated as the percentage of the total 16S rRNA gene copy number (defined as the sum of the gene copy numbers obtained by bacterial and archaeal primer sets) in the DNA extracts. With respect to sample AZM13, extra PCR products were observed by using the archaeal primer set and the copy numbers could not be obtained. The 16S rRNA genes of cluster 1 of the OP5 phylum were detected in some hot-spring and microbial-mat samples, and the relative abundance in the prokaryotic 16S rRNA gene was between 0.2 and 1.4%.
DISCUSSION
A novel chemoheterotrophic bacterium was isolated from a hot spring in Otari, Japan, and the isolate, named AZM16c01, was found to belong to an uncultured lineage, the candidate phylum OP5. Our phylogenetic analyses indicated that the OP5 lineage including strain AZM16c01 is independent at the phylum level (Fig. 3), which is consistent with previous reports (11, 12, 29). The OP5 phylum was originally proposed as one of the candidate phyla based on short partial 16S rRNA gene sequences obtained from the OP of Yellowstone National Park (12). Since then, environmental clone sequences belonging to this phylum have been detected in various environments, such as the microbial mat of a hot spring (32), submarine hydrothermal systems (14, 34), a meromictic soda lake (13), a hypersaline microbial mat (18), a hydrocarbon- and chlorinated-solvent-contaminated aquifer (6), and a fluidized-bed reactor (15) (Fig. 3). In addition to these environmental clone sequences, we tried to search for related sequences in public databases, which resulted in a total of 18 authentic sequences of the OP5 phylum. In our results, the 16S rRNA genes of the OP5 phylum were detected in the range of 0.2 to 1.4% of the prokaryotic 16S rRNA genes from the hot-spring samples (Table 2). Considering these results, it may be that bacteria of the OP5 phylum inhabit unique environments such as hot springs rather than being ubiquitous.
By the real-time PCR analysis using the OP5-specific primer, the 16S rRNA gene fragments of the OP5 phylum were detected only in some hot-spring samples in the study area (Table 2). Although the appearance of the OP5 phylum is likely to be controlled by geographical distribution, it is noteworthy that cluster 1 of the OP5 phylum was not detected in AZM11, AZM12, and AZM13, where sulfate was depleted in the hot-spring water. Anaerobic and sulfur compound-rich conditions may be important elements for the inhabitation of bacteria of the OP5 phylum. Factors defining their existence remain to be probed further, along with the chemical and physical characteristics, geological properties, microbial communities, and seasonal changes of individual sites.
While strain AZM16c01 was a strictly chemoheterotrophic bacterium and used yeast extract for its growth, it could not utilize organic compounds as the sole substrate, such as sugars, alcohols, and short-chain fatty acids in the presence of 0.1 g liter−1 yeast extract (Table 3). In the present study, we did not specify which component(s) of yeast extract is necessary for the growth of strain AZM16c01. However, because the growth was not observed in the absence of yeast extract, it seems reasonable to assume that strain AZM16c01 is a scavenger in nature. Based on chemical composition and the isotopic compositions of oxygen and hydrogen, the AZM16 hot-spring water is considered to originate mainly from meteoric water that circulates within a sedimentary layer consisting of sandstone, shale, and dacite tuff, and to involve a small contribution of a deeply sourced fluid that has reacted with a serpentine-rich volcanic layer (21). The nutrient sources for the inhabitants of the AZM16 site are organic components that are transported by the meteoric water.
On the other hand, strain AZM16c01 was found to require sulfur compounds for its growth (Table 3). Although we did not know whether this anaerobic sulfur respiration was a characteristic of strain AZM16c01 or the phylogenetic group as a whole, bacteria of the OP5 phylum may be significant contributors to the sulfur cycle in nature. There is considerable validity to this notion in view of the following points: (i) Koksonen et al. reported that 9.1% of the total clones were detected as OP5 clones in the fluidized-bed reactor in which ethanol-utilizing sulfate-reducing bacteria were dominant (15), and (ii) some clones were retrieved from relatively sulfur-rich environments such as hot springs (12, 32) and hydrothermal vent sites (14, 34). At any rate, further analyses of various fields as well as characterizations of other isolates belonging to the OP5 phylum are needed.
In conclusion, through the isolation of a representative belonging to the OP5 phylum for the first time, we made it clear that bacteria belonging to this phylum may play a part in the sulfur cycle in nature. The isolation of additional novel representatives of the phylum will be important for establishing a detailed image of the phylum as well as the genetic resources held by its members.
Acknowledgments
We thank the staff of GERD and the Otari village office for collecting the hot-spring samples. We also thank Kuniko Shimamura for technical support and Katsumi Isono for critical reading of the manuscript. We gratefully acknowledge helpful discussions with Atsushi Yamazoe, Shinzo Mayuzumi, and Fumiyoshi Okazaki on several aspects of the paper.
Footnotes
Published ahead of print on 5 September 2008.
REFERENCES
- 1.Adachi, J., and M. Hasegawa. 1995. Improved dating of the human/chimpanzee separation in the mitochondrial DNA tree: heterogeneity among amino acid sites. J. Mol. Evol. 40:622-628. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casamayor, E. O., R. Massana, S. Benlloch, L. Ovreas, B. Diez, V. J. Goddard, J. M. Gasol, I. Joint, F. Rodriguez-Valera, and C. Pedros-Alio. 2002. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 4:338-348. [DOI] [PubMed] [Google Scholar]
- 5.Cho, J. C., K. L. Vergin, R. M. Morris, and S. J. Giovannoni. 2004. Lentisphaera araneosa gen. nov., sp. nov, a transparent exopolymer producing marine bacterium, and the description of a novel bacterial phylum, Lentisphaerae. Environ. Microbiol. 6:611-621. [DOI] [PubMed] [Google Scholar]
- 6.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gieskes, J. M., T. Gamo, and H. Brumsack. 1991. Chemical methods for interstitial water analysis aboard JOIDES Resolution. Technical note 15. Ocean Drilling Program, Texas A&M University, College Station.
- 8.Hasegawa, M., H. Kishino, and T. A. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160-174. [DOI] [PubMed] [Google Scholar]
- 9.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 10.Hugenholtz, P. 2002. Exploring prokaryotic diversity in the genomic era. Genome Biol. 3:reviews0003.1-0003.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humayoun, S. B., N. Bano, and J. T. Hollibaugh. 2003. Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inagaki, F., M. M. Kuypers, U. Tsunogai, J. Ishibashi, K. Nakamura, T. Treude, S. Ohkubo, M. Nakaseama, K. Gena, H. Chiba, H. Hirayama, T. Nunoura, K. Takai, B. B. Jorgensen, K. Horikoshi, and A. Boetius. 2006. Microbial community in a sediment-hosted CO2 lake of the southern Okinawa Trough hydrothermal system. Proc. Natl. Acad. Sci. USA 103:14164-14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaksonen, A. H., J. J. Plumb, P. D. Franzmann, and J. A. Puhakka. 2004. Simple organic electron donors support diverse sulfate-reducing communities in fluidized-bed reactors treating acidic metal- and sulfate-containing wastewater. FEMS Microbiol. Ecol. 47:279-289. [DOI] [PubMed] [Google Scholar]
- 16.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-176. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 17.Levin, J. D., D. Fiala, M. F. Samala, J. D. Kahn, and R. J. Peterson. 2006. Position-dependent effects of locked nucleic acid (LNA) on DNA sequencing and PCR primers. Nucleic Acids Res. 34:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ley, R. E., J. K. Harris, J. Wilcox, J. R. Spear, S. R. Miller, B. M. Bebout, J. A. Maresca, D. A. Bryant, M. L. Sogin, and N. R. Pace. 2006. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl. Environ. Microbiol. 72:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miroshnichenko, M. L., N. A. Kostrikina, N. A. Chernyh, N. V. Pimenov, T. P. Tourova, A. N. Antipov, S. Spring, E. Stackebrandt, and E. A. Bonch-Osmolovskaya. 2003. Caldithrix abyssi gen. nov., sp. nov., a nitrate-reducing, thermophilic, anaerobic bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent, represents a novel bacterial lineage. Int. J. Syst. Evol. Microbiol. 53:323-329. [DOI] [PubMed] [Google Scholar]
- 21.Miyoshi, Y., S. Kawamura, J. Ishibashi, M. Noto, T. Okabe, M. Sato, M. Sunamura, K. Yanagawa, and T. Urabe. 2008. Geochemical study of geothermal fluid collected with a high temperature borehole fluid sampler, abstr. V170-P007. Abstr. JPN Geosci. Union Meet. 2008. Chiba, Japan.
- 22.Mori, K., H. Kim, T. Kakegawa, and S. Hanada. 2003. A novel lineage of sulfate-reducing microorganisms: Thermodesulfobiaceae fam. nov., Thermodesulfobium narugense, gen. nov., sp. nov., a new thermophilic isolate from a hot spring. Extremophiles 7:283-290. [DOI] [PubMed] [Google Scholar]
- 23.Mori, K., A. Maruyama, T. Urabe, K.-I. Suzuki, and S. Hanada. 2008. Archaeoglobus infectus sp. nov., a novel thermophilic, chemolithoheterotrophic archaeon isolated from a deep-sea rock collected at Suiyo Seamount, Izu-Bonin Arc, western Pacific Ocean. Int. J. Syst. Evol. Microbiol. 58:810-816. [DOI] [PubMed] [Google Scholar]
- 24.Mori, K., H. Yamamoto, Y. Kamagata, M. Hatsu, and K. Takamizawa. 2000. Methanocalculus pumilus sp. nov., a heavy-metal-tolerant methanogen isolated from a waste-disposal site. Int. J. Syst. Evol. Microbiol. 50:1723-1729. [DOI] [PubMed] [Google Scholar]
- 25.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.NBRC. 2005. NBRC catalogue of biological resources: microorganisms, genomic DNA clones, and cDNAs, 1st ed. National Institute of Technology and Evaluation, Chiba, Japan.
- 27.Olsen, G. J., D. J. Lane, S. J. Giovannoni, N. R. Pace, and D. A. Stahl. 1986. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40:337-365. [DOI] [PubMed] [Google Scholar]
- 28.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 9:1-55. [Google Scholar]
- 29.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 30.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 31.Saitou, N., and M. Nei. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 32.Skirnisdottir, S., G. O. Hreggvidsson, S. Hjorleifsdottir, V. T. Marteinsson, S. K. Petursdottir, O. Holst, and J. K. Kristjansson. 2000. Influence of sulfide and temperature on species composition and community structure of hot spring microbial mats. Appl. Environ. Microbiol. 66:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sunamura, M., Y. Higashi, C. Miyako, J. Ishibashi, and A. Maruyama. 2004. Two Bacteria phylotypes are predominant in the Suiyo seamount hydrothermal plume. Appl. Environ. Microbiol. 70:1190-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teske, A., K. U. Hinrichs, V. Edgcomb, A. de Vera Gomez, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, H., Y. Sekiguchi, S. Hanada, P. Hugenholtz, H. Kim, Y. Kamagata, and K. Nakamura. 2003. Gemmatimonas aurantiaca gen. nov., sp. nov., a gram-negative, aerobic, polyphosphate-accumulating microorganism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 53:1155-1163. [DOI] [PubMed] [Google Scholar]