Abstract
The hide and carcass hygiene of cull cattle at slaughter in four geographically distant regions of the United States was examined from July 2005 to April 2006 by measuring the aerobic plate counts (APC) and the prevalences and loads of Salmonella and Escherichia coli O157:H7. The geometric mean log10 APC CFU/100 cm2 levels on hides and preevisceration and postintervention carcasses ranged from 6.17 to 8.19, 4.24 to 6.47, and 1.46 to 1.96, respectively, and were highest in the summer (P < 0.0001). The average prevalences of Salmonella on hides and preevisceration and postintervention carcasses were 89.6% (95% confidence interval [CI], 85.1 to 94.0), 50.2% (95% CI, 40.9 to 59.5), and 0.8% (95% CI, 0.18 to 1.42), respectively. The prevalences of E. coli O157:H7 were 46.9% (95% CI, 37.3 to 56.6) and 16.7% (95% CI, 9.8 to 23.6) on hides and preevisceration carcasses, respectively. Examination of the concomitant incidence of Salmonella and E. coli O157:H7 showed that, on average, 33.3% (95% CI, 15.9 to 69.8) of cattle hide and 4.1% (95% CI, 0.98 to 17.3) of preevisceration carcass samples were contaminated with both pathogens. The pathogen prevalence on hides and carcasses was not significantly affected by the season; however, significant differences were observed between plants with respect to the incoming pathogen load and the ability to mitigate hide-to-carcass transfer. In spite of these differences, postintervention carcass contamination was significantly reduced (P < 0.001), likely as a result of the use of one or more of the processing interventions employed at each of the four processing plants examined.
Salmonella and Escherichia coli O157:H7 are estimated to cause 1.3 million and 62,000 cases of gastroenteritis, respectively, each year in the United States (30). Agriculture production environments and animals, including swine, poultry, and cattle, are noted reservoirs of Salmonella and E. coli O157:H7 (1, 37). Ground beef has been implicated as a mode of transmission for these pathogens in a number of food-borne disease outbreaks (13-15). Beef from cull cattle represents approximately 15% of total U.S. beef production, and a majority of this is marketed as ground beef (38, 39). Often, cattle shedding these pathogens in their feces do so without showing any clinical signs of disease (18, 23, 29), a factor that complicates efforts geared toward preventing pathogen entry into the U.S. food supply. Many studies on the microbiological hygiene of cattle at slaughter have shown that hide contamination is strongly correlated with carcass contamination, which is likely the result of cross-contamination (inter- and/or intrahide-to-carcass contamination) during processing (3, 11, 21, 28, 33). The majority of these studies have examined fed-cattle hygiene at slaughter, while fewer have investigated pathogen prevalence associated with cull cattle at slaughter. Increasing our understanding of the variation in the prevalence and load of Salmonella and E. coli O157:H7 contamination present on the hides and carcasses of cull cattle during processing is an important prerequisite for risk analysis and process control point assessment for meat entering the ground-beef supply.
One aspect of determining accurate point assessments of pathogen prevalence is the use of sensitive detection methods. Over the past 2 decades, detection methods for Salmonella and E. coli O157:H7 have undergone substantial improvements. Specifically, the introduction of immunomagnetic separation (IMS) has substantially improved the ability to detect the presence of these pathogens at relatively low levels from a variety of matrices (16, 17, 26, 27). While a number of studies have described the prevalence of E. coli O157:H7 on cattle hides and carcasses at slaughter using IMS in the process of pathogen isolation (2, 4, 22, 35), relatively few have examined Salmonella prevalence in this manner (8, 23, 29). Similarly, few studies have examined the levels (CFU/100 cm2) of these pathogens on cattle hides and carcasses and the extent to which cattle are contaminated with either or both pathogens prior to and during the slaughter process. Here, we report on the prevalence and load of Salmonella and E. coli O157:H7 on cull cattle hides and carcasses presented for slaughter in four geographically distant regions of the United States sampled in four seasons. Both pathogens were isolated using IMS, and the pathogen load was evaluated using rapid direct plating enumeration techniques (12). Also presented is an examination of the levels of aerobic bacterial contamination in hide and carcass samples and the extents to which they were found to be contaminated with either or both Salmonella and E. coli O157:H7.
MATERIALS AND METHODS
Sample collection.
Four processing plants that mainly slaughter cull cows and bulls or dairy cattle in four geographically distant regions of the United States were sampled every 3 months (July, October, January, and April) in a 10-month period from 2005 to 2006. Samples were collected over 2 days on each trip, totaling 8 sampling days per plant. On each sampling day, 95 hide and preevisceration (prior to any intervention) and postintervention (after receiving the full complement of all processing interventions and having been chilled for no more than 2 hours) carcass samples were collected, resulting in a total of 3,040 for each sample type. The hides and carcasses were tagged prior to being sampled so that the samples were matched, and samples were collected consecutively. All samples were shipped in coolers with ice packs and were received and processed at the U.S. Meat Animal Research Center within 24 h of collection.
Hide samples were obtained by swabbing approximately 1,000 cm2 with a sterile sponge (Whirl Pak; Nascom, Fort Atkinson, WI) prewetted with 20 ml sterile Difco buffered peptone water (Beckton Dickinson, Sparks, MD). Hide samples were collected from the brisket plate region of animals on the line, after stunning and exsanguination and prior to hide removal. Carcass samples were obtained by swabbing approximately 8,000 cm2 of carcass with two sterile sponges (Nasco), each prewetted with 10 ml buffered peptone water as previously described (4).
Culture media, aerobic plate counts (APC), enrichment, and enumeration.
Hide and preevisceration and postintervention carcass sample enrichments were analyzed for the presence of Salmonella, while only hide and preevisceration carcass enrichments were analyzed for the presence of E. coli O157:H7. Sponge samples were enriched as previously described (4, 10). Briefly, Difco trypticase soy broth (Beckton Dickinson, Sparks, MD) was added to the sponge samples in a 1:5 ratio and incubated at 25°C for 2 h and at 42°C for 6 h and then held at 4°C until the samples were processed the next day. E. coli O157:H7 or Salmonella was isolated from culture enrichments using IMS as previously described (31).
For Salmonella isolation, IMS beads were placed into 3 ml of Rappaport-Vassiliadis soya peptone broth (Oxoid, Basingstoke, United Kingdom) and incubated at 42°C for 18 to 20 h. These enrichments were swabbed onto Difco Hektoen enteric medium (Beckton Dickinson) with novobiocin at a concentration of 5 mg liter−1 and Difco Brilliant Green agar with sulfidiazine at 80 mg liter−1 (Beckton Dickinson) and then streaked for isolation and incubated at 37°C for 18 to 20 h. Putative Salmonella isolates were confirmed by PCR for the Salmonella-specific portion of the invA gene (32, 34).
For E. coli O157:H7 isolation, IMS beads were plated directly onto ntChrom-O157 agar (DRG International, Mountainside, NJ) containing 5 mg liter−1 novobiocin and 2.5 mg liter−1 potassium tellurite and ctSMac (Difco sorbitol MacConkey agar; Beckton Dickinson) with cefixime at 0.05 mg liter−1 and tellurite at 2.5 mg liter−1. The plates were incubated at 37°C for 18 to 20 h, and putative E. coli O157:H7 colonies were tested for the O157 antigen using the DrySpot agglutination test kit (Oxoid) and further genotypically confirmed as being the O157:H7 serotype with a multiplex PCR (25).
Enumeration of Salmonella and E. coli O157:H7 was performed as previously described (12). Hide samples collected in all four sampling seasons were examined using enumeration methods, while for carcass samples, only those collected in winter and spring were evaluated using enumeration methods. Briefly, for pathogen enumeration from hide samples, a 50-μl aliquot of each 20-ml sponge sample was spiral plated onto the appropriate selective medium and incubated as described below. For pathogen enumeration from carcass samples, 500 μl (for Salmonella) or 300 μl (for E. coli O157:H7) of carcass sponge sample was added to 7 ml of phosphate-buffered saline with 1% (vol/vol) Tween 80 (Sigma), and these samples were analyzed by hydrophobic grid membrane filtration using Isogrid membranes (Neogen) and a FiltaFlex spread filter apparatus (FiltaFlex, Ltd.), and the membranes were then placed on the appropriate selective medium.
Salmonella enumeration was performed on XLDtnc medium (xylose lysine desoxycholate medium; Oxoid, Remel) with 4.6 ml liter−1 tergitol (also known as niaproof; Sigma), 15 mg liter−1 novobiocin, and 10 mg liter−1 cefsulodin). The plates were incubated at 37°C for 18 to 20 h and then for an additional 18 to 20 h at room temperature (23 to 25°C) (12). Up to 10 presumptive Salmonella isolates per plate were tested by PCR as described above (32, 34). The CFU counts for confirmed Salmonella isolates were adjusted for the percentage of verified isolates per positive sample and then reported as CFU per 100 cm2.
Enumeration of E. coli O157:H7 was performed on ntChrom-O157 agar (12). The plates were incubated at 42°C for 18 to 20 h and then inspected for the presence of E. coli O157:H7, and putative E. coli O157:H7 isolates were tested using the DrySpot agglutination test kit as described above. Colonies that gave a positive agglutination reaction were then subcultured to ctSMac and confirmed to be the O157:H7 serotype by PCR as described above. The CFU counts for confirmed E. coli O157:H7 isolates were adjusted and reported as described for Salmonella above.
APC for hide and preevisceration carcass samples were determined by impedance measurements using a Bactometer (bioMérieux, Hazelwood, MO), with a subset of each day's samples also examined in parallel with APC Petrifilm (3 M Microbiology, St. Paul, MN) to construct an APC standard curve. APC values for postintervention carcasses were determined using only APC Petrifilm. Hide samples were serially diluted out to 10−3 or 10−4, and then 0.1 ml of the diluted sample was added to 0.9 ml of General Purpose Medium-Plus (bioMérieux) supplemented with 1.8% (wt/vol) dextrose for Bactometer analysis. Preevisceration carcass samples were diluted to 10−1 or 10−2 prior to dilution in General Purpose Medium-Plus. Hide and preevisceration carcass samples that were evaluated in parallel, using both the Bactometer and APC Petrifilm, were serially diluted out to 10−4, 10−5, and 10−6, and then 1 ml of each dilution was plated on APC Petrifilm and incubated according to the manufacturer's instructions. Initial detection times were converted to CFU per milliliter using a standard curve generated from representative samples examined in parallel with APC Petrifilm. The APC values for postintervention carcass samples were determined by plating 1 ml each of the 100 and the 10−2 dilutions onto APC Petrifilm. The Petrifilms were incubated at 35°C for 20 to 48 h and were counted manually. All APC values were corrected for their dilution factor, converted to their log10 values, and reported as CFU per 100 cm2.
Statistics.
The APC data are reported as the geometric mean log10 CFU/100 cm2 with the 95% confidence interval (CI). Data were analyzed by season (8 sample days per season; 2 days at each of four plants) or by plant (8 sample days per plant; 2 days in each season). Salmonella and E. coli O157:H7 prevalence values were calculated by dividing the number of culture-positive samples by the total number of samples collected (n = 95 per sample type per day). The percent prevalence on hide and carcass samples was determined for each sample day and reported as the mean and standard deviation (SD) of 8 sample days per season or per plant. The percentage of hide and carcass samples found to be enumeration positive was also analyzed by season or by plant and reported as the mean percent enumerable (±SD), the median value (CFU/100 cm2), and the minimum and maximum values observed for the enumerable samples. Comparisons of seasonal or plant prevalence values, percent enumerable, and arithmetic mean APC values were examined using a one-way analysis of variance (ANOVA) and the Bonferroni multiple-comparison posttest. Comparisons of median values of enumeration data sets were made using the Kruskal-Wallis test for nonparametric data and Dunn's multiple-comparison posttest. Comparisons of data sets with only two groups of values were made using either a two-tailed unpaired t test or the Mann-Whitney U test for nonparametric data. Interactions between plant and season hide prevalence values were examined using a 4-by-4-by-2 ANOVA factorial analysis. Data were analyzed using Prism 4 GraphPad software, and P values of less than 0.05 were considered significantly different.
In order to examine the extent of concomitant contamination with Salmonella and E. coli O157:H7, samples were evaluated for the presence of each pathogen using both the enrichment and the enumeration techniques described above, and the results were then scored using the following value definitions. Hide samples that were enumeration and enrichment negative were assigned an arbitrarily low value of 0.1 CFU/100 cm2 (equivalent to ≤1 CFU/1,000 cm2, a value that would likely result in a sample enrichment testing negative) so that they could be evaluated in context with the other numerical data. Hide samples that were enumeration negative but enrichment positive were assigned a value of 5 CFU/100 cm2 (equivalent to 50 CFU/1,000 cm2, which is below the likely enumerable range). Data from enumeration-positive samples, regardless of pathogen, were binned (CFU/100 cm2) as follows: 40 to 400, 440 to 800, 840 to 1,600, 1,640 to 2,400, 2,440 to 3,200, and > 3,240. The geometric mean of each bin and the percentage of samples in each bin were then calculated. Carcass samples that were negative for both enumeration and enrichment were assigned a value of 0.01 CFU/100 cm2 (equivalent to <1 CFU/8,000 cm2). Those that were enumeration negative but enrichment positive were assigned a value of 0.2 CFU/100 cm2 (equivalent to 16 CFU/8,000 cm2). Samples that were enumeration positive were binned (CFU/100 cm2) as follows: for Salmonella, the ranges were 0.5 to 5.0, 5.5 to 10.0, 10.5 to 20.0, 20.5 to 30.0, 30.5 to 40.0, and > 40.5; for E. coli O157:H7, the ranges were 0.83 to 8.3, 9.1 to 16.6, 17.4 to 33.2, 43.0 to 49.8, 50.6 to 66.4, and >67.2. The geometric mean of each bin was calculated as described above.
RESULTS
Aerobic microbial loads on hides and carcasses.
Examination of the aerobic microbial loads on the hides of cattle at slaughter showed that the geometric mean APC was highest in the summer (P < 0.001) and ranged from log10 6.17 to 8.19 CFU/100 cm2 (Table 1). The geometric mean of APC counts for preevisceration carcasses in the dressing process ranged from log10 4.24 to 6.47 CFU/100 cm2 and also was highest in the summer (P < 0.001). An estimate of the hide-to-carcass transfer (HTCT) of bacterial contamination was calculated by determining the ratio of APC contamination present on preevisceration carcasses (which should theoretically be sterile) to the APC level determined on hides. The results of this analysis showed that average HTCT values were lowest in winter, when the APC levels on hides were generally lower. All plants sampled used a combination of multiple-hurdle interventions, including steam vacuum, hot water, and a final organic-acid wash. The use of one or more of these interventions resulted in a consistent and generally low APC level on postintervention carcasses, which ranged from log10 1.46 to 1.96 CFU/100 cm2.
TABLE 1.
Log10 geometric mean APC load for each sample type in each season or plant
| Season or plant | Log10 geometric mean APC load (95% CI)a
|
% HTCTb | ||
|---|---|---|---|---|
| Hide | Carcass
|
|||
| Preevisceration | Postintervention | |||
| Seasons | ||||
| Summer | 8.19 (8.13-8.24)B | 6.47 (6.42-6.55)B | 1.96 (1.91-2.04)B | 3.4 ± 3.6 |
| Fall | 6.53 (6.57-6.67)A | 4.70 (4.62-4.76)A | 1.55 (1.49-1.59)A | 2.7 ± 2.5 |
| Winter | 6.17 (6.12-6.22)A | 4.24 (4.19-4.30)A | 1.46 (1.42-1.51)A | 1.4 ± 0.7 |
| Spring | 6.54 (6.49-6.60)A | 4.94 (4.88-5.01)A | 1.49 (1.45-1.54)A | 3.9 ± 3.9 |
| Plants | ||||
| A | 7.07 (7.00-7.16)B | 5.25 (5.18-5.32)A | 1.74 (1.69-1.79)A | 1.9 ± 1.4 |
| B | 6.72 (6.64-6.80)A | 5.39 (5.29-5.50)B | 1.52 (1.48-1.56)A | 5.8 ± 3.7 |
| C | 6.92 (6.85-6.99)A | 4.55 (4.47-4.64)A | 1.73 (1.67-1.80)B | 0.5 ± 0.3 |
| D | 6.71 (6.75-6.90)A | 5.16 (5.06-5.22)A | 1.47 (1.43-1.52)A | 3.3 ± 2.1 |
Each season or plant log10 value represents the geometric mean of 760 samples. Values that do not have a common uppercase letter following them are significantly different as determined by a one-way ANOVA and Bonferroni's multiple-comparison posttest (P < 0.05).
Percent HTCT calculated by dividing the geometric mean APC of the pre-evisceration carcass samples calculated for each season or each plant, by the geometric mean APC determined for the paired hide samples. HTCT values are shown as the mean and standard deviation (± SD) calculated either by season or by plant.
Analysis of the APC data collected from each plant over the course of the study showed that the arithmetic mean load on cattle hides was significantly higher at plant A than at the other three plants (P < 0.001) and the mean load on preevisceration carcasses was significantly higher for plant B (P < 0.001), while the arithmetic mean APC on postintervention carcasses was significantly higher in plant C (P < 0.001) (Table 1). Examination of the log10 geometric mean APC values present on preevisceration carcasses in relation to those present on hides showed that plant C appeared to have the lowest HTCT values (0.5% ± 0.3%), while plant B had the highest (5.8% ± 3.7%). This difference was consistent with the significantly higher average APC value on preevisceration carcass samples from plant B (Table 1).
Salmonella prevalences and loads on hides and carcasses.
No significant seasonal effect was detected for the prevalence of Salmonella on hides (P = 0.67), which ranged from 86 to 94% (Table 2). The percentages of hide samples yielding enumeration results (those with a Salmonella load of ∼40 CFU/100 cm2 or greater) ranged from 9 to 21% across seasons and also were not significantly different (P = 0.24). However, the median value of enumerable samples was significantly lower in the spring than in the other three seasons (P < 0.0001). Preevisceration carcass prevalences were consistent and ranged from 44 to 55%, and the percentages of preevisceration carcass samples in the winter and spring that were in the enumeration range (∼0.5 CFU/100 cm2 or greater) varied from 5 to 13%. The median loads of Salmonella in these enumerable samples were 1.0 and 0.5 CFU/100 cm2, respectively (Table 2).
TABLE 2.
Mean percent prevalence and load of Salmonella on cull cattle hides and carcasses analyzed by season
| Salmonella parameter | Valuea
|
P value | |||
|---|---|---|---|---|---|
| Summer | Fall | Winter | Spring | ||
| Hide | |||||
| Prevalence (%) (±SD) | 93.8 ± 10.2 | 88.5 ± 13.1 | 89.8 ± 8.4 | 86.1 ± 16.9 | 0.6680b |
| Load (CFU/100 cm2) | |||||
| Enumeration positive (%) (±SD) | 17.1 ± 17.4 | 9.4 ± 10.7 | 21.3 ± 14.0 | 10.7 ± 7.5 | 0.2363b |
| n | 130 | 71 | 162 | 81 | |
| Median | 120A | 120A | 80A | 40B | <0.0001c |
| Minimum | 40 | 40 | 40 | 40 | |
| Maximum | 2.1 × 103 | 3.4 × 104 | 2.8 × 104 | 5.2 × 102 | |
| Preevisceration carcass | |||||
| Prevalence (%) (±SD) | 49.4 ± 26.3 | 51.8 ± 28.9 | 55.3 ± 31.5 | 44.2 ± 19.2 | 0.8670b |
| Load (CFU/100 cm2)d | |||||
| Enumeration positive (%) (±SD) | ND | ND | 13.0 ± 14.6 | 5.1 ± 4.4 | 0.1656e |
| n | ND | ND | 99 | 39 | |
| Median | ND | ND | 1.0 | 0.5 | 0.6536f |
| Minimum | ND | ND | 0.5 | 0.5 | |
| Maximum | ND | ND | 3.8 × 102 | 4.9 × 102 | |
| Postintervention carcass | |||||
| Prevalence (%) (±SD) | 1.3 ± 2.2 | 0.1 ± 0.4 | 0.7 ± 0.8 | 1.1 ± 2.6 | 0.5602b |
Values that do not have a common uppercase letter following them are significantly different (P < 0.05). ND, not determined.
Comparisons of mean percent prevalence or percentages of samples that were enumerable were made using a one-way ANOVA and Bonferroni's multiple comparison post-test.
Comparisons of enumeration median values were made using the Kruskal-Wallis test for nonparametric data and Dunn's multiple-comparison posttest.
Enumeration values were determined in the winter and spring sampling seasons only.
Comparisons of percentages of samples that were enumerable were made using a two-tailed unpaired t test (t = 1.463).
Comparisons of enumeration median values were made using a two-tailed Mann-Whitney U test.
Analysis of Salmonella prevalence and enumeration data by plant showed that the hide prevalences of animals entering processing facilities were similar (85 to 93%), and the percentages of samples with a Salmonella load high enough to be enumerated were not significantly different between plants (P = 0.89). However, the median value for enumerable hide samples was significantly lower in plant D than in plant A (P = 0.02). Examination of average preevisceration carcass prevalence values for Salmonella also revealed significant differences between plants (Table 3). Specifically, the percentage of preevisceration carcasses found to be contaminated with Salmonella was significantly lower at plant C than at plant B (P = 0.01). These data are consistent with the APC data and the calculated HTCT values summarized in Table 1.
TABLE 3.
Mean percent prevalence and Salmonella load on cull cattle hides and carcasses analyzed by plant
| Salmonella parameter | Value for planta:
|
P value | |||
|---|---|---|---|---|---|
| A | B | C | D | ||
| Hide | |||||
| Prevalence (%) (±SD) | 88.0 ± 18.0 | 92.1 ± 4.9 | 93.1 ± 9.1 | 84.9 ± 13.8 | 0.5454b |
| Load (CFU/100 cm2) | |||||
| Enumeration positive (%) (±SD) | 15.9 ± 14.7 | 11.8 ± 8.9 | 13.9 ± 13.8 | 16.7 ± 16.5 | 0.8974b |
| n | 121 | 90 | 106 | 127 | |
| Median | 120A | 80AB | 80AB | 80B | 0.0185c |
| Minimum | 40 | 40 | 40 | 40 | |
| Maximum | 3.4 × 104 | 4.3 × 103 | 2.1 × 103 | 2.8 × 104 | |
| Preevisceration carcass | |||||
| Prevalence (%) (±SD) | 52.2 ± 24.2AB | 67.2 ± 23.4A | 26.9 ± 12.5B | 54.4 ± 26.4AB | 0.0097b |
| Load (CFU/100 cm2)d | |||||
| Enumeration positive (%) (±SD) | 4.7 ± 3.6AB | 8.1 ± 6.8AB | 1.3 ± 1.9A | 22.1 ± 15.1B | 0.0239b |
| n | 18 | 31 | 5 | 84 | |
| Median | 0.5 | 0.5 | 2.5 | 1.0 | 0.0526c |
| Minimum | 0.5 | 0.5 | 1.0 | 0.5 | |
| Maximum | 2.5 | 4.9 × 102 | 8.5 | 3.8 × 102 | |
| Postintervention carcass | |||||
| Prevalence (%) (±SD) | 1.3 ± 2.2 | 0.4 ± 0.6 | 0.1 ± 0.4 | 1.3 ± 2.6 | 0.3938b |
Values that do not have a common uppercase letter following them are significantly different (P < 0.05).
Comparisons of mean percentage prevalence or percentages of samples that were enumerable were made using a one-way ANOVA and Bonferroni's multiple-comparison posttest.
Comparisons of enumeration median values were made using the Kruskal-Wallis test for nonparametric data and Dunn's multiple-comparison posttest.
Enumeration values were determined in the winter and spring sampling seasons only.
The prevalence of Salmonella on postintervention carcasses was significantly decreased by one or more of the processing interventions employed (P < 0.001) and was found to vary from 0.1 to 1% (Tables 2 and 3). In the course of this study, only one postintervention carcass was found to have Salmonella present at a level that could be detected with the enumeration methods employed. This sample was identified at plant D in winter, and the calculated Salmonella concentration was in the range of 0.01 to 0.07 CFU/100 cm2.
E. coli O157:H7 prevalences and loads on hides and carcasses.
The prevalence of E. coli O157:H7 on hides and carcasses was lower than that of Salmonella, ranging from 39 to 56%, and no significant seasonal effect was detected. The prevalences on preevisceration carcasses were consistent and ranged from 14 to 20% (Table 4). The percentages of hide samples found to contain E. coli O157:H7 at enumerable levels ranged from 0.4 to 6%, while those of preevisceration carcass samples in the winter and spring were approximately 3 and 2%, respectively (Table 4), and the median value of these enumerable carcass samples was found to be significantly lower in spring than in winter (P = 0.008).
TABLE 4.
Mean percent prevalence and load of E. coli O157:H7 on cull cattle hides and carcasses analyzed by season
| E. coli O157:H7 parameter | Valuea
|
P value | |||
|---|---|---|---|---|---|
| Summer | Fall | Winter | Spring | ||
| Hide | |||||
| Prevalence (%) (±SD) | 55.7 ± 22.6 | 38.7 ± 24.6 | 50.4 ± 37.8 | 43.2 ± 20.7 | 0.6107b |
| Load (CFU/100 cm2) | |||||
| Enumeration positive (%) (±SD) | 5.8 ± 7.2 | 0.4 ± 0.6 | 3.7 ± 6.4 | 1.5 ± 2.5 | 0.1626b |
| n | 44 | 3 | 28 | 11 | |
| Median | 80 | 80 | 40 | 40 | 0.1817c |
| Minimum | 40 | 40 | 40 | 40 | |
| Maximum | 9.8 × 103 | 80 | 7.2 × 102 | 1.0 × 103 | |
| Preevisceration carcass | |||||
| Prevalence (%) (±SD) | 18.3 ± 16.6 | 14.9 ± 13.6 | 19.5 ± 31.2 | 14.2 ± 13.2 | 0.9407b |
| Load (CFU/100 cm2)d | |||||
| Enumeration positive (%) (±SD) | ND | ND | 3.2 ± 7.1 | 1.6 ± 2.5 | 0.5615e |
| n | ND | ND | 24 | 12 | |
| Median | ND | ND | 1.6A | 0.8B | 0.0082f |
| Minimum | ND | ND | 0.8 | 0.8 | |
| Maximum | ND | ND | 45.6 | 5.6 | |
Values that do not have a common uppercase letter following them are significantly different (P < 0.05). ND, not determined.
Comparisons of mean percent prevalence or percentages of samples that were enumerable were made using a one-way ANOVA and Bonferroni's multiple-comparison posttest.
Comparisons of enumeration median values were made using the Kruskal-Wallis test for nonparametric data and Dunn's multiple-comparison posttest.
Enumeration values were determined in the winter and spring sampling seasons only.
Comparisons of percentages of samples that were enumerable were made using a two-tailed unpaired t test (t = 0.5948).
Comparisons of enumeration median values were made using a two-tailed Mann-Whitney U test.
Analysis of the E. coli O157:H7 prevalence in each plant showed significant differences between plants in the incoming load of E. coli O157:H7, as seen by plants C and D exhibiting a higher percent hide prevalence than plants A and B (P = 0.0005) (Table 5). The prevalences on preevisceration carcasses ranged from 7 to 42% (Table 5), and in keeping with the results for APC and Salmonella levels, plant C was found to have one of the lowest average percent prevalence values for E. coli O157:H7 on preevisceration carcasses, despite having one of the higher average hide prevalence values (Table 5). The percentages of hide samples that carried E. coli O157:H7 at enumerable levels ranged from 0.3 to 9%, with plant D having a significantly higher percentage of enumerable hides (P = 0.001). The hide prevalence and load in plant D were reflected in a significantly higher percent preevisceration prevalence value (P = 0.0001), as summarized in Table 5.
TABLE 5.
Mean percent prevalence and load of E. coli O157:H7 on cull cattle hides and carcasses analyzed by plant
| E. coli O157:H7 parameter | Value for planta:
|
P value | |||
|---|---|---|---|---|---|
| A | B | C | D | ||
| Hide | |||||
| Prevalence (%) (±SD) | 29.9 ± 20.2A | 28.0 ± 15.9A | 64.0 ± 23.8B | 65.9 ± 21.6B | 0.0005b |
| Load (CFU/100 cm2) | |||||
| Enumeration positive (%) (±SD) | 0.9 ± 1.4A | 0.3 ± 0.5A | 1.6 ± 1.6A | 8.6 ± 8.0B | 0.0013b |
| n | 7 | 2 | 12 | 65 | |
| Median | 120 | 80 | 60 | 80 | 0.7125c |
| Minimum | 40 | 80 | 40 | 40 | |
| Maximum | 4.0 × 102 | 80 | 6.0 × 102 | 9.8 × 103 | |
| Preevisceration carcass | |||||
| Prevalence (%) (±SD) | 11.5 ± 12.8A | 6.9 ± 4.6A | 6.9 ± 5.8A | 41.5 ± 21.6B | <0.0001b |
| Load (CFU/100 cm2)d | |||||
| Enumeration positive (%) (±SD) | 0.8 ± 1.0 | 8.7 ± 7.8 | 0.0927e | ||
| n | 0 | 0 | 3 | 33 | |
| Median | 0.8 | 1.6 | 0.5281f | ||
| Minimum | 0.8 | 0.8 | |||
| Maximum | 5.6 | 45.6 | |||
Values that do not have a common uppercase letter following them are significantly different (P < 0.05).
Comparisons of mean percent prevalence or percentages of samples that were enumerable were made using a one-way ANOVA and Bonferroni's multiple-comparison posttest.
Comparisons of enumeration median values were made using the Kruskal-Wallis test for nonparametric data and Dunn's multiple-comparison posttest.
Enumeration values were determined in the winter and spring sampling seasons only.
Comparisons of percentages of samples that were enumerable were made using a two-tailed unpaired t test (t = 1.998).
Comparisons of enumeration median values were made using a two-tailed Mann-Whitney U test.
Concomitant contamination of hides and carcasses with Salmonella and E. coli O157:H7.
Analysis of paired hide and carcass samples collected from all four plants in the winter and spring of 2006 (n = 1,520) showed that on average 33.3% (95% CI, 15.9% to 69.8%) and 4.1% (95% CI, 0.98% to 17.3%) of cattle hides and carcasses, respectively, were contaminated with both Salmonella and E. coli O157:H7. A substantial proportion of samples were found to be contaminated with Salmonella alone, as 36.6% (95% CI, 20.0% to 67.0%) of hides and 35.9% (95% CI, 25.2% to 51.3%) of carcasses were found to be contaminated with Salmonella but not E. coli O157:H7. This was in contrast with the small percentage of samples found to be contaminated only with E. coli O157:H7, 1.1% (95% CI, 0.2% to 6.7%) and 2.7% (95% CI, 1.1% to 6.7%) for hides and carcasses, respectively. Hides found not to be contaminated with either pathogen were uncommon (3.7% [95% CI, 0.5% to 29.2%]), while carcasses were frequently determined not to be contaminated with either pathogen (35.3% [95% CI, 16.5% to 75.3%]).
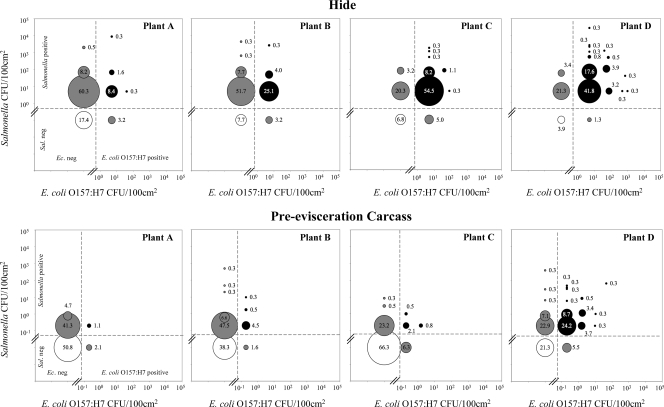
In order to further examine the incidence of contamination with either or both Salmonella and E. coli O157:H7, the load of each pathogen (CFU/100 cm2) and the percentage of hides and carcasses found to be contaminated with neither, either, or both pathogens at the indicated levels were plotted (Fig. 1). These plots show the considerable variation that was observed between plants in terms of the incoming pathogen load and concomitant HTCT. As seen in Fig. 1, cattle hide samples in plants A and B were predominantly found to be contaminated with Salmonella and to a lesser extent with E. coli O157:H7, while hide samples in plants C and D were contaminated to a greater extent with both pathogens. Presenting the data in this way also demonstrates the correlation between enumerable pathogen levels on hides and those on preevisceration carcasses, a likely result of processing cross-contamination, as seen most prominently in the graph of the data from plant D (Fig. 1).
FIG. 1.
Examination of coincident contamination with Salmonella and E. coli O157:H7 on cattle hides and carcasses at slaughter. A total of 380 hide and carcass samples were collected from each of four plants in the winter and spring (95 samples each day; 2 days each season). The samples were evaluated for the presence of each pathogen using both the enrichment and the enumeration techniques described, and the results were scored using the definitions described in Materials and Methods. The samples were binned according to the outcome, and the geometric mean and percentage of samples in each bin were then calculated. The circles represent the percentages of cattle hide and carcass samples at each plant that were found to contain neither (indicated by white circles), either (indicated by gray circles), or both (indicated by black circles) Salmonella and E. coli O157:H7, and the geometric mean pathogen load is indicated by the position in the plot of E. coli O157:H7 CFU/100 cm2 versus Salmonella CFU/100 cm2. Ec., E. coli; Sal, Salmonella; neg, negative.
DISCUSSION
Studies describing the hide and carcass hygiene of cull cattle at slaughter are few and yet are important with respect to food safety, as a large portion of the meat obtained from these animals is processed into ground beef. Ultimately, the hygiene of cull cattle at slaughter has an important impact on the incidence of food-borne disease outbreaks that are attributed to the consumption of undercooked ground beef. In this study, we examined the hide and carcass hygiene of cull cattle at slaughter and found that on average, approximately 1% of the aerobic load on hides appeared to be transferred to preevisceration carcasses in the dressing process. Use of one or more of the multiple-hurdle processing interventions employed effectively reduced this contamination, however, by an average 3.5 log units.
Numerous studies have reported seasonal effects on the prevalence of Salmonella and E. coli O157:H7 (1, 19, 20, 35). In this study, we found no statistically significant differences between seasonal prevalence values. However, significant differences were observed for the median enumeration values determined in each season. Specifically, Salmonella contamination levels on hides and E. coli O157:H7 levels on carcasses were both found to be lowest in the spring (P < 0.0001 and P = 0.0082, respectively). The lack of a seasonal effect on the pathogen prevalence values observed in this experiment may have been due in part to differences in the geographical locations of the packing plants sampled compared to those studied in our evaluation of the effects of seasonality in fed-beef plants (8), where the cattle sampled were predominantly from one region of the United States (the Midwest). Additionally, plants that slaughter cull cows and bulls tend to obtain animals from a broader area than fed-beef plants. For each of the plants studied in this experiment, the cattle sampled included a number of animals that were sourced from markets or operations located over 1,200 km from the packing plant. For three of the four plants, less than half of the animals sampled originated from the same state as the plant where they were processed. The cattle sampled in this experiment originated from 33 of the 48 continental United States, with 6 to 14 states represented in each of the plants sampled. Therefore, at any time of the year, cattle entering cull beef plants may have originated from a variety of environments, and as a consequence, seasonal effects would be muted.
Another factor likely to have influenced the prevalence values determined in this study is that Salmonella and E. coli O157:H7 prevalence is typically higher on cattle hides at slaughter than on the hides of cattle sampled on farms, in part because of transportation and lairage effects (1). Arthur et al. have shown that the E. coli O157:H7 hide prevalences of the same animals increased from 50.3% as measured at the feedlot to 94.4% at slaughter (2). Barham et al. have demonstrated that the average cattle hide prevalence for Salmonella increased from 6% to 89% as measured before transport and after slaughter (7). Similarly, Fluckey et al. showed Salmonella prevalence to increase from 35%, as measured on animal hides at the feedlot, to 84% on the hides of the same animals at slaughter (24). In this study, we found the average Salmonella hide prevalence at slaughter to be 89.6%, while that of E. coli O157:H7 was 46.9%. Given the relatively high level of contamination observed on cattle hides at slaughter and that IMS was used for pathogen isolation from culture enrichments, it is not surprising that a seasonal effect was not observed. It is also noteworthy that the prevalence values determined in this study were from hides sampled at one site (1,000 cm2 of the brisket plate region). Given the nonuniform distribution of pathogen contamination on hides, sampling at multiple hide sites would likely show even higher prevalence values.
A unique aspect of the data collected in this study regarding the levels of Salmonella and E. coli O157:H7 on cull cattle hides and carcasses centers on the analysis and presentation of the enumeration data. Using the rapid and direct methods described, enumeration data were collected for 18.5% and 11.4% of 1,520 hide and carcass samples (samples found with pathogen levels in the enumeration range), respectively. Analysis of these data allows not only an examination of the extent to which cattle hides and carcasses were coincidently contaminated with Salmonella and E. coli O157:H7, but more importantly, a description of the observed variation in both the incoming load and the resultant transfer to carcasses via cross-contamination during the dressing process (Fig. 1). It should be noted that the reported enumeration values are likely underestimates of the actual pathogen levels. This is in part due to the sampling error associated with the enumeration methods employed, especially when pathogen levels are near the limits of detection of the methods (12), but perhaps to a greater extent because of the selective media used in evaluating the pathogen load. As a result, only a fraction of the pathogens present grow, due to either injury or other phenotypic determinants. However, in spite of these caveats, the use of direct plating enumeration methods can provide valuable baseline information. The results of this study are in keeping with those reported from other research laboratories, which have previously described the efficacy of direct plating enumeration methods (26, 36). Large-scale enumeration data, generated using methods such as those employed in this study, are important for process modeling and risk assessment analyses. Moreover, the prevalence and enumeration data collected in this study allow the correlation between hide and carcass contamination to be visualized in a way not previously described (Fig. 2).
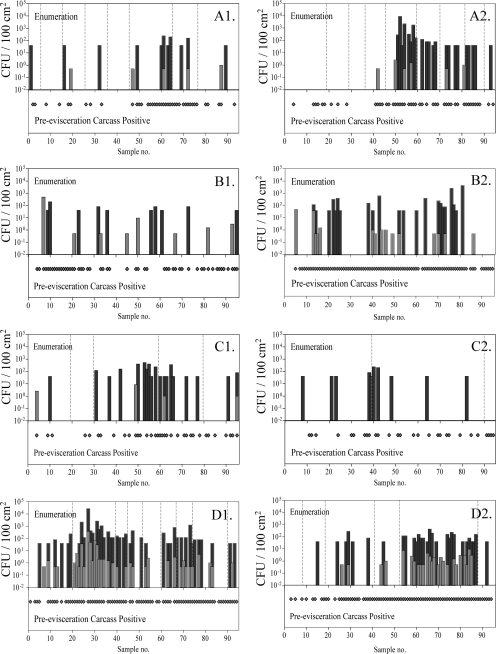
FIG. 2.
Comparison of hide and preevisceration carcass sample enumeration data for Salmonella (CFU/100 cm2) with paired preevisceration carcass prevalence data. The data presented were collected on two different days at each of the four plants sampled (plant A [A1 and A2], plant B [B1 and B2], plant C [C1 and C2], and plant D [D1 and D2]). Samples were collected consecutively, and lot break information, when available, is indicated by gray dashed lines. The dark-gray bars and light-gray bars indicate enumerable hide and preevisceration carcass samples and the Salmonella loads in those samples, while the light-gray diamonds indicate preevisceration carcass samples that were positive for Salmonella by enrichment.
Many studies have shown that cattle hide contamination is transferred to carcasses in the processing environment (2, 5, 9, 21). These analyses typically involved examination of the pulsed-field gel electrophoresis patterns of isolates collected from hide and carcass samples, but little information was available to model the cross-contamination process. In this study, hides and carcasses were tagged so that they were matched and were sampled consecutively. As a consequence of this sampling scheme, it was possible to plot the prevalence and enumeration results obtained from these samples consecutively. Plotting the data in this fashion demonstrated that enumerable pathogen levels on hides and carcasses often clustered on the processing line and that these clusters were frequently correlated with lots (Fig. 2). Further examination of the comparison of enumeration and prevalence data from paired hide and carcass samples showed that animals entering the processing line with enumerable pathogen levels on their hides frequently coincided with preevisceration carcasses that were also contaminated (Fig. 2). These data not only demonstrate the considerable variation in the incoming pathogen load on cattle hides, but also illustrate the differences between plants in their abilities to prevent cross-contamination between hides and carcasses in the dressing process. As a case in point, the Salmonella hide prevalence in the data set depicted in Fig. 2C1 was 93.7%, similar to that in Fig. 2D2, which was 98.9%, and yet the resulting preevisceration carcass prevalence for the data set depicted in Fig. 2C1 was 38.9% while that in Fig. 2D2 was 84.2%.
Previous studies have shown that multiple-hurdle processing interventions effectively reduce carcass contamination (6), conclusions that are supported by this study. However, ultimately the utility of carcass-processing interventions is dependent upon the level of pathogens entering the plant. As shown here, the interventions employed at each of the four abattoirs substantially decreased carcass contamination with Salmonella an average of 98.4% (95% CI, 97.6% to 99.7%). It is intuitive that lower pathogen levels on hides at slaughter should result in lower pathogen levels on carcasses, but as the slaughter process is multifaceted, so are the factors that affect the extent of contaminated postintervention carcasses, such as the effective use of multiple-hurdle interventions and the skill, training, and experience of plant personnel. In summary, the results presented here provide data on hide and carcass pathogen levels during the harvesting process that have previously been unavailable. These data indicate that the continued development and implementation of intervention strategies that reduce hide pathogen levels are key elements in the effort to decrease the incidence of postintervention carcass contamination with Salmonella or E. coli O157:H7.
Acknowledgments
This project was funded in part by the Beef Checkoff.
We thank the plant personnel for their kind assistance and cooperation. We also thank Julie Dyer, Frank Reno, Bruce Jasch, Greg Smith, Kim Kucera, Jade Franklin, Troy Worth, and Jessica Ott for their technical support and Debbie Kummer for administrative assistance.
The mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 22 August 2008.
REFERENCES
- 1.Armstrong, G. L., J. Hollingsworth, and J. G. Morris, Jr. 1996. Emerging foodborne pathogens: Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol. Rev. 18:29-51. [DOI] [PubMed] [Google Scholar]
- 2.Arthur, T. M., J. M. Bosilevac, D. M. Brichta-Harhay, M. N. Guerini, N. Kalchayanand, S. D. Shackelford, T. L. Wheeler, and M. Koohmaraie. 2007. Transportation and lairage environment effects on prevalence, numbers, and diversity of Escherichia coli O157:H7 on hides and carcasses of beef cattle at processing. J. Food Prot. 70:280-286. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, T. M., J. M. Bosilevac, D. M. Brichta-Harhay, N. Kalchayanand, S. D. Shackelford, T. L. Wheeler, and M. Koohmaraie. 2007. Effects of a minimal hide wash cabinet on the levels and prevalence of Escherichia coli O157:H7 and Salmonella on the hides of beef cattle at slaughter. J. Food Prot. 70:1076-1079. [DOI] [PubMed] [Google Scholar]
- 4.Arthur, T. M., J. M. Bosilevac, X. Nou, S. D. Shackelford, T. L. Wheeler, M. P. Kent, D. Jaroni, B. Pauling, D. M. Allen, and M. Koohmaraie. 2004. Escherichia coli O157 prevalence and enumeration of aerobic bacteria, Enterobacteriaceae, and Escherichia coli O157 at various steps in commercial beef processing plants. J. Food Prot. 67:658-665. [DOI] [PubMed] [Google Scholar]
- 5.Aslam, M., F. Nattress, G. Greer, C. Yost, C. Gill, and L. McMullen. 2003. Origin of contamination and genetic diversity of Escherichia coli in beef cattle. Appl. Environ. Microbiol. 69:2794-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacon, R. T., K. E. Belk, J. N. Sofos, R. P. Clayton, J. O. Reagan, and G. C. Smith. 2000. Microbial populations on animal hides and beef carcasses at different stages of slaughter in plants employing multiple-sequential interventions for decontamination. J. Food Prot. 63:1080-1086. [DOI] [PubMed] [Google Scholar]
- 7.Barham, A. R., B. L. Barham, A. K. Johnson, D. M. Allen, J. R. Blanton, Jr., and M. F. Miller. 2002. Effects of the transportation of beef cattle from the feedyard to the packing plant on prevalence levels of Escherichia coli O157 and Salmonella spp. J. Food Prot. 65:280-283. [DOI] [PubMed] [Google Scholar]
- 8.Barkocy-Gallagher, G. A., T. M. Arthur, M. Rivera-Betancourt, X. Nou, S. D. Shackelford, T. L. Wheeler, and M. Koohmaraie. 2003. Seasonal prevalence of Shiga toxin-producing Escherichia coli, including O157:H7 and non-O157 serotypes, and Salmonella in commercial beef processing plants. J. Food Prot. 66:1978-1986. [DOI] [PubMed] [Google Scholar]
- 9.Barkocy-Gallagher, G. A., T. M. Arthur, G. R. Siragusa, J. E. Keen, R. O. Elder, W. W. Laegreid, and M. Koohmaraie. 2001. Genotypic analyses of Escherichia coli O157:H7 and O157 nonmotile isolates recovered from beef cattle and carcasses at processing plants in the Midwestern states of the United States. Appl. Environ. Microbiol. 67:3810-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barkocy-Gallagher, G. A., K. K. Edwards, X. Nou, J. M. Bosilevac, T. M. Arthur, S. D. Shackelford, and M. Koohmaraie. 2005. Methods for recovering Escherichia coli O157:H7 from cattle fecal, hide, and carcass samples: sensitivity and improvements. J. Food Prot. 68:2264-2268. [DOI] [PubMed] [Google Scholar]
- 11.Bell, R. G. 1997. Distribution and sources of microbial contamination on beef carcasses. J. Appl. Microbiol. 82:292-300. [DOI] [PubMed] [Google Scholar]
- 12.Brichta-Harhay, D. M., T. M. Arthur, J. M. Bosilevac, M. N. Guerini, N. Kalchayanand, and M. Koohmaraie. 2007. Enumeration of Salmonella and Escherichia coli O157:H7 in ground beef, cattle carcass, hide and faecal samples using direct plating methods. J. Appl. Microbiol. 103:1657-1668. [DOI] [PubMed] [Google Scholar]
- 13.CDC. 2002. Multistate outbreak of Escherichia coli O157:H7 infections associated with eating ground beef—United States, June-July 2002. MMWR Morb. Mortal. Wkly. Rep. 51:637-639. [PubMed] [Google Scholar]
- 14.CDC. 2006. Multistate outbreak of Salmonella typhimurium infections associated with eating ground beef—United States, 2004. MMWR Morb. Mortal. Wkly. Rep. 55:180-182. [PubMed] [Google Scholar]
- 15.CDC. 2002. Outbreak of multidrug-resistant Salmonella newport—United States, January-April 2002. MMWR Morb. Mortal. Wkly. Rep. 51:545-548. [PubMed] [Google Scholar]
- 16.Chapman, P. A., A. T. Malo, C. A. Siddons, and M. Harkin. 1997. Use of commercial enzyme immunoassays and immunomagnetic separation systems for detecting Escherichia coli O157 in bovine fecal samples. Appl. Environ. Microbiol. 63:2549-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coleman, D. J., K. J. Nye, K. E. Chick, and C. M. Gagg. 1995. A comparison of immunomagnetic separation plus enrichment with conventional Salmonella culture in the examination of raw sausages. Lett. Appl. Microbiol. 21:249-251. [DOI] [PubMed] [Google Scholar]
- 18.Cray, W. C., Jr., and H. W. Moon. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:1586-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dargatz, D. A., P. J. Fedorka-Cray, S. R. Ladely, C. A. Kopral, K. E. Ferris, and M. L. Headrick. 2003. Prevalence and antimicrobial susceptibility of Salmonella spp. isolates from US cattle in feedlots in 1999 and 2000. J. Appl. Microbiol. 95:753-761. [DOI] [PubMed] [Google Scholar]
- 20.Edrington, T. S., T. R. Callaway, S. E. Ives, M. J. Engler, M. L. Looper, R. C. Anderson, and D. J. Nisbet. 2006. Seasonal shedding of Escherichia coli O157:H7 in ruminants: a new hypothesis. Foodborne Pathog Dis. 3:413-421. [DOI] [PubMed] [Google Scholar]
- 21.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laegreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fegan, N., G. Higgs, P. Vanderlinde, and P. Desmarchelier. 2005. An investigation of Escherichia coli O157 contamination of cattle during slaughter at an abattoir. J. Food Prot. 68:451-457. [DOI] [PubMed] [Google Scholar]
- 23.Fegan, N., P. Vanderlinde, G. Higgs, and P. Desmarchelier. 2005. A study of the prevalence and enumeration of Salmonella enterica in cattle and on carcasses during processing. J. Food Prot. 68:1147-1153. [DOI] [PubMed] [Google Scholar]
- 24.Fluckey, W. M., W. G. Loneragan, R. Warner, and M. M. Brashears. 2007. Antimicrobial drug resistance of Salmonella and Escherichia coli isolates from cattle feces, hides, and carcasses. J. Food Prot. 70:551-556. [DOI] [PubMed] [Google Scholar]
- 25.Hu, Y., Q. Zhang, and J. C. Meitzler. 1999. Rapid and sensitive detection of Escherichia coli O157:H7 in bovine faeces by a multiplex PCR. J. Appl. Microbiol. 87:867-876. [DOI] [PubMed] [Google Scholar]
- 26.LeJeune, J. T., D. D. Hancock, and T. E. Besser. 2006. Sensitivity of Escherichia coli O157 detection in bovine feces assessed by broth enrichment followed by immunomagnetic separation and direct plating methodologies. J. Clin. Microbiol. 44:872-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maciorowski, K. G., P. Herrera, F. T. Jones, S. D. Pillai, and S. C. Ricke. 2006. Cultural and immunological detection methods for Salmonella spp. in animal feeds: a review. Vet. Res. Commun. 30:127-137. [DOI] [PubMed] [Google Scholar]
- 28.McEvoy, J. M., A. M. Doherty, M. Finnerty, J. J. Sheridan, L. McGuire, I. S. Blair, D. A. McDowell, and D. Harrington. 2000. The relationship between hide cleanliness and bacterial numbers on beef carcasses at a commercial abattoir. Lett. Appl. Microbiol. 30:390-395. [DOI] [PubMed] [Google Scholar]
- 29.McEvoy, J. M., A. M. Doherty, J. J. Sheridan, I. S. Blair, and D. A. McDowell. 2003. The prevalence of Salmonella spp. in bovine faecal, rumen and carcass samples at a commercial abattoir. J. Appl. Microbiol. 94:693-700. [DOI] [PubMed] [Google Scholar]
- 30.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nou, X., T. M. Arthur, J. M. Bosilevac, D. M. Brichta-Harhay, M. N. Guerini, N. Kalchayanand, and M. Koohmaraie. 2006. Improvement of immunomagnetic separation for Escherichia coli O157:H7 detection by the PickPen magnetic particle separation device. J. Food Prot. 69:2870-2874. [DOI] [PubMed] [Google Scholar]
- 32.Nucera, D. M., C. W. Maddox, P. Hoien-Dalen, and R. M. Weigel. 2006. Comparison of API 20E and invA PCR for identification of Salmonella enterica isolates from swine production units. J. Clin. Microbiol. 44:3388-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Omisakin, F., M. MacRae, I. D. Ogden, and N. J. C. Strachan. 2003. Concentration and prevalence of Escherichia coli O157 in cattle feces at slaughter. Appl. Environ. Microbiol. 69:2444-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahn, K., S. A. De Grandis, R. C. Clarke, S. A. McEwen, J. E. Galan, C. Ginocchio, R. Curtiss III, and C. L. Gyles. 1992. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 6:271-279. [DOI] [PubMed] [Google Scholar]
- 35.Rivera-Betancourt, M., S. D. Shackelford, T. M. Arthur, K. E. Westmoreland, G. Bellinger, M. Rossman, J. O. Reagan, and M. Koohmaraie. 2004. Prevalence of Escherichia coli O157:H7, Listeria monocytogenes, and Salmonella in two geographically distant commercial beef processing plants in the United States. J. Food Prot. 67:295-302. [DOI] [PubMed] [Google Scholar]
- 36.Robinson, S. E., E. J. Wright, N. J. Williams, C. A. Hart, and N. P. French. 2004. Development and application of a spiral plating method for the enumeration of Escherichia coli O157 in bovine faeces. J. Appl. Microbiol. 97:581-589. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez, A., P. Pangloli, H. A. Richards, J. R. Mount, and F. A. Draughon. 2006. Prevalence of Salmonella in diverse environmental farm samples. J. Food Prot. 69:2576-2580. [DOI] [PubMed] [Google Scholar]
- 38.Thrift, T. A. 2000. Realizing more value for cull cattle: results of the 1999 national market cow and bull quality audit. Texas and Southwestern Cattle Raisers Association, Ft. Worth, TX. http://www.thecattlemanmagazine.com/issues/2000/05-00/realizingMore.asp.
- 39.Wells, S. J., P. J. Fedorka-Cray, D. A. Dargatz, K. Ferris, and A. Green. 2001. Fecal shedding of Salmonella spp. by dairy cows on farm and at cull cow markets. J. Food Prot. 64:3-11. [DOI] [PubMed] [Google Scholar]